Abstract
Nitric oxide (NO) is a well-known active site ligand and inhibitor of respiratory terminal oxidases. Here, we investigated the interaction of NO with a purified chimeric bcc-aa3 supercomplex composed of Mycobacterium tuberculosis cytochrome bcc and Mycobacterium smegmatis aa3-type terminal oxidase. Strikingly, we found that the enzyme in turnover with O2 and reductants is resistant to inhibition by the ligand, being able to metabolize NO at 25 °C with an apparent turnover number as high as ≈303 mol NO (mol enzyme)−1 min−1 at 30 µM NO. The rate of NO consumption proved to be proportional to that of O2 consumption, with 2.65 ± 0.19 molecules of NO being consumed per O2 molecule by the mycobacterial bcc-aa3. The enzyme was found to metabolize the ligand even under anaerobic reducing conditions with a turnover number of 2.8 ± 0.5 mol NO (mol enzyme)−1 min−1 at 25 °C and 8.4 µM NO. These results suggest a protective role of mycobacterial bcc-aa3 supercomplexes against NO stress.
Keywords: enzyme, ligand binding, enzyme inhibition, nitric oxide, respiratory chain complexes, mycobacteria, bcc-aa3 supercomplex
1. Introduction
Nitric oxide (NO) is a gaseous free radical that, while exerting physiological functions at low concentrations, can have deleterious effects on the cell at high levels, being able itself or in combination with reactive oxygen species to damage proteins, lipids and nucleic acids. NO production by activated macrophages through the inducible form of the enzyme NO synthase (iNOS) is indeed a common host defence mechanism against infections. Mycobacterium (M.) tuberculosis, like several other microorganisms, has evolved the ability to sense this ligand and elicit defensive mechanisms [1], such as the downregulation of the host iNOS expression [2,3], the degradation of proteins damaged by NO and reactive nitrogen species [4] and the production of enzymes resisting and/or metabolizing the ligand. Two types of enzymes are mainly involved in bacterial NO detoxification, the NO-reductases (NORs) and the NO-dioxygenases (NODs). The former reductively metabolize NO to dinitrogen oxide (N2O) under anaerobic conditions, whereas the latter degrade NO to nitrate (NO3−) using O2 as co-substrate. Most NODs belong to the globin superfamily, such as flavohaemoglobins (flavoHb) and truncated haemoglobins (trHb) that detoxify NO with high efficiency in many microbes. Genes encoding flavoHb and trHb have been found in mycobacteria too. In M. tuberculosis, a truncated single domain haemoglobin (HbN) was found to effectively oxidise NO to harmless nitrate and protect the microorganism from nitrosative stress [5,6]. In M. smegmatis, a similar role is played by a flavoHb [7].
One of the main targets of NO is the respiratory chain that generates the proton motive force across the bacterial or mitochondrial membrane needed by ATP synthase for ATP production. Among the respiratory enzymes, terminal oxidases are the preferential targets of the ligand [8]. At nanomolar concentrations and in competition with O2, NO rapidly inhibits the activity of both prokaryotic and eukaryotic terminal oxidases, leading to energy deficiency and redox imbalance due to enhanced production of reactive oxygen species. The inhibition occurs rapidly and is reversible. Interestingly, if the ligand is removed from solution, the bacterial bd type-terminal oxidase recovers the activity faster compared to the studied haem copper oxidases, suggesting that bd oxidases can enhance bacterial tolerance to NO and related nitrosative stress [9].
As an obligate aerobe, M. tuberculosis possesses a flexible, branched electron transport chain sustaining O2 reduction [10,11]. Electrons pass through type II NADH dehydrogenase or other dehydrogenases to the menaquinone pool, and then to O2 through either a supercomplex formed by cytochrome bcc and aa3-type cytochrome c oxidase (bcc-aa3) or a bd-type terminal menaquinol oxidase [12,13,14]. Mycobacteria have no soluble cytochrome c, but the bcc complex displays a dihaem c-type cytochrome playing the role of the cytochromes c and c1 in canonical electron transfer chains. Therefore, formation of the bcc-aa3 supercomplex is necessary to mediate the direct electron transfer from menaquinol to O2 [15]. The bcc-aa3 is composed of the two different transmembrane complexes bcc and aa3. The bcc complex encoded by the qcrCAB operon transfers electrons from menaquinol to the aa3-type cytochrome c oxidase. It comprises cytochrome b (QcrB) containing two b haem groups, a Rieske-type high potential Fe2S2 iron-sulfur protein (QcrA), and a dihaem c-type cytochrome (QcrC). The bcc is a homologue of the mitochondrial cytochrome bc1 (Complex III) and the chloroplast b6f complexes. Like the bc1 and b6f complexes, the mycobacterial bcc complex is proposed to utilize the Q-cycle mechanism to build an electrochemical proton gradient. The second component of the bcc-aa3, the aa3-type cytochrome c oxidase, is encoded by the ctaBCDE operon. Of interest, the genes ctaD and ctaC (but not ctaB and ctaE) are in close proximity to the qcrCAB operon [13]. The oxidase carries four redox-active metal centers: CuA, haem a, haem a3 and CuB. The CuA center, composed of two copper atoms, is located on CtaC (subunit II or COX2) and is probably the immediate electron acceptor from c-type dihaem in QcrC. The low-spin haem a possibly accepts electrons from CuA and is located on CtaD (subunit I or COX1). Ultimately, the electrons are transferred to the binuclear center (also located on CtaD), composed of the magnetically coupled high-spin haem a3 and CuB ion, where O2 reduction takes place. CtaE (subunit III or COX3) seems to hold no redox cofactors. Intriguingly, the cryo-EM structure of the bcc-aa3 respiratory supercomplex from M. smegmatis has revealed the association of the copper superoxide dismutase (SOD) SodC, present as a dimer on the periplasmic side of the membrane, on the top of bcc dimer and in contact with the QcrA and/or QcrC cytochrome cc subunit [16,17]. Sod C was suggested to provide protection against superoxide, locally generated by the respiratory chain, similarly to mitochondrial SOD-2, which is found associated with the respiratory supercomplex CI–CIII2–CIV in Caenorhabditis elegans [18]. Like other haem-copper terminal oxidases [19,20,21,22,23,24,25,26,27], the mycobacterial bcc-aa3 supercomplex is proposed to act as a proton pump to generate the proton-motive force [16]. It is worth mentioning that there is a kinetic advantage associated with the formation of a supercomplex composed of complexes III and IV, as reported recently [28]. Importantly, genetic deletion by homologous recombination of bcc-aa3 is lethal for mycobacteria [29], pointing to a central role in energy metabolism of the bcc-aa3 branch of the electron transport chain.
Recently, a purification protocol was reported, yielding a stable and active chimeric supercomplex (bcc-aa3) consisting of the cytochrome bcc from M. tuberculosis and the aa3-type cytochrome c oxidase from M. smegmatis [30]. Here, we studied the interaction of such a purified supercomplex with NO. Surprisingly, we found that the enzyme not only resists inhibition by the ligand, but also rapidly metabolizes it.
2. Results
2.1. Cytochrome bcc-aa3 is Resistant to NO Inhibition
The effect of NO on the dithiothreitol (DTT)/menadione (MD)-sustained O2-reductase activity of bcc-aa3 was tested oxygraphically at 25 °C by simultaneously monitoring the concentration of O2 and NO in solution (Figure 1). Surprisingly, NO (even at relatively high concentrations) has little or no effect on the enzyme activity. As shown in Figure 1, only a small, transient decline in the O2 consumption is caused by NO additions, followed by quick and full restoration of the control O2-reductase activity of bcc-aa3.
Figure 1.
Effect of NO on O2 consumption by purified mycobacterial cytochrome bcc-aa3 supercomplex. The addition of cytochrome bcc-aa3 (67 nM) to the oxygraphic chamber containing the reducing system DTT/MD results in an O2 reductase activity of 144 e− min−1. The effect of NO on the enzymatic O2 consumption is tested by sequentially adding to the chamber increasing volumes of NO-saturated water. Following O2 depletion and sample reoxygenation, O2 consumption re-starts and the activity increases up to 600 e− min−1. Sequential additions to the 1.5-mL reaction chamber: 9.5 μL of 800 mM DTT (5 mM); 4 μL of 100 mM MD (0.26 mM); 10 μL of 10 μM cytochrome bcc-aa3 (67 nM); 3, 10 and 25 μL of 1.8 mM NO (3.6, 12 and 30 μM, respectively).
The activity recovery occurs as NO levels decline, the NO decay in the presence of the enzyme being faster than expected. Following O2 exhaustion and subsequent sample reoxygenation, the O2-reductase activity of bcc-aa3 is enhanced (Figure 1). This phenomenon resembles the so-called ‘pulsing effect’ initially reported for the mammalian cytochrome c oxidase [31] and more recently documented for Escherichia coli cytochrome bd-I too [32]. It should be noted that after reoxygenation the enzyme retains its resistance to NO inhibition (Figure 1).
2.2. Cytochrome bcc-aa3 in Turnover Metabolizes NO
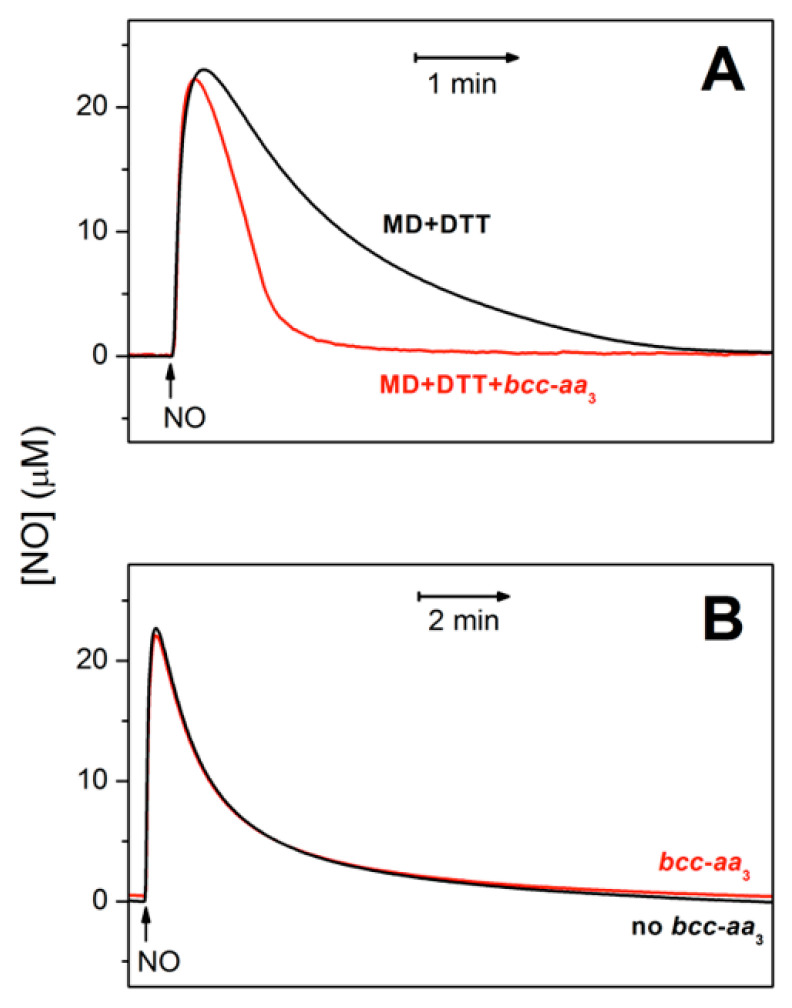
Figure 2A shows that bcc-aa3 in turnover with excess reductants and O2 can metabolize NO. In the presence of the supercomplex, the rate of NO decay is indeed significantly higher than measured in the absence of bcc-aa3 under otherwise identical experimental conditions (Figure 2A, red trace vs. black trace). At an initial NO concentration of 30 µM, bcc-aa3 displays a maximal NO-metabolizing activity of ≈303 mol NO (mol bcc-aa3)−1 min−1 at 25 °C, as estimated from initial rate analysis. The supercomplex can catalyze the aerobic degradation of NO only under turnover conditions. Indeed, as shown in Figure 2B, in the absence of reductants, the kinetics of NO decay with and without the enzyme in aerobic solution are virtually the same.
Figure 2.
Purified mycobacterial cytochrome bcc-aa3 supercomplex in turnover conditions metabolizes NO. NO traces in the presence (red) and absence (black) of the supercomplex. (A) Cytochrome bcc-aa3 (67 nM) in turnover with the reducing system DTT (5 mM) and MD (0.26 mM) accelerates the decomposition of NO (30 µM) added to the chamber at [O2] ≈ 130 µM. At an initial NO concentration of 30 µM, the NO-consuming activity of the enzyme was ≈ 303 mol NO (mol bcc-aa3)−1 min−1. In the absence of the enzyme (black line) the NO degradation mainly occurs due to the reaction of NO with O2. (B) In the absence of DTT/MD, i.e., under non-turnover conditions, cytochrome bcc-aa3 (200 nM) does not accelerate the decomposition of added NO (30 µM).
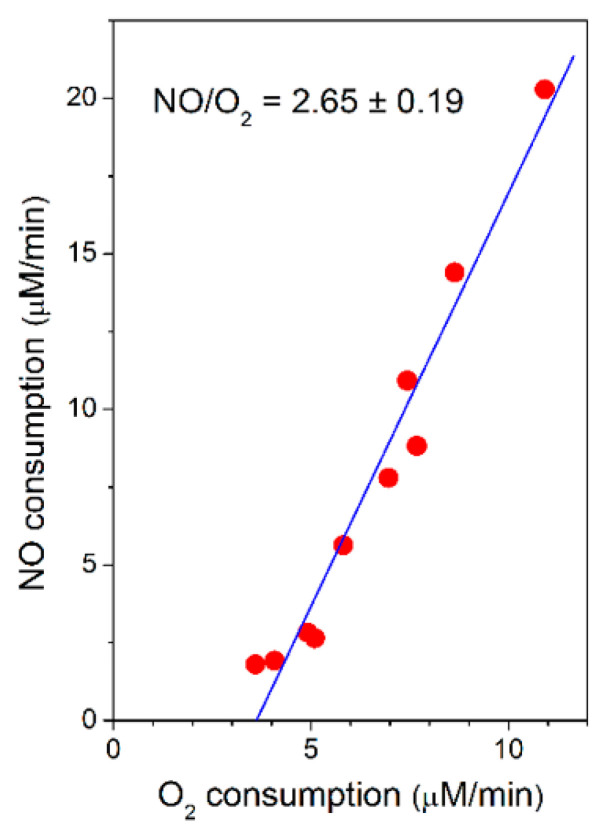
Taking advantage of the spontaneous increase in the O2-reductase activity of bcc-aa3 observed along each oxygraphic assay (Figure 1), the rate of NO consumption by bcc-aa3 could be measured at different actual rates of O2 consumption and the former was found to be proportional to the latter, according to a NO/O2 ratio of 2.65 ± 0.19 (Figure 3).
Figure 3.
Rate of NO consumption plotted as a function of the rate of O2 consumption by purified mycobacterial cytochrome bcc-aa3 supercomplex. NO (30 µM) was added to cytochrome bcc-aa3 in the presence of DTT (5 mM) and MD (0.26 mM) at [O2] ≈ 130 µM. Conditions as in Figure 1. Linear regression analysis (solid line) of experimental data points (red circles: NO consumption rate) gives a NO/O2 ratio of 2.65 ± 0.19 (mean ± standard deviation).
2.3. Cytochrome bcc-aa3 Possesses NO-Reductase Activity
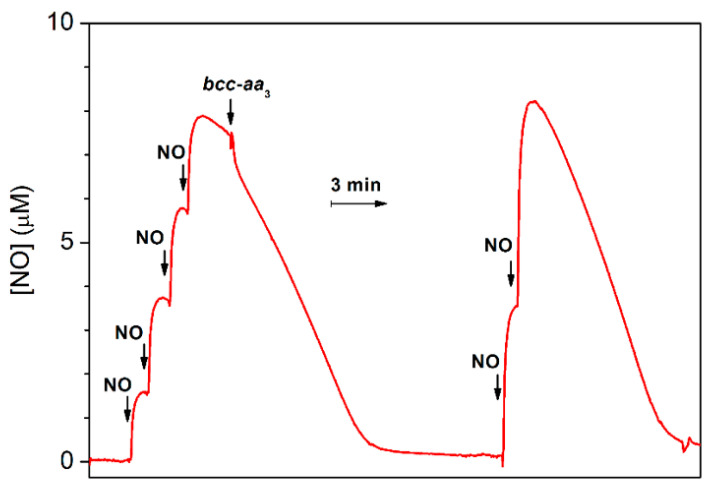
The ability of bcc-aa3 to degrade NO was also tested under anaerobic reducing conditions. In this assay, an aliquot of pre-reduced supercomplex was anaerobically added to an O2-free solution of NO containing excess reductants (DTT and MD), and the NO concentration in solution was then monitored using a NO-selective electrode. As shown in Figure 4, prior to enzyme addition, in the presence of excess DTT and MD, a slow decay of NO is observed due to reaction of NO with the reductants. The addition of bcc-aa3 clearly accelerates the decomposition of NO (Figure 4). The fast drop in the NO concentration observed immediately after addition of bcc-aa3 can be explained, at least partially, by NO binding to the reduced of bcc-aa3. The NO decay observed subsequently is consistent with a substantial catalytic NO reductase activity of bcc-aa3. After NO depletion, if the gas is re-added, the NO-consuming activity of bcc-aa3 is observed again (Figure 4). A notably slower NO consumption was observed if the same volume of aerobic buffer was added instead of the supercomplex in control experiments conducted under otherwise identical conditions (Supplementary Figure S1). The enzymatic NO reductase activity value was obtained from the slope of the trace with the enzyme after subtraction of the background non-enzymatic NO-reduction rate. Under anaerobic conditions, at [NO] = 8.4 µM, the estimated NO-reductase activity of bcc-aa3 proved to be 2.8 ± 0.5 mol NO (mol of bcc-aa3)−1 min−1 at 25 °C.
Figure 4.
NO reductase activity of purified mycobacterial cytochrome bcc-aa3 supercomplex. Four aliquots of 2.1 µM NO (1.8 μL of 1.8 mM NO each) were sequentially added to degassed buffer (buffer composition as in Figure 1) containing DTT (5 mM), MD (0.26 mM) as the reducing system, and glucose (5 mM) and glucose oxidase (16 units/mL) to scavenge residual O2 in the 1.5-mL reaction chamber. Then, 30 μL of 10 μM cytochrome bcc-aa3 (200 nM), pre-reduced with DTT (5 mM) and MD (0.33 mM) in the presence of catalase (520 units/mL) and SOD (60 units/mL), was added to the chamber. The NO decay observed before addition of the supercomplex is likely due to the reaction with the reductants. Addition of pre-reduced cytochrome bcc-aa3 accelerates the NO decay due to catalytic NO consumption. The initial fast drop in the NO concentration observed just after addition of the supercomplex is likely due at least partly to NO binding to cytochrome bcc-aa3. Following NO depletion from solution, two aliquots of 4.2 µM NO (3.5 μL of 1.8 mM NO each) were sequentially added and catalytic NO consumption further observed. The activity of bcc-aa3 at [NO] = 8.4 µM was 2.8 ± 0.5 mol NO (mol bcc-aa3)−1 min−1. The calculated activity is expressed as mean ± standard deviation.
3. Discussion
Clinical resistance of M. tuberculosis antibiotics represents an increasing threat to public health globally, preventing effective treatments against tuberculosis, one of the top 10 causes of death worldwide [33]. Interestingly, a correlation between resistance to first-line anti-TB drugs and reduced NO susceptibility has been found in clinical strains of M. tuberculosis [34]. Understanding how this bacterial pathogen resists the host immune system attack and identifying novel drug targets are therefore crucial for the successful cure of this infectious disease. The respiratory complexes of M. tuberculosis, including the bcc-aa3 supercomplex, are attractive targets for the development of new antitubercular agents.
Kim et al. [30] recently succeeded in the purification of an untagged hybrid bcc-aa3 supercomplex with M. tuberculosis cytochrome bcc and M. smegmatis cytochrome aa3. The supercomplex is stable and remains active during protein extraction and purification in the presence of the non-ionic detergent dodecyl-β-d-maltoside (DDM) [30]. This allowed us to use these bcc-aa3 preparations for functional studies. Assays were meant to test whether bcc-aa3 could contribute to mechanisms subverting, suppressing or evading the host immune response. In particular, we sought to gain insight into the reactivity of bcc-aa3 with NO, a harmful species produced by the host as part of the immune response to fight microbial infections. To this purpose, we investigated the effect of NO on the O2 reductase activity of bcc-aa3 and the ability of the supercomplex to metabolize NO under different conditions.
First, we assayed whether the O2-reductase activity of bcc-aa3 is affected by NO, which is known to bind with high affinity to the active site of respiratory terminal oxidases, resulting in a potent inhibition (reviewed in [35,36,37,38,39,40,41]). Surprisingly, we found that the mycobacterial supercomplex is resistant to NO inhibition, even at the highest concentration of the gas tested (30 µM). Such insensitivity to NO is unprecedented for a terminal oxidase. Indeed, the oxidases thus far assayed for NO inhibition, such as mitochondrial cytochrome c oxidase [42,43,44], the aa3 oxidase from Rhodobacter sphaeroides [45], the cbb3 oxidases from Rhodobacter sphaeroides and Vibrio cholerae [45] and the cytochromes bd from Escherichia coli and Azotobacter vinelandii [32,41,46], were found to be strongly and reversibly inhibited by NO with IC50 values in the nanomolar range.
Figure 1 (blue trace) shows that the addition of 3.6 µM NO has no detectable effect on the O2 consumption by bcc-aa3 in the presence of excess respiratory substrates. At the highest concentration of NO added, there is only a small decline in the O2 consumption followed by a relatively fast (≈1–2 min after the NO addition) and complete recovery of enzyme activity. The activity recovery occurs as NO disappears from solution (Figure 1, red trace). Careful comparison of the NO traces acquired in the presence and absence of bcc-aa3 shows that the enzyme drastically accelerates the kinetics of NO decay (Figure 2A). The maximum NO-consuming activity of bcc-aa3 detected at 30 µM NO and ≈130 µM O2 was ≈303 mol NO (mol bcc-aa3)−1 min−1. Thus, one can conclude that the apparent resistance of the supercomplex to NO inhibition results from its ability to directly metabolize NO, likely to nitrite (see below). NO consumption by the supercomplex requires turnover conditions, as bcc-aa3 in the same aerobic buffer but without the DTT/MD reducing system proved to be unable to metabolize NO (Figure 2B).
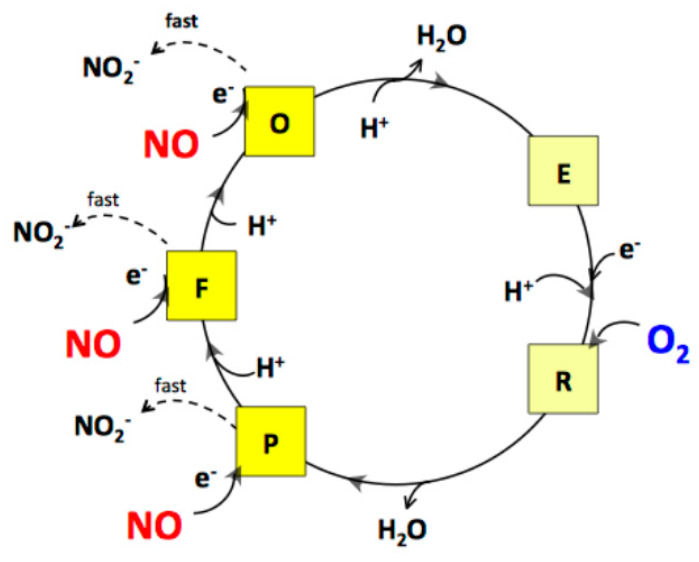
The rates of NO and O2 consumption catalysed by bcc-aa3 are proportional, yielding a NO/O2 ratio of 2.65 ± 0.19 (Figure 3). In the bovine mitochondrial cytochrome c oxidase (mtCcOX), unlike O2 that binds only to the fully reduced (R) active site of the enzyme, NO was previously reported to bind and react also with the catalytic intermediates O (with the fully oxidized haem a3-CuB site), P (peroxy) and F (ferryl), each according to a 1:1 stoichiometry [47]. The reaction with the intermediates O, P and F is accompanied by injection of 1 electron into the enzyme from NO [47]. Donation of such an electron, while causing NO oxidation to nitrite, leads to conversion of an O2 intermediate into the succeeding one along the catalytic cycle (Figure 5). By reacting with NO, intermediates O, P and F are therefore respectively converted into the intermediates E (with the single electron-reduced haem a3-CuB site), F and O [47]. In mtCcOX, the nitrite formed from NO binds with relatively high affinity to the oxidized haem a3 in the active site, impairing the Fe reduction and, thus, its ability to react with O2. Consequently, the reaction with O2 stops. At lower reductive pressure on the enzyme, when intermediates O, P, F are more populated at steady-state, this inhibition pathway leading to the haem a3 Fe3+-NO2− adduct prevails over the so-called nitrosyl pathway, that is instead favoured at higher reductive pressure and occurs via NO binding to reduced haem a3, leading to formation of a Fe2+-NO adduct (reviewed in [48]). Under the assumption that the mycobacterial bcc-aa3 catalytic intermediates share a similar reactivity with NO, the observed >1 NO/O2 stoichiometry indicates that in the case of the mycobacterial enzyme NO can also react with more than one catalytic intermediate, but possibly with the notable difference (compared to the mitochondrial enzyme) that the newly formed nitrite does not bind to the haem a3 moiety with high affinity and, therefore, is ejected into the bulk phase from the enzyme without impairing its catalytic O2-reductase activity (Figure 5).
Figure 5.
Proposed mechanism for the interaction of NO with the catalytic intermediates of the mycobacterial bcc-aa3 supercomplex under aerobic conditions. As shown for mtCcOX, by reacting with the intermediates O, P and F, NO is suggested to lead to formation of nitrite. However, at variance from the mammalian enzyme, in the case of the mycobacterial supercomplex, the observed NO/O2 stoichiometry = 2.65 ± 0.19 suggests that nitrite does not bind with high affinity to oxidized haem a3 and is rapidly ejected from the enzyme, without affecting its catalytic activity.
Interestingly, bcc-aa3 was found to be capable of consuming NO also under anaerobic reducing conditions (Figure 4), with an estimated NO-reductase activity of 2.8 ± 0.5 mol NO (mol bcc-aa3)−1 min−1 (at [NO] = 8.4 µM and 25 °C), most likely leading to nitrous oxide (N2O) as the reaction product, similarly to other terminal oxidases. Indeed, the ability to catalyze the reduction of NO to N2O was previously documented under similar experimental conditions and setup (amperometric measurements under anaerobic reducing conditions at [NO] = 5–10 µM) in several haemcopper oxidases, namely in the ba3 and caa3 oxidases from Thermus thermophilus [49], the cbb3 oxidases from Pseudomonas stutzeri [50] and Rhodobacter sphaeroides [51], and the bo3 oxidase from Escherichia coli [52], but not in the mitochondrial beef heart cytochrome c oxidase [49,53] or the cytochromes bd from Escherichia coli and Azotobacter vinelandii [46]. Notably, the NO reductase activities of all terminal oxidases tested so far are significantly lower than that displayed by bacterial NORs. For instance, NOR purified from Paracoccus denitrificans shows the activity as high as ≈1400 mol NO (mol enzyme)−1 min−1 [50] (see also [54,55]).
Taken together, the novel findings reported here, particularly the high resistance to NO inhibition of the mycobacterial bcc-aa3 supercomplex and its unexpected NO-metabolizing activity, suggest a role for this enzymatic complex in the defence against NO, a major effector in the immune response to M. tuberculosis infection [56]. The resistance of bcc-aa3 to NO inhibition may help the pathogen survive acute NO stress before transcriptional and late phase responses take place [57]. All in all, this study reveals unexpected properties of the mycobacterial bcc-aa3 supercomplex, further pointing to this protein as a valuable drug target.
4. Materials and Methods
4.1. Supercomplex Purification and Concentration Determination
The cytochrome bcc-aa3 supercomplex was purified from a strain of M. smegmatis expressing the hybrid supercomplex, as described by Kim et al. [30] with modifications. Cells were collected by centrifugation. Membranes were prepared by sonication of cells in 50 mM K-Pi pH 7.4, 0.5 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF), followed by differential centrifugation, resuspended to 35 g/L protein in the same buffer, and frozen. For extraction, membranes were treated with DDM at a concentration of 10 g/L protein, 5 g/L detergent in 50 mM KPi pH 7.4, 100 mM NaCl, 0.1 mM PMSF. Insoluble material was separated from the extract by centrifugation. In this case the initial extract contained little soluble supercomplex but did have a large amount of soluble protein containing flavin and cytochrome b557. Therefore, the pellet was resuspended in buffer without detergent, protein content was measured, and the material was re-extracted as before but with 1 g/(g protein) of added DDM. This extract was diluted with ¼ volume of water, applied to DEAE-Sepharose CL6B column, and eluted with a gradient from 100 to 500 mM NaCl in 5-mM KPi, 0.5 mM EDTA, and 0.1 g/L DDM. Fractions containing bcc-caa3 were identified by UV-visible spectra and haem-stained gels [58,59]. The pooled fractions were concentrated by ultrafiltration and layered on glycerol density step-gradients with 60, 40, and 20% w/w glycerol in 20 mM K-MPOS, 100 mM NaCl, and 0.5 mM EDTA, pH 7.2. The gradients were centrifuged at 30,000 rpm in the Beckman SW-32.Ti rotor for 72 h. Fractions containing bcc-aa3 were pooled and chromatographed on Sepharose CL-6B size-exclusion medium equilibrated with 20 mM K MOPS, 100 mM NaCl, 0.5 mM EDTA, and 0.1 g/L DDM (pH 7.2). Fractions containing pure supercomplex, as assessed by SDS-PAGE (Supplementary Figure S2), were pooled and concentrated. Concentration of the supercomplex was estimated using the extinction coefficient for the oxidized form Δε413-371 nm = 485 mM−1 cm−1.
4.2. Catalytic Assays
Oxygraphic measurements were performed at 25 °C with a high-resolution respirometer (Oxygraph-2k, Oroboros Instruments) equipped with two 1.5-mL gas-tight thermostated chambers. The assays were carried out at 25 °C in 20 mM K/MOPS buffer (pH 7.3) containing 100 mM NaCl, 0.5 mM EDTA and 0.01% dodecyl-β-d-maltoside. Changes in the O2 and NO levels in solution were recorded in real time simultaneously. The NO concentration was recorded with the aid of a NO-selective electrode (World Precision Instruments, Sarasota FL, USA). The electrode was calibrated by sequential additions of NO from a stock solution, prepared by equilibrating degassed water at room temperature with pure NO (Air Liquide, Paris, France) at 1 atm. The O2 reductase activity of purified mycobacterial cytochrome bcc-aa3 supercomplex was measured with an excess of the reductants DTT (5 mM) and MD (0.26 mM). Mycobacterial bcc-aa3 preparations are indeed active with menadiol as substrate, which has a higher solubility in water solutions compared to menaquinol [60]. DTT was used to keep MD in the reduced state during the oxygraphic assays (Supplementary Figure S3). In the presence of DDT and MD, 520 units/mL catalase and 60 units/mL SOD were added to the solution to reduce the non-enzymatic background oxygen consumption.
4.3. Data Analysis
The catalytic NO-consuming activity was obtained by subtracting from the rate of NO consumption measured after addition of bcc-aa3 from that of the non-enzymatic NO consumption. The latter was obtained by analyzing either the same trace prior to the enzyme addition (anaerobic reducing conditions with excess DTT and MD) or a trace independently collected under identical experimental conditions in the absence of bcc-aa3 (aerobic conditions with excess DTT and MD). Origin software (OriginLab Corporation) was used for data analysis and figure preparation.
Acknowledgments
The strain of M. smegmatis expressing the supercomplex was provided by Jichan Jang of the Institut Pasteur, Korea.
Abbreviations
| NO | nitric oxide |
| TB | tuberculosis |
| iNOS | inducible NO synthase |
| NOD | NO-dioxygenase |
| NOR | NO reductase |
| flavoHb | flavohaemoglobin |
| trHb | truncated haemoglobin |
| SOD | superoxide dismutase |
| DTT | dithiothreitol |
| MD | menadione |
| mtCcOX | bovine mitochondrial cytochrome c oxidase |
| PMSF | phenylmethylsulfonyl fluoride |
| DDM | dodecyl β-d-maltoside |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/22/8521/s1. Figure S1. Control NO trace acquired under anaerobic conditions in the absence of the supercomplex but in the presence of DTT/MD, Figure S2. SDS-PAGE analysis of the purified chimeric bcc-aa3 supercomplex composed of M. tuberculosis cytochrome bcc and M. smegmatis aa3-type terminal oxidase Figure S3. Scheme illustrating the electron flow from DTT to O2 via MD and the cytochrome bcc-aa3 supercomplex
Author Contributions
Conceptualization, E.F., A.G. and V.B.B.; data curation, V.B.B. and E.F.; supercomplex purification, E.A.B. and L.-s.H.; writing—original draft preparation, V.B.B., E.F. and A.G.; data discussion, writing—review and editing, V.B.B., E.F., A.G., E.A.B and L.-s.H.; funding acquisition, V.B.B., E.F. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Russian Foundation for Basic Research—http://www.rfbr.ru/rffi/eng—research project № 19-04-00094 (to V.B.B.), by Ministero dell’Istruzione, della Ricerca e dell’Università of Italy, PNR-CNR Aging Program 2012–2014 (to A.G.) and by Sapienza grant № RM11715C7F529A09 (to E.F.)
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jamaati H., Mortaz E., Pajouhi Z., Folkerts G., Movassaghi M., Moloudizargari M., Adcock I.M., Garssen J. Nitric oxide in the pathogenesis and treatment of tuberculosis. Front. Microbiol. 2017;8:2008. doi: 10.3389/fmicb.2017.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Queval C.J., Song O.-R., Deboosère N., Delorme V., Debrie A.-S., Iantomasi R., Veyron-Churlet R., Jouny S., Redhage K., Deloison G., et al. STAT3 represses nitric oxide synthesis in human macrophages upon Mycobacterium tuberculosis infection. Sci. Rep. 2016;6:29297. doi: 10.1038/srep29297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat K.H., Srivastava S., Kotturu S.K., Ghosh S., Mukhopadhyay S. The PPE2 protein of Mycobacterium tuberculosis translocates to host nucleus and inhibits nitric oxide production. Sci. Rep. 2017;7:39706. doi: 10.1038/srep39706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darwin K.H., Ehrt S., Gutierrez-Ramos J.C., Weich N., Nathan C.F. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 5.Pathania R., Navani N.K., Gardner A.M., Gardner P.R., Dikshit K.L. Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN in Escherichia coli. Mol. Microbiol. 2002;45:1303–1314. doi: 10.1046/j.1365-2958.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 6.Arya S., Sethi D., Singh S., Hade M.D., Singh V., Raju P., Chodisetti S.B., Verma D., Varshney G.C., Agrewala J.N., et al. Truncated hemoglobin, HbN, is post-translationally modified in Mycobacterium tuberculosis and modulates host-pathogen interactions during intracellular infection. J. Biol. Chem. 2013;288:29987–29999. doi: 10.1074/jbc.M113.507301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakur N., Gupta S., Hade M.D., Dikshit K.L. Type I flavohemoglobin of mycobacterium smegmatis is a functional nitric oxide dioxygenase. IUBMB Life. 2014;66:396–404. doi: 10.1002/iub.1275. [DOI] [PubMed] [Google Scholar]
- 8.Sarti P., Forte E., Giuffrè A., Mastronicola D., Magnifico M.C., Arese M. The chemical interplay between nitric oxide and mitochondrial cytochrome c oxidase: Reactions, effectors and pathophysiology. Int. J. Cell Biol. 2012;2012:1–11. doi: 10.1155/2012/571067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forte E., Borisov V.B., Vicente J.B., Giuffrè A. Cytochrome bd and gaseous ligands in bacterial physiology. Adv. Microb. Physiol. 2017;71:171–234. doi: 10.1016/bs.ampbs.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Cook G.M., Hards K., Vilchèze C., Hartman T., Berney M. Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.MGM2-0015-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook G.M., Greening C., Hards K., Berney M. Energetics of pathogenic bacteria and opportunities for drug development. Adv. Microb. Physiol. 2014;65:1–62. doi: 10.1016/bs.ampbs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Bald D., Villellas C., Lu P., Koul A. Targeting energy metabolism in Mycobacterium tuberculosis, a new paradigm in antimycobacterial drug discovery. MBio. 2017;8:e00272-17. doi: 10.1128/mBio.00272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook G.M., Hards K., Dunn E., Heikal A., Nakatani Y., Greening C., Crick D.C., Fontes F.L., Pethe K., Hasenoehrl E., et al. Oxidative phosphorylation as a target space for tuberculosis: Success, caution, and future directions. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.TBTB2-0014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hards K., Cook G.M. Targeting bacterial energetics to produce new antimicrobials. Drug Resist. Updat. 2018;36:1–12. doi: 10.1016/j.drup.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Megehee J.A., Hosler J.P., Lundrigan M.D. Evidence for a cytochrome bcc–aa3 interaction in the respiratory chain of Mycobacterium smegmatis. Microbiology. 2006;152:823–829. doi: 10.1099/mic.0.28723-0. [DOI] [PubMed] [Google Scholar]
- 16.Wiseman B., Nitharwal R.G., Fedotovskaya O., Schäfer J., Guo H., Kuang Q., Benlekbir S., Sjöstrand D., Ädelroth P., Rubinstein J.L., et al. Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nat. Struct. Mol. Biol. 2018;25:1128–1136. doi: 10.1038/s41594-018-0160-3. [DOI] [PubMed] [Google Scholar]
- 17.Gong H., Li J., Xu A., Tang Y., Ji W., Gao R., Wang S., Yu L., Tian C., Li J., et al. An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science. 2018;362 doi: 10.1126/science.aat8923. [DOI] [PubMed] [Google Scholar]
- 18.Suthammarak W., Somerlot B.H., Opheim E., Sedensky M.M., Morgan P.G. Novel interactions between mitochondrial superoxide dismutases and the electron transport chain. Aging Cell. 2013;12:1132–1140. doi: 10.1111/acel.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira M.M., Gomes C.M., Teixeira M. Plasticity of proton pathways in haem-copper oxygen reductases. FEBS Lett. 2002;522:14–18. doi: 10.1016/S0014-5793(02)02920-4. [DOI] [PubMed] [Google Scholar]
- 20.Wikström M., Krab K., Sharma V. Oxygen activation and energy conservation by cytochrome c oxidase. Chem. Rev. 2018;118:2469–2490. doi: 10.1021/acs.chemrev.7b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarti P., Forte E., Mastronicola D., Giuffrè A., Arese M. Cytochrome c oxidase and nitric oxide in action: Molecular mechanisms and pathophysiological implications. Biochim. Biophys. Acta. 2012;1817:610–619. doi: 10.1016/j.bbabio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Siletsky S.A., Borisov V.B., Mamedov M.D. Photosystem II and terminal respiratory oxidases molecular machines operating in opposite directions. Front. Biosci. 2017;22:1379–1426. doi: 10.2741/4550. [DOI] [PubMed] [Google Scholar]
- 23.von Ballmoos C., Ädelroth P., Gennis R.B., Brzezinski P. Proton transfer in ba3 cytochrome c oxidase from Thermus thermophilus. Biochim. Biophys. Acta Bioenerg. 2012;1817:650–657. doi: 10.1016/j.bbabio.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Melin F., Xie H., Meyer T., Ahn Y.O., Gennis R.B., Michel H., Hellwig P. The unusual redox properties of C-type oxidases. Biochim. Biophys. Acta Bioenerg. 2016;1857:1892–1899. doi: 10.1016/j.bbabio.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Rich P.R. Mitochondrial cytochrome c oxidase: Catalysis, coupling and controversies. Biochem. Soc. Trans. 2017;45:813–829. doi: 10.1042/BST20160139. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa S., Shimada A. Reaction mechanism of cytochrome c oxidase. Chem. Rev. 2015;115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 27.Papa S., Capitanio G., Papa F. The mechanism of coupling between oxido-reduction and proton translocation in respiratory chain enzymes. Biol. Rev. Camb. Philos. Soc. 2018;93:322–349. doi: 10.1111/brv.12347. [DOI] [PubMed] [Google Scholar]
- 28.Stuchebrukhov A., Schäfer J., Berg J., Brzezinski P. Kinetic advantage of forming respiratory supercomplexes. Biochim. Biophys. Acta Bioenerg. 2020;1861:148193. doi: 10.1016/j.bbabio.2020.148193. [DOI] [PubMed] [Google Scholar]
- 29.Matsoso L.G., Kana B.D., Crellin P.K., Lea-Smith D.J., Pelosi A., Powell D.R., Dawes S.S., Rubin H., Coppel R.L., Mizrahi V. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J. Bacteriol. 2005;187:6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M.-S., Jang J., Ab Rahman N.B., Pethe K., Berry E.A., Huang L.-S. Isolation and characterization of a hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis cytochrome bcc and Mycobacterium smegmatis cytochrome aa3. J. Biol. Chem. 2015;290:14350–14360. doi: 10.1074/jbc.M114.624312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonini E., Brunori M., Colosimo A., Greenwood C., Wilson M.T. Oxygen “pulsed” cytochrome c oxidase: Functional properties and catalytic relevance. Proc. Natl. Acad. Sci. USA. 1977;74:3128–3132. doi: 10.1073/pnas.74.8.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borisov V.B., Forte E., Siletsky S.A., Sarti P., Giuffrè A. Cytochrome bd from Escherichia coli catalyzes peroxynitrite decomposition. Biochim. Biophys. Acta Bioenerg. 2015;1847:182–188. doi: 10.1016/j.bbabio.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization . Global Tuberculosis Report 2019. World Health Organization; Geneva, Switzerland: 2019. [Google Scholar]
- 34.Idh J., Mekonnen M., Abate E., Wedajo W., Werngren J., Ängeby K., Lerm M., Elias D., Sundqvist T., Aseffa A., et al. first-line anti-TB drugs is associated with reduced nitric oxide susceptibility in Mycobacterium tuberculosis. PLoS ONE. 2012;7:e39891. doi: 10.1371/journal.pone.0039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper C.E. Nitric oxide and cytochrome oxidase: Substrate, inhibitor or effector? Trends Biochem. Sci. 2002;27:33–39. doi: 10.1016/S0968-0004(01)02035-7. [DOI] [PubMed] [Google Scholar]
- 36.Sarti P., Arese M., Bacchi A., Barone M.C., Forte E., Mastronicola D., Brunori M., Giuffrè A. Nitric oxide and mitochondrial complex IV. IUBMB Life. 2003;55:605–611. doi: 10.1080/15216540310001628726. [DOI] [PubMed] [Google Scholar]
- 37.Forte E., Borisov V.B., Konstantinov A.A., Brunori M., Giuffrè A., Sarti P. Cytochrome bd, a key oxidase in bacterial survival and tolerance to nitrosative stress. Ital. J. Biochem. 2007;56:265–269. [PubMed] [Google Scholar]
- 38.Borisov V.B., Gennis R.B., Hemp J., Verkhovsky M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta Bioenerg. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giuffrè A., Borisov V.B., Mastronicola D., Sarti P., Forte E. Cytochrome bd oxidase and nitric oxide: From reaction mechanisms to bacterial physiology. FEBS Lett. 2012;586:622–629. doi: 10.1016/j.febslet.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 40.Giuffrè A., Borisov V.B., Arese M., Sarti P., Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim. Biophys. Acta Bioenerg. 2014;1837:1178–1187. doi: 10.1016/j.bbabio.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Borisov V.B., Forte E., Siletsky S.A., Arese M., Davletshin A.I., Sarti P., Giuffrè A. Cytochrome bd protects bacteria against oxidative and nitrosative stress: A potential target for next-generation antimicrobial agents. Biochemistry. 2015;80:565–575. doi: 10.1134/S0006297915050077. [DOI] [PubMed] [Google Scholar]
- 42.Brown G.C., Cooper C.E. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 43.Torres J., Darley-Usmar V., Wilson M.T. Inhibition of cytochrome c oxidase in turnover by nitric oxide: Mechanism and implications for control of respiration. Biochem. J. 1995;312:169–173. doi: 10.1042/bj3120169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giuffrè A., Sarti P., D’Itri E., Buse G., Soulimane T., Brunori M. On the mechanism of inhibition of cytochrome c oxidase by nitric oxide. J. Biol. Chem. 1996;271:33404–33408. doi: 10.1074/jbc.271.52.33404. [DOI] [PubMed] [Google Scholar]
- 45.Arjona D., Wikström M., Ädelroth P. Nitric oxide is a potent inhibitor of the cbb3-type heme-copper oxidases. FEBS Lett. 2015;589:1214–1218. doi: 10.1016/j.febslet.2015.03.033. [DOI] [PubMed] [Google Scholar]
- 46.Borisov V.B., Forte E., Konstantinov A.A., Poole R.K., Sarti P., Giuffrè A. Interaction of the bacterial terminal oxidase cytochrome bd with nitric oxide. FEBS Lett. 2004;576:201–204. doi: 10.1016/j.febslet.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 47.Giuffrè A., Barone M.C., Mastronicola D., D’Itri E., Sarti P., Brunori M. Reaction of nitric oxide with the turnover intermediates of cytochrome c oxidase: Reaction Pathway and Functional Effects. Biochemistry. 2000;39:15446–15453. doi: 10.1021/bi000447k. [DOI] [PubMed] [Google Scholar]
- 48.Brunori M., Forte E., Arese M., Mastronicola D., Giuffrè A., Sarti P. Nitric oxide and the respiratory enzyme. Biochim. Biophys. Acta Bioenerg. 2006;1757:1144–1154. doi: 10.1016/j.bbabio.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 49.Giuffrè A., Stubauer G., Sarti P., Brunori M., Zumft W.G., Buse G., Soulimane T. The heme-copper oxidases of Thermus thermophilus catalyze the reduction of nitric oxide: Evolutionary implications. Proc. Natl. Acad. Sci. USA. 1999;96:14718–14723. doi: 10.1073/pnas.96.26.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forte E., Urbani A., Saraste M., Sarti P., Brunori M., Giuffrè A. The cytochrome cbb 3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur. J. Biochem. 2001;268:6486–6491. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 51.Huang Y., Reimann J., Lepp H., Drici N., Ädelroth P. Vectorial proton transfer coupled to reduction of O2 and NO by a heme-copper oxidase. Proc. Natl. Acad. Sci. USA. 2008;105:20257–20262. doi: 10.1073/pnas.0805429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler C., Forte E., Scandurra F.M., Arese M., Giuffrè A., Greenwood C., Sarti P. Cytochrome bo(3) from Escherichia coli: The binding and turnover of nitric oxide. Biochem. Biophys. Res. Commun. 2002;296:1272–1278. doi: 10.1016/S0006-291X(02)02074-0. [DOI] [PubMed] [Google Scholar]
- 53.Stubauer G., Giuffrè A., Brunori M., Sarti P. Cytochrome c oxidase does not catalyze the anaerobic reduction of NO. Biochem. Biophys. Res. Commun. 1998;245:459–465. doi: 10.1006/bbrc.1998.8457. [DOI] [PubMed] [Google Scholar]
- 54.Girsch P., De Vries S. Purification and initial kinetic and spectroscopic characterization of NO reductase from Paracoccus denitrificans. Biochim. Biophys. Acta Bioenerg. 1997;1318:202–216. doi: 10.1016/S0005-2728(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 55.Hendriks J., Warne A., Gohlke U., Haltia T., Ludovici C., Lübben M., Saraste M. The active site of the bacterial nitric oxide reductase is a dinuclear iron center. Biochemistry. 1998;37:13102–13109. doi: 10.1021/bi980943x. [DOI] [PubMed] [Google Scholar]
- 56.Singh R., Manjunatha U., Boshoff H.I.M., Ha Y.H., Niyomrattanakit P., Ledwidge R., Dowd C.S., Lee I.Y., Kim P., Zhang L., et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cortes T., Schubert O.T., Banaei-Esfahani A., Collins B.C., Aebersold R., Young D.B. Delayed effects of transcriptional responses in Mycobacterium tuberculosis exposed to nitric oxide suggest other mechanisms involved in survival. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-08306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas P.E., Ryan D., Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 59.Francis R.T., Jr., Becker R.R. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 1984;136:509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 60.Lemma E., Schägger H., Kröger A. The menaquinol oxidase of Bacillus subtilis W23. Arch. Microbiol. 1993;159:574–578. doi: 10.1007/BF00249037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.