Abstract
Liquid biopsy is a rapidly emerging field due to an increasing number of oncogenic drivers and a better understanding of resistance mechanisms to targeted therapies in non-small cell lung cancer (NSCLC). The sensitivity of the most widely used blood-based assays is, however, limited in particular in cases of low tumor volume where shed of tumor-derived material can be limited. A negative result thus requires biopsy confirmation using minimally invasive sampling procedures that can result in small specimens, which are often not suitable for genotyping. Liquid biopsy is not limited to plasma, and tumor DNA circulating in other body fluids such as urine, pleural fluid, cerebrospinal fluid, or cytology specimen-derived supernatant can be exploited. In comparison to cell blocks, these fluids in close contact to the tumor may contain a more abundant and less analytically demanding tumor DNA. In this review, we discuss the potential applications of circulating tumor DNA derived from cytology samples in NSCLC, from early stage (screening, nodule characterization) to metastatic disease.
Keywords: genotyping, targeted therapy, screening, urine, fine-needle aspiration, cerebrospinal fluid, liquid biopsy, circulating tumor DNA, lung cancer
1. Introduction
Mechanisms of oncogenesis in lung cancer have been largely deciphered over the past 20 years. Lung adenocarcinoma, unlike other histological subtypes, can now be considered as a cluster of discrete molecular subtypes, the majority being defined by a single alteration of an oncogenic driver. In addition to EGFR and KRAS mutations and ALK rearrangements, new molecular targets, such as BRAF, MET, or HER2 mutations, and ROS1, NTRK, and RET rearrangements, have been recently highlighted. Multiplex genotyping and high-throughput genomic profiling by next-generation sequencing (NGS) is thus increasingly refining molecular diagnoses [1]. The evolution of tumors bearing a molecular alteration is usually dependent on a single mechanism following the principle of oncogenic addiction, which has been described as the dependence of tumor cells on the specific activity of an activated oncogene [2]. These tumors respond to genotype-directed therapies but inevitably progress usually through the emergence of genomic alterations that confer resistance to first-line targeted agents, requiring iterative assessment of the molecular profile of the tumor. In parallel, sampling methods are also evolving. There is currently a paradox between the need to obtain a significant amount of tumor tissue to test an increasing number of biomarkers and the development of bronchoscopic minimally invasive techniques, resulting in small tissue samples with limited amounts of DNA [1]. Bronchoscopy currently constitutes the preferred approach for tumor sampling as it is less invasive than radio-guided biopsies [3]. It is however frequent (10 to 20% of cases) [4,5] that these cytologic samples, when only the cell block is considered, are rejected for genotyping after time consuming processing steps because of limited tumor content.
Liquid biopsy, and in particular plasma circulating tumor DNA (ctDNA) genotyping to detect driver and resistance mutations, is a rapidly emerging field in non-small cell lung cancer (NSCLC) [6]. While tissue only offers a snapshot of the tumor at a given time and location, liquid biopsy has the potential to overcome both spatial and temporal tumor heterogeneity and can non-invasively interrogate the molecular landscape of a tumor, taking into account different clones present within all metastatic sites, and can follow subclonal evolution through iterative blood draws. However, the most sensitive cell-free DNA (cfDNA) genotyping platforms still have a sensitivity of only 70–80% for advanced diseases [7,8,9] and below 50% for early stages [6,10], such that a negative result requires biopsy confirmation. This poses a clinical challenge because false negative plasma genotyping is usually associated with limited metastatic spread and lower tumor burden [9], and biopsy of these patients may be more challenging. Blood sampling strategies at the tumor draining vein could be a solution to increase recovery rate, however such a strategy has so far only been applied to circulating tumor cell (CTC) detection and necessitates additional steps during surgery [11,12].
However, liquid biopsy is not limited to plasma. Tumor nucleic acids floating (alone or in exosomes) in other body fluids such as urine, pleural fluid, cerebrospinal fluid, or cytology specimen-derived supernatant can be exploited.
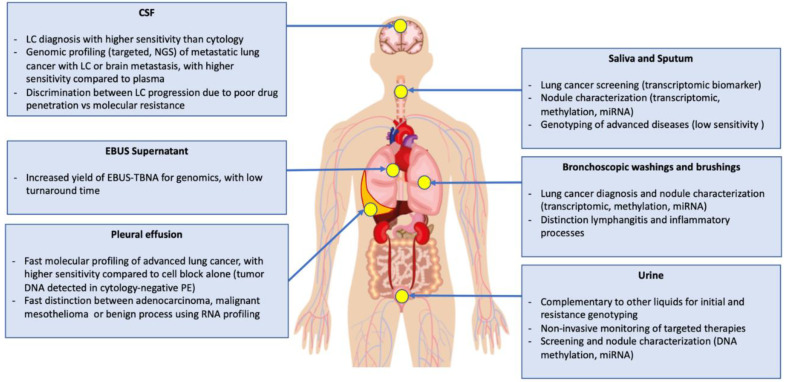
A fluid close to a tumor/metastasis may contain more abundant and less diluted tumor DNA (tDNA) with higher concentrations/mutant allelic fraction (MAF), derived from active secretion and/or cell death, compared to what can be isolated from the bloodstream. Non-invasive collection of multiple fluids could be complementary and increase sensitivity compared to plasma alone and may better characterize spatial tumor heterogeneity. We report herein the main “non-blood” biological fluids that have been studied for liquid biopsy tests in the field of NSCLC and their potential advantages (summarized in Table 1 and Figure 1). We studied the main publications reported in journals indexed on PubMed (using the terms: Pleural effusion cfDNA, urine cfDNA, CSF cfDNA, ascitic fluid cfDNA, supernatant cfDNA, brushings cfDNA, washings cfDNA) on the topic in the past years.
Table 1.
Diagnostic accuracy, advantages and potential future applications of free-floating DNA derived from body fluids.
| Sample | State of the Art Advantages | Challenges Potential Future Applications |
|---|---|---|
| Plasma [5,6,7,8,9] | Non-invasive, rapid Sensitivity 70 to 80%, below 50% for stage I Early detection of acquired resistance (e.g., EGFR T790M) Addresses challenges of tumour heterogeneity |
Integration of plasma NGS in routine clinical care for initial and resistance genotyping Minimal Residual Disease/Follow up Improve sensitivity in early diagnosis/Screening |
| FNA supernatant [12,13,14,15,16,17] |
Rich and immediate source of tumor DNA Increases the yield of a low tumour content biospecimen for genomics and maybe diagnosis |
Prospective validation of correlation with tissue, improvement in turnaround time and genomics feasibility Ultimately use supernatant for genomics and save cell block for pathology Improve the yield of bronchoscopy for nodule characterization |
| Brushing washing [18,19,20,21,22] |
Increases the yield for cancer diagnosis Sensitivity of 88% for EGFR detection Prescence of lung cancer biomarkers |
Fast discrimination between inflammatory pneumonitis (after radiation therapy, under immune therapy) and carcinomatous lymphangitis Lung cancer screening and nodule characterization (cfDNA, met-cfDNA, miRNAs) |
| Pleural and ascitic fluids [23,24,25,26,27,28,29,30,31,32,33,34] | Minimally invasive Good ratio tDNA/fDNA Reliable correlation with cell pellet Low turnaround time compared to tissue Increases the yield of low tumour content biospecimens |
Integration of PE-ctDNA NGS in routine for initial and resistance genotyping Fast discrimination between benign and malignant PE (e.g., after surgery) |
| Urine [35,36,37,38,39,40] | Non-invasive, easy to collect Sensitivity around 70% Low turnaround time Can address tumour heterogeneity if trans-renal clearance of cfDNA (trDNA) Complimentary with plasma and tissue (T790M) |
NGS: Wider range of genotype coverage Monitoring of response under targeted therapy Lung cancer screening and nodule characterization (cfDNA, met-cfDNA, miRNAs) |
| Saliva/sputum [41,42,43,44,45] | Non-invasive, easy to collect Potential for early lung cancer detection (miRNA, methylation) low sensitivity of EGFR mutation detection (42% for EGFR in sputum) |
Lung cancer screening and nodule characterization (cfDNA, met-cfDNA, miRNAs) |
| Cerebrospinal fluid [46,47,48,49,50,51,52] | Minimally invasive High ctDNA MAF Increases the yield of a low tumour content biospecimen: diagnosis of leptomeningeal carcinomatosis (LC) in pauci or acellular samples |
Discrimination between relapse and post-radiation necrosis after radiation therapy for brain metastasis Resistance analysis in CNS progression and distinction with poor drug penetration in CNS Monitoring of response for metastases and LC |
BW: bronchial washing, PE: pleural effusion, CSF: cerebrospinal fluid, LC: leptomeningeal carcinomatosis, EBUS-TBNA: endobronchial ultrasonography transbronchial needle aspiration, NGS: next generation sequencing, cfDNA: cell free DNA, ctDNA: circulating tumor DNA, CNS: Central Nervous System, met-cfDNA: methylated cfDNA, trDNA: transrenal DNA.
Figure 1.
Easily available body fluids and their potential applications. BW: bronchial washing, PE: pleural effusion, CSF: cerebrospinal fluid, LC: leptomeningeal carcinomatosis, EBUS-TBNA: endobronchial ultrasonography transbronchial needle aspiration, NGS: next generation sequencing.
2. Fine-Needle Aspiration (FNA) Supernatant
The supernatant of fine-needle aspiration (FNA) specimens (Computed tomography (CT)-guided or EBUS-TBNA (endobronchial ultrasonography transbronchial needle aspiration) in the context of NSCLC) is usually discarded. However, it is now clear that this supernatant is rich in cell free DNA. In patients with pancreatic cancer, the yield of FNA supernatant matches, and in a subset of patients, exceed cellblock [13]. Roy-Chowdhuri et al. reported in FNA supernatant mean and median DNA yields of 445 ng and 176.4 ng (i.e., much higher than what is usually found in plasma), respectively. Tumor-associated mutations were detected (via NGS) in all 25 FNAs from patients with solid tumors and none of the 10 benign controls [14].
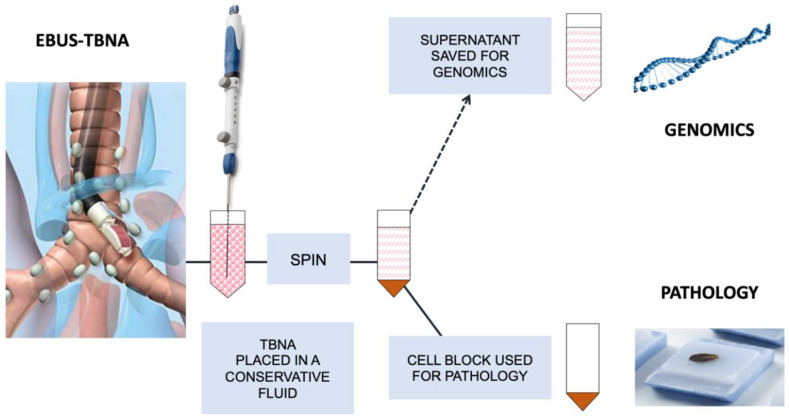
In the context of lung cancer, several studies have also demonstrated the feasibility and accuracy of NGS-based genotyping of cfDNA derived from FNA supernatant [14,15,16,53]. In particular for EBUS-TBNA, in which a specimen of low tumor cell content is rejected from genotyping in up to 20% of cases [17], mutations of interest could be detected in all supernatant-derived cfDNA from a 17 patient cohort [15]. Genomics is usually performed after conventional time-consuming diagnostic steps on the cell block (e.g., tumor adequacy review, formalin fixed paraffin embedded (FFPE) preparations, immunostaining). Supernatant, an immediately available biospecimen, could significantly improve the overall yield of these pauci-cellular samples for genomics and decrease the turnaround time [18]. This is particularly appealing in the context of tyrosine kinase inhibitors (TKI) resistance where histology is less needed since, with the exception of the rare small cell transformation, most resistance mechanisms are genetic alterations. The example of a potential alternative handling of EBUS-TBNA is summarized in Figure 2.
Figure 2.
Alternative handling of cytology specimens for genomics using Fine-Needle Aspiration (FNA) supernatant: example of EBUS-TBNA. EBUS-TBNA: endobronchial ultrasonography transbronchial needle aspiration.
3. Bronchoscopy Cytology Specimen Supernatant
With the increase in lung cancer screening, the management of peripheral nodules is becoming a highly prevalent and challenging situation for pulmonologists. Because most nodules are benign and biopsies are difficult and dangerous (CT-guided biopsies are complicated with pneumothorax in more than 20% of cases), less invasive approaches are urgently needed to discriminate benign from malignant nodules. Sensitivity of plasma genotyping remains poor for early stage NSCLC because of inconsistent/low DNA shed [10]. Bronchoscopy, a minimally invasive procedure, appears to be a good compromise. Cytology examination of bronchoalveolar lavage has poor sensitivity to detect lung cancer but its yield could be improved using new biomarkers, such as exosome-derived miRNA [19] or free DNA methylation (e.g., targeting SHOX2) [20]. Ruy et al. demonstrated that DNA extracted from bronchial washing (BW) supernatant has potential to increase the yield of BW for early-stage lung cancer detection [21].
The free DNA derived from bronchoscopy brushings and washings has also been investigated for advanced lung cancer genotyping by using a TaqMan PCR assay limited to common EGFR mutations. The sensitivity reached 88% across 74 specimens with no false positives [22]. EGFR genotyping in bronchoalveolar lavage fluid could be improved by the use of extracellular vesicles (EV) as demonstrated in a study of 137 patients [23]. The sensitivity and specificity of EV-based EGFR genotyping were 76% and 87%, respectively, with high sensitivity irrespective of the stage (79% for stage I, 100% for stage II, 74% for stage III, and 92% for stage IV).
New bronchoscopic approaches to better target peripheral nodules have been developed such as electromagnetic navigation or radial EBUS, but these samples are sometimes of low tumor content and sensitivity is low for the small lesions (below 70% for nodules < 20 mm) [3]. Following a similar approach as the one reported in the previous chapter, it is very likely that the supernatant derived from the cytology samples collected during these procedures (brushing, rinses) are of high free floating DNA content and could be used to help characterize nodules (using DNA methylation analysis, for example).
4. Pleural, Pericardial, and Ascitic Fluids
Pleural, pericardial, and ascitic fluids are ultrafiltrates from the blood deprived of peripheral blood cells but that can be enriched in cfDNA. Malignant pleural effusion (MPE) is a common complication of lung cancer. Pleural effusion (PE) supernatant contains cell free tumor DNA (PE-cfDNA) with high MAF (comparable to tissue). A study of 102 NSCLC patients showed a good correlation between supernatant-cfDNA and cell block molecular profile (NGS) in MPE [24]. Tong et al. recently demonstrated in 63 lung cancer patients that PE-cfDNA outperforms plasma and cell block for genotyping, with some samples being cytologically negative but rich in free-floating tumor DNA [25]. Reliable detection of EGFR mutations in cytology-negative pleural effusion has been demonstrated in another study [26]. Thus, NGS genotyping of PE-cfDNA appears to be a very appealing approach in cases where no tissue is left. Additionally, it may have a lower turnaround time and a higher sensitivity in cases with adequate tissue. Moreover, tumor mutational burden in PE-cfDNA and in tumor tissue DNA are well correlated [27]. This rich source of tumor DNA can be used to quickly detect driver and resistance EGFR mutations using digital droplet PCR (ddPCR), with high concordance between supernatant and cell pellet [28,29]. Other supernatant-derived material can be used to provide genomic information (EGFR T790M mutations in particular [30]), such as extracellular vesicle-derived DNA (EV-DNA). DNA concentration in EV-DNA extracted from pleural effusion supernatant was significantly higher than that of cfDNA. Moreover, EV-DNA demonstrated 100% agreement with tissue for EGFR genotyping, with higher specificity compared to cytology [30].
After surgery for early stages, liquid biopsy of pleural effusion could help discriminate a metastatic PE from a benign process using RNA profiling (miR-200 and LCN2 expression) [31]. miR-130A quantification could help distinguish lung adenocarcinoma and malignant mesothelioma [32].
Similarly to pleural fluid, metastatic ascites contains free-floating DNA that can be used for mini-invasive and fast initial or resistance genotyping, as demonstrated by the detection of copy number variations (CNVs) in cancer-associated genes in a small series of 6 metastatic non lung cancer patients [33]. Another prospective study has reported that ascitic fluid in patients with peritoneal carcinomatosis (n = 8, among other liquids) is more sensitive than plasma for the detection of clinically relevant mutations in melanoma and NSCLC [34].
Finally, the feasibility of genotyping pericardial effusion supernatant, when a drainage is necessary, has also been reported in 3 patients for the detection of EGFR driver [35] or T790M resistance [36] mutations.
5. Urine
Urine also contains free-floating tumor DNA and is particularly easy to collect repeatedly, giving it great potential for longitudinal follow up throughout treatment. Two fractions of urinary cfDNA have been described: high molecular weight nucleic acids originating from urinary tract and endothelial cells, and low molecular weight [50–250 base pairs] circulating DNA fragments excreted into urine following glomerular filtration by the kidneys. Urine cfDNA, derived from local tumor shed, is already an attractive tool to monitor the molecular profile of urological malignancies [37]. In other cancers, urine cfDNA, can serve as an alternative to plasma genotyping. In 63 patients with advanced EGFR-mutant NSCLC, sensitivities of tissue, plasma and urine were 73%, 82%, and 75%, respectively for T790M detection, these specimens being complementary [38]. In the same study, a significant decrease in T790M MAF in urine was observed in 9 patients treated with rociletinib, highlighting a potential for follow-up (cfDNA being extracted from the same volume). These findings were confirmed in another study investigating ctDNA kinetics in 8 patients treated with osimertinib, in whom the early kinetics of ctDNA shed into the urine correlated with tumor response [39]. Another prospective study in advanced NSCLC demonstrated a good correlation and complementarity between genomic profiles of cfDNA extracted from plasma, sputum, and urine compared to tissue [40]. In earlier stages, the analysis of DNA methylation at cancer-specific loci [41] in urine could help characterize nodules after screening via computed tomography (CT).
6. Saliva and Sputum
Like urine, saliva is another great example of an easy-to-access and potentially cost-effective alternative biospecimen. Data suggest it could be adequate for genotyping [42]. In advanced diseases, EGFR mutation detection is feasible in saliva [43], but with low sensitivity (46.2%) likely due to a low tumor content in tumors not proximal to airways [44].
A study combining five transcriptomic markers and a miRNA signature to discriminate cancer patients (n = 32) from controls (n = 64) reported a sensitivity of 94% but with a specificity of 83% [45]. This approach could be useful to help characterize nodules and guide potentially harmful biopsy procedures.
Sputum might be enriched with tumor-derived material compared to saliva and could help with lung cancer prediction in patients with a positive CT screen, using DNA hypermethylation of genes of interest [54]. The sensitivity and specificity of the combination of three genes (TAC1, HOXA17, and SOX17) in sputum was 93% and 89%, respectively, with a corresponding ROC AUC of 0.89 (95% CI: 0.80–0.98). In a validation cohort from the NELSON trial (subsequently diagnosed with cancer vs. control patients), this targeted methylation analysis approach remained specific (99.3%) but sensitivity was very low (17% for RASSF1A, 28% for the panel RASSF1A, 3OST2, and PRDM14) [55]. Combining CT lung cancer screening and DNA methylation marker research on CT positive patients would relieve the problem of false positive scans and the unnecessary invasive procedures that result [46].
7. Cerebrospinal Fluid (CSF)
The cerebrospinal fluid (CSF) is another challenging cytology specimen. Exploiting CSF-derived cell-free DNA (CSF-cfDNA) is particularly appealing to avoid invasive surgical biopsies in primary brain tumors, but also in cases of brain metastases, because the number of tumor cells in CSF is usually low and DNA shed from central nervous system (CNS) tumors into the blood can be limited. Additionally, due to the blood brain barrier, the proportion of ctDNA is much higher in CSF than in blood, because normal DNA is sparsely released in CSF. The detection of CSF-cfDNA is feasible, more likely in cases of tumors adjacent to CSF reservoirs [47,48,49]. In NSCLC, EGFR mutation detection in CSF-cfDNA by ARMS-PCR is feasible (sensitivity 67%) but limited by a suboptimal concordance with tissue (specificity 82%) [50]. Through iterative hybrid-capture NGS of CSF-cfDNA, De Mattos-Aruda et al. demonstrated a good correlation between tumoral DNA load kinetics in CSF and response to local or systemic therapy in 6 patients, including 2 with NSCLC [48]. In contrast to the cytology study of CSF, the analysis of CSF-cfDNA could be very useful to identify molecular mechanisms of resistance to tyrosine kinase inhibitors (TKI) from a progression solely due to a poor central nervous system penetration of the drug. In patients with ALK-rearranged NSCLC with leptomeningeal carcinomatosis, liquid biopsy of CSF is more sensitive than plasma cfDNA to detect driver or resistance mutations and monitoring tumor response [51], even though repeating lumbar punctures seems invasive. CSF-cfDNA provided a comprehensive profiling of driver and resistance genes in 26 patients with leptomeningeal carcinomatosis. Driver genes were detected, by NGS, in 100%, 84.6%, and 73.1% of CSF-cfDNA, CSF precipitates, and plasma, respectively. Overall, 92.3% of patients had a much higher MAF in CSF-cfDNA compared to cells and plasma [52]. Another study of 26 NSCLC patients demonstrated a higher sensitivity of CSF-cfDNA compared to cytology (100% vs. 71%) for the diagnosis of leptomeningeal carcinomatosis [56].
8. Discussion, Perspectives, and Conclusions
The sensitivity of plasma genotyping can be limited, in particular when tumor burden is limited, and tissue biopsy in these patients is also challenging. Two approaches could address this issue: (i) considering other body fluids that contain tumor-derived free floating DNA at high concentrations and that outperform cytology for most of these pauci-cellular samples, with cfDNA being detected in cytologically negative samples [34]; (ii) improving methods to maximize the yield of biopsy procedures because invasive biopsy remains an integral part of any diagnostic strategy. The supernatant of cytology specimens is also a rich source of immediately available tumor DNA that appears to be highly complementary with tissue.
The interest of non-blood liquid biopsy is thus increasing, and multiple studies have demonstrated that cfDNA extracted from diverse body fluids can be a reliable source of genomic information, obtained consistently in a far less invasive manner than tissue biopsy.
Free-floating DNA in easy-to-access liquids, such as urine, sputum or bronchoscopy cytology samples’ supernatant could be very useful tools to discriminate malignant from benign nodules in the upcoming lung cancer screening era. CT scan is very sensitive but of poor specificity and, in addition, most blood-based screening tests have shown low sensitivity [6,10,57]. Exploiting liquids in direct contact to the tumor could represent a more reliable approach, even though most data regarding these biospecimens concern advanced stages.
In more advanced disease, body fluids collected near the tumor or metastases are more concentrated in cfDNA compared to blood, offering increased amounts of genetic material. A superiority of non-blood free DNA over cytology has not been clearly demonstrated but suggested by some reports, where ctDNA is detected in the supernatant of apparently negative cytology specimens (EBUS-TBNA [15] or pleural effusion [25]). Both are probably highly complementary, the limited cell block being preserved for diagnosis, and the immediately available supernatant for genomics. These high MAF may decrease the risk of false positive calls due to potential sequencing artefact of NGS. CfDNA may thus increase the overall yield of cytology specimens for tumor genotyping. Moreover, the turnaround time could be dramatically shortened, as free DNA is immediately available compared to the conventional approach where DNA can only be extracted after time-consuming diagnosis steps on FFPE blocks. This will be particularly interesting in the context of acquired resistance to targeted therapy where genomics is much more needed than histology. Cell -free DNA has also shown to be useful for prognostication, either by the analysis of tumor mutation burden [58] or by targeting alterations known to be associated with response or resistance to these agents [59] and to assess response to immune checkpoint inhibitors [59,60,61]. These potential applications could be extrapolated to the non-blood approaches described here.
In addition, these biospecimens could improve specificity by avoiding the detection of mutations linked to clonal hematopoiesis that can be detected in blood [62].
In conclusion, liquid biopsy should not be limited to blood. Tumor-derived materials are released in many other fluids in higher amounts when in direct contact with the tumor. Non-blood free-floating DNA could be of high value at all lung cancer stages, from screening and nodule characterization to genotyping patients with metastatic disease both at diagnosis and progression. Commercially available and clinically validated plasma genotyping platforms must be tested in the context of these additional samples before this approach can be translated into routine clinical use, as most of the studies reported here were performed with “in-house” sequencing platforms within academic research programs.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Calvayrac O., Pradines A., Pons E., Mazieres J., Guibert N. Molecular biomarkers for lung adenocarcinoma. Eur. Respir. J. 2017;49:1601734. doi: 10.1183/13993003.01734-2016. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein I.B. CANCER: Addiction to Oncogenes—The Achilles Heal of Cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 3.Steinfort D.P., Khor Y.H., Manser R.L., Irving L.B. Radial probe endobronchial ultrasound for the diagnosis of peripheral lung cancer: Systematic review and meta-analysis. Eur. Respir. J. 2010;37:902–910. doi: 10.1183/09031936.00075310. [DOI] [PubMed] [Google Scholar]
- 4.Guisier F., Salaün M., Lachkar S., Lamy A., Piton N., Obstoy B., Sabourin J.-C., Thiberville L. Molecular analysis of peripheral non-squamous non-small cell lung cancer sampled by radial EBUS. Respirology. 2016;21:718–726. doi: 10.1111/resp.12737. [DOI] [PubMed] [Google Scholar]
- 5.Good W.R., Christensen P.M., Herath S., Dawkins P., Yap E. Radial-probe endobronchial ultrasound outcomes in the investigation of peripheral pulmonary lesions: A New Zealand perspective. Intern. Med. J. 2018;48:1481–1487. doi: 10.1111/imj.14070. [DOI] [PubMed] [Google Scholar]
- 6.Guibert N., Pradines A., Favre G., Mazieres J. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur. Respir. Rev. 2020;29:190052. doi: 10.1183/16000617.0052-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oxnard G.R., Thress K.S., Alden R.S., Lawrance R., Paweletz C.P., Cantarini M., Yang J.C.-H., Barrett J.C., Jänne P.A. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2016;34:3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacher A.G., Alden R.S., Oxnard G.R. Early Intervention in Lung Cancers with Rapid Plasma Genotyping for EGFR and KRAS Mutations—Reply. JAMA Oncol. 2016;2:1096–1097. doi: 10.1001/jamaoncol.2016.1963. [DOI] [PubMed] [Google Scholar]
- 9.Oxnard G.R., Paweletz C.P., Kuang Y., Mach S.L., O’Connell A., Messineo M.M., Luke J.J., Butaney M., Kirschmeier P., Jackman D.M., et al. Noninvasive Detection of Response and Resistance in EGFR-Mutant Lung Cancer Using Quantitative Next-Generation Genotyping of Cell-Free Plasma DNA. Clin. Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbosh C., Birkbak N.J., Swanton C. Early stage NSCLC—Challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018;15:577–586. doi: 10.1038/s41571-018-0058-3. [DOI] [PubMed] [Google Scholar]
- 11.Crosbie P.A., Shah R., Krysiak P., Zhou C., Morris K., Tugwood J., Booton R., Blackhall F., Dive C. Circulating Tumor Cells Detected in the Tumor-Draining Pulmonary Vein Are Associated with Disease Recurrence after Surgical Resection of NSCLC. J. Thorac. Oncol. 2016;11:1793–1797. doi: 10.1016/j.jtho.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemi F., Rothwell D.G., McGranahan N., Gulati S., Abbosh C., Pearce S.P., Zhou C., Wilson G.A., Jamal-Hanjani M., Brikbak N., et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019;25:1534–1539. doi: 10.1038/s41591-019-0593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein S.D., Bibbo M., Kowalski T.E., Loren D.E., Siddiqui A.A., Solomides C., Ellsworth E. Mutational analysis of cytocentrifugation supernatant fluid from pancreatic solid mass lesions. Diagn. Cytopathol. 2013;42:719–725. doi: 10.1002/dc.23048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy-Chowdhuri S., Mehrotra M., Bolivar A.M., Kanagal-Shamanna R., Barkoh B.A., Hannigan B., Zalles S., Ye W., Duose D., Broaddus R., et al. Salvaging the supernatant: Next generation cytopathology for solid tumor mutation profiling. Mod. Pathol. 2018;31:1036–1045. doi: 10.1038/s41379-018-0006-x. [DOI] [PubMed] [Google Scholar]
- 15.Guibert N., Tsukada H., Hwang D.H., Chambers E., Cibas E.S., Bale T., Supplee J., Ulrich B., Sholl L.M., Paweletz C.P., et al. Liquid biopsy of fine-needle aspiration supernatant for lung cancer genotyping. Lung Cancer. 2018;122:72–75. doi: 10.1016/j.lungcan.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Janaki N., Harbhajanka A., Michael C.W., Do P.B., Wasman J., Bs M.A., Bs K.M., Alouani D., Sadri N. Comparison of cytocentrifugation supernatant fluid and formalin-fixed paraffin-embedded tissue for targeted next-generation sequencing. Cancer Cytopathol. 2019;127:297–305. doi: 10.1002/cncy.22126. [DOI] [PubMed] [Google Scholar]
- 17.Malapelle U., Bellevicine C., De Luca C., Salatiello M., De Stefano A., Rocco D., De Rosa N., Vitiello F., Russo S., Pepe F., et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol. 2013;121:552–560. doi: 10.1002/cncy.21322. [DOI] [PubMed] [Google Scholar]
- 18.Gokozan H.N., Harbhajanka A., Do P.B., Michael C.W., Sadri N. Use of cytology centrifuged supernatants improves cost and turnaround time for targeted next generation sequencing. Diagn. Cytopathol. 2020 doi: 10.1002/dc.24548. [DOI] [PubMed] [Google Scholar]
- 19.Kim J.E., Eom J.S., Kim W.-Y., Jo E.J., Mok J., Lee K., Kim K.U., Park H.-K., Lee M.K., Kim M.-H. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: A pilot study. Thorac. Cancer. 2018;9:911–915. doi: 10.1111/1759-7714.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt B., Liebenberg V., Dietrich D., Schlegel T., Kneip C., Seegebarth A., Flemming N., Seemann S., Distler J., Lewin J., et al. SHOX2 DNA Methylation is a Biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. doi: 10.1186/1471-2407-10-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu J.-S., Lee M.K., Lee S.J. Feasibility of bronchial washing fluid-based approach to early-stage lung cancer diagnosis. Lung Cancer. 2019;24:e603–e606. doi: 10.1634/theoncologist.2019-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara A., Ct C.F., Taira T., Abe H., Ct Y.T., Murata K., Yamaguchi T., Azuma K., Ishii H., Takamori S., et al. Epidermal growth factor receptor mutation status in cell-free DNA supernatant of bronchial washings and brushings. Cancer Cytopathol. 2015;123:620–628. doi: 10.1002/cncy.21583. [DOI] [PubMed] [Google Scholar]
- 23.Hur J.Y., Lee J.S., Kim I.A., Kim H.J., Kim W.S., Lee K.Y. Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl. Lung Cancer Res. 2019;8:1051–1060. doi: 10.21037/tlcr.2019.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Li X., Wang W., Shao Y., Zhang Y., Song Z. Gene Alterations in Paired Supernatants and Precipitates from Malignant Pleural Effusions of Non-Squamous Non-Small Cell Lung Cancer. Transl. Oncol. 2020;13:100784. doi: 10.1016/j.tranon.2020.100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong L., Ding N., Tong X., Li J., Zhang Y., Wang X., Xu X., Ye M., Li C., Wu X., et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9:5532–5541. doi: 10.7150/thno.34070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Z., Wang W., Li M., Liu J., Zhang Y. Cytological-negative pleural effusion can be an alternative liquid biopsy media for detection of EGFR mutation in NSCLC patients. Lung Cancer. 2019;136:23–29. doi: 10.1016/j.lungcan.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Goodman A.M., Kato S., Bazhenova L., Patel S.P., Frampton G.M., Miller V., Stephens P.J., Daniels G.A., Kurzrock R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hummelink K., Muller M., Linders T.C., Van Der Noort V., Nederlof P.M., Baas P., Burgers S., Smit E.F., Meijer G.A., Heuvel M.M.V.D., et al. Cell-free DNA in the supernatant of pleural effusion can be used to detect driver and resistance mutations, and can guide tyrosine kinase inhibitor treatment decisions. ERJ Open Res. 2019;5:00016-2019. doi: 10.1183/23120541.00016-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soh J., Toyooka S., Aoe K., Asano H., Ichihara S., Katayama H., Hiraki A., Kiura K., Aoe M., Sano Y., et al. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int. J. Cancer. 2006;119:2353–2358. doi: 10.1002/ijc.22190. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.S., Hur J.Y., Kim I.A., Kim H.J., Choi C.M., Lee J.C., Kim W.S., Lee K.Y. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: A comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer. 2018;18:1236. doi: 10.1186/s12885-018-5138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hydbring P., De Petris L., Zhang Y., Brandén E., Koyi H., Novak M., Kanter L., Hååg P., Hurley J., Tadigotla V., et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer. 2018;124:45–52. doi: 10.1016/j.lungcan.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Cappellesso R., Galasso M., Nicolè L., Dabrilli P., Volinia S., Fassina A. miR-130A as a diagnostic marker to differentiate malignant mesothelioma from lung adenocarcinoma in pleural effusion cytology. Cancer Cytopathol. 2017;125:635–643. doi: 10.1002/cncy.21869. [DOI] [PubMed] [Google Scholar]
- 33.Husain H., Nykin D., Bui N., Quan D., Gomez G., Woodward B., Venkatapathy S., Duttagupta R., Fung E., Lippman S.M., et al. Cell-Free DNA from Ascites and Pleural Effusions: Molecular Insights into Genomic Aberrations and Disease Biology. Mol. Cancer Ther. 2017;16:948–955. doi: 10.1158/1535-7163.MCT-16-0436. [DOI] [PubMed] [Google Scholar]
- 34.Villatoro S., Mayo-De-Las-Casas C., Jordana-Ariza N., Viteri-Ramírez S., Garzón-Ibañez M., Moya-Horno I., García-Peláez B., González-Cao M., Malapelle U., Balada-Bel A., et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol. Oncol. 2019;13:2633–2645. doi: 10.1002/1878-0261.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P., Wu X., Tang M., Nie X., Lin L. Detection of EGFR gene mutation status from pleural effusions and other body fluid specimens in patients with lung adenocarcinoma. Thorac. Cancer. 2019;10:2218–2224. doi: 10.1111/1759-7714.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provencio M., Torrente M., Calvo V., Gutiérrez L., Pérez-Callejo D., Pérez-Barrios C., Barquín M., Royuela A., Rodríguez-Alfonso B., Sotelo M., et al. Dynamic circulating tumor DNA quantificaton for the individualization of non-small-cell lung cancer patients treatment. Oncotarget. 2017;8:60291–60298. doi: 10.18632/oncotarget.20016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Meo A., Bartlett J., Cheng Y., Pasic M.D., Mac-Way F. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer. 2017;16:80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reckamp K.L., Melnikova V.O., Karlovich C., Sequist L.V., Camidge D.R., Wakelee H., Perol M., Oxnard G.R., Kosco K., Croucher P., et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J. Thorac. Oncol. 2016;11:1690–1700. doi: 10.1016/j.jtho.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 39.Husain H., Melnikova V.O., Kosco K., Woodward B., More S., Pingle S.C., Weihe E., Park B.H., Tewari M., Erlander M.G., et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin. Cancer Res. 2017;23:4716–4723. doi: 10.1158/1078-0432.CCR-17-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z., Yang Z., Li C.S., Zhao W., Liang Z.X., Dai Y., Zhu Q., Miao K.L., Cui D.H., Chen L. Differences in the genomic profiles of cell-free DNA between plasma, sputum, urine, and tumor tissue in advanced NSCLC. Cancer Med. 2019;8:910–919. doi: 10.1002/cam4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B., Filho J.R., Mallisetty A., Villani C., Kottorou A.E., Rodgers K.P., Chen C., Ito T., Holmes K., Gastala N., et al. Detection of Promoter DNA Methylation in Urine and Plasma Aids the Detection of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020;26:4339–4348. doi: 10.1158/1078-0432.CCR-19-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., Ehli E.A., Nelson K., Bohlen K., Lynch C., Huizenga P., Kittlelsrud J., Soundy T.J., Davies G.E. Genotyping Performance between Saliva and Blood-Derived Genomic DNAs on the DMET Array: A Comparison. PLoS ONE. 2012;7:e33968. doi: 10.1371/journal.pone.0033968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei F., Lin C.-C., Joon A., Feng Z., Troche G., Lira M.E., Wong D.T. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am. J. Respir. Crit. Care Med. 2014;190:1117–1126. doi: 10.1164/rccm.201406-1003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z., Zhang L., Li L., Li X., Xu Y., Wang M., Du J. Sputum Cell-Free DNA: Valued Surrogate Sample for Detection of EGFR Mutation in Patients with Advanced Lung Adenocarcinoma. J. Mol. Diagn. 2020;22:934–942. doi: 10.1016/j.jmoldx.2020.04.208. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L., Xiao H., Zhou H., Santiago S., Lee J.M., Garon E.B., Yang J., Brinkmann O., Yan X., Akin D., et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell. Mol. Life Sci. 2012;69:3341–3350. doi: 10.1007/s00018-012-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hulbert A., Jusue-Torres I., Stark A., Chen C., Rodgers K., Lee B., Griffin C., Yang A., Huang P., Wrangle J., et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin. Cancer Res. 2017;23:1998–2005. doi: 10.1158/1078-0432.CCR-16-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Springer S., Zhang M., McMahon K.W., Kinde I., Dobbyn L., Ptak J., Brem H., Chaichana K.L., Gallia G.L., et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl. Acad. Sci. USA. 2015;112:9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Mattos-Arruda L., Mayor R., Ng C.K.Y., Weigelt B., Martínez-Ricarte F., Torrejon D., Oliveira M., Arias A., Raventos C., Tang J., et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pentsova E.I., Shahiba O., Tang J., Boire A., You D., Briggs S., Omuro A., Lin X., Fleisher M., Grommes C., et al. Evaluating Cancer of the Central Nervous System Through Next-Generation Sequencing of Cerebrospinal Fluid. J. Clin. Oncol. 2016;34:2404–2415. doi: 10.1200/JCO.2016.66.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H., Cai L., Zhang Y., Tan H., Deng Q., Zhao M., Xu X. Sensitive Detection of EGFR Mutations in Cerebrospinal Fluid from Lung Adenocarcinoma Patients with Brain Metastases. J. Mol. Diagn. 2014;16:558–563. doi: 10.1016/j.jmoldx.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Zheng M.-M., Li Y.-S., Jiang B.-Y., Tu H.-Y., Tang W.-F., Yang J.-J., Zhang X.-C., Ye J.-Y., Yan H.-H., Su J., et al. Clinical Utility of Cerebrospinal Fluid Cell-Free DNA as Liquid Biopsy for Leptomeningeal Metastases in ALK-Rearranged NSCLC. J. Thorac. Oncol. 2019;14:924–932. doi: 10.1016/j.jtho.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Li Y., Jiang B., Yang J., Zhang X.-C., Zhang Z., Ye J., Zhong W., Tu H., Chen H., Wang Z., et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: A new medium of liquid biopsy. Ann. Oncol. 2018;29:945–952. doi: 10.1093/annonc/mdy009. [DOI] [PubMed] [Google Scholar]
- 53.Hannigan B., Ye W., Mehrotra M., Lam V., Bolivar A., Zalles S., Barkoh B., Duose D., Hu P., Broaddus R., et al. Liquid biopsy assay for lung carcinoma using centrifuged supernatants from fine-needle aspiration specimens. Ann. Oncol. 2019;30:963–969. doi: 10.1093/annonc/mdz102. [DOI] [PubMed] [Google Scholar]
- 54.Hubers A.J., Heideman D.A.M., Burgers S.A., Herder G.J.M., Sterk P.J., Rhodius R.J., Smit H.J., Krouwels F., Welling A., Witte B.I., et al. DNA hypermethylation analysis in sputum for the diagnosis of lung cancer: Training validation set approach. Br. J. Cancer. 2015;112:1105–1113. doi: 10.1038/bjc.2014.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hubers A.J., Heideman D.A.M., Duin S., Witte B.I., De Koning H.J., Groen H.J.M., Prinsen C.F.M., Bolijn A.S., Wouters M., Van Der Meer S.E., et al. DNA hypermethylation analysis in sputum of asymptomatic subjects at risk for lung cancer participating in the NELSON trial: Argument for maximum screening interval of 2 years. J. Clin. Pathol. 2016;70:250–254. doi: 10.1136/jclinpath-2016-203734. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y., He J., Zou Y., Guo X.-S., Cui J.-Z., Guo L., Bu H. Evaluating the cerebrospinal fluid ctDNA detection by next-generation sequencing in the diagnosis of meningeal Carcinomatosis. BMC Neurol. 2019;19:331. doi: 10.1186/s12883-019-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marquette C.-H., Boutros J., Benzaquen J., Ferreira M., Pastre J., Pison C., Padovani B., Bettayeb F., Fallet V., Guibert N., et al. Circulating tumour cells as a potential biomarker for lung cancer screening: A prospective cohort study. Lancet Respir. Med. 2020;8:709–716. doi: 10.1016/S2213-2600(20)30081-3. [DOI] [PubMed] [Google Scholar]
- 58.Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y., Rittmeyer A., Fehrenbacher L., Otto G., Malboeuf C., et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018;24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 59.Guibert N., Jones G., Beeler J.F., Plagnol V., Morris C., Mourlanette J., Pradines A. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer. 2019;137:1–6. doi: 10.1016/j.lungcan.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Cabel L., Riva F., Servois V., Livartowski A., Daniel C., Rampanou A., Lantz O., Romano E., Milder M., Buecher B., et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: A proof-of-concept study. Ann. Oncol. 2017;28:1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 61.Goldberg S.B., Narayan A., Kole A.J., Decker R.H., Teysir J., Carriero N.J., Lee A., Nemati R., Nath S.K., Mane S.M., et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018;24:1872–1880. doi: 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Y., Ulrich B.C., Supplee J., Kuang Y., Lizotte P.H., Feeney N.B., Guibert N.M., Awad M.M., Wong K.-K., Jänne P.A., et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin. Cancer Res. 2018;24:4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]