Abstract
Humans have always been encountered to big infectious diseases outbreak throughout the history. In December 2019, novel coronavirus (COVID-19) was first noticed as an agent causing insidious pneumonia in Wuhan, China. COVID-19 was spread rapidly from Wuhan to the rest of the world. Until late June 2020, it infected more than 10,000,000 people and caused more than 500,000 deaths in almost all of countries in the world, creating a global crisis worse than all previous epidemics and pandemics. In the current review, we gathered and summarized the results of various studies on characteristics, diagnosis, treatment, and prevention of this pandemic crisis.
Keywords: COVID-19, infection, pandemic, severe acute respiratory syndrome coronavirus-2
INTRODUCTION
Humans have always been afflicted by epidemics and pandemics of infectious diseases. The outbreaks have been around for thousands of years. Even in this modern era, infection outbreaks have ravaged humanity until it causes people to despair. In the meantime, viruses have always posed significant challenges that have left deadly epidemics and pandemics.
In December 2019, novel coronavirus (CoV) was first noticed as an agent causing insidious pneumonia in Wuhan, China. CoVs are a group of enveloped single-stranded ribonucleic acid (RNA) viruses that are notorious for developing severe outbreaks in the last two decades. The first one was severe acute respiratory syndrome coronavirus (SARS-CoV) that occurred in 2002–2003 with 8096 cases and 774 deaths, which was successfully controlled.[1] The second CoV-caused outbreak, Middle East respiratory syndrome coronavirus (MERS-CoV), occurred in 2012, which is still under investigation.[2,3] COVID-19 is spreading rapidly from Wuhan to the rest of the world. By the end of June 2020, it infected more than 10,000,000 people and caused more than 500,000 deaths in almost all countries, creating a global crisis worse than all previous epidemics and pandemics.[4] It can infect people of all ages, but older adults and people with certain underlying medical conditions are at higher risk and may develop more severe disease. The virus spreads 1–2 m via droplets by coughing and sneezing. In an appropriate condition, it could maintain viable potential on the surfaces up to several days. People who touch these surfaces and then touch their face, mouth, nose, and eyes could get contaminated. Fecal-oral transmission is also possible but not proved. Lung angiotensin-converting enzyme 2 (ACE2) is the potential receptor for the virus.[5,6] Patients usually experience symptoms similar to an upper respiratory infection. However, a common presentation of infection is fever (88%), and patients may be afebrile in the first 1–2 days.[7] Other common symptoms are dry cough, sore throat, upper airways obstruction, shortness of breath, headache, fatigue, myalgia, and arthralgia.[8,9] The disease could progress in some patients with more severe manifestations, such as severe dyspnea, pneumonia, respiratory failure, renal failure,[10,11] and death within a week. Up to 30% of patients may require intensive unit care (ICU) during hospitalization. Moreover, complications such as acute respiratory distress syndrome (ARDS), acute lung, or kidney injuries are probable consequences. The severity of the disease is worse in elders and patients with comorbidities (hypertension, chronic heart disease, chronic obstructive pulmonary disease, diabetes).[10]
Despite some reported evidence about COVID-19, we still have some gap knowledge in characteristics, diagnosis, and treatment of this novel virus. Hence, in this narrative review, our team summarized and established a feature for COVID-19 centered on the history, symptoms, pathogenesis, diagnosis, and treatment of this disease.
CORONAVIRUS CHARACTERISTICS, STRUCTURE, AND FUNCTION
CoVs are enveloped nonsegmented positive-sense large RNA structure viruses belonging to the Nidovirales order, which includes Coronaviridae, Arteriviridae, Mesoniviridae, and Roniviridae families.[11,12,13] Nidovirales order has conserved genomic organizing, with a large replicase gene preceding structural and supplementary genes; expression of many nonstructural genes by ribosomal frameshifting; various unique or uncommon enzymatic activities encoded within the large replicase–transcriptase polyprotein; and expression of downstream genes by the synthesis of 3′ nested subgenomic mRNAs. The important distinctions inside the Nidovirus families are in the number, type, and sizes of the structural proteins. These differences lead to important changes in the structure and morphology of the nucleocapsids and virions.[11,14] All viruses in the Nidovirales order contain large genomes, and some viruses have the largest recognized RNA genomes containing until 33.5 kb genomes.[14] The Coronaviridae is a monogeneric family of a large family of mammalian and avian pathogens that were first described in 1966.[12,14]
Four subgroups of CoV family, namely alpha-, beta-, gamma-, and delta-CoVs exist.[15,16] Alpha- and beta-CoVs originate from mammals, especially from bats, while gamma and delta viruses originate from pigs and birds.[17] There are several CoVs discovered that cause infection in humans. Four of which are distinguished to only cause the common cold or other mild symptoms, including HCoV-229E (in 1966), HCoV-NL63 (in 2004), HCoV-OC43 (in 1967), and HCoV-HKU1 (in 2005), the first two cases are alpha-CoVs, and the last two are beta-CoVs.[18,19]
The three beta-CoVs, SARS-CoV discovered in 2002, MERS-CoV discovered in 2012, and SARS-CoV-2 (2019-nCoV), were originally from animals, eventually infected human, and resulted in severe health problems. SARS-CoV originated from bats with a mortality rate of 10%, and MERS-CoV was isolated from camel with a mortality rate of 37%.[7,16] Although bats are possible reservoir hosts for SARS-CoV-2, the intermediate host that facilitates the transmission of this virus to humans is still not clear. Recently, Lam et al. identified CoVs in Malayan pangolins (Manis javanica) which are related to SARS-CoV-2. By metagenomic sequencing and identifying the similarity of these CoVs to SARS-CoV-2, they suggested Malayan pangolins as possible intermediate host for SARS-CoV-2.[20]
Similar to SARS-CoV and MERS-CoV, the genome of SARS-CoV-2 (2019-nCoV) encodes nonstructural proteins (such as 3-chymotrypsin-like protease, papain-like protease, helicase, and RNA-dependent RNA polymerase) as key enzymes and structural proteins (such as spike glycoprotein) for virus–cell receptor interactions during viral entry in the viral life cycle. These proteins were recognized as the interesting targets to improve antiviral agents against SARS-CoV and MERS-CoV.[21] The sequence analysis of SARS-CoV-2 has a structure typical to that of other CoVs, and its genome has been likened to the SARS outbreak.[22]
CoVs are minute in size (65–125 nm in diameter) and contain a nucleic material, with size ranging from 26 to 33 kb in length.[16] The club-shaped spike protrusions flow out from the surface of the virion. These spikes are a shaping feature of the virion and give them the appearance of a solar corona, prompting the name, CoVs (corona = crown). CoVs have helically symmetric nucleocapsids, which are unusual among positive-sense RNA viruses, but more usual for negative-sense RNA viruses.[14,23] The CoVs can infect humans and may cause severe disease and deaths.[17]
CoV particles contain four principal structural proteins, including the spike (S), a small membrane protein (M), envelope (E), and nucleocapsid (N) proteins, all of which are encoded within the 3′ end of the viral genome. The S glycoprotein (~150 kDa) is a class of fusion protein and constitutes the special spike structure on the surface of the virus, which facilitates binding to the host receptor [Figure 1].[14,24] The small M protein (~25–30 kDa) is the most abundant structural protein in the virion and gives the virion its shape. The M protein is dimer and helps membrane curvature besides to bind to the nucleocapsid.[14] The E protein (~8–12 kDa) is found in small amounts in the virion and is a transmembrane protein with anion channel activity, which is required for pathogenesis.[14,25] The N protein is heavily phosphorylated and triggers a structural change increasing the affinity for viral RNA, a key component of the replicase complex.[14,17] An additional membrane glycoprotein (HE) is found in the beta-CoVs, and the whole genome of SARS CoV-2 is exactly alike (96%) with a bat CoV.[17]
Figure 1.
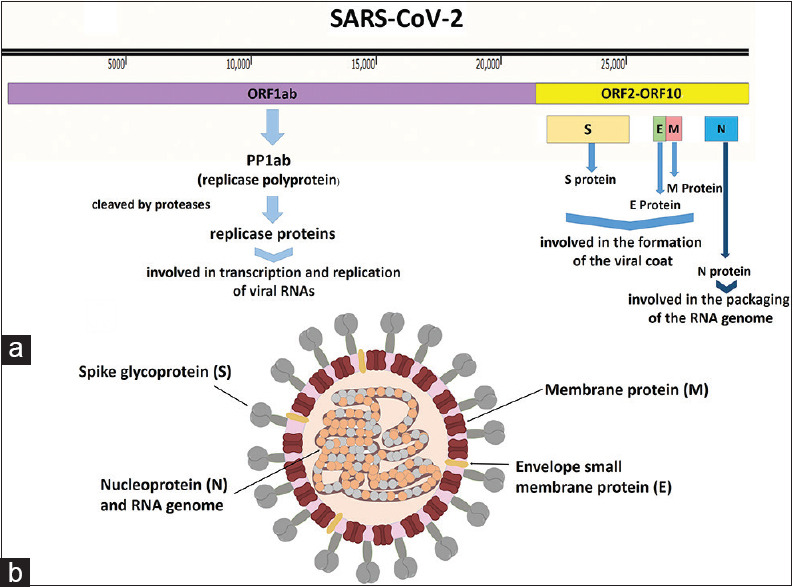
(a) Ribonucleic acid genome and proteins of severe acute respiratory syndrome coronavirus-2. (b) Schematic representation of severe acute respiratory syndrome coronavirus-2 structure
SYMPTOMS, PATHOGENESIS, AND MECHANISM OF DISEASE
Clinical studies show that the symptoms in COVID-19 patients include fever, shortness of breath, nonproductive cough, headache, dyspnea, myalgia, fatigue, lymphopenia, and radiographic evidence of pneumonia and acute lung injury. This acute lung injury often leads to hypoxemic respiratory failure, ARDS, and ultimately death (in 2.8% of cases). Most of these symptoms are similar to SARS-CoV and MERS-CoV infections. Based on the latest study, the incubation of this disease is estimated to be 5.2 days, and the period time from disease onset to death is 6–41 days,[26] in which the clinical manifestations are completely dependent on the age and immune system of the patient. This period in patients over 70 years of age and patients with underlying diseases or immunodeficiency disorders is shorter.[27,28] In most patients, the distribution of ground-glass opacities (GGOs) in both lungs is revealed on the chest CT imaging that likely triggers local and systemic immune responses and enhances inflammation.[26]
The exact pathogenesis of COVID-19 is not clear at present; however, the similar mechanisms of SARS-CoV and MERS-CoV can provide us numerous information to elucidate the pathogenicity and mechanism of COVID-19.[7] SARS-CoV-2, like SARS-CoV, uses ACE2 as a specific cellular receptor to go through the target cell,[29] and this binding is mediated by CoV spike (S) glycoprotein.[30] In addition to binding to the target cell, the S protein functions in the cell tropism and pathogenesis are the main target of the host immune system.[31] Brielle et al. analyzed the binding of the SARS-CoV-2 spike protein to the human ACE2 receptor and compared the SARS-CoV-2–ACE2 complex with that of other pathogenic CoVs using molecular dynamic simulations. They found that the SARS-CoV-2–ACE2 complex contains a higher number of contacts, a larger interface area, and decreased interface residue fluctuations relative to SARS-CoV.[32] Sigrist et al., by sequence analyzing and PROSITE (a protein database) scanning, suggested that integrins may be alternative receptors for SARS-CoV-2. Their results showed that SARS-CoV-2 binds to integrins through the RGD (Arg Gly Asp) motif in the receptor binding domain of the S protein. This motif is not present in other CoVs.[31]
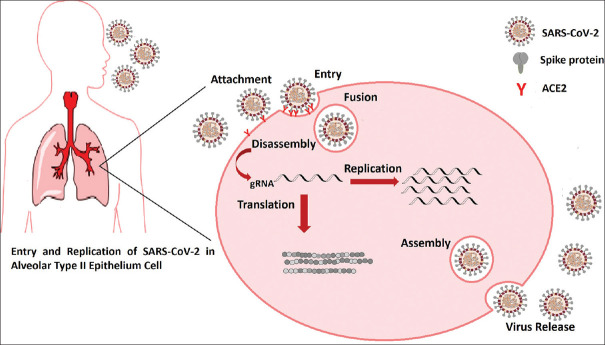
Following virus attachment and entry into the target cell by fusion between the virus envelope and cell membrane, the viral genome is uncoated into the cytoplasm and translated into viral proteins. Then, the genomic RNA replicates and the viral particles were assembled by the combination of viral genome and proteins, and newly formed viruses release by exocytosis [Figure 2]. Based on previous studies of SARS-CoV infection, rapid replication of SARS-CoV-2 can lead to massive epithelial and endothelial cell apoptosis and also may cause pyroptosis in macrophages and lymphocytes.[33] The results of these events can cause symptoms such as peripheral blood lymphopenia, as seen in most SARS-CoV-2 patients.[34] Similar to other viral infectionsafter virus entrance and antigen presentation, the host's humoral and cellular immunity will be stimulated. By humoral immune response against viral infections, SARS-CoV-2-specific B-cell will produce a specific antibody against this pathogen. In SARS-CoV infection, a typical mold of SARS-specific IgM and IgG construction is reported in the virus-specific antibody profile of SARS-CoV patients. Unlike IgM, which disappears after 12 weeks from the onset of illness, SARS-specific IgG can stay for a long time.[35]
Figure 2.
Attachment, entry, and replication of severe acute respiratory syndrome coronavirus-2 in an angiotensin-converting enzyme 2 expressing cell
Cellular immunity mediated by T-lymphocytes, especially CD4+ T-cells and CD8+ T-cells, plays an essential role in defense against virus infections and viral clearance. Many studies on cellular immunity against CoVs indicate that MHC I molecules play a main role in the antigen presentation of SARS-CoV to cytotoxic T-lymphocytes and numerous HLA polymorphisms correlated to the susceptibility of SARS-CoV have been identified.[7] In the acute phase of SARS-CoV infection, the number of CD4+ T-cells and CD8+ T-cells substantially decreased.[36] Similarly, Xu et al. investigated the pathological characteristics of a patient who died from a severe infection of SARS-CoV-2 and reported that the number of peripheral CD4+ and CD8+ T-cells was significantly reduced.[37]
Fan et al. analyzed the cellular immune response of 21 recovered SARS patients and showed that both memory CD4+ T-cells and CD8+ T-cells specific for SARS-CoV could be maintained for 4 years and gradually declined in the absence of SARS-CoV.[36]
The common immunopathological event and the main cause of death in COVID-19 patients are ARDS. The main mechanism that leads to ADRS is the cytokine storm (or hypercytokinemia), resulting from the uncontrolled produce and release of substantial quantity of pro-inflammatory cytokines and chemokines by immune effector. Studies show that severe cases of SARS-CoV and MERS-CoV increase serum concentrations of interferon (IFN)-α, interleukin (IL)-6 and CXCL8, CCL5, and CXCL-10 compared to mild–moderate cases of disease.[38] High blood levels of pro-inflammatory cytokines such as IL-2, IL-7, tumor necrosis factor-α (TNF-α) and MCP1, and MIP1α were reported in some of the SARS-CoV-2 patients who were admitted to the ICU.[7]
In addition to ARDS, the cytokine storm could trigger the immune system attacking the whole body and lead to loss of lung function, multiple organ disorder, shock, and finally death in critical cases of COVID-19. However, it seems that the virus enters the peripheral blood and attacks the other organs such as the heart that express ACE2.[39]
The clinical course of COVID-19 infection noticeably varies due to various factors such as genetic changes in the virus or host factors.[40]
The severity of COVID-19 could be divided into four conditions:
Mild disease: people with mild disease (approximately 40%) are symptomatic patients without evidence of viral pneumonia or hypoxia
Moderate disease: around 40% of patients show clinical signs of patients with clinical signs of pneumonia (fever, cough, dyspnea, and fast breathing) but no signs of severe pneumonia, including SpO2 ≥ 90% on room air
Severe disease: patients who require oxygen support (approximately 15%)
Critical disease: approximately 5% of patients have complications such as respiratory failure, ARDS, sepsis and septic shock, thromboembolism, and/or multiorgan failure.[40]
DIAGNOSIS OF COVID-19
Clinical identification of SARS-CoV-2 is principally based on the epidemiological history, clinical manifestations and some examinations, such as CT scan, nucleic acid detection, and immune identification technology (point-of-care [POC] testing of IgM/IgG, enzyme-linked immunosorbent assay).[41] As in SARS and MERS, the diagnosis of 2019-nCoV infection is based on a history of detailed contact and travel and precise laboratory testing.[42]
Clinical diagnosis
The clinical signs and symptoms of individuals infected with SARS-CoV-2 are highly atypical, counting fever (the typical symptom), fatigue, dry cough, and dyspnea. Some patients have symptoms such as nasal congestion, runny nose, sore throat, diarrhea, or other upper respiratory symptoms.[43] A few patients only have a low fever, mild fatigue, and no pneumonia. In severe cases, the infection can cause viral pneumonia, shortness of breath, and breathing difficulties occurring more than 1 week after infection.[7]
Laboratory tests
In the early stage of the disease, the total number of leukocytes in the peripheral blood is decreased or may be normal, the lymphocyte count is decreased, and some patients show increases in the liver enzymes, muscle enzymes, and myoglobin.[7,43] The C-reactive protein and erythrocyte sedimentation rate are increased in most patients, and procalcitonin is normal.[7] However, in severe cases, the neutrophil count, D-dimer, blood urea, and creatinine levels were significantly increased, and the lymphocytes were progressively decreased.[7,44] In addition, inflammatory factors (IL-6, IL-10, and TNF-α) increase, indicating the immune status of patients.[44] CoV nucleic acid can be detected from the upper respiratory tract (oropharyngeal and nasopharyngeal) and lower respiratory tract (endotracheal aspirate, expectorated sputum or bronchoalveolar lavage, blood, and stool).[7,43] Diagnostic criteria for COVID-19 are virus isolation and viral nucleic acid detection.[43]
Nucleic acid detection technology
Rapid collection and testing of appropriate specimens in suspected patients to COVID-19 are priority for the clinical management and outbreak control and should be guided by a laboratory specialist.[45]
Molecular methods such as reverse transcription polymerase chain reaction (RT-PCR) or real-time quantitative polymerase chain reaction (RT-qPCR) and high-throughput sequencing are the most common diagnostic methods, which are made using RNA from respiratory samples.[41,42] These rapid techniques could be more verified by next-generation sequencing. However, the application of this technology in clinical diagnosis is limited because of its equipment dependency and high cost.[41]
Moreover, RT-qPCR is more sensitive than the conventional RT-PCR assay, which helps much for the diagnosis in early infection.[46] Up to now, the golden clinical diagnosis method of SARS-CoV-2 is the TaqMan-based real-time RT-PCR assay in the lower respiratory tract samples that can offer significantly higher viral load and genome fraction than upper respiratory tract samples.[42,44,46]
Serological testing
In the early phase of the disease, infected patients may not have tested positive for viral RNA but retroactively can be shown to have an immune response, and serology can be performed as an auxiliary diagnostic test.[47]
Combining PCR with serology tests would allow more confirmation of infection.[48] High serological tests for SARS-CoV-2 are becoming more widely available for determining of asymptomatic and recovered cases.[45,48] Enzyme immunoassays on high-throughput automated platforms and rapid, POC tests are becoming more widely available, which are subject to many interferences, including specific binding and cross-reaction with other CoVs. It depends on the type of antigen used to coat the plates and may produce false-positive results.[41]
Therefore, emerging other sensitive and specific assisting methods include the fully automated serology test developed to detect SARS-CoV-2 IgG antibodies against both the S1 and S2 domain of the spike protein. This increases the quality assurance and the specificity of the test and prevents cross-reaction and false-positive results due to other CoVs.[41]
Viral sequencing
Besides confirming the presence of the virus, ordinary sequencing of clinical specimens can be useful. Virus whole-genome sequencing can also help molecular epidemiology studies.[45]
Newly developed methods
The RNA-targeting CRISPR-associated enzyme Cas13 has recently been adapted for multiplexable, portable, rapid, and quantitative detection platform of nucleic acids.[46] Cas13 can be programmed to target and demolish the genomes of diverse mammalian single-stranded RNA viruses.[46] A platform was developed termed SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) that combined isothermal preamplification with Cas13 to detect single molecules of RNA or DNA.[42,49] A protocol for the detection of COVID-19 using CRISPR diagnostics has been reported on the website (https://broad.io/sherlockprotocol), which may provide some reference points for researchers interested in further progressing of this diagnostics system.[46]
Computed tomography imaging examination
SARS-CoV-2 causes inflammatory lesions in the lungs such as pneumonia. Chest computed tomography (CT) is the main examination for lung lesions and plays a main function in the clinical diagnosis.[50] The imaging findings change with the patient's age, immunity condition, disease stage at the time of scanning, underlying diseases, and drug treatment. In the early stage of pneumonia cases, chest images show multiple small patchy shadows and interstitial changes notable in the lung periphery.[43] Severe cases can further develop to bilateral multiple GGO, infiltrating shadows, and segmental consolidation in the bilateral lungs, with infrequent pleural effusion. Chest CT scan pulmonary lesions are shown more clearly by CT than X-ray examination, and the high-resolution CT for the chest is essential for early diagnosis and rating of disease severity of patients with SARS-CoV-2.[41,43]
If there are typical imaging characteristics of CoV pneumonia, a nucleic acid test is not necessary to confirm the diagnosis.[51]
Although RT-qPCR is specific for the detection of COVID-19, its false-negative rate cannot be neglected. Due to severe outcomes of missed diagnosis, for high clinically suspicious patients, chest CT scan proposed as a necessary auxiliary diagnostic method.[41]
Due to inadequate availability of RT-PCR kits and the potential of false-negative results, some guidelines have suggested diagnosis based on the clinical and chest CT findings.[52] Quick diagnosis of COVID-19 is critical for patient isolation and control of this communicable disease. Chest CT scan is a first-line imaging modality for the management of highly suspected people for COVID-19 with negative results of a RT-PCR assay.[53] Furthermore, it has been proposed that in comparison with initial RT-PCR, chest CT scan might have higher sensitivity to detect patients with COVID-19. In a large study among 1014 patients in China, chest CT has been reported to have a sensitivity of 97%, a specificity of 25%, and an accuracy of 68% and has been proposed as the primary tool for the detection of COVID-19.[54] However, normal CT has been reported in 56% of early patients (>2 days after symptom onset) and could not be a single reliable tool to rule out COVID-19 infection.[55]
TREATMENT STRATEGY
Chloroquine and hydroxychloroquine
In the 1940s, chloroquine was first introduced as an antimalarial drug. Nowadays, it is also a major medication for rheumatic disease and systemic lupus erythematosus.[30,56] In the 2000s, during SARS-CoV epidemic, the antiviral effects of this drug were noticed. The mechanism of this effect is the glycosylation of virus–cell receptors and inhibition of cell fusion by increasing endomysial pH.[57,58,59] The therapeutic effects of chloroquine on the novel CoV was first declared by Wang et al. The outcome of their in vitro study demonstrated the positive effect of the drug on SARS-CoV-2;[7] following these results, multiple clinical studies have been conducted. A multicenter study by the Department of Science and Technology of Guangdong Province showed that adding chloroquine into patients' drug regimen can shorten the hospitalization duration and enhance the outcome of the treatment. Further, the optimum dosage recommended was administering 500 mg of chloroquine phosphate twice a day for 10 days with no contraindication.[8]
Hydroxychloroquine is a drug similar to chloroquine with a slightly molecular difference. In an in vitro study which compared these two, hydroxychloroquine demonstrated greater potency against SARS-CoV-2 activity.[52,60,61] Therefore, Colson et al. recommended prescription of it as the first choice in SARS-CoV-2 treatment with a dose of 600 mg/day.[62] Another research recommends administering the hydroxychloroquine 200 mg/day for 5–20 days based on the patients' condition.[59] A recent study in France showed that adding azithromycin (500 mg as a loading dose followed by 250 mg/day) will enhance the result of the treatment.[63] However, there are some studies with different results, e.g., Borba et al. discontinued early when preliminary results showed higher rates of mortality among the patients taking the high dose of chloroquine.[64] In addition, FDA cautions about the use of chloroquine or hydroxychloroquine to treat the patients with COVID-19 and recommend monitoring the patient for adverse effects.[65]
Angiotensin-converting enzyme 2
ACE2 is a cleavage enzyme taking part in the renin–angiotensin cascade. This protein is expressed in various organs throughout the body, especially in type 2 pneumocytes.[66,67] The structure of this enzyme provides an extracellular domain that works as a receptor for SARS-CoV-2 virus.[68] An in vitro study showed that recombinant ACE immunoglobulin is a great neutralizer of the virus, and it can be prescribed for treatment and even prophylaxis.[69] Gurwitz et al. suggested soluble ACE2 medication as a competitive component to neutralize the virus. Furthermore, angiotensin receptor blockers such as losartan and telmisartan are suggested to be used to prevent acute respiratory syndrome. However, the possibility of hypotension exacerbation should be noticed.[70,71,72,73]
Since, angiotensin-converting enzyme inhibitor (ACEI) and angiotensin II receptor blocker (ARB) have been shown to upregulate the expression of ACE2 or prevent the loss of ACE2 in the heart;[74,75] they may have similar effect in the lung tissue. Hence, it could be supposed that ACEI and ARB prescription in COVID-19 patient may make them vulnerable due to additional virus entrance and replication in the type II alveolar cells.[75]
However, multiple regulatory association has recommended neither to stop taking ACEI or ARB in patients who are suffering from hypertension nor discontinue these drugs COVID-19 patients who were previously taking ACEIs.[75]
Antiviral drugs
Remdesivir is a broad-spectrum antiviral medicine which was synthesized for Ebola in 2017.In vitro studies proved the inhibitory effect of this medicine on MERS and SARS-CoV.[76] Huang et al. confirmed the same results for SARS-CoV-2.[7] Several studies suggest adding remdesivir into drug regimen.[77,78] At the time of preparing this paper, clinical trials are being conducted, and on May 1, 2020, remdesivir has been approved by the FDA for COVID-19. In an ongoing random clinical trial by Zhu et al., remdesivir is administered to the patient 200 mg on the 1st day following by 100 mg/day for 9 days.[79]
Lopinavir is an antiviral drug that was used for SARS in 2003. Therefore, it was assumed that it could be a beneficial treatment for SARS-CoV-2.[80] Liu et al. claimed that using lopinavir 400 mg/12 h is advantageous by decreasing the viral load and improvement in clinical symptoms. However, it is important to notice to the risk of hypokalemia and the adverse digestive effect of the drug.[81] Several other studies also have been reported the therapeutic effects using lopinavir/ritonavir.[82] Prescribing lopinavir/ritonavir (400–100 mg/12 h) plus arbidol 200 mg/8 h causes a significant improvement in patients.[75] In a study in Singapore, lopinavir/ritonavir (200–100 mg/12 h for 14 days) was used. Although this treatment was effective in several patients, gastrointestinal side effects (nausea, vomiting, and diarrhea) were developed in the majority of cases.[83]
Arbidol is another antiviral drug that can inhibit COVID-19 infections. As mentioned, some studies used this medicine in combination with lopinavir and ritonavir.[75,84] Wang et al. reported that 400 mg arbidol three times a day can reduce the mortality rate and duration of hospitalization.[85] However, in another study, the standard administered dosage has been reported 200 mg three times a day for less than 10 days.[86]
During the epidemic of SARS and MERS, ribavirin was a therapeutic medicine, and it was administered 2 g intravenous followed by 400–600 mg/8 h. Therefore, it is assumed that it has the same impact in SARS-CoV-2 infection.[87] However, in COVID-19 infections, it is recommended to use 500 mg every 8–12 h via intravenous infusion and in combination with either lopinavir and ritonavir or INF-α.[86,88] INF-α is a wide-spectrum antiviral medicine. Vapor inhalation of 5 million U is helpful in SARS-CoV-2 disease, and it is preferred to be used in combination with other drugs.[86]
Common antiviral drugs such as ganciclovir, acyclovir, and neuraminidase inhibitors (oseltamivir [Tamiflu], peramivir, and zanamivir) are not recommended.[89,90,91]
Corticosteroids
Proper corticosteroid therapy in severe ARDS patients can alleviate their symptoms and improve patients' well-being. However, it will not reduce the duration of hospitalization. This therapy can be done using 40–80 mg methylprednisolone daily (not more than 2 mg/kg/day) for patients suffering from SARS.[92,93] However, a recent study claims that corticosteroid does not have benefit SARS-CoV treatment.[94,95]
Other drugs
The concentration of ILs during SARS-CoV infection is higher than healthy people. Tocilizumab is an anti-IL-6 monoclonal antibody. The anti-inflammatory effects of this drug can improve patients' symptoms.[96]
Olaparib and mefuparib hydrochloride (CVL218) are inhibitors of poly ADP-ribose polymerase 1.In vitro study shows that these drugs (especially CVL218) inhibit SARS-CoV-2 replication.[97]
Teicoplanin is an antibiotic that has shown therapeutic effects against MERS, Ebola, SARS-CoV.[98] Studies show that this drug can be helpful in SARS-CoV-2 management. 400 mg/day teicoplanin will provide blood concentration above the therapeutic level.[99,100,101]
Further, herbal-based medicines can be effective, but further studies are needed.[102]
NUTRITION AND SUPPLEMENT
The high morbidity and mortality of COVID-19 virus highlight the need for additional strategies to support the optimal function of the immune system to reduce the impact of the infection.[103] Now, the effects of nutrition on immune function are well established. Some of the nutrients such as fatty acids and omega-3, vitamins including vitamins A, C, D, E, B6, B12, and B9, and minerals, including iron, selenium, magnesium, zinc, and copper play important roles in supporting both the adaptive and innate immune systems[104] and so might impact on the treatment of COVID-19. The effects of these micronutrients in supporting immune function are suggested via the production of antimicrobial proteins, development of physical barriers, phagocytic activities of macrophages and neutrophils, and promotion of inflammation.[105]
The micronutrients with the strongest evidence for immune support against COVID-19 are Vitamin A, Vitamin C, Vitamin D, omega-3, selenium, and zinc. However, there is no proved therapy by nutrition/supplements for this novel virus, so far.
Vitamin A
Vitamin A is a fat-soluble vitamin, and β-carotene is its plant-derived precursor. Some studies demonstrated that Vitamin A supplementation diminishes morbidity and mortality in a variety of infectious diseases and protects against the complications of life-threatening infections such as respiratory diseases.[106] It had been reported that the diet with low Vitamin A might compromise the inactivated bovine CoV vaccine's effectiveness.[107] Thus, Vitamin A could be a promising option for the treatment of COVID-19 and lung infection prevention.[108]
Vitamin C
Vitamin C is a water-soluble vitamin that has an important role in the synthesis of collagen, supporting immune functions and integrity of the epithelial barriers. In addition, Vitamin C acts as an antioxidant to protect cell membranes from damage caused by free radicals.[109] The evidence shows the therapeutic use of Vitamin C to prevent inflammatory hyperactivation in the lymphoid and myeloid cells. Some of the clinical trials have reported the effects of Vitamin C to protect against infection caused by a CoV and incidence of the respiratory tract infections.[110] As COVID-19 causes lower respiratory tract infection, Vitamin C could be one of the effective choices in the treatment strategies of this disease. According to a recent study, the high-dose infusion of Vitamin C might be a timely choice for the treatment of the COVID-19-related ARDS.[111]
Vitamin D
Vitamin D is a fat-soluble vitamin and also a hormone essential for life, which has a role in maintaining bone integrity and stimulates the maturation of immune cells. Vitamin D regulates antimicrobial proteins and protects the lungs against infection as well.[112] A high prevalence of Vitamin D deficiency is observed in adults and elderly people, especially in the dark months of winter. It has been reported that decreased Vitamin D in calves causes the infection of bovine CoV. A significant body of studies suggests that the dietary recommendation allowance for Vitamin D is unlikely to raise the serum levels needed for adequate function of the immune system, and it needs to supplementation.[113,114] Moreover, Vitamin D regulates the renin-angiotensin system that is involved in binding the virus to the host cells. Decreased Vitamin D is along with the increased activity of this system.[115] Low-to-moderate evidence suggests the potential benefits of Vitamin D supplementation with pulmonary exacerbations, including tuberculosis, or lower/upper respiratory infection.[116] Vitamin D can involve in the elimination of viruses and bacteria through regulation of the pathogen recognition receptors, production of antimicrobial peptides, and modulation the adaptive immune system.[117] In observational studies, associations between low serum concentrations of 25(OH) D and acute respiratory infection have been reported.[118] Now, Vitamin D is an effective potential candidate to treat or prevent COVID-19.
Omega-3 polyunsaturated fatty acids
Omega-3 and omega-6 polyunsaturated fatty acids promote anti-inflammatory effects. In addition, the omega-3 polyunsaturated fatty acids could markedly attenuate influenza virus replication via machinery RNA export.[108] Omega-3 served as a novel antiviral drug and so could be considered for one of the potential interventions of COVID-19.
Selenium
Selenium is an essential trace element that plays a crucial role in the defense against infectious diseases.[119] Selenium deficiency impacts the immune response and viral pathogens. Oxidative stress that is caused by selenium deficiency can alter the viral genome, and the pathogenic virus can become highly virulent.[108] Besides, deficiency in the selenium induces rapid mutation of benign variants of RNA viruses to virulence. It was reported that selenium could induce an immune response to CoV vaccine in chickens.[120] Hence, selenium supplementation could be a right choice in the therapeutic strategies of COVID-19.
Zinc
Zinc is a trace element that helps maintain the integrity of skin and mucosal membrane and is important for the development of immunity. Zinc maintains or enhances NK cell cytotoxic activity, differentiation, and turnover of immune cells and improves the phagocytic capacity of monocytes.[105] Marginal zinc deficiency can impact immunity and increase susceptibility to infectious diseases.[121] Studies demonstrated that supplementation by zinc could impair the replication of a variety of RNA viruses and reduce morbidity and mortality caused by lower respiratory tract infections.[105,122] It has been reported that the combination of zinc and pyrithione inhibits the replication of SARS-CoV as well.[123] Therefore, a zinc supplement may have an inhibitory effect on COVID-19 and its symptoms.
COVID-19 POTENTIAL VACCINE CANDIDATES
According to the World Health Organization (WHO) advises and other reports, vaccination strategy against COVID-19 is one of the most effective and main ways to prevent/therapeutic of disease, and it gained a priority for governments and public health organizations around the world. Vaccination against COVID-9 will minimize the size of the virus spread and reduce global morbidity and mortality and so helps health systems with the opportunity to scale up and respond sufficiently in outbreaks.
Further, based on the “WHO Draft Landscape of COVID-19 Candidate Vaccines on June 18, 2020,” “Regulatory Affairs Professionals Society on March 23” as well as scientific reports, more than 140 potential preventive/therapeutic vaccines candidates projects are ongoing and are in different stages of preclinical (127 candidates) and human clinical trials (13 candidates) and exploiting different technologies and platforms currently ongoing in different parts of the world.
Furthermore, in “Coronavirus disease 2019 (COVID-19) Situation Report-65” on March 25, 2020, the WHO has activated the Research and Development blueprint to accelerate diagnostics, vaccines, and therapeutics.
Until now, there is no certified vaccine against COVID-19 (WHO Draft Landscape, 2020), although multiple attempts are in progress to develop effective vaccines. In February 2020, the WHO announced that the COVID-19 vaccine will not be available for at least the next 1.5 years, and most vaccine candidates are still in the early stage of developments.[7] However, in comparison, vaccines funded by the Coalition for Epidemic Preparedness Innovations (CEPI) and FDA or their methodology already approved can be available and certify in less time.[124]
Although many vaccine candidates for CoVs tested in animal models, totally, near 600 unique studies on SARS-CoV-2, SARS-CoV and MERS-CoV have advanced to the phase I and II of clinical trials testing. Still, no vaccine has not yet passes all human clinical trials and filed marketing authorization.[124,125,126,127,128] If these vaccines were available, they could use and probably provide acceptable immunity against COVID-19.[129]
Initially, before the virus has spread to different countries and made it available to research organizations and companies, many companies and organizations tried to design COVID-19 vaccines through the first Chinese published genome in Genbank. In addition, three vaccines projected being supported by the CEPI, include projects by the biotechnology companies of Moderna, Inovio Pharmaceuticals, and the University of Queensland.
Current target antigen selection and vaccine platforms in COVID-19 are designed mostly based on SARS-CoV and MERS-CoV previous vaccines studies, which may help the quick development of successful vaccines.[130] Most immunoresponses are against the structural proteins (spike glycoprotein, envelope, membrane, and nucleocapsid), so these proteins are used as main vaccine protein candidates in the present vaccine developments.[131]
In case of COVID-19, live-attenuated virus mutants are not suitable choices for vaccination because of virus recombination (especially homologous recombination) and reversion to a virulent strain risk, requiring cold chain circulation and the probability of susceptibility of the population such as infants, immunocompromised individuals, or elderly individuals. Base on the WHO Draft Landscape in June 2020, Moderna/NIAID Messenger RNA (LNP-encapsulated mRNA) vaccines are first and frequent vaccines that have entered to different human clinical phase 1, 2, and 3 (to evaluate their safety and immunogenicity).
In addition, immunoinformatics and virus genome sequences in databases can be used to design and develop new generations of vaccines.[132,133]
NONPHARMACEUTICAL INTERVENTIONS FOR COVID-19
National planning and public health action are crucial to minimize the spread of the COVID-19, the most severe seen viral respiratory disease since the 1918 H1N1 influenza pandemic in the population. Prevention of COVID-19 is extremely challenging because of many infected individuals without or with mild symptoms who have viral shedding.[134] Therefore, well-organized surveillance is essential for preventing transmission to moderate the growth rate of this epidemic, and the increased demand of healthcare services, as well as reduce the total number of infected patients.[135]
Nonpharmaceutical (NP) interventions developed for lowering contact rates and transmission of the infection are crucial for the management of COVID-19 because there are no approved vaccines or treatment.[136]
Control of COVID-19 relies on containment of three main routes including sources of infection, routes of transmission, and susceptible hosts.
The main goal of NP measures is to reduce the impact of an outbreak by reducing the number of contacts that result in disease transmission. Depending on their implementation, NP measures can postpone the time to epidemic peak and reduce the overall number of cases.[137]
NP interventions can be implemented at all stages of an epidemic based on the containment and mitigation phases. These measures encompass from personal protective measures, such as hand, respiratory, and environmental hygiene, to activities demanding the engagement of communities and the involvement of national authorities, such as social distancing and travel restrictions.
During a widespread epidemic, a combined approach may increase the efficacy of NP measures and mitigate the disease impact, as well as reduce socioeconomic costs. NP measures need to be implemented according to their necessity, acceptability, and feasibility.[137]
Finding the best time is a critical determinant of success or failure, as outbreak and outbreak response could be mentioned as time contest. Therefore, the correct and on-time application of NP measures is necessary for their effectiveness during the epidemic phases.
It is worth noting that digital technologies can provide advantageous tools to effective and quick implementation of NP interventions. They include the telecommunication networks; big-data analysis, internet of things; and artificial intelligence (AI).[138]
Telemedicine platforms not only provide good care for confined people but also allow health workers to protect themselves by performing initial screening through teleconsultations. It can also improve public health education and risk communication. Mathematical models using big-data and AI can help us understand how the pandemic could spread across the world and find how control methods mitigate transmission.
In conclusion, the expeditious implementation of NP measures to tackle COVID-19 epidemic suppression is the only worthwhile strategy at the current time.
CHALLENGES AND PROMISING FUTURE
Despite the researchers' effort to better understanding COVID-19, a lot of issues remain unclear regarding treatment in different comorbidities, prevention, transmission, and other aspects.[136,137,138,139,140] There are some documents that show the presence of the virus in patient stools.[139,140,141] However, the fecal-oral transmission of COVID-19 remains unclear.[135,142]
There are no approved drugs for the management of the disease, and it is the main challenge for the control of the pandemic infection. Protease inhibitors, neuraminidase inhibitors, nucleoside analogs, RNA synthesis inhibitors, anti-inflammatory drugs, ACE2, arbidol, and remdesivir are found to be effective for the disease.[90,139]
Similar to MERS and SARS, SARS-CoV-2 infection does not have a unique clinical manifestation, and symptoms mostly overlap with other severe respiratory infections. As a result, a definite diagnosis is difficult to reach. Delay in the diagnosis of the disease can lead to new cases. In addition, many cases are asymptomatic, and it is hard to estimate when the disease will peak and what problems will be in the epidemic peak. Protecting healthcare personals is another serious challenge.[143,144] Further, there is currently no specific vaccine for COVID-19.[145]
Finally, sometimes, it is difficult to diagnose COVID-19 because the laboratory tests and radiographic images are not fit with the patient's clinical symptoms. False-negative cases are discharged from the hospital and contact with other people, while false-positive cases with pneumonia or fever are incorrectly isolated with other patients in a medical ward.[134]
The most promising treatments and vaccine are the leading priority for control of this novel CoV and more rigorous evidence is needed in this area.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The present study was conducted in the Infectious Diseases and Tropical Medicine Research Center of Isfahan University of Medical Sciences. We would like to thank the Research Center for supporting of this research.
REFERENCES
- 1.Muller MP, McGeer A. Severe acute respiratory syndrome (SARS) coronavirus. Semin Respir Crit Care Med. 2007;28:201–12. doi: 10.1055/s-2007-976492. [DOI] [PubMed] [Google Scholar]
- 2.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirani K, Sheikhbahaei E, Torkpour Z, Ghadiri Nejad M, Kamyab Moghadas B, Ghasemi M, et al. A narrative review of COVID-19: The new pandemic disease. Iran J Med Sci. 2020;45:233–49. doi: 10.30476/ijms.2020.85869.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronavirus Outbreak. 2020. Jun 20, Available from: https://wwwworldometersinfo/coronavirus/
- 5.Cheng ZJ, Shan J. 2019 Novel coronavirus: Where we are and what we know. Infection. 2020;48:155–63. doi: 10.1007/s15010-020-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–6. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan KW, Wong VT, Tang SC. COVID-19: An update on the epidemiological, clinical, preventive and therapeutic evidence and guidelines of integrative Chinese-Western Medicine for the management of 2019 novel coronavirus disease. Am J Chin Med. 2020;48:737–62. doi: 10.1142/S0192415X20500378. [DOI] [PubMed] [Google Scholar]
- 10.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J Pediatrics. 2020;13:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddell SG, Anderson R, Cavanagh D, Fujiwara K, Klenk HD, Macnaughton MR, et al. Coronaviridae. Intervirology. 1983;20:181–9. doi: 10.1159/000149390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–53. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehr AR, Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102–8. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–8. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velavan TP, Meyer CG. The COVID-19 Epidemic.Tropical medicine & international health. 2020;25(3):278. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Zhang Y, Liu S, Peng H, Mackey V. Coronaviruses and the associated potential therapeutics for the viral infections. J Infect Dis Ther. 2020;8:2. [Google Scholar]
- 20.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–5. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 21.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–50. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 22.Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, Al-Jabir A, et al. World health organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–6. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song D, Park B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–75. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10:e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Tang J, Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol. 2020;92:441–7. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: Findings of 487 cases outside Wuhan. Crit Care. 2020;24:108. doi: 10.1186/s13054-020-2833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–4. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–92. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigrist CJ, Bridge A, Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brielle ES, Schneidman-Duhovny D, Linial M. The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses. 2020;12(5):497. doi: 10.3390/v12050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M. CellPpyroptosis, a Potential Pathogenic Mechanism of 2019-nCoV Infection. 2020 [Google Scholar]
- 34.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020 [Google Scholar]
- 35.Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508–9. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 36.Fan YY, Huang ZT, Li L, Wu MH, Yu T, Koup RA, et al. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch Virol. 2009;154:1093–9. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–2. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min CK, Cheon S, Ha NY, Sohn KM, Kim Y, Aigerim A, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep. 2016;6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–9. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. Clinical Management of COVID-19. 2020:62. [PubMed] [Google Scholar]
- 41.Li T, Lu H, Zhang W. Clinical observation and management of COVID-19 patients. Emerg Microbes Infect. 2020;9:687–90. doi: 10.1080/22221751.2020.1741327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahin AR, Erdogan A, Agaoglu PM, Dineri Y, Cakirci AY, Senel ME, et al. 2019 Novel coronavirus (COVID-19) outbreak: A review of the current literature. EJMO. 2020;4:1–7. [Google Scholar]
- 43.Wu D, Wu T, Liu Q, Yang Z. The SARS-CoV-2 outbreak: What we know. Int J Infect Dis. 2020;94:44–8. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-An update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.World Health Organization. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance, 2 March, 2020 WHO. 2020 [Google Scholar]
- 46.Shen M, Zhou Y, Ye J, Abdullah Al-Maskri AA, Kang Y, Zeng S, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020;10:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections-The state of the art. Emerg Microbes Infect. 2020;9:747–56. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh GC, Hoenig H. How should the rehabilitation community prepare for 2019-nCoV? Arch Phys Med Rehab. 2020;101(6):1068–1071. doi: 10.1016/j.apmr.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–44. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, Liu B, Yu Y, Wang X, Du Y, Gu J, et al. The characteristics and clinical value of chest CT images of novel coronavirus pneumonia. Clin Radiol. 2020;75:335–40. doi: 10.1016/j.crad.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forouzesh M, Rahimi A, Valizadeh R, Dadashzadeh N, Mirzazadeh A. Clinical display, diagnostics and genetic implication of novel coronavirus (COVID-19) epidemic. Eur Rev Med Pharmacol Sci. 2020;24:4607–15. doi: 10.26355/eurrev_202004_21047. [DOI] [PubMed] [Google Scholar]
- 53.Huang P, Liu T, Huang L, Liu H, Lei M, Xu W, et al. Use of Chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295:22–3. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology. 2020;296:E32–40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, et al. Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection. Radiology. 2020;295:685–91. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Touret F, de Lamballerie X. Of chloroquine and COVID-19. Antiviral Res. 2020;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu W, Zhu HL, Duan Y. Effective chemicals against novel coronavirus (COVID-19) in China. Curr Top Med Chem. 2020;20:603–5. doi: 10.2174/1568026620999200305145032. [DOI] [PubMed] [Google Scholar]
- 59.Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Critical Care. 2020;57:279–83. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–9. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valizadeh R, Dadashzadeh N, Zakeri R, James Kelllner S, Rahimi M. Drug therapy in hospitalized patients with very severe symptoms following COVID-19. J Nephropharmacol. 2020;9:e21. [Google Scholar]
- 62.Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020;55:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raoult D, Philippe Gautret JC, Parola P, Hoang VT, Meddeb L, Morgane Mailhe BD, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Borba MG, Val FF, Sampaio VS, Alexandre MA, Melo GC, Brito M, et al. Effect of high vs.low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: A randomized clinical trial. JAMA Network Open. 2020;3:e208857. doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.FDA. FDA Cautions Against use of Hydroxychloroquine or Chloroquine for COVID-19 Outside of the Hospital Setting or a Clinical Trial Due to Risk of Heart Rhythm Problems. 2020. [[Last accessed on 2020 May 08]]. Available from: https://wwwfdagov/drugs/drug-safety-and-availability/fda-cautions-against-use-hydroxychloroquine-or-chloroquine -covid-19-outside-hospital-setting-or .
- 66.Serfozo P, Wysocki J, Gulua G, Schulze A, Ye M, Liu P, et al. Ang II (Angiotensin II) conversion to angiotensin-(1-7) in the circulation Is POP (prolyloligopeptidase)-dependent and ACE2 (angiotensin-converting enzyme 2)-independent. Hypertension. 2020;75:173–82. doi: 10.1161/HYPERTENSIONAHA.119.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus.A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: A potential approach for coronavirus infection therapy? Clin Sci (Lond) 2020;134:543–5. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 69.Lei C, Fu W, Qian K, Li T, Zhang S, Ding M, et al. Potent Neutralization of 2019 Novel Coronavirus by Recombinant ACE2-Ig. 2020 doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Develop Res. 2020 doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mubarak M, Nasri N. COVID-19 nephropathy; an emerging condition caused by novel coronavirus infection. J Nephropathol. 2020;9:e21. [Google Scholar]
- 72.Tolouian R, Zununi Vahed S, Ghiyasvand S, Tolouian A, Ardalan M. COVID-19 interactions with angiotensin-converting enzyme 2 (ACE2) and the kinin system; Looking at a potential treatment. J Renal Inj Prev. 2020;9:e19. [Google Scholar]
- 73.Valizadeh R, Baradaran A, Mirzazadeh A, Bhaskar L. Coronavirus-nephropathy; renal involvement in COVID-19. J Renal Inj Prev. 2020;9:e18. [Google Scholar]
- 74.Ocaranza MP, Godoy I, Jalil JE, Varas M, Collantes P, Pinto M, et al. Enalapril attenuates downregulation of Angiotensin-converting enzyme 2 in the late phase of ventricular dysfunction in myocardial infarcted rat. Hypertension. 2006;48:572–8. doi: 10.1161/01.HYP.0000237862.94083.45. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Chen X, Lu Y, Chen F, Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 2020;14:64–8. doi: 10.5582/bst.2020.01030. [DOI] [PubMed] [Google Scholar]
- 76.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–5. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko WC, Rolain JM, Lee NY, Chen PL, Huang CT, Lee PI, et al. Arguments in favor of remdesivir for treating SARS-CoV-2 infections. Int J Antimicrob Agents. 2020;55(4):105933. doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Al-Tawfiq JA, Al-Homoud AH, Memish ZA. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med Infect Dis. 2020;34:101615. doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu RF, Gao RI, Robert SH, Gao JP, Yang SG, Zhu C. Systematic review of the registered clinical trials of coronavirus diseases 2019 (COVID-19) medRxiv. 2020;18(1):274. doi: 10.1186/s12967-020-02442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JY. Letter to the editor: Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35:e88. doi: 10.3346/jkms.2020.35.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu F, Xu A, Zhang Y, Xuan W, Yan T, Pan K, et al. Patients of COVID-19 may benefit from sustained Lopinavir-combined regimen and the increase of Eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis. 2020;95:183–91. doi: 10.1016/j.ijid.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, et al. Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–94. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deng L, Li C, Zeng Q, Liu X, Li X, Zhang H, et al. Arbidol combined with LPV/r versus LPV/r alone against corona virus disease 2019: A retrospective cohort study. J Infect. 2020;81:e1–e5. doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769–77. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 87.Zeng YM, Xu XL, He XQ, Tang SQ, Li Y, Huang YQ, et al. Comparative effectiveness and safety of ribavirin plus interferon-alpha, lopinavir/ritonavir plus interferon-alpha and ribavirin plus lopinavir/ritonavir plus interferon-alphain in patients with mild to moderate novel coronavirus disease 2019: study protocol. Chin Med J. 2020;133(9):1132–4. doi: 10.1097/CM9.0000000000000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Du B, Qiu H, Zhan X, Wang Y, Kang H, Li X, et al. Pharmacotherapeutics for the new coronavirus pneumonia. Chin J Tubercul Res Dis. 2020;43:E012. doi: 10.3760/cma.j.issn.1001-0939.2020.0012. [DOI] [PubMed] [Google Scholar]
- 89.Li H, Wang Y, Xu J, Cao B. Potential antiviral therapeutics for 2019 novel coronavirus. Chin J Tubercul Res Dis. 2020;43:E002. doi: 10.3760/cma.j.issn.1001-0939.2020.0002. [DOI] [PubMed] [Google Scholar]
- 90.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 91.Dadashzadeh N, Farshid S, Valizadeh R, Nanbakhsh M, Rahimi M. Acute respiratory distress syndrome in COVID-19 disease. Immunopathol Persa. 2020;6(2):e16. [Google Scholar]
- 92.Zhao ZW, Zhang FC, Xu M, Huang K, Zhong WN, Cai WP, et al. Clinical analysis of 190 cases of outbreak with atypical pneumonia in Guangzhou in spring, 2003. Zhonghua Yi Xue Za Zhi. 2003;83:713–8. [PubMed] [Google Scholar]
- 93.Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu K, Fang YY, Deng Y, Liu W, Wang MF, Ma JP, et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl) 2020;133:1025–31. doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–5. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai X. An Insight of Comparison between COVID-19 (2019-nCoV) and SARS-CoV in Pathology and Pathogenesis. 2020 [Google Scholar]
- 97.Ge Y, Tian T, Huang S, Wan F, Li J, Li S, et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19.bioRxiv. 2020 doi: 10.1038/s41392-021-00568-6. doi: https://doiorg/101101/20200311986836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baron SA, Devaux C, Colson P, Raoult D, Rolain JM. Teicoplanin: An alternative drug for the treatment of COVID-19? Int J Antimicrob Agents. 2020;55:105944. doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang J, Ma X, Yu F, Liu J, Zou F, Pan T, et al. Teicoplanin potently blocks the cell entry of 2019-nCoV. bioRxiv. 2020 doi: https://doiorg/101101/20200205935387. [Google Scholar]
- 100.Ghelichi-Ghojogh M, Allah Kalteh E, Fararooei M. Coronavirus disease 2019; epidemiology and recommendations. J Prev Epidemiol. 2020;5:e01. [Google Scholar]
- 101.Hamidian Jahromi A, Mazloom S, Ballard D. What the European and American health care systems can learn from China COVID-19 epidemic; action planning using purpose designed medical telecommunication, courier services, home-based quarantine, and COVID-19 walk-in centers. Immunopathol Persa. 2020;6:e17. [Google Scholar]
- 102.Sriwijitalai W, Wiwanitkit V. Herbs that might be effective for the management of COVID-19: A bioinformatics analysis on anti-tyrosine kinase property. J Res Med Sci. 2020;25:44. doi: 10.4103/jrms.JRMS_312_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vetter P, Eckerle I, Kaiser L. COVID-19: A puzzle with many missing pieces. BMJ. 2020;368:m627. doi: 10.1136/bmj.m627. [DOI] [PubMed] [Google Scholar]
- 104.Calder PC, Carr AC, Gombart AF, Eggersdorfer M. Optimal nutritional status for a well-functioning immune system is an important factor to protect against viral infections. Nutrients. 2020;12(4):1181. doi: 10.3390/nu12041181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gombart AF, Pierre A, Maggini S. A review of micronutrients and the immune system-working in harmony to reduce the risk of infection. Nutrients. 2020;12(1):236. doi: 10.3390/nu12010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Villamor E, Mbise R, Spiegelman D, Hertzmark E, Fataki M, Peterson KE, et al. Vitamin A supplements ameliorate the adverse effect of HIV-1, malaria, and diarrheal infections on child growth. Pediatrics. 2002;109:E6. doi: 10.1542/peds.109.1.e6. [DOI] [PubMed] [Google Scholar]
- 107.Jee J, Hoet AE, Azevedo MP, Vlasova AN, Loerch SC, Pickworth CL, et al. Effects of dietary vitamin A content on antibody responses of feedlot calves inoculated intramuscularly with an inactivated bovine coronavirus vaccine. Am J Vet Res. 2013;74:1353–62. doi: 10.2460/ajvr.74.10.1353. [DOI] [PubMed] [Google Scholar]
- 108.Zhang L, Liu Y. Potential interventions for novel coronavirus in China: A systematic review. J Med Virol. 2020;92:479–90. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carr AC, Maggini S. Vitamin C and immune function. Nutrients. 2017;9(11):1211. doi: 10.3390/nu9111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hemilä H. Vitamin C and infections. Nutrients. 2017;9:339. doi: 10.3390/nu9040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carr AC. A new clinical trial to test high-dose vitamin C in patients with COVID-19. Crit Care. 2020;24(1):133. doi: 10.1186/s13054-020-02851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clark A, Mach N. Role of vitamin D in the hygiene hypothesis: The interplay between Vitamin D, Vitamin D receptors, gut microbiota, and immune response. Front Immunol. 2016;7:627. doi: 10.3389/fimmu.2016.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xiao L, Xing C, Yang Z, Xu S, Wang M, Du H, et al. Vitamin D supplementation for the prevention of childhood acute respiratory infections: A systematic review of randomised controlled trials. Br J Nutr. 2015;114:1026–34. doi: 10.1017/S000711451500207X. [DOI] [PubMed] [Google Scholar]
- 114.Vuichard Gysin D, Dao D, Gysin CM, Lytvyn L, Loeb M. Effect of Vitamin D3 supplementation on respiratory tract infections in healthy individuals: A systematic review and meta-analysis of randomized controlled trials. PLoS One. 2016;11:e0162996. doi: 10.1371/journal.pone.0162996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ajabshir S, Asif A, Nayer A. The effects of Vitamin D on the renin-angiotensin system. J Nephropathol. 2014;3:41–3. doi: 10.12860/jnp.2014.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Das RR, Singh M, Naik SS. Vitamin D as an adjunct to antibiotics for the treatment of acute childhood pneumonia. Cochrane Database Syst Rev. 2018;7:CD011597. doi: 10.1002/14651858.CD011597.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hejazi ME, Modarresi-Ghazani F, Entezari-Maleki T. A review of Vitamin D effects on common respiratory diseases: Asthma, chronic obstructive pulmonary disease, and tuberculosis. J Res Pharm Pract. 2016;5:7–15. doi: 10.4103/2279-042X.176542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maes K, Serré J, Mathyssen C, Janssens W, Gayan-Ramirez G. Targeting Vitamin D deficiency to limit exacerbations in respiratory diseases: Utopia or strategy with potential? Calcif Tissue Int. 2020;106:76–87. doi: 10.1007/s00223-019-00591-4. [DOI] [PubMed] [Google Scholar]
- 119.Rayman MP. Selenium intake, status, and health: A complex relationship. Hormones (Athens) 2020;19:9–14. doi: 10.1007/s42000-019-00125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lipinski B. Redox-active selenium in health and disease: A conceptual review. Mini Rev Med Chem. 2019;19:720–6. doi: 10.2174/1389557517666161104125022. [DOI] [PubMed] [Google Scholar]
- 121.Cuevas LE, Koyanagi A. Zinc and infection: A review. Ann Trop Paediatr. 2005;25:149–60. doi: 10.1179/146532805X58076. [DOI] [PubMed] [Google Scholar]
- 122.Mayo-Wilson E, Junior JA, Imdad A, Dean S, Chan XH, Chan ES, et al. Zinc supplementation for preventing mortality, morbidity, and growth failure in children aged 6 months to 12 years of age. Cochrane Database Syst Rev. 2014;5:CD009384. doi: 10.1002/14651858.CD009384.pub2. [DOI] [PubMed] [Google Scholar]
- 123.te Velthuis A, van den Worm S, Sims A, Baric R, Snijder E. Zn2+Inhibits Coronavirus and Arterivirus RNA Polymerase Activity In vitro. 2010 doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pang J, Wang MX, Ang IY, Tan SH, Lewis RF, Chen JI, et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): A systematic review. J Clin Med. 2020;9(3):623. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao W, Tamin A, Soloff A, D'Aiuto L, Nwanegbo E, Robbins PD, et al. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–6. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim E, Okada K, Kenniston T, Raj VS, AlHajri MM, Farag EA, et al. Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine. 2014;32:5975–82. doi: 10.1016/j.vaccine.2014.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jiang S, Lu L, Du L. Development of SARS vaccines and therapeutics is still needed. Future Virol. 2013;8:1–2. doi: 10.2217/fvl.12.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yong CY, Ong HK, Yeap SK, Ho KL, Tan WS. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 131.Shin HS, Kim Y, Kim G, Lee JY, Jeong I, Joh JS, et al. Immune responses to Middle East respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin Infect Dis. 2019;68:984–92. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ranjbar MM, Ebrahimi MM, Shahsavandi S, Farhadi T, Mirjalili A, Tebianian M, et al. Novel applications of immuno-bioinformatics in vaccine and bio-product developments at research institutes. Arch Razi Inst. 2019;74:219–33. doi: 10.22092/ari.2018.122523.1224. [DOI] [PubMed] [Google Scholar]
- 133.Robson B. Computers and viral diseases.Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput Biol Med. 2020;119:103670. doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92(6):568–76. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Heymann DL, Shindo N. COVID-19: What is next for public health? Lancet. 2020;395:542–5. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eurosurveillance Editorial Team. Latest updates on COVID-19 from the European Centre for Disease Prevention and Control. Euro Surveill. 2020;25(6):2002131. doi: 10.2807/1560-7917.ES.2020.25.6.2002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee VJ, Lye DC, Wilder-Smith A. Combination strategies for pandemic influenza response-A systematic review of mathematical modeling studies. BMC Med. 2009;7:76. doi: 10.1186/1741-7015-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ting DS, Carin L, Dzau V, Wong TY. Digital technology and COVID-19. Nat Med. 2020;26:459–61. doi: 10.1038/s41591-020-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–36. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tang A, Tong ZD, Wang HL, Dai YX, Li KF, Liu JN, et al. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg Infect Dis. 2020;26:1337–9. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92:680–2. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5:335–7. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arabi YM, Murthy S, Webb S. COVID-19: A novel coronavirus and a novel challenge for critical care. Intensive Care Med. 2020;46(5):833–6. doi: 10.1007/s00134-020-05955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barry M, Al Amri M, Memish ZA. COVID-19 in the shadows of MERS-CoV in the Kingdom of Saudi Arabia. J Epidemiol Glob Health. 2020;10:1–3. doi: 10.2991/jegh.k.200218.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tang B, Bragazzi NL, Li Q, Tang S, Xiao Y, Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect Dis Model. 2020;5:248–55. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]