Abstract
Neuropilins are transmembrane coreceptors expressed by endothelial cells and neurons. NRP1 and NRP2 bind a variety of ligands, by which they trigger cell signaling, and are important in the development of lymphatic valves and lymphatic capillaries, respectively. This study focuses on identifying rare variants in the NRP1 and NRP2 genes that could be linked to the development of lymphatic malformations in patients diagnosed with lymphedema. Two hundred and thirty-five Italian lymphedema patients, who tested negative for variants in known lymphedema genes, were screened for variants in NRP1 and NRP2. Two probands carried variants in NRP1 and four in NRP2. The variants of both genes segregated with lymphedema in familial cases. Although further functional and biochemical studies are needed to clarify their involvement with lymphedema and to associate NRP1 and NRP2 with lymphedema, we suggest that it is worthwhile also screening lymphedema patients for these two new candidate genes.
Keywords: NRP1, NRP2, NGS, lymphedema, genetic diagnostics
1. Introduction
Neuropilins 1 and 2 are transmembrane coreceptor proteins encoded by the NRP1 and NRP2 genes. They were first identified in the nervous system of Xenopus laevis embryos [1,2]. Although they map to different chromosomes (10p11.22 and 2q33.3, respectively; see Table 1), they have 45–50% amino acid sequence homology [3]. The two neuropilins are implicated in angiogenesis, and both are expressed by endothelial cells and neurons [4].
Table 1.
Comparison of NRP1 and NRP2.
| Characteristics | NRP1 | NRP2 |
|---|---|---|
| Localization | 10p11.22 | 2q33.3 |
| Gene function | Protein coding | Protein coding |
| Expression in lymphatic system | Predominantly lymphatic valves | Small lymphatic vessels and capillaries |
| Lethality in mouse model | Embryo death at E8.5 (Nrp1−/−/Nrp2−/−) Embryo death at E10-E10.5 (Nrp1+/−/Nrp2−/−; Nrp1−/−/Nrp2+/−) |
|
| Lymphatic phenotype in mouse model | Nrp1−/− malformation of valve precursor LECs [18] |
Nrp2−/− reduction or loss of small lymphatic vessels and capillaries [19] Nrp2+/− and Vegfr3+/− abnormal lymphatic development and reduced lymphatic vessel branching [13] |
The two neuropilins have a very short intracellular domain (only about 40 amino acids long) and a longer extracellular region. Both neuropilins complex a large variety of ligands, by which they take part in cell signaling. These transmembrane proteins have a critical role in the regulation of vascular and neural development through binding vascular endothelial growth factors (VEGFs) and semaphorins [5]. The major neuropilin ligands are vascular endothelial growth factors VEGF-A, VEGF-C and VEGF-D [4,6]. While VEGF-A plays a primary role in angiogenesis, VEGF-C and VEGF-D are important for normal development of the lymphatic system. NRP1 and NRP2 also bind other ligands—for instance, hepatocyte growth factor (HGF) [7], EGF receptor [8] and an adhesion receptor L1-CAM [9]. Although they mediate cell signaling by ligand binding, it is unclear whether they also transduce signals on their own. In any case, they take part in many biological processes, and changes in their function may modulate tumor growth and metastasis, affecting vascular and lymphatic biology [10]. This can give rise to pathologies such as cancer and autoimmune disorders [11].
While NRP1 is predominantly expressed in the arterial endothelium and lymphatic valves, NRP2 mainly functions in venous and lymphatic endothelial cells (LECs) [12]. This indicates that NRP1 acts as a semaphorin receptor in the valve endothelium throughout lymphatic vessel development [12]. In contrast, NRP2 is implicated in VEGF-C–driven lymphatic vessel growth [13], while double-heterozygous nrp2+/− vegfr3+/− mice show a decrease in lymphatic vessel sprouting in adult organs (Table 1) [13].
Most information about neuropilin function comes from studies with mice. Early studies of Nrp1−/− mice showed deficient neuronal vascularization, aortic arch malformations and impaired yolk sac vascularization [14,15]. Abnormal blood vessel formation due to overexpression of the Nrp1 gene was also observed [16], although the authors of the study did not investigate the lymphatic phenotype.
Nrp1 was recently implicated in lymphatic valve development via Sema3a/Nrp1 signaling [12]. Sema3a is a ligand bound by Nrp1. Deficiency or blockage of Nrp1 bound to Sema3a leads to malformations of a specific type of LEC that later migrates from the wall of lymphatic vessels to give rise to lymphatic valves. Defects in this process lead to abnormal lymphatic valve development [13,17,18].
In comparison, the Nrp2−/− mouse model provided evidence of the selective role of Nrp2 in the development of lymphatic vessels. Nrp2-deficient mice showed significantly fewer or a complete absence of small lymphatic vessels and capillaries in many organs. The small number of vessels that did develop were wrongly positioned, and in some cases, small vessels and capillaries were larger in size. A decrease in LEC proliferation was also observed, indicating a role of Nrp2 in the proliferation of LECs in selected types of lymphatic vessels and implying the control of positional guidance by Nrp2 [19].
Knocking out Nrp1 and Nrp2 separately has evident effects on the mouse phenotype, and the joint deletion of Nrp1 and Nrp2 has much graver consequences. Takashima et al. [20] reported that Nrp1−/−/Nrp2−/− mice died at an early stage of embryogenesis (E8.5) and displayed severely defective blood vessel development in the yolk sac. In the study, the authors observed that double-knockout mice had severer vascular malformations than Nrp1- or Nrp2-deleted animals. Surprisingly, heterozygous mice with Nrp1+/−/Nrp2−/− or Nrp1−/−/Nrp2+/− genotypes did not survive embryo development either but lived longer than double-knockout mice (E10–E10.5) [20]. Although the authors did not study the development of lymphatics in these double-knockout mice, the premature death of the embryos and the fact that Nrp1 and Nrp2 deletions caused severe lymphatic system malformations suggested that a double knockout also results in impaired lymphatic system development.
The development of small lymphatic vessels, capillaries and valves is important, and its impairment may lead to an overaccumulation of lymph in the tissues and lymphedema. Lymphedema is a pathological condition characterized by swelling, usually at the extremities, accompanied by pain and inflammation [21]. Lymphatic capillaries, the development of which is regulated by NRP2, are indispensable for normal drainage of interstitial fluid into the lymphatic system. Lymphatic capillaries lack a basement membrane and feature endothelial cell junctions that grant the passage of interstitial fluid into the lymphatic system [22]. When lymphatic capillaries and small vessels are malformed, interstitial fluid cannot enter lymphatic collecting vessels and accumulates in the tissues. The proper functioning of valves is also crucial to ensure retrograde lymph flow. Valve defects cause lymph flow obstruction and fluid accumulation [22]. The blockage of lymph flow and lymph accumulation are the primary causes of lymphedema.
In our study, a cohort of 235 Italian lymphedema patients, who tested negative for variants in known lymphedema-associated genes, was screened by a next-generation sequencing (NGS)-targeted panel for variants the in NRP1 and NRP2 genes. A segregation analysis in familial lymphedema cases added evidence that NRP1 and NRP2 qualify as candidate genes to include in the genetic testing of lymphedema patients.
2. Materials and Methods
2.1. Clinical Evaluation
We retrospectively enrolled 235 Caucasian lymphedema patients in this study, which was conducted in accordance with the Declaration of Helsinki, and whose protocol was approved by the Ethics Committee of Azienda Sanitaria dell’Alto Adige, Italy (Approval No. 94-2016). No consanguinity was reported in any family. Clinical diagnosis of lymphedema was made in hospitals across Italy, according to generally accepted criteria. All subjects gave their informed consent for inclusion before they participated in the study.
2.2. Genetic Analysis
Genetic testing was performed on genomic DNA extracted from saliva or peripheral blood of probands, as previously described [23]. Briefly, a custom-made oligonucleotide probe library was designed to capture all coding exons and flanking exon/intron boundaries (±15 bp) of genes known to be associated with lymphedema [24]. We added the candidate genes NRP1 and NRP2 to our panel. DNA from probands was analyzed for germline variants. Variants were confirmed by bidirectional Sanger sequencing on a CEQ8800 Sequencer (Beckman Coulter, Brea, CA, USA).
We searched the international Single Nucleotide Polymorphism Database (dbSNP) and the Human Gene Mutation Database professional for all nucleotide changes. In-silico evaluation of the pathogenicity of sequence changes in NRP1 and NRP2 was performed using the Variant Effect Predictor tool and MutationTaster. Minor allele frequencies were checked in the Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/). All variants were evaluated according to the American College of Medical Genetics and Genomics guidelines [25]. Detailed pretest genetic counseling was provided to all subjects, who were then invited to sign informed consent to use of their anonymized genetic results for research.
2.3. In-Silico Analysis
The primary amino acid sequences of NRP1 and NRP2 in FASTA format (Table 2) were used as targets to search template libraries (i.e., the SWISS-MODEL Template Library (version 2019-10-24) and the Protein Data Bank (release 2019-10-18)) [26] for matching evolution-related structures by means of BLAST [27] and HHBlits [28], as previously described [23]. Briefly, models were based on target-template alignment using ProMod3 of the SWISS-MODEL server [29]. An alternative model was obtained with ProMod-II in those cases in which loop modeling failed with ProMod3 [30]. Coordinates conserved between the target and the template were copied from the template to the model. Insertions and deletions were remodeled using a fragment library. Side chains were then rebuilt. Finally, the geometry of the resulting model was regularized with the CHARMM27 force field [31]. Global and per-residue model quality was assessed using the QMEAN scoring function [32]. BioVia Discovery Studio Visualizer (version 17.2) [33] was used to visualize the modeled protein, to vary the targeted amino acids and to analyze interactions at the molecular level.
Table 2.
NRP1 and NRP2 primary amino acid sequences. Primary amino acid sequences used to search for templates to build models for NRP1 and NRP2 proteins.
| NRP1 | MERGLPLLCAVLALVLAPAGAFRNDKCGDTIKIESPGYLTSPGYPHSYHPSEKCEWLIQAPDPYQRIMINFNPHFDLEDRDCKYDYVEVFDGENENGHFRGKFCGKIAPPPVVSSGPFLFIKFVSDYETHGAGFSIRYEIFKRGPECSQNYTTPSGVIKSPGFPEKYPNSLECTYIVFVPKMSEIILEFESFDLEPDSNPPGGMFCRYDRLEIWDGFPDVGPHIGRYCGQKTPGRIRSSSGILSMVFYTDSAIAKEGFSANYSVLQSSVSEDFKCMEALGMESGEIHSDQITASSQYSTNWSAERSRLNYPENGWTPGEDSYREWIQVDLGLLRFVTAVGTQGAISKETKKKYYVKTYKIDVSSNGEDWITIKEGNKPVLFQGNTNPTDVVVAVFPKPLITRFVRIKPATWETGISMRFEVYGCKITDYPCSGMLGMVSGLISDSQITSSNQGDRNWMPENIRLVTSRSGWALPPAPHSYINEWLQIDLGEEKIVRGIIIQGGKHRENKVFMRKFKIGYSNNGSDWKMIMDDSKRKAKSFEGNNNYDTPELRTFPALSTRFIRIYPERATHGGLGLRMELLGCEVEAPTAGPTTPNGNLVDECDDDQANCHSGTGDDFQLTGGTTVLATEKPTVIDSTIQSEFPTYGFNCEFGWGSHKTFCHWEHDNHVQLKWSVLTSKTGPIQDHTGDGNFIYSQADENQKGKVARLVSPVVYSQNSAHCMTFWYHMSGSHVGTLRVKLRYQKPEEYDQLVWMAIGHQGDHWKEGRVLLHKSLKLYQVIFEGEIGKGNLGGIAVDDISINNHISQEDCAKPADLDKKNPEIKIDETGSTPGYEGEGEGDKNISRKPGNVLKTLDPILITIIAMSALGVLLGAVCGVVLYCACWHNGMSERNLSALENYNFELVDGVKLKKDKLNTQSTYSEA |
| NRP2 | MDMFPLTWVFLALYFSRHQVRGQPDPPCGGRLNSKDAGYITSPGYPQDYPSHQNCEWIVYAPEPNQKIVLNFNPHFEIEKHDCKYDFIEIRDGDSESADLLGKHCGNIAPPTIISSGSMLYIKFTSDYARQGAGFSLRYEIFKTGSEDCSKNFTSPNGTIESPGFPEKYPHNLDCTFTILAKPKMEIILQFLIFDLEHDPLQVGEGDCKYDWLDIWDGIPHVGPLIGKYCGTKTPSELRSSTGILSLTFHTDMAVAKDGFSARYYLVHQEPLENFQCNVPLGMESGRIANEQISASSTYSDGRWTPQQSRLHGDDNGWTPNLDSNKEYLQVDLRFLTMLTAIATQGAISRETQNGYYVKSYKLEVSTNGEDWMVYRHGKNHKVFQANNDATEVVLNKLHAPLLTRFVRIRPQTWHSGIALRLELFGCRVTDAPCSNMLGMLSGLIADSQISASSTQEYLWSPSAARLVSSRSGWFPRIPQAQPGEEWLQVDLGTPKTVKGVIIQGARGGDSITAVEARAFVRKFKVSYSLNGKDWEYIQDPRTQQPKLFEGNMHYDTPDIRRFDPIPAQYVRVYPERWSPAGIGMRLEVLGCDWTDSKPTVETLGPTVKSEETTTPYPTEEEATECGENCSFEDDKDLQLPSGFNCNFDFLEEPCGWMYDHAKWLRTTWASSSSPNDRTFPDDRNFLRLQSDSQREGQYARLISPPVHLPRSPVCMEFQYQATGGRGVALQVVREASQESKLLWVIREDQGGEWKHGRIILPSYDMEYQIVFEGVIGKGRSGEIAIDDIRISTDVPLENCMEPISAFAGENFKVDIPEIHEREGYEDEIDDEYEVDWSNSSSATSGSGAPSTDKEKSWLYTLDPILITIIAMSSLGVLLGATCAGLLLYCTCSYSGLSSRSCTTLENYNFELYDGLKHKVKMNHQKCCSEA |
3. Results
3.1. Clinical and Genetic Evaluation
DNA from 235 Caucasian patients diagnosed with lymphedema, from different Italian hospitals, previously testing negative for variants in known lymphedema and lymphatic malformation-causing genes, was screened for variants in new candidate genes. Here, we report the results for NRP1 and NRP2. No consanguinity was reported in their families.
NGS screening detected two different variants in NRP1 in two families and four different variants in NRP2 in four families. All the variants were heterozygous. The clinical features of the probands and family members are shown in Table 3. A segregation analysis performed on family members carrying variants in NRP1 are shown in Figure 1. The pedigrees of families with variants in NRP2 are shown in Figure 2.
Table 3.
Clinical features of probands (grey background) and family members with variants in the NRP1 and NRP2 genes. Abbreviations: M = male, F = female and ACMG = American College of Medical Genetics and Genomics. VUS: variant of unknown significance.
| Gene | Family | Pedigree | Sex | Age | Clinical Features | Age of Onset | Familial Case | Variant Nomenclature (NRP1: NM_003873.6; NP_001019799.1; NRP2: NM_003872.2; NP_003863.2) |
dbSNP id | ACMG Classification | Frequency in GnomAD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NRP1 | 1 | Proband | M | 68 | lymphedema of the lower limbs | Juvenile | NO | c.2557A > C; p.Ile853Leu | rs760388137 | Likely benign | 0.0001178 |
| NRP1 | 2 | Proband | F | 54 | edema of left foot and ankle | 27 | YES | c.1655G > A; p.Arg552Gln | rs757990959 | VUS | 0.00004873 |
| NRP1 | 2 | Daughter | F | 21 | lymphedema of left lower limb | 10 | YES | c.1655G > A; p.Arg552Gln | |||
| NRP2 | 1 | Proband | F | 18 | edema of left hand | Congenital | NO | c.580T > G; p.Phe194Val | rs755679361 | VUS | 0.000008122 |
| NRP2 | 1 | Mother | F | 44 | Healthy | / | NO | - | |||
| NRP2 | 1 | Father | M | 46 | Apparently healthy but with subclinical symptoms | / | NO | c.580T > G; p.Phe194Val | |||
| NRP2 | 2 | Proband | M | 9 | edema of left foot, ankle and heel since vaccination | 3 months | NO | c.1748T > C; p.Ile583Thr | rs746130411 | VUS | 0.000004068 |
| NRP2 | 3 | Proband | F | 45 | edema of right foot | 32 | YES | c.838C > T; p.Pro280Ser | rs79750907 | Likely benign | 0.001783 |
| NRP2 | 3 | Son | M | 15 | edema of left foot | 9 | YES | c.838C > T; p.Pro280Ser | |||
| NRP2 | 4 | Proband | F | 50 | swelling of lower limbs | 42 | NO | c.1000C > T; p.Arg334Cys | rs114144673 | Likely benign | 0.001525 |
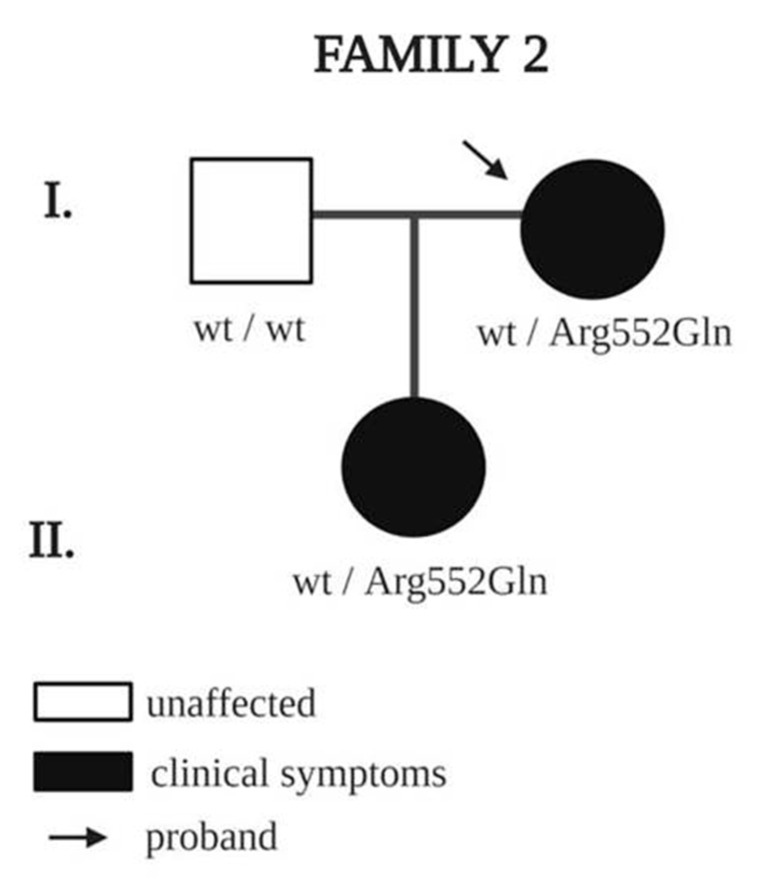
Figure 1.
Pedigree of family 2 with a rare NRP1 variant. Square indicates male, and circles indicate females; black indicates clinical symptoms of lymphedema. Arrows indicate probands, and wt indicates wildtype amino acid.
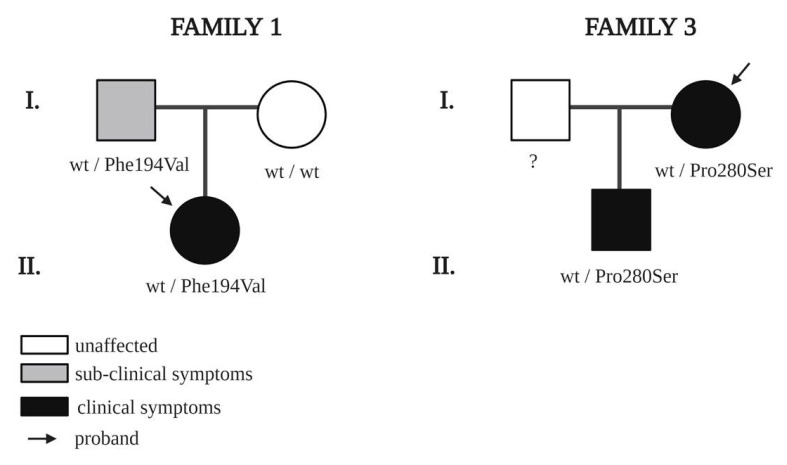
Figure 2.
Pedigrees of families with rare NRP2 variants. Squares indicate males and circles, females; black indicates clinical symptoms of lymphedema; grey indicates subclinical symptoms confirmed by lymphoscintigraphy. Arrows indicate probands. ? means “not screened”, and wt indicates wildtype amino acid.
Regarding variants in the NRP1 gene, the first proband, male, 68 years, has had lymphedema of the lower limbs since his youth. The case is sporadic, and no other family members of the proband were tested. The proband carries a missense single-nucleotide variation c.2557A > C (dbSNP rs760388137) with a GnomAD-reported frequency of 0.0001178.
The second proband with an NRP1 variant, female, 54 years, has stage 2 lymphedema of the left foot and ankle, diagnosed at age 27. The case is familial: the proband’s daughter also has stage 2 lymphedema of the lower limbs (Figure 1). Both carry the same heterozygous missense variant c.1655G > A (dbSNP ID rs757990959) with a frequency of 0.00004873 (GnomAD) classified variant of unknown significance (VUS) by the ACMG [34].
Regarding variants in the NRP2 gene, we identified four families with rare variants. In the first family, the proband, female, 18 years, has congenital edema of the left hand (lymphedema stage 2). Indeed, upper limb lymphoscintigraphy failed to visualize the left axillary lymph nodes. She carries a missense variant c.580T > C (rs755679361), which has an allele frequency in healthy control populations of 0.000008122, according to GnomAD. The segregation analysis in available family members showed that the proband inherited the variant from her father. The father was apparently healthy; however, lymphoscintigraphy showed clear bilateral lymph transport problems, especially on the left side (Figure 3).
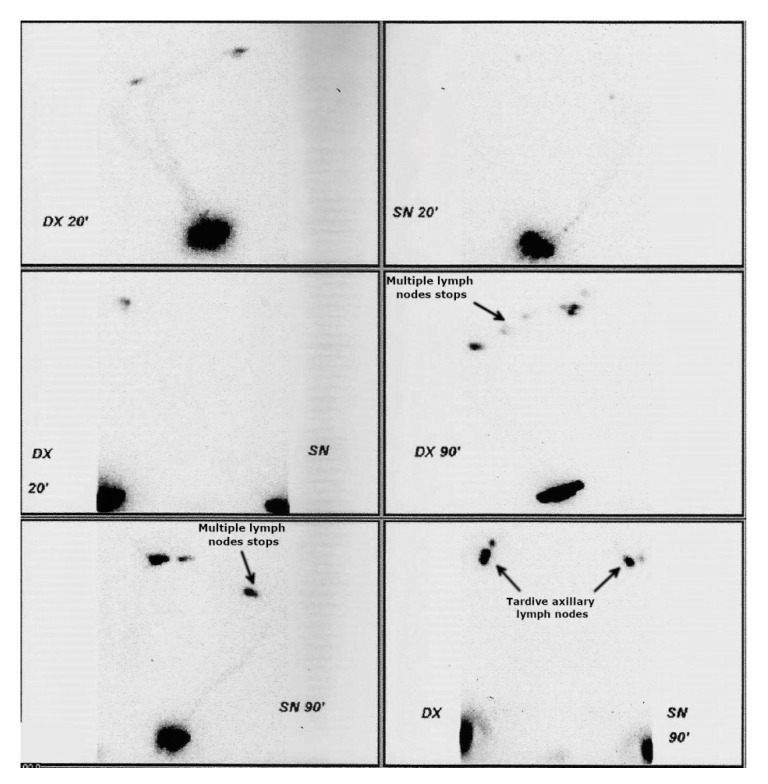
Figure 3.
Lymphoscintigraphy of father of the proband with c.580T > C variants in NRP2 (family 1). Although phenotypically healthy, lymphoscintigraphy highlighted subclinical anomalies (arrows): radiotracer accumulates in multiple lymph nodes of the left (panel 4) and right arm (panel 5). After 90 min of tracer administration, there is reduced tardive visualization of the right axillary lymph nodes (right axillary packet hypogenesis) and even more reduced tardive visualization of the left axillary lymph nodes (left axillary packet hypogenesis) (arrows in panel 6).
The second proband with an NRP2 variant, male, nine years, developed edema of the left foot, ankle and heel after vaccination at age three months. The case is sporadic, and no other family members were tested. The missense variant is classified as VUS (c.1748T > C) (dbSNP rs746130411), with a GnomAD frequency of 0.000004068.
The third proband, female, 45 years, was diagnosed with edema of the right foot at age 32. The heterozygous missense variant is c.838C >T (dbSNP rs79750907) and has a frequency of 0.001783 (GnomAD). This is a familial case, since the proband’s son was also diagnosed with left foot lymphedema at age nine. The son was found to carry the same variant as the proband.
The fourth proband, female, 50 years, was diagnosed with swelling of the lower limbs at age 42. She carries a heterozygous single-nucleotide missense variant c.1000C > T (dbSNP rs114144673), with a frequency of 0.001525 (GnomAD). This is a sporadic case without family history, and no other family members were tested.
3.2. In Silico Analysis, Template Selection and Building
In an attempt to study the possible role of the variants found in our cohort compared to the wildtype, we performed an in-silico study. A wildtype template (Table 2) search with BLAST and HHBlits against the SWISS-MODEL template library (last update: 2019-10-24; last included PDB release: 2019-10-18) produced a total of 267 and 254 templates that matched the NRP1 and NRP2 genes, respectively, with different sequence identity and quality percentages. Details of the top 10 templates are shown in Table 4.
Table 4.
Best models for 3D modeling of the NRP1 and NRP2 structures. Abbreviation: QSQE= quaternary structure quality estimate.
| Template | Seq Identity | Oligo State | QSQE | Found by | Method | Resolution | Seq Similarity | Coverage | Description | |
|---|---|---|---|---|---|---|---|---|---|---|
| NRP1 | 4gz9.1.A | 91.50 | monomer | - | BLAST | X-ray | 2.70Å | 0.60 | 0.61 | Neuropilin-1 |
| 4gz9.1.A | 91.50 | monomer | - | HHBlits | X-ray | 2.70Å | 0.60 | 0.61 | Neuropilin-1 | |
| 2qql.1.A | 51.95 | Homodimer | 0.37 | HHBlits | X-ray | 3.10Å | 0.46 | 0.61 | Neuropilin-2 | |
| 2qql.1.A | 53.14 | Homodimer | 0.35 | BLAST | X-ray | 3.10Å | 0.47 | 0.60 | Neuropilin-2 | |
| 2qqk.1.A | 51.95 | Monomer | - | HHBlits | X-ray | 2.75Å | 0.48 | 0.61 | Neuropilin-2 | |
| 2qqk.1.A | 53.14 | Monomer | - | BLAST | X-ray | 2.75Å | 0.47 | 0.60 | Neuropilin-2 | |
| 2qqm.1.A | 99.78 | monomer | - | BLAST | X-ray | 2.00Å | 0.62 | 0.48 | Neuropilin-1 | |
| 2qqm.1.A | 99.78 | monomer | - | HHBlits | X-ray | 2.00Å | 0.62 | 0.48 | Neuropilin-1 | |
| 2qqo.1.A | 51.36 | monomer | - | HHBlits | X-ray | 2.30Å | 0.46 | 0.48 | Neuropilin-2 | |
| 2qqo.1.A | 52.51 | monomer | - | BLAST | X-ray | 2.30Å | 0.46 | 0.47 | Neuropilin-2 | |
| NRP2 | 2qql.1.A | 100.00 | Homo-monomer | 0.51 | BLAST | X-ray | 3.10 Å | 0.62 | 0.62 | Neuropilin-2 |
| 2qqk.1.A | 100.00 | monomer | - | BLAST | X-ray | 2.75Å | 0.62 | 0.62 | Neuropilin-2 | |
| 2qql.1.A | 100.00 | Homodimer | 0.51 | HHBlits | X-ray | 3.10Å | 0.62 | 0.61 | Neuropilin-2 | |
| 2qqk.1.A | 100.00 | monomer | - | HHBlits | X-ray | 2.75Å | 0.62 | 0.61 | Neuropilin-2 | |
| 2qqo.1.A | 100.00 | Monomer | - | HHBlits | X-ray | 2.30Å | 0.62 | 0.48 | Neuropilin-2 | |
| 2qqm.1.A | 51.81 | Monomer | - | HHBlits | X-ray | 2.00Å | 0.46 | 0.47 | Neuropilin-1 | |
| 2qqm.1.A | 53.05 | monomer | - | BLAST | X-ray | 2.00Å | 0.46 | 0.48 | Neuropilin-1 | |
| 2qqj.1.A | 100.00 | monomer | - | BLAST | X-ray | 1.95Å | 0.62 | 0.34 | Neuropilin-2 | |
| 2qqj.1.A | 100.00 | monomer | - | HHBlits | X-ray | 1.95Å | 0.62 | 0.34 | Neuropilin-2 | |
| 2qqi.1.A | 51.77 | monomer | - | BLAST | X-ray | 1.80Å | 0.46 | 0.33 | Neuropilin-1 |
Based on the percentage of sequence identity, similarity and best quality square, the 4gz9.1.A and 2qql.1.A chains were selected to align the template and query sequences in order to build models of NRP1 and NRP2 (Figure 4 and Figure 5).
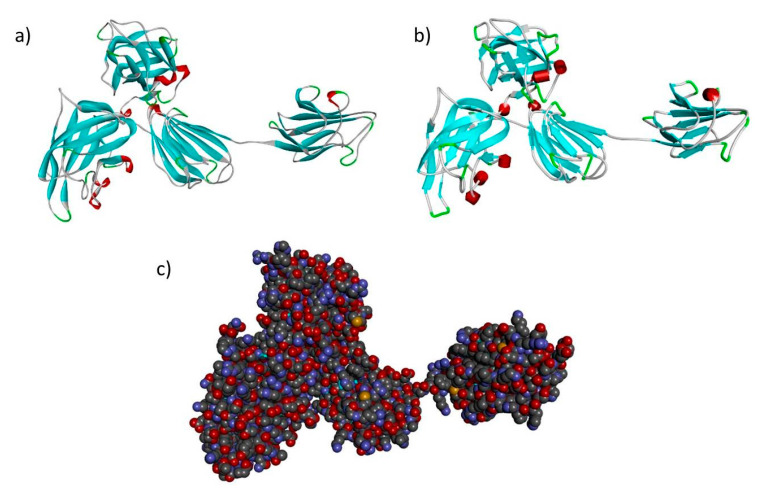
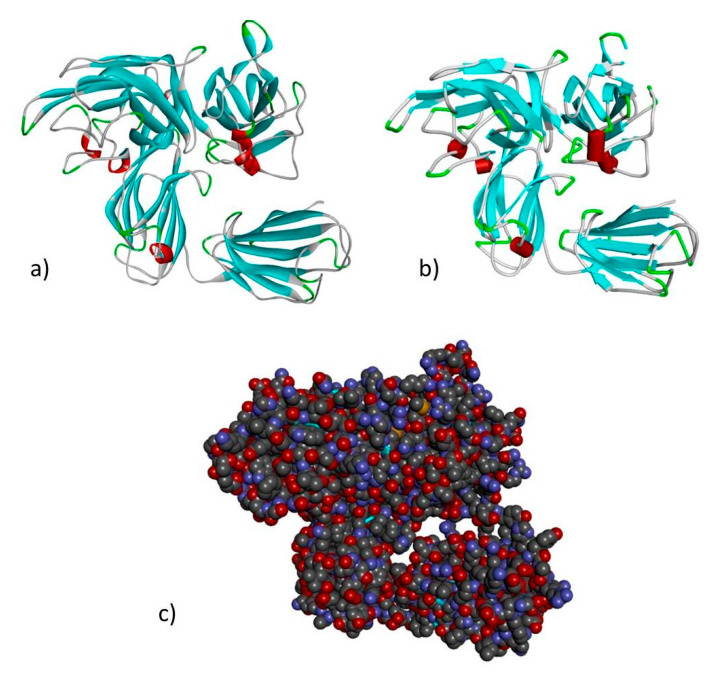
Figure 4.
Modeled structure of the NRP1 gene in (a) ribbon (b) schematic and (c) CPK views. Cyan regions are β sheets, white represents loops and red represents α helices.
Figure 5.
Modeled structure of the NRP2 gene in (a) ribbon (b) schematic and (c) CPK views. Cyan regions are β sheets, white represents loops and red represents α helices.
Then, we entered the model in the Discovery studio visualizer to generate versions of the NRP1 structure with Arg552Gln and versions of the NRP2 structure with Phe194Val, Pro280Ser, Arg334Cys and Ile583Thr. Unfortunately, the template to generate the model for Ile853 was not available, so we could not study its interaction with adjacent residues. A molecular-level interaction analysis between native/variant residues was performed (Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10 show snapshots). Details of the residues involved in the interactions, the types of bond they formed and bond lengths in angstrom units are shown in Table 5 and Table 6.
Figure 6.
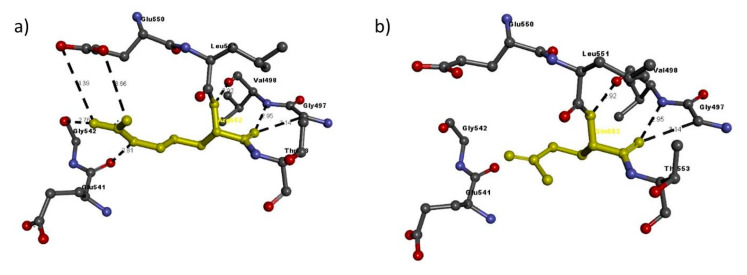
Molecular interactions of (a) Arg552 and (b) Gln552 (in yellow) of the modeled NRP1 protein with adjacent residues.
Figure 7.
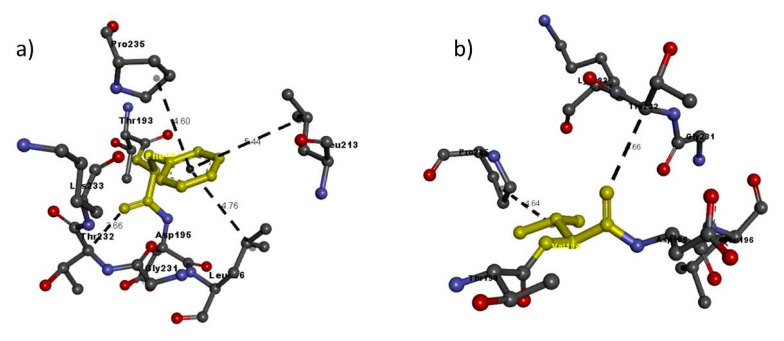
Molecular interactions of (a) Phe194 and (b) Val194 (in yellow) of the modeled NRP2 protein with adjacent residues.
Figure 8.
Molecular interactions of (a) Pro280 and (b) Ser280 (in yellow) of the modeled NRP2 protein with adjacent residues.
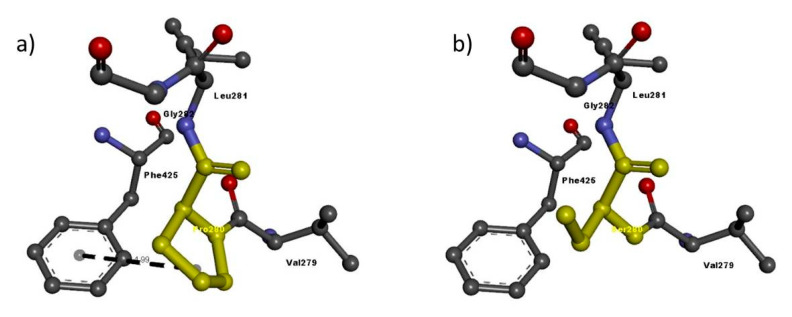
Figure 9.
Molecular interactions of (a) Arg334 and (b) Cys334 (in yellow) of the modeled NRP2 protein with adjacent residues.
Figure 10.
Molecular interactions of (a) Ile583 and (b) Thr583 (in yellow) of the modeled NRP2 protein with adjacent residues.
Table 5.
Details of molecular interactions with adjacent residues of wildtype Arg552 or variant Gln552 of the modeled NRP1 protein in family 2.
| Variant | Amino Acid | Molecular Interactions | Bond Length in Angstroms | Bond Type | |
|---|---|---|---|---|---|
| NRP1 FAM2 |
Arg552Gln | Arg552 | Arg552: N–Glu550: O | 3.66 | H-bond |
| Arg552: N–Glu550: O | 4.39 | H-bond | |||
| Val498: N–Arg552: O | 2.95 | H-bond | |||
| Arg552: N–Val498: O | 2.92 | H-bond | |||
| Arg552: N–Glu541: O | 2.81 | H-bond | |||
| Arg552: N–Gly542: O | 2.76 | H-bond | |||
| Gly497: C–Arg552: O | 3.14 | H-bond | |||
| Gln552 | Val498: N–Gln552: O | 2.95 | H-bond | ||
| Gln552: N–Val498: O | 2.92 | H-bond | |||
| Gly497: C–Gln552: O | 3.14 | H-bond |
Table 6.
Details of molecular interactions with adjacent residues of the wildtype or variant residue in the modeled NRP2 protein in family 1 (Phe194Val), family 3 (Pro280Ser), family 4 (Arg334Cys) and family 2 (Ile583Thr).
| Variant | Amino Acid | Molecular Interactions | Bond Length in Angstroms | Bond Type | |
|---|---|---|---|---|---|
| NRP2 FAM 1 | Phe194Val | Phe194 | Thr232: C–Phe194: O | 3.66 | H-bond |
| Phe194: A–Phe196 | 4.76 | H-bond | |||
| Phe1941: A–Phe213 | 5.44 | Pi interaction | |||
| Phe194: A–Pro235 | 4.60 | Pi interaction | |||
| Val194 | Thr232: C–Val194: O | 3.66 | H-bond | ||
| Val194: A–Pro235 | 4.64 | Hydrophobic interaction | |||
| NRP2 FAM 3 | Pro280Ser | Pro280 | Phe425–Pro280 | 4.99 | Pi interaction |
| Ser280 | No bonds | - | - | ||
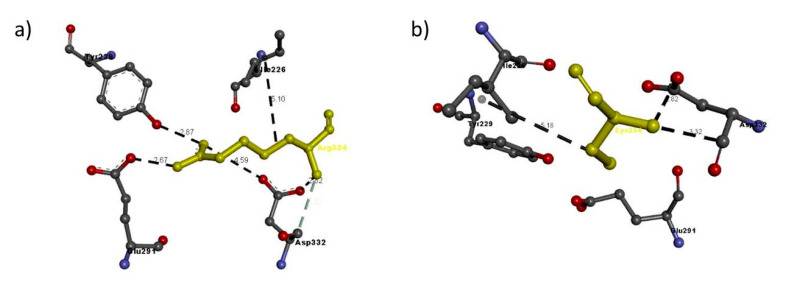
| NRP2 FAM 4 | Arg334Cys | Arg334 | Arg334: N–Glu291: O | 2.67 | H-bond |
| Arg334: N–Asp332: O | 4.59 | H-bond | |||
| Arg334: N–Asp332: O | 2.82 | H-bond | |||
| Arg334: N–Tyr229: O | 2.87 | H-bond | |||
| Asp332: C–Arg334: N | 3.32 | H-bond | |||
| Arg334–Ile226 | 5.10 | Hydrophobic interaction | |||
| Cys334 | Cys334: N - Asp332: O | 2.82 | H-bond | ||
| Asp332: C–A:Cys334:N | 3.32 | H-bond | |||
| A:Cys334–A:Ile226 | 5.18 | Hydrophobic interaction | |||
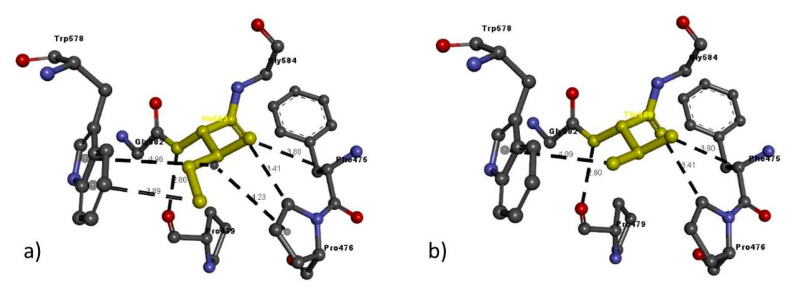
| NRP2 FAM 2 | Ile583Thr | Ile583 | Ile583: N–Pro479: O | 2.80 | H-bond |
| Phe475: C–Ile583: O | 3.80 | H-bond | |||
| Pro476: C–Ile583: O | 3.41 | H-bond | |||
| Ile583: C–Trp578 | 3.89 | Pi interactions | |||
| Pro476: A–Ile583 | 4.23 | Pi interactions | |||
| Trp578: A–Ile583 | 4.91 | Pi interactions | |||
| Thr583 | Thr583: N–Pro479: O | 2.80 | H-bond | ||
| Phe475: C–Thr583: O | 3.80 | H-bond | |||
| Pro476: C–Thr583: O | 3.41 | H-bond | |||
| Thr583: O–Trp578 | 4.09 | Pi interactions |
Briefly, for all the residues analyzed, there is a change in the number or length of the bonds that are formed in the variant with respect to the modeled wildtype structure. Indeed, the in-silico analysis showed that NRP1 encoded with Arg552 has slightly different stability from the Gln552 variant, because it has seven interactions with adjacent residues, whereas the variant has only three that exactly match three of the Arg552 interactions (Table 5 and Figure 6).
By contrast, NRP2 encoded with Phe194 has slightly different stability from the Val194 variant due to four interactions with adjacent residues, whereas the variant has only two, similar to Phe194 but slightly different in bond length (Table 6 and Figure 7).
In the case of Pro280Ser, there is a shift from a nonpolar residue (Pro) to a polar residue (Ser). Differences in the amino acid side chain greatly alter the interaction of the residues with the phenylalanine at residue 425, leading to a loss of Pi interaction. These results suggest that the overall protein structure is altered by these different interactions with nearby residues and is functionally defective (Table 6 and Figure 8).
In the case of Arg334Cys, Arg334 has six interactions with adjacent residues, whereas Cys334 has only three similar to those of Arg334, and the interaction with Ile226 has a different hydrophobic interaction bond length (Table 6 and Figure 9).
Likewise, in the case of Ile583Thr, Ile583 has six interactions with adjacent residues, whereas the Thr583 variant has only four that are the same, but the Pi interaction with Trp578 varies slightly in bond length (Table 6 and Figure 10).
4. Discussion
Lymphedema is a progressive disease caused by lymphatic system malformations and impaired lymph flow. A panel consisting of 29 known lymphedema genes is currently used for the genetic testing of patients [24] but only detects relevant variants in 25–30% of patients [35,36]. This highlights the need for new target genes to screen. Here, we report our findings from screening a cohort of 235 patients, negative for variants in the said 29 genes, for variants in the neuropilin-coding genes NRP1 and NRP2.
Neuropilins are highly conserved transmembrane receptors that perform various functions in vessels, neurons and tumors, because they bind structurally to different ligands and coreceptors. Although NRP1 and NRP2 share basic structural features, they are implicated in different biological processes, such as the development of blood and lymphatic endothelial cells [37]. They have a role in the development of lymphatic valves [13,17,18], lymphatic vessels and capillaries [12,19,22].
Our screening detected two (2/235; 0.852%) and four (4/235; 1.702%) distinct probands with missense variants in NRP1 and NRP2, respectively. In four out of six cases, the proband was a female.
Overall, our results showed rare variants in NRP1 and NRP2 in patients who tested negative for variants in the 29 genes already associated with lymphedema. In three out of six cases, family members were screened, highlighting segregation of the variant with lymphedema or with subclinical signs of abnormal lymph flow. Bioinformatic modeling added evidence of a possible impact of the variant on the stability of the wildtype protein. Further biochemical studying or in-vitro studying is necessary to verify the altered function of coreceptors carrying the variant. In addition, other studies aiming at evaluating the effect of the interaction of the altered NRP1 with its principal ligand SEMA3A, or of the altered NRP2 with its coreceptor VEGFR3, could be important to define whether the variants also have a role in such signaling. Moreover, the loss of SEMA3A function by the specific inhibition of its interaction with NRP1 leads to impairment of lymphatic valve development [12], while variants in VEGFR3, a tyrosine kinase receptor important in the development and establishment of the lymphatic system, are known to cause Milroy disease, a form of primary lymphedema [38].
Additional studies could shed light on altered pathways, leading to a better understanding of the mechanism of action of the alteration; in particular, it could help determine whether the phenotype seen in our patients is due to haploinsufficiency or to a dominant negative effect of the coreceptor carrying the variant.
Although these results are not sufficient to associate the two genes, known to be involved in the development of lymphatic capillaries and valves, with lymphedema, they suggest that it would be worthwhile screening a larger cohort of patients for variants in NRP1 and NRP2, especially in large families with more than one affected subject, and performing a further functional analysis of the variants identified by us in in vitro and in vivo models to confirm their involvement in the development of lymphedema.
5. Conclusions
A comprehensive review of the literature strongly suggests that neuropilins NRP1 and NRP2 play a role in the development of lymphatic capillaries and lymphatic valves, crucial components of the lymphatic system. If these components do not function normally, the lymph does not drain, and fluid may accumulate in tissues, eventually leading to lymphedema. Given their functional role and our findings, we suggest that NRP1 and NRP2 should be considered candidate genes for inclusion in the gene panel for genetic testing of lymphedema patients.
Acknowledgments
We thank Helen Ampt for English language editing.
Abbreviations
| NGS | Next-Generation Sequencing |
| NRP1/2 | Neuropilin 1/2 |
| LECs | Lymphatic endothelial cells |
Author Contributions
Conceptualization, M.B. (Matteo Bertelli); data curation, D.K., M.B. (Mirko Baglivo), E.M., M.D., J.K., S.H.B., S.P. and M.B. (Matteo Bertelli); funding acquisition, M.B. (Matteo Bertelli); investigation, S.M., B.A. (Bruno Amato), M.R., R.S. and A.D.; project administration, M.B. (Matteo Bertelli); resources, S.M., B.A. (Bruno Amato), M.R. and M.B. (Matteo Bertelli); visualization, M.B. (Mirko Baglivo), S.H.B. and S.P.; writing—original draft, S.K. and D.V.; and writing—review and editing, S.M., B.A. (Bruno Amato), M.R., D.K., M.B. (Mirko Baglivo), E.M., M.D., J.K., S.H.B., R.S., G.A.D.M., B.A. (Barbara Aquilanti), G.M., V.V., L.G., A.D. and M.B. (Matteo Bertelli) All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Provincia Autonoma di Trento in the framework of LP 6/99 (dgp 1045/2017).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kolodkin A.L., Levengood D.V., Rowe E.G., Tai Y.T., Giger R.J., Ginty D.D. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/S0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 2.He Z., Tessier-lavigne M. Neuropilin Is a Receptor for the Axonal Chemorepellent Semaphorin III. Cell. 1997;90:739–751. doi: 10.1016/S0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 3.Giger R.J., Urquhart E.R., Gillespie S.K.H., Levengood D.V., Ginty D.D., Kolodkin A.L. Neuropilin-2 is a receptor for semaphorin IV: Insight into the structural basis of receptor function and specificity. Neuron. 1998;21:1079–1092. doi: 10.1016/S0896-6273(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 4.Gluzman-Poltorak Z., Cohen T., Herzog Y., Neufeld G. Neuropilin-2 and neuropilin-1 are receptors for the 165-amino acid form of vascular endothelial growth factor (VEGF) and of placenta growth factor-2, but only neuropilin-2 functions as a receptor for the 145-amino acid form of VEGF. J. Biol. Chem. 2000;275:18040–18045. doi: 10.1074/jbc.M909259199. [DOI] [PubMed] [Google Scholar]
- 5.Raimondi C., Ruhrberg C. Neuropilin signalling in vessels, neurons and tumours. Semin. Cell Dev. Biol. 2013;24:172–178. doi: 10.1016/j.semcdb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Kärpänen T., Heckman C.A., Keskitalo S., Jeltsch M., Ollila H., Neufeld G., Tamagnone L., Alitalo K. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J. 2006;20:1462–1472. doi: 10.1096/fj.05-5646com. [DOI] [PubMed] [Google Scholar]
- 7.Sulpice E., Plouët J., Bergé M., Allanic D., Tobelem G., Merkulova-Rainon T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood. 2008;111:2036–2045. doi: 10.1182/blood-2007-04-084269. [DOI] [PubMed] [Google Scholar]
- 8.Rizzolio S., Rabinowicz N., Rainero E., Lanzetti L., Serini G., Norman J., Neufeld G., Tamagnone L. Neuropilin-1-dependent regulation of EGF-receptor Signaling. Cancer Res. 2012;72:5801–5811. doi: 10.1158/0008-5472.CAN-12-0995. [DOI] [PubMed] [Google Scholar]
- 9.Castellani V., De Angelis E., Kenwrick S., Rougon G. Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 2002;21:6348–6357. doi: 10.1093/emboj/cdf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bagri A., Tessier-Lavigne M., Watts R.J. Neuropilins in tumor biology. Clin. Cancer Res. 2009;15:1860–1864. doi: 10.1158/1078-0432.CCR-08-0563. [DOI] [PubMed] [Google Scholar]
- 11.Neufeld G., Kessler O. The Neuropilins: Role and Function in Health and Disease. Springer International Publishing; Basel, Switzerland: 2017. [Google Scholar]
- 12.Bouvrée K., Brunet I., Del Toro R., Gordon E., Prahst C., Cristofaro B., Mathivet T., Xu Y., Soueid J., Fortuna V., et al. Semaphorin3A, Neuropilin-1, and PlexinA1 are required for lymphatic valve formation. Circ. Res. 2012;111:437–445. doi: 10.1161/CIRCRESAHA.112.269316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Yuan L., Mak J., Pardanaud L., Caunt M., Kasman I., Larrivée B., Del Toro R., Suchting S., Medvinsky A., et al. Neuropilin-2 mediates VEGF-C-induced lymphatic sprouting together with VEGFR3. J. Cell Biol. 2010;188:115–130. doi: 10.1083/jcb.200903137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawasaki T., Kitsukawa T., Bekku Y., Matsuda Y., Sanbo M., Yagi T., Fujisawa H. A requirement for neuropilin-1 in embryonic vessel formation. Development. 1999;126:4895–4902. doi: 10.1242/dev.126.21.4895. [DOI] [PubMed] [Google Scholar]
- 15.Kitsukawa T., Shimizu M., Sanbo M., Hirata T., Taniguchi M., Bekku Y., Yagi T., Fujisawa H. Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron. 1997;19:995–1005. doi: 10.1016/S0896-6273(00)80392-X. [DOI] [PubMed] [Google Scholar]
- 16.Kitsukawa T., Shimono A., Kawakami A., Kondoh H., Fujisawa H. Overexpression of a membrane protein, neuropilin, in chimeric mice causes anomalies in the cardiovascular system, nervous system and limbs. Development. 1995;121:4309–4318. doi: 10.1242/dev.121.12.4309. [DOI] [PubMed] [Google Scholar]
- 17.Jurisic G., Maby-El Hajjami H., Karaman S., Ochsenbein A.M., Alitalo A., Siddiqui S.S., Ochoa Pereira C., Petrova T.V., Detmar M. An Unexpected Role of Semaphorin3A–Neuropilin-1 Signaling in Lymphatic Vessel Maturation and Valve Formation. Circ. Res. 2012;111:426–436. doi: 10.1161/CIRCRESAHA.112.269399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W., Aspelund A., Alitalo K. Lymphangiogenic factors, mechanisms, and applications. J. Clin. Investig. 2014;124:878–887. doi: 10.1172/JCI71603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L., Moyon D., Pardanaud L., Bréant C., Karkkainen M.J., Alitalo K., Eichmann A. Abnormal lymphatic vessel development in neuropilin 2 mutant mice. Development. 2002;129:4797–4806. doi: 10.1242/dev.129.20.4797. [DOI] [PubMed] [Google Scholar]
- 20.Takashima S., Kitakaze M., Asakura M., Asanuma H., Sanada S., Tashiro F., Niwa H., Miyazaki J.I., Hirota S., Kitamura Y., et al. Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michelini S., Cardone M., Maltese P., Bruson A., Fiorentino A., Bertelli M. Primary lymphedema and genetic implications. EuroBiotech J. 2017;1:144–146. [Google Scholar]
- 22.Petrova T.V., Karpanen T., Norrmén C., Mellor R., Tamakoshi T., Finegold D., Ferrell R., Kerjaschki D., Mortimer P., Ylä-Herttuala S., et al. Defective valves and abnormal mural cell recruitment underlie lymphatic vascular failure in lymphedema distichiasis. Nat. Med. 2004;10:974–981. doi: 10.1038/nm1094. [DOI] [PubMed] [Google Scholar]
- 23.Michelini S., Ricci M., Veselenyiova D., Kenanoglu S., Kurti D., Baglivo M., Fiorentino A., Basha S.H., Priya S., Serrani R., et al. TIE1 as a candidate gene for lymphatic malformations with or without lymphedema. Int. J. Mol. Sci. 2020;21:6780. doi: 10.3390/ijms21186780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michelini S., Paolacci S., Manara E., Eretta C., Mattassi R., Lee B.-B., Bertelli M. Genetic tests in lymphatic vascular malformations and lymphedema. J. Med. Genet. 2018;55:222–232. doi: 10.1136/jmedgenet-2017-105064. [DOI] [PubMed] [Google Scholar]
- 25.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remmert M., Biegert A., Hauser A., Söding J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30:162–173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 31.Mackerell A.D., Feig M., Brooks C.L. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulation. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 32.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studio B.D. Discovery Studio Modeling Environment [Internet] Dassault Systèmes BIOVIA; San Diego, CA, USA: 2016. [Google Scholar]
- 34.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E., The Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 35.Maltese P.E., Michelini S., Ricci M., Maitz S., Fiorentino A., Serrani R., Lazzerotti A., Bruson A., Paolacci S., Benedetti S., et al. Increasing evidence of hereditary lymphedema caused by CELSR1 loss-of-function variants. Am. J. Med. Genet. 2019;179:1718–1724. doi: 10.1002/ajmg.a.61269. [DOI] [PubMed] [Google Scholar]
- 36.Brouillard P., Boon L., Vikkula M. Genetics of lymphatic anomalies. J. Clin. Investig. 2014;124:898–904. doi: 10.1172/JCI71614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz Q., Ruhrberg C. Neuropilin, you gotta let me know: Should I stay or should I go? Cell Adhes. Migr. 2010;4:61–66. doi: 10.4161/cam.4.1.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connell F.C., Ostergaard P., Carver C., Brice G., Williams N., Mansour S., Mortimer P.S., Jeffery S. Lymphoedema Consortium Analysis of the coding regions of VEGFR3 and VEGFC in Milroy disease and other primary lymphoedemas. Hum. Genet. 2009;124:625–631. doi: 10.1007/s00439-008-0586-5. [DOI] [PubMed] [Google Scholar]