Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19 (coronavirus disease-19), represents a far more serious threat to public health than SARS and MERS coronaviruses, due to its ability to spread more efficiently than its predecessors. Currently, there is no worldwide-approved effective treatment for COVID-19, urging the scientific community to intense efforts to accelerate the discovery and development of prophylactic and therapeutic solutions against SARS-CoV-2 infection. In particular, effective antiviral drugs are urgently needed. With few exceptions, therapeutic approaches to combat viral infections have traditionally focused on targeting unique viral components or enzymes; however, it has now become evident that this strategy often fails due to the rapid emergence of drug-resistant viruses. Targeting host factors that are essential for the virus life cycle, but are dispensable for the host, has recently received increasing attention. The spike glycoprotein, a component of the viral envelope that decorates the virion surface as a distinctive crown (“corona”) and is essential for SARS-CoV-2 entry into host cells, represents a key target for developing therapeutics capable of blocking virus invasion. This review highlights aspects of the SARS-CoV-2 spike biogenesis that may be amenable to host-directed antiviral targeting.
Keywords: Antiviral, Coronavirus, COVID-19, ERp57, Furin, N-Glycosylation
1. Introduction
SARS-CoV-2 coronavirus entered human history near the end of 2019. Starting from China, SARS-CoV-2 rapidly spread globally with more than 40 million people testing positive worldwide, and causing more than 1.100.000 deaths as of October 20, 2020 (https://coronavirus.jhu.edu/map.html).
SARS-CoV-2 is a member of the Coronaviridae family that comprises a large number of enveloped, positive-sense single-stranded RNA viruses causing respiratory, enteric and neurological diseases of varying severity in domestic and wild animals, as well as in humans [1].
On the basis of their phylogenetic relationships and genomic structures, coronaviruses (CoV) are subdivided in four genera: alpha-, beta-, gamma- and delta-coronavirus; among these, only alpha- and beta-CoVs can infect humans [1,2]. Human coronaviruses (HCoV) include four globally distributed viruses (HCoV-NL63, HCoV-229E, HCoV-OC43 and HCoV-HKU1) that generally cause mild upper respiratory tract diseases in immunocompetent hosts, and three highly pathogenic (HP) viruses that have emerged since the beginning of this century [[1], [2], [3], [4]]. HP-HCoV include, in addition to SARS-CoV-2, the lineage B beta-CoV SARS coronavirus that emerged in China and Hong Kong in 2002–2003, causing more than 8.000 cases worldwide with a death rate of approximately 10% (https://www.who.int/csr/sars/country/table2004_04_21/en/), and the lineage C beta-CoV MERS (Middle East Respiratory Syndrome) coronavirus that emerged in 2012 in the Arabian Peninsula, causing over 2.500 confirmed cases and a case-fatality rate higher than 34% (https://www.who.int/csr/don/24-february-2020-mers-saudi-arabia/en/).
SARS-CoV and MERS-CoV infections can result in acute respiratory distress syndrome (ARDS), which may lead to long-term reduction in lung function and death [5]; infection with SARS-CoV-2 in humans manifests as coronavirus disease-2019 (COVID-19), a spectrum of diseases ranging from asymptomatic infection to respiratory symptoms that, in a subset of patients, may progress to pneumonia, ARDS, multi organ dysfunction and death [6,7].
Given the sudden appearance and rapid spread of SARS-CoV-2, there is no current validated vaccine or SARS-CoV-2-specific targeting therapy that is clinically approved, although steroids, heparin and statins look promising for lowering fatality rates, and the antiviral remdesivir has been proven to reduce the duration of symptomatic disease presentation [8,9].
Despite the extraordinary worldwide commitment, efforts to defeat COVID-19 are hampered by lack of information on several important aspects of this new coronavirus, ranging from SARS-CoV-2 biology to its interaction with the host response.
At the genomic level SARS-CoV-2 is highly similar (nearly 80% identical) to SARS-CoV [10,11] and to RaTG13-CoV (>95% identical) circulating in bats, the natural reservoir host for multiple coronaviruses [[10], [11], [12], [13]]. Similarly to other coronaviruses, it harbors a large (∼30 kb) nonsegmented RNA genome, with the replicase-transcriptase gene encoded within the 5′-end and the structural proteins encoded in the 3′-end, following the CoV invariant gene order: 5′- S (spike) - E (envelope) - M (membrane) - N (nucleocapsid)-3’; numerous small open reading frames, encoding accessory proteins, are distributed among the structural genes [2,14].
The spike protein, together with the M and E proteins, is anchored into the viral envelope, decorating the virion surface as a distinctive crown (“corona”) (Fig. 1 A), and is essential for viral entry into target cells [15,16]. As it induces neutralizing antibody responses, S-protein is also an important target for vaccine development [17,18], and therefore it has been studied extensively [[19], [20], [21]].
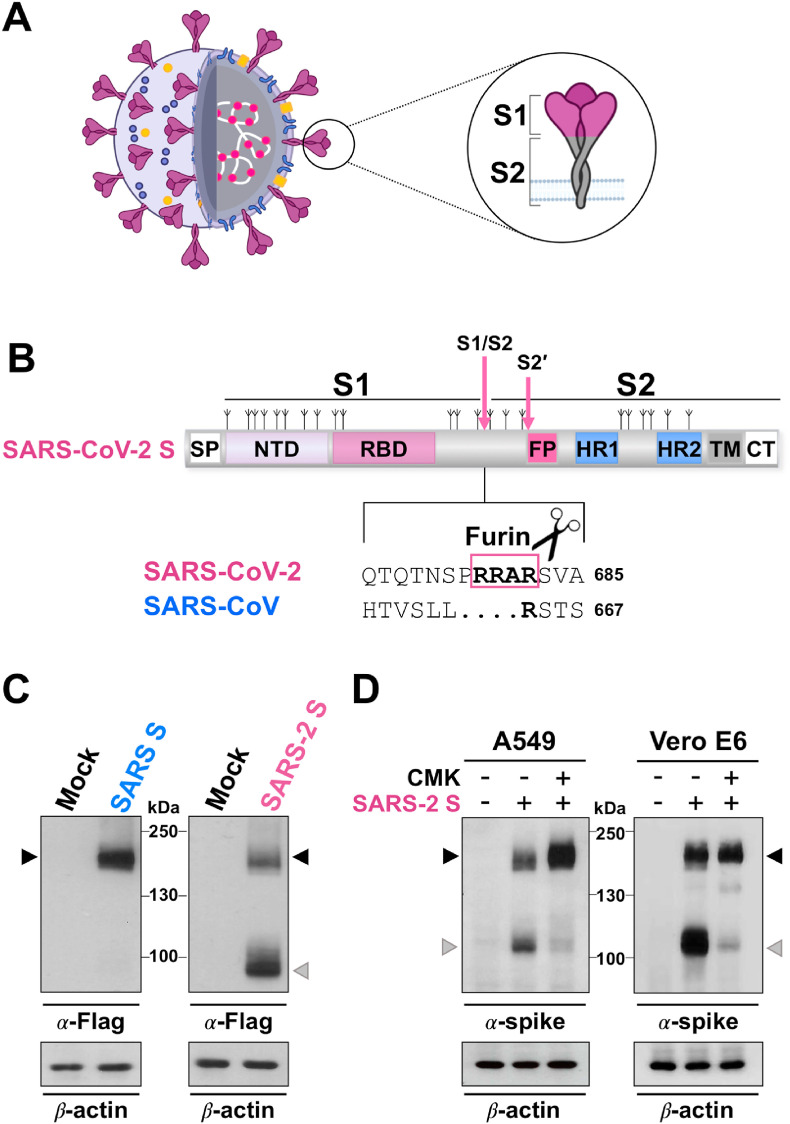
Fig. 1.
Furin-mediated S1/S2 cleavage during SARS-CoV-2 spike biogenesis. (A) Schematic representation of the SARS-CoV-2 virus and the spike glycoprotein. The lipid bilayer comprising the spike protein (S, violet), the membrane protein (M, blue) and the envelope protein (E, orange), and the viral RNA (white) associated with the nucleocapsid protein (N, pink) are shown. The spike protein S1 and S2 subunits are indicated in the zoom. (B) Schematic representation of the SARS-CoV-2 S glycoprotein. The positions of N-linked glycosylation sequons are shown as branches. S1, receptor-binding subunit; S2, membrane fusion subunit. The S1/S2 and S2’ protease cleavage sites are indicated by arrows. Protein domains are illustrated: SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Sequence comparison of the S1/S2 cleavage site of SARS-CoV-2 and SARS-CoV spike proteins, and the putative furin cleavage site (RRAR residues in the box) in SARS-CoV-2 are shown. GenBank accession numbers are QHD43416.1 for SARS-CoV-2 spike and AFR58740.1 for SARS-CoV spike [104]. (C) Human A549 alveolar type II-like epithelial cells (ATCC) transfected with Flag-tagged SARS-CoV-2 spike (SARS-2 S, C-terminal Flag VG40590-CF, Sino Biological) or Flag-tagged SARS-CoV spike (SARS S, C-terminal Flag VG40150-CF, Sino Biological) constructs, or empty vector (Mock) were analyzed by immunoblot (IB) using anti-Flag antibodies (Cell Signaling). (D) IB for SARS-CoV-2 spike protein using anti-SARS-CoV-2 spike antibody (α-spike, Sino Biological) in whole cell extracts from A549 and Vero E6 cells (ATCC) transfected with the SARS-CoV-2 spike construct (SARS-2 S, +) or empty vector (−), and treated with the furin inhibitor decanoyl-RVKR-CMK (CMK, Sigma-Aldrich) or vehicle (−). In panels C and D, black arrowheads indicate bands corresponding to uncleaved S protein, whereas gray arrowheads indicate the cleaved S fragment.
1.1. The SARS-CoV-2 spike glycoprotein
The SARS-CoV-2 spike is a trimeric class I fusion protein. As most viral fusion proteins, each monomer is synthesized as a fusogenically-inactive precursor of about 180 kDa, which assembles into an inactive homotrimer and is endoproteolytically cleaved by cellular proteases, giving rise to a metastable complex of two functional subunits: S1 (bulb) responsible for receptor binding and the membrane-anchored S2 (stalk) that contains the fusion machinery (Fig. 1A).
The structure of the SARS-CoV-2 S precursor has been characterized and it consists of a signal peptide (SP) located at the N-terminus, followed by the S1 and S2 subunits (Fig. 1B). The S1 subunit contains the N-terminal domain (ND) and the receptor-binding domain (RBD), with the receptor-binding motif (RBM), responsible for recognition and attachment to the host angiotensin-converting enzyme 2 (ACE2) receptor (Fig. 1B) [20]. The attachment process starts with the recognition of the RBD 394 glutamine residue by the lysine 31 residue on the human ACE2 [22]. Interestingly, the SARS-CoV-2 S protein has a 10- to 20-fold higher affinity for ACE2 than the S protein of SARS-CoV [19].
S2 harbors the fusion peptide (FP), a short segment of 15–20 conserved mainly hydrophobic amino acids, which anchors to target membranes and plays an essential role in mediating membrane fusion by disrupting and connecting lipid bilayers of the host cell. The FP is followed by two heptapeptide repeat sequences HR1 and HR2, the transmembrane anchor domain (TM), and a short cytoplasmic tail (CT) (Fig. 1B) [20]. Similarly to other viral fusion proteins, when the S1 subunit recognizes its receptor on human cells, the HR1 and HR2 domains are exposed to interact with each other to form a six-helical bundle (6-HB), consequently bringing viral and cellular membranes into close proximity to permit lipid bilayer fusion [23]. Fusion inhibitors derived from the SARS-CoV-2 HR2 domain and pan-CoV fusion inhibitors have been developed that are able to bind to S-HR1-trimers forming heterologous 6-HBs, thus inhibiting viral/cell membrane fusion [23].
As most viral glycoproteins that become incorporated into the envelope membrane bilayer, SARS-CoV-2 S protein is extensively decorated with N-linked glycans at both the S1 and S2 subunits, possessing a total of 22 N-linked glycan sites (as compared with 23 on SARS) (Fig. 1B) [20,24,25]. Glycosylation plays an essential role in establishing viral spike proteins bioactive conformation and stability, for shaping viral tropism and has effects on virus antigenic properties, receptor binding and fusion activity [[26], [27], [28]]. Using a site-specific mass spectrometric analysis, Watanabe et al. have recently revealed the glycan structures on a recombinant SARS-CoV-2 S immunogen, mapping the glycan-processing states across the trimeric viral spike [24].
There is still little information on the molecular processes involved in SARS-CoV-2 spike protein glycosylation and maturation; however, due to the similarity with other CoV spike proteins, such as the SARS-CoV S, the knowledge obtained on these viruses can be used to identify elements of the secretory pathway essential for virus morphogenesis that could represent appealing targets for novel anti-coronavirus drugs [3,29,30].
1.2. Coronavirus spike glycosylation
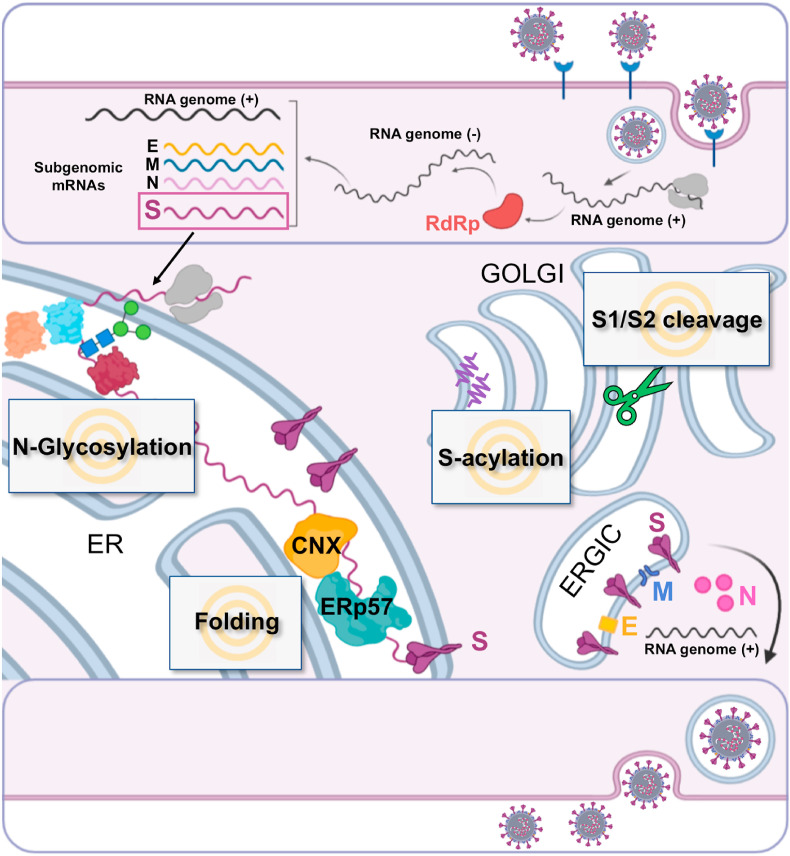
Like other enveloped viruses, coronaviruses exploit the ability of the host cell to produce, properly fold and transport glycoproteins to the correct cellular location [29,31]. For SARS-CoV-2 and other CoVs, these steps take place in organelles of the early secretory pathway, the endoplasmic reticulum (ER), the ER-Golgi intermediate compartment (ERGIC), and the Golgi apparatus, and efficient virus spread critically depends on hijacking the host secretory machinery (Fig. 2 ) [29,31].
Fig. 2.
Schematic representation of SARS-CoV-2 spike glycoprotein biogenesis. Different steps of SARS-CoV-2 replication cycle are illustrated in the cartoon, including binding to the ACE2 receptor (blue), virus entry, viral RNA replication, sub-genomic RNA transcription and translation. RdRp, RNA-dependent RNA polymerase; E, envelope; M, membrane; N, nucleoprotein; S, spike; CNX, Calnexin; ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartment. Gray text boxes highlight host-cell processes implicated in SARS-CoV-2 spike biogenesis that might represent potential targets for host-directed antiviral drugs.
Co-translational N-glycosylation was shown to be essential for CoV spike proteins to fold properly and to exit the ER. Interestingly the S protein of some coronaviruses, including S proteins of transmissible gastroenteritis virus (TGEV) [32,33] and SARS-CoV-2 (Santoro et al., unpublished results), was shown to aggregate when glycosylation was inhibited; furthermore growth of coronaviruses in the presence of the N-glycosylation inhibitor tunicamycin resulted in the production of spikeless, noninfectious virions [[34], [35], [36]].
In addition, tunicamycin and α-glucosidases I and II inhibitors, which hamper the removal of terminal glucose residues from N-glycan chains attached to the nascent glycoprotein in the ER, such as N-methyl-1-deoxynojirimycin and castanospermine, were shown to influence the intracellular transport as well as the antigenic properties of MHV (mouse hepatitis virus) coronavirus spike glycoprotein [37], and to cause both a delay in the surface expression of the glycoprotein and a drastic reduction in progeny virion formation. Castanospermine and the α-glucosidase inhibitor N-butyl-deoxynojirimycin (NB-DNJ) were also found to alter SARS S protein maturation and to decrease incorporation of the spike into virus particles [38,39].
Potential use of α-glucosidase inhibitors, including imino sugars, as host-directed antiviral agents have been largely investigated [40], and potential candidates for treatment of COVID-19, including miglitol, celgosivir and miglustat, have been recently reviewed [41]. It should be noted that, in the case of N-linked glycans, more than 30 enzymes, located in the cytosol, ER and Golgi apparatus, are required to generate, attach and process oligosaccharides, and may then represent potential targets for S protein biogenesis inhibitors; however, the molecular events participating in the protein glycosylation process have been largely described [29,40,42] and will not be discussed here.
1.3. CoV spike glycoprotein folding and disulfide bond formation
Like most viral glycoproteins, during its biosynthesis the S polypeptide, while it is co-translationally N-glycosylated in the ER, interacts with the cellular molecular chaperones calnexin and/or calreticulin, after which the carbohydrates are processed in the ER and in the Golgi apparatus [27,43]. In the case of the SARS coronavirus it has been shown that the binding of the S protein to calnexin is essential for correct folding of the glycosylated spike protein, and that this ER chaperone plays a critical role in the infectious ability of progeny viruses and consequently on SARS-CoV infection [39].
In addition to the calnexin/calreticulum folding machine, post-translational formation of disulfide bonds plays a decisive role in the generation of the final glycoprotein architecture. As an example, disulfide bond formation was shown to be essential for the correct folding, trafficking and trimerization of the spike protein of the MHV coronavirus [44]. In the case of SARS-CoV-2 S protein, nine cysteine residues were found in the S1 receptor binding domain, eight of which form four pairs of disulfide bonds [45]. Among these four pairs, three (Cys336–Cys361, Cys379–Cys432 and Cys391–Cys525) were found to stabilize the β sheet structure, whereas the fourth (Cys480–Cys488) connects the loops in the distal end of the RBM [45]. These intra-molecular disulfide bonds are believed to contribute to the stereospecific orientations of the ACE2-interacting amino acid residues of the spike protein, and therefore to play a relevant role in the binding of the RBM to the receptor. It has been hypothesized that the perturbation of the spike functionally active conformation through the reduction of accessible disulfide bonds may be a feasible strategy to disintegrate the spike protein from the ACE2 receptor, preventing infection [46]. In silico studies suggest that N-acetyl cysteine (NAC), a drug used as antioxidant and mucolytic agent, may bind in the close proximity of a solvent-accessible disulfide bond (Cys391-Cys525); reduction of this disulfide bond via thiol/disulfide exchange, followed by covalent conjugation of NAC, is predicted to perturb the stereospecific orientations of interacting key residues of the S glycoprotein, weakening the binding affinity of the spike with ACE2 receptor [46].
In addition, mutations in SARS-CoV-2 spike that allow the production of thermostable, disulfide-bonded S-protein trimers that are trapped in the closed, prefusion state, have been recently described, and potential applications of a designed, thermostable S trimer as a reagent for serology/virology and as an immunogen have been recently suggested [47].
A critical role for the correct dynamics of disulfide bond formation of viral glycoproteins, such as the influenza hemagglutinin (HA) and the parainfluenza fusion (F) protein in the early secretory pathway is played by the calnexin/calreticulin associated co-chaperone ERp57 [48]. ERp57, also known as GRP58 (glucose-regulated protein-58) is a member of the protein disulfide-isomerase (PDI) family coded by the PDIA3 gene, mostly, but not exclusively, localized in the ER [48,49]. Like other PDIs, ERp57 is characterized by the presence of two catalytically active thioredoxin-like domains (TLD) containing a cys-gly-his-cys sequence, termed a and a′, which provide ERp57 with its redox properties, and two inactive TLD, termed b and b′. Crystallography studies demonstrate that the four TLD form a twisted “U”-shape structure [50].
ERp57 is a multifunctional protein acting both as oxidoreductase and isomerase, helping ER-glycoproteins to obtain native disulfides by rearranging nonnative linkages. The catalytically-active domains are located at the C- and N-termini at the top of the U, while the two noncatalytic domains are localized to the inside surface providing the binding sites for calnexin/calreticulin; in particular, ERp57-b’ needs to associate with calreticulin/calnexin that position lectin-bound misfolded glycoproteins allowing ERp57-mediated catalysis [50,51]. Interestingly ERp57 was recently shown to be the target of the broad-spectrum anti-infective agent nitazoxanide [52].
Nitazoxanide, a thiazolide originally developed as an antiprotozoal agent and used in clinical practice for the treatment of infectious gastroenteritis [53,54], has recently emerged as broad-spectrum antiviral drug [55,56]. We and others have previously reported that nitazoxanide and its active circulating-metabolite tizoxanide are effective against a broad range of RNA viruses, including influenza and parainfluenza viruses, hepatitis C and rotavirus infection in-vitro as well as in clinical studies [52,[57], [58], [59], [60], [61]].
Rather than affecting viral targets, thiazolides act through a cell-mediated mechanism. In the case of influenza and parainfluenza viruses nitazoxanide was found to act by a novel mechanism, impairing terminal glycosylation and intracellular trafficking of class-I viral fusion glycoproteins influenza HA and paramyxovirus F spike proteins [52,57], an effect mediated by the inhibition of ERp57 [52].
Interestingly nitazoxanide was found to also be effective against different animal and human coronaviruses in vitro [55,62,63], as well as in clinical studies (Rossignol et al., submitted). Recent studies have also reported the antiviral activity of nitazoxanide against SARS-CoV-2 in cell culture assays [64,65]. We are currently investigating the possibility that, as for the influenza HA and parainfluenza F glycoproteins, nitazoxanide may act by affecting the biogenesis of SARS-CoV-2 spike protein, impairing its correct folding, maturation and fusion activity (Santoro et al., unpublished results).
1.4. Furin and SARS-CoV-2 spike cleavage
Previous studies on different coronaviruses have shown that activation of the CoV spike protein is a complex process involving several host proteases and multiple cleavage events at distinct sites of the glycoprotein [66]. A variety of host proteases, including cell surface TMPRSS2 (transmembrane serine protease 2) proteases, trypsin and furin, and endosomal cathepsins, have been shown to direct S protein cleavage during CoVs entry or viral protein biogenesis, depending on their distribution in host cells. In addition to controlling virus entry, S protein cleavage may also influence host tropism and pathogenesis of CoV infection [16,[67], [68], [69]].
The SARS-CoV-2 spike is cleaved at the boundary between the S1 and S2 subunits that remain non-covalently bound in the prefusion conformation [[70], [71], [72], [73]]. Similarly to other viral fusion glycoproteins, including influenza hemagglutinin and parainfluenza fusion F-protein, S cleavage at the S1/S2 boundary is essential for membrane-fusion activity [[71], [72], [73]]. As for all CoVs, a second cleavage by host proteases occurring at a different site termed S2’ located immediately upstream of the fusion peptide has been proposed to activate the protein for membrane fusion via extensive conformational changes [67,70,[74], [75], [76]].
In the case of SARS-CoV-2 early characterizations of the genome revealed the existence of an exposed loop at the S1/S2 site harboring a polybasic furin-like cleavage (FLC) site containing multiple arginine residues (RRAR) (Fig. 1B) [15,19,77,78]; a similar multibasic cleavage site was previously found in MERS-CoV, but not in other lineage B beta-CoVs, including SARS-CoV and SARS-related coronaviruses (SARSr-CoV) [77].
Furin, a protease belonging to the proprotein convertase (PC) family, is ubiquitously expressed in the Golgi apparatus of all cells, with some cell types showing enhanced expression or altered intracellular distribution [79,80]. To date, furin and/or PC-mediated cleavage is considered to represent a key event in activating fusion activity of envelope glycoproteins of several evolutionarily diverse virus families, including, besides Coronaviridae, Flaviviridae, Filoviridae, Retroviridae, Orthomyxoviridae and Paramyxoviridae; however, both subcellular localization and timing of furin-mediated activation may vary considerably depending on the virus family [80,81].
Interestingly, in the case of SARS-CoV-2, the FLC site is processed during S protein biogenesis; in fact, differently from SARS-CoV, the spike of SARS-CoV-2 is mainly detected as the cleaved S1/S2 subunits when exogenously expressed in different types of cells (Fig. 1B–D) [15,77].
The furin-like cleavage site, promoting S-protein “priming” intracellularly during morphogenesis, facilitates virus progeny entry into the target cells and may be responsible for SARS-CoV-2 high infectivity and transmissibility, providing a gain-of-function to the new coronavirus for efficient spreading in the human population compared to other lineage B beta-CoVs [15,22]. More importantly, the acquisition of similar polybasic cleavage sites is associated with increased pathogenicity in different viruses, including avian influenza A viruses (AIVs) [82,83]. In the case of AIVs, a multibasic cleavage site in the virus surface glycoprotein, the hemagglutinin (HA), is considered a critical virulence factor [82,83]: the HA cleavage site motif of low-pathogenicity avian influenza viruses (LPAIV) typically contains one or two basic amino acid residues that are cleaved by trypsin and trypsin-like proteases with monobasic specificity, confining LPAIV replication to trypsin-expressing epithelial cells of the respiratory and gastrointestinal tracts. On the other hand, the HA cleavage site motif of highly pathogenic viruses (HPAIV) contains multiple basic amino acids, facilitating cleavage by ubiquitously expressed proteases with polybasic specificity, most notably furin, thus allowing HPAIV to replicate in multiple tissues, including the vascular endothelium. Therefore, the FLC site may be implicated in the ability of SARS-CoV-2 to invade different tissues and organs, not restricting virus replication to the respiratory and gastrointestinal tracts. In fact, although SARS-CoV-2 infection predominantly causes substantial respiratory pathology, it can also result in several extrapulmonary manifestations, including, beside gastrointestinal symptoms, myocardial dysfunction and arrhythmia, thrombotic complications, kidney and hepatocellular injury, neurologic illnesses and ocular symptoms [84].
As indicated above, similarly to SARS-CoV, the cellular receptor of SARS-CoV-2 is the angiotensin-converting enzyme 2. ACE2 is a transmembrane protein acting as a carboxypeptidase negatively regulating the renin–angiotensin system [85]. In addition to the respiratory system, ACE2 expression was observed is several human organs and cell types, including enterocytes, cardiomyocytes and renal tubules [86]. Since ACE2 is expressed in multiple extrapulmonary tissues, direct viral tissue damage is considered a possible mechanism of injury [84].
On this basis, in addition of the well studied TMPRSS2 inhibitors [16,67], furin inhibitors might be considered as a treatment option for COVID-19. Since furin, unlike TMPRSS2, is required for normal cell function [87], blocking this enzyme for prolonged time periods might cause unwanted toxic effects; however, short treatments might be well tolerated and lead to a therapeutic benefit [88,89].
In recent years, several small molecule and peptide-based inhibitors targeting furin/PCs have been developed as putative antiviral agents, which may inhibit maturation and fusion activity of viral envelope glycoproteins [80,81,90]. Among these, the peptide inhibitor decanoyl-RVKR-chloromethylketone (CMK) has proven to be particularly effective in inhibiting SARS-CoV-2 spike cleavage during biogenesis in different types of cells (Fig. 1D).
Interestingly, it was recently reported that the loss of the furin cleavage site in the spike of a SARS-CoV-2 mutant (ΔPRRA) resulted in reduced infection in human respiratory cells (but not in Vero E6 cells) and ablated disease in a hamster pathogenesis model of SARS-CoV-2, demonstrating a critical role for the FLC in SARS-CoV-2 replication and pathogenesis [91].
1.5. Coronavirus spike protein S-acylation
Protein S-acylation, a post-translational modification that involves linkage of a fatty acid chain predominantly to a cysteine amino acid in a thioester bond, was first described by Schmidt and Schlesinger in the VSV (Vesicular Stomatitis Virus) glycoprotein G [92], and subsequently found to be ubiquitous and highly conserved from yeast to human [93,94]. Since the fatty acid molecule is predominantly palmitate, the term ‘palmitoylation’ is also used, but other fatty acids both saturated (e.g., myristic and stearic) and unsaturated (e.g., oleic and arachidonic) can also form modifications.
S-acylation affects protein trafficking, protein-protein and protein-membrane interactions, and, being coupled to membrane fusion or virus assembly, is known to influence viral replication and pathogenesis.
Coronavirus S protein palmitoylation was initially identified in cells infected with MHV coronavirus [43]. Treatment of palmitoyl acyltransferase inhibitor 2-bromopalmitate was found to decrease MHV S protein palmitoylation, and caused a significant reduction in MHV infectivity [[95], [96], [97]]. Reduction of S palmitoylation was also found to impair S association with the M protein with subsequent exclusion of the spike protein from virions. In addition, MHV mutants harboring mutations in the putative palmitoylation sites exhibited reduced infectivity, further supporting the importance of palmitoylation in virion assembly and infectivity [[95], [96], [97]].
Alpha-coronavirus TGEV S protein is also modified by palmitoylation, and inhibition of palmitoylation by 2-bromopalmitate treatment reduced TGEV replication in cell culture [98].
In the case of SARS-CoV, the cytoplasmic portion of the spike protein contains four cysteine-rich clusters, two of which (clusters I and II) were found to be modified by palmitoylation; S-mediated cell fusion was markedly reduced by mutations in these cysteine clusters as compared with wild-type protein, suggesting that palmitoylation in the endodomain may be required for the fusogenic activity of SARS-CoV S protein [99]. In a different study, using a recombinant nonpalmitoylated SARS-CoV S protein mutant, it was shown that similarly to MHV S protein, palmitoylation of the SARS-CoV S protein was required for its partitioning into membranes and for cell–cell fusion [100]. However, differently from MHV S protein, SARS-CoV spike palmitoylation was not required for S–M interaction [100]. Finally, treatment with nitric oxide or its derivatives was found to reduce SARS-CoV S protein palmitoylation, which affected the spike binding to the ACE2 receptor [101]. Therefore, depending on the type of coronavirus, palmitoylation may differentially affect the folding, fusogenic activity and/or protein–protein interaction of the S protein.
Very little is currently known about palmitoylation of the SARS-CoV-2 spike protein. Analysis of the alignment of SARS-CoV and SARS-CoV-2 S proteins has revealed that all 9 putative palmitoylation sites in SARS-CoV are conserved in SARS-CoV-2, and it has been speculated that palmitoylation could contribute to nascent SARS-CoV-2 spikes targeting to GM1 lipid rafts in the producing cells [102]. Characterizing SARS-CoV-2 spike glycoprotein S-acylation, and understanding the molecular mechanisms involved may uncover new targets for antiviral therapy during S protein biogenesis.
1.6. Perspectives
As obligatory parasites, viruses are directly dependent upon their host cell environment for replication, protein expression and assembly of progeny particles. With few exceptions, therapeutic approaches to combat viral infections have traditionally focused on targeting unique viral components or enzymes; however, it has now become evident that this strategy often fails due to the rapid emergence of drug-resistant viruses. Targeting host factors that are essential for the virus life cycle, rather than pathogen components directly, has recently received increasing attention. Genome-wide approaches have been used successfully to identify cellular factors that are critical for virus replication, but are dispensable for the host, and can thus serve as novel targets for antiviral drug development [103].
The SARS-CoV-2 spike, which is essential for virus invasion, represents a key target for developing therapeutics capable of blocking viral entry and inhibiting membrane fusion. Characterizing the components of the molecular machines operating in the biogenesis of SARS-CoV-2 spike protein, together with their tissue specificity and redundancy, will provide opportunities in the search for unique targets for novel host-directed antiviral drugs for therapeutic intervention in SARS-COV-2 infections.
Declaration of competing interest
All authors declare no conflict of interest.
Acknowledgments
The authors thank the Italian Ministry of University and Scientific Research (PRIN project N 2010PHT9NF-006) for research support.
References
- 1.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung T.S., Liu D.X. Human Coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 3.Lim Y.X., Ng Y.L., Tam J.P., et al. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang X., Wu C., Li X., et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Wit E., van Doremalen N., Falzarano D., et al. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiersinga W.J., Rhodes A., Cheng A.C., et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 7.Tay M.Z., Poh C.M., Rénia L., et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders J.M., Monogue M.L., Jodlowski T.Z., et al. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a Review. J. Am. Med. Assoc. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 9.Beigel J.H., Tomashek K.M., Dodd L.E., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong N.S., Zheng B.J., Li Y.M., et al. February, 2003, Lancet. vol. 362. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China; pp. 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020;37:2699–2705. doi: 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D., Lee J.Y., Yang J.S., et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Park Y.J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 18.Dagotto G., Yu J., Barouch D.H. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe. 2020;28:364–370. doi: 10.1016/j.chom.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y., Zhang J., Xiao T., et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florindo H.F., Kleiner R., Vaskovich-Koubi D., et al. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia S., Zhu Y., Liu M., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein, Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y., Allen J.D., Wrapp D., et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe Y., Berndsen Z.T., Raghwani J., et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander S., Elder J.H. Carbohydrate dramatically influences immune reactivity of antisera to viral glycoprotein antigens. Science. 1984;226:1328–1330. doi: 10.1126/science.6505693. [DOI] [PubMed] [Google Scholar]
- 27.Braakman I., van Anken E. Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic. 2000;1:533–539. doi: 10.1034/j.1600-0854.2000.010702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulze I.T. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 1997;176:S24–S28. doi: 10.1086/514170. [DOI] [PubMed] [Google Scholar]
- 29.Sicari D., Chatziioannou A., Koutsandreas T., et al. Role of the early secretory pathway in SARS-CoV-2 infection. J. Cell Biol. 2020;219 doi: 10.1083/jcb.202006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vigant F., Santos N.C., Lee B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su S., Wong G., Shi W., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delmas B., Laude H. Carbohydrate-induced conformational changes strongly modulate the antigenicity of coronavirus TGEV glycoproteins S and M. Virus Res. 1991;20:107–120. doi: 10.1016/0168-1702(91)90103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes K.V., Doller E.W., Sturman L.S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rottier P.J., Horzinek M.C., van der Zeijst B.A. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J. Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern D.F., Sefton B.M. Coronavirus proteins: structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J. Virol. 1982;44:804–812. doi: 10.1128/jvi.44.3.804-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Repp R., Tamura T., Boschek C.B., et al. The effects of processing inhibitors of N-linked oligosaccharides on the intracellular migration of glycoprotein E2 of mouse hepatitis virus and the maturation of coronavirus particles. J. Biol. Chem. 1985;260:15873–15879. doi: 10.1016/S0021-9258(17)36339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie G., Harvey D.J., Feldmann F., et al. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology. 2010;399:257–269. doi: 10.1016/j.virol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukushi M., Yoshinaka Y., Matsuoka Y., et al. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J. Virol. 2012;86:11745–11753. doi: 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dwek R.A., Butters T.D., Platt F.M., et al. Targeting glycosylation as a therapeutic approach. Nat. Rev. Drug Discov. 2002;1:65–75. doi: 10.1038/nrd708. [DOI] [PubMed] [Google Scholar]
- 41.Williams S.J., Goddard-Borger E.D. α-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem. Soc. Trans. 2020;48:1287–1295. doi: 10.1042/BST20200505. [DOI] [PubMed] [Google Scholar]
- 42.Helenius A., Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 43.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opstelten D.J., de Groote P., Horzinek M.C., et al. Disulfide bonds in folding and transport of mouse hepatitis coronavirus glycoproteins. J. Virol. 1993;67:7394–7401. doi: 10.1128/jvi.67.12.7394-7401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 46.Debnath U., Dewaker V., Prabhakar Y.S., et al. ChemRxiv; 2020. Conformational perturbation of SARS-CoV-2 spike protein using N-Acetyl Cysteine, a molecular scissor: a probable strategy to combat COVID-19. [DOI] [PubMed] [Google Scholar]
- 47.Xiong X., Qu K., Ciazynska K.A., et al. A thermostable, closed SARS-CoV-2 spike protein trimer. Nat. Struct. Mol. Biol. 2020;27:934–941. doi: 10.1038/s41594-020-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellgaard L., Ruddock L.W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jessop C.E., Chakravarthi S., Garbi N., et al. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian G., Xiang S., Noiva R., et al. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 51.Hebert D.N., Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 52.Piacentini S., La Frazia S., Riccio A., et al. Nitazoxanide inhibits paramyxovirus replication by targeting the Fusion protein folding: role of glycoprotein-specific thiol oxidoreductase ERp57. Sci. Rep. 2018;8:10425. doi: 10.1038/s41598-018-28172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossignol J.F., Ayoub A., Ayers M.S. Treatment of diarrhea caused by Giardia intestinalis and Entamoeba histolytica or E. dispar: a randomized, double-blind, placebo-controlled study of nitazoxanide. J. Infect. Dis. 2001;184:381–384. doi: 10.1086/322038. [DOI] [PubMed] [Google Scholar]
- 54.Rossignol J.F., Kabil S.M., el-Gohary Y., et al. Effect of nitazoxanide in diarrhea and enteritis caused by Cryptosporidium species. Clin. Gastroenterol. Hepatol. 2006;4:320–324. doi: 10.1016/j.cgh.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 55.Rossignol J.F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antivir. Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 57.Rossignol J.F., La Frazia S., Chiappa L., et al. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J. Biol. Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossignol J.F., Elfert A., El-Gohary Y., et al. Improved virologic response in chronic hepatitis C genotype 4 treated with nitazoxanide, peginterferon, and ribavirin. Gastroenterology. 2009;136:856–862. doi: 10.1053/j.gastro.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 59.La Frazia S., Ciucci A., Arnoldi F., et al. Thiazolides, a new class of antiviral agents effective against rotavirus infection, target viral morphogenesis, inhibiting viroplasm formation. J. Virol. 2013;87:11096–11106. doi: 10.1128/JVI.01213-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossignol J.F., Abu-Zekry M., Hussein A., Santoro M.G. Effect of nitazoxanide for treatment of severe rotavirus diarrhoea: randomised double-blind placebo-controlled trial. Lancet. 2006;368:124–129. doi: 10.1016/S0140-6736(06)68852-1. [DOI] [PubMed] [Google Scholar]
- 61.Haffizulla J., Hartman A., Hoppers M., et al. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 2014;14:609–618. doi: 10.1016/S1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao J., Forrest J.C., Zhang X. A screen of the NIH Clinical Collection small molecule library identifies potential anti-coronavirus drugs. Antivir. Res. 2015;114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M., Cao R., Zhang L., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Son J., Huang S., Zeng Q., et al. bioRxiv; 2020. Nitazoxanide and JIB-04 have broad-spectrum antiviral activity and inhibit SARS-CoV-2 replication in cell culture and Coronavirus pathogenesis in a pig model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hulswit R.J.G., de Haan C.A.M., Bosch B.-J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park J.E., Li K., Barlan A., et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U.S.A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Q., Qiu Y., Li J.Y., et al. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol. Sin. 2020;35:337–339. doi: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coutard B., Valle C., de Lamballerie X., et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burkard C., Verheije M.H., Wicht O., et al. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peacock T.P., Goldhill D.H., Zhou J., et al. bioRxiv; 2020. The furin cleavage site of SARS-CoV-2 spike protein Is a key determinant for transmission due to enhanced replication in airway cells. [Google Scholar]
- 73.Kirchdoerfer R.N., Cottrell C.A., Wang N., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madu I.G., Roth S.L., Belouzard S., et al. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walls A.C., Tortorici M.A., Snijder J., et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jaimes J.A., Millet J.K., Whittaker G.R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 Site. iScience. 2020;23 doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shapiro J., Sciaky N., Lee J., et al. Localization of endogenous furin in cultured cell lines. J. Histochem. Cytochem. 1997;45:3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- 80.Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunology. 2019;8 doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses. 2019;11:837. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luczo J.M., Stambas J., Durr P.A., et al. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev. Med. Virol. 2015;25:406–430. doi: 10.1002/rmv.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Horimoto T., Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 84.Gupta A., Madhavan M.V., Sehgal K., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tipnis S.R., Hooper N.M., Hyde R., et al. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 86.Hikmet F., Méar L., Edvinsson Å., et al. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16 doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roebroek A.J., Umans L., Pauli I.G., et al. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125:4863–4876. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- 88.Sarac M.S., Cameron A., Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect. Immun. 2002;70:7136–7139. doi: 10.1128/IAI.70.12.7136-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sarac M.S., Peinado J.R., Leppla S.H., et al. Protection against anthrax toxemia by hexa-D-arginine in vitro and in vivo. Infect. Immun. 2004;72:602–605. doi: 10.1128/IAI.72.1.602-605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng Y.W., Chao T.L., Li C.L., et al. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33:108254. doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson B.A., Xie X., Kalveram B., et al. bioRxiv; 2020. Furin cleavage site Is key to SARS-CoV-2 pathogenesis. [Google Scholar]
- 92.Schmidt M.F., Schlesinger M.J. Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell. 1979;17:813–819. doi: 10.1016/0092-8674(79)90321-0. [DOI] [PubMed] [Google Scholar]
- 93.Anderson A.M., Ragan M.A. Palmitoylation: a protein S-acylation with implications for breast cancer. NPJ Breast Cancer. 2016;2:16028. doi: 10.1038/npjbcancer.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chamberlain L.H., Shipston M.J. The physiology of protein S-acylation. Physiol. Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thorp E.B., Boscarino J.A., Logan H.L., et al. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006;80:1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shulla A., Gallagher T. Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 2009;284:32725–32734. doi: 10.1074/jbc.M109.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang J., Lv J., Wang Y., et al. Replication of murine coronavirus requires multiple cysteines in the endodomain of spike protein. Virology. 2012;427:98–106. doi: 10.1016/j.virol.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gelhaus S., Thaa B., Eschke K., et al. Palmitoylation of the Alphacoronavirus TGEV spike protein S is essential for incorporation into virus-like particles but dispensable for S-M interaction. Virology. 2014;464–465:397–405. doi: 10.1016/j.virol.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petit C.M., Chouljenko V.N., Iyer A., et al. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McBride C.E., Machamer C.E. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology. 2010;405:139–148. doi: 10.1016/j.virol.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akerström S., Gunalan V., Keng C.T., et al. Dual effect of nitric oxide on SARS-CoV replication: viral RNA production and palmitoylation of the S protein are affected. Virology. 2009;395:1–9. doi: 10.1016/j.virol.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H., Yuan Z., Pavel M.A., et al. bioRxiv; 2020. The role of high cholesterol in age-related COVID19 lethality. [Google Scholar]
- 103.Kaufmann S.H.E., Dorhoi A., Hotchkiss R.S., et al. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018;17:35–56. doi: 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shang J., Wan Y., Luo C., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]