Abstract
SARS-CoV-2 papain-like protease is considered as an important potential target for anti-SARS-CoV-2 drug discovery due to its crucial roles in viral spread and innate immunity. Here, we have utilized an in silico molecular docking approach to identify the possible inhibitors of the SARS-CoV-2 papain-like protease, by screening 21 antiviral, antifungal and anticancer compounds. Among them, Neobavaisoflavone has the highest binding energy for SARS-CoV-2 papain-like protease. These molecules could bind near the SARS-CoV-2 papain-like protease crucial catalytic triad, ubiquitination and ISGylation residues: Trp106, Asn109, Cys111, Met208, Lys232, Pro247, Tyr268, Gln269, His272, Asp286 and Thr301. Because blocking the papain-like protease is an important strategy in fighting against viruses, these compounds might be promising candidates for therapeutic intervention against COVID-19.
Keywords: SARS-CoV-2, COVID-19, Papain-like protease, Molecular docking
Abbreviations: COVID-19, Corona Virus Disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; PLpro, papain-like protease; nsP3, non-structural protein 3; UBL, ubiquitin-like; DS, Discovery Studio; -Se-S-, selenylsulfide
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by the new severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) poses an unprecedented challenge [1,2]. Globally, as of October 21, 2020, more than 40.6 million cases have been reported to the World Health Organization, and 1,121,843 deaths have occurred in more than 235 countries. The scale of the pandemic and the rapidly spread make COVID-19 one of the most serious diseases facing mankind so far.
At present, there is no vaccine or approved antiviral treatment for SARS-CoV-2. In the extensive research on the development of new antiviral drugs for COVID-19, main protease of SARS-CoV-2, which plays an important role in blocking viral infection, has been identified as a potential target [3]. Another attractive antiviral target is papain-like protease (PLpro) non-structural protein 3 (nsP3), an essential component of the replicase-transcriptase complex [4]. Its topological structure is divided into ubiquitin-like (UBL), thumb, palm, and fingers [5]. It process the viral polyprotein by recognizing the tetrapeptide Leu-X-Gly-Gly motif, and functions as de-ubiquitinating and de-ISGylating enzyme to suppresses inflammation and antiviral signaling [6,7]. Therefore, inhibition of PLpro activity can prevent virus replication and destroy its role in host immune response evasion, making it a promising target for antiviral drugs. Recently, an inhibitor for PLpro GRL-0617 has been proved to be an effective therapeutic agent for SARS-CoV PLpro [8,9].
In current study, the inhibitory effects towards PLpro for 21 compounds, including azvudine, aminoethyl, bavachinin, corylifol A, chromen, darunavir, disulfiram, ebselen, efavirenz, famciclovir, GRL-0617, isobavachalcone, mercaptopurine, neobavaisoflavone, oseltamivir, papyriflavonol A, psoralidin, ribavirin, sofosbuvir, tioguanine and 4′-O-methylbavachalcone were investigated by molecular docking. These compounds are antiviral, antifungal or anticancer drugs. They have been proved to be highly effective against HIV, HCV, influenza virus, coronavirus, antifungal and anticancer agents [[10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]]. The focus of our study is on the repurposing and identification of compounds, aiming to accelerate the identification of potential drugs for COVID-19 treatment.
2. Materials and methods
2.1. Optimization of the 3D coordinates of the ligands
The 3D coordinates of the 21 drug ligands, azvudine (CSID: 24717759), aminoethyl (CSID: 29361441), bavachinin (CSID: 8512670), corylifol A (CSID: 22913562), chromen (CSID: 57578897), darunavir (CSID: 184733), disulfiram (CSID: 3005), ebselen (CSID: 3082), efavirenz (CSID: 57715), famciclovir (CSID: 3207), GRL-0617 (Chemical Book ID: CB74667969), isobavachalcone (CSID: 4444667), mercaptopurine (CSID: 580869), neobavaisoflavone (CSID: 4478223), oseltamivir (CSID: 58540), papyriflavonol A (CSID: 8518529), psoralidin (CSID: 4445118), ribavirin (CSID: 34439), sofosbuvir (CSID: 26286922), tioguanine (CSID: 2005804), and 4′-O-methylbavachalcone (CSID: 24845874) were obtained from ChemSpider (http://www.chemspider.com/) and Chemical Book (https://www.chemicalbook.com/) as a .mol file. The code within the parenthesis signifies the ChemSpider ID and ChemicalBook Number of the respective molecules. The secondary structure shown is the predicted by DSSP for SARS-CoV-2 (PDB: 7JIW), Rhinolophus affinis (GenBank: KF569996.1), SARS (PDB: 4OW0), MERS (PDB: 4RF1) and MHV (PDB: 5WFI) PLpro. Similarity and alignment calculations were performed using MEGA-X and ClustalW.
2.2. Drug-like properties of the ligands
The drug likeliness of a molecule is indicated by the Lipinski’s rule of five parameters (molecular weight ranges from 152.18 Da to 547.66 Da, no more than 5 hydrogen bond donors, no. of hydrogen bond acceptors should be less than 10 and logP should not be greater than 5). The Lipinski’s rule of five parameters was obtained from the SWISSADME server (www.swissadme.ch/index.php). The chemical structures, chemical formula and the Lipinski’s rule parameters of the ligands are listed in Table 1 .
Table 1.
Candidate drugs properties screened and dockscore through LibDock in the study.
| No. | Compounds | Structure | Treatment | Mass | Dockscore |
|---|---|---|---|---|---|
| 1 | Azvudine | 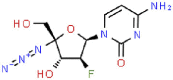 |
HIV | 286.22 | 83.52 |
| 2 | Aminoethyl | 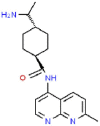 |
HIV | 312.41 | 65.25 |
| 3 | Bavachinin | 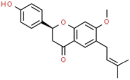 |
SARS | 338.40 | 82.65 |
| 4 | Corylifol A | 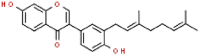 |
SARS | 390.47 | 89.15 |
| 5 | Chromen | 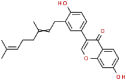 |
H1N1, H3N2 | 390.47 | 88.00 |
| 6 | Darunavir | 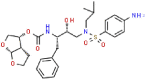 |
HIV | 547.66 | 99.38 |
| 7 | Disulfiram | 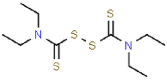 |
HIV, MERS, SARS | 296.54 | 36.89 |
| 8 | Ebselen | 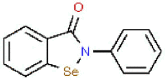 |
HIV | 274.18 | 69.10 |
| 9 | Efavirenz | 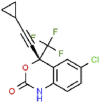 |
HIV | 315.68 | 69.73 |
| 10 | Famciclovir | 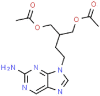 |
VZV | 321.33 | 75.77 |
| 11 | GRL-0617 | 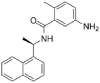 |
SARS | 304.39 | 67.70 |
| 12 | Isobavachalcone | 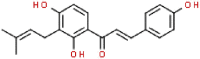 |
SARS | 324.37 | 52.81 |
| 13 | Mercaptopurine | 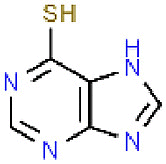 |
SARS | 152.18 | 60.27 |
| 14 | Neobavaisoflavone | 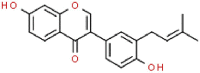 |
SARS | 322.35 | 110.53 |
| 15 | Oseltamivir | 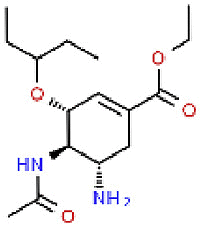 |
AIV, H1N1, IBV | 312.41 | 94.09 |
| 16 | Papyriflavonol A | 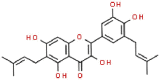 |
Antifungal | 438.47 | 56.56 |
| 17 | Psoralidin | 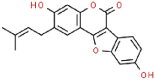 |
Anticancer | 336.34 | 120.99 |
| 18 | Ribavirin | 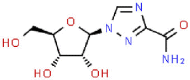 |
HCV | 244.21 | 71.04 |
| 19 | Sofosbuvir | 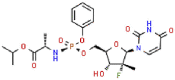 |
HCV | 529.45 | 124.29 |
| 20 | Tioguanine | 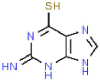 |
Anticancer | 167.19 | 37.43 |
| 21 | 4′-O-methylbavachalcone |  |
SARS | 352.42 | 82.85 |
2.3. Structure and preparation of PLpro for docking studies
SARS-CoV-2 papain-like protease (PDB: 6W9C, chains A, B, C) was used as the template for molecular docking. The 6W9C is solved by cryo-electron microscopy with 2.70 Å resolution, which is a trimer of three homologous amino acid chains (A, B, and C). Water molecules and ligands were removed from the PBD using Pymol.
2.4. Molecular docking analyses and visualization
Blind dockings were conducted based on docking ligands into a protein target using Discovery Studio (BIOVIA, 2016). In the key steps of the dockings, the protein remained rigid, while the ligand was fully flexible. Energy minimization of the protein was performed using the DS with CHARMM force field to relax the structure and remove the steric overlap. Then, polar hydrogen atoms were added. The centroid of the site model was used to define the active site box required by LibDock. The SARS-CoV-2 PLpro-binding site in the X-ray crystal structure of ligand complex was determined by co-crystallization (X: -46.0103, Y: 14.2997, and Z: 29.9464), with a radius of 13.8 Å. The ligand conformation generation method was selected as “best”, the number of binding site hotspots was set to 100, and other parameters were set by default. After each compound was docked, the best conformations and LibDock score were obtained. Considering their interaction energy and binding free energy, the compounds were screened. The binding modes (3D or 2D ligand-receptor interaction simulation map) of the selected compounds were analyzed.
3. Results and disscussion
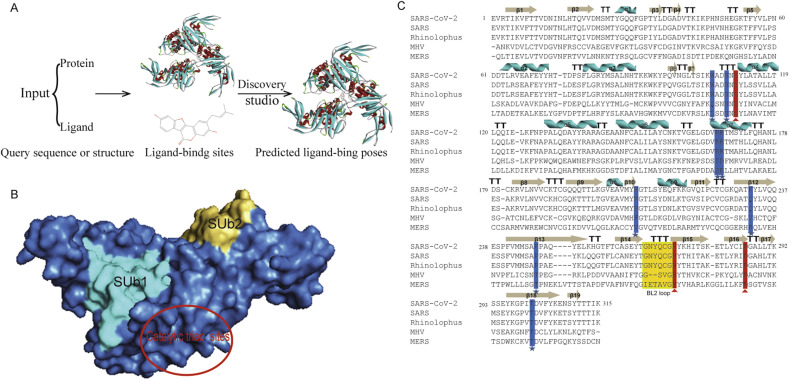
Docking and scoring software is used widely to enhance the drug design and predict the interaction between drugs and macromolecules in the pharmaceutical products. The docking method involves searching for binding sites on the whole macromolecular surface. Therefore, the papain-like protease of COVID-19, SARS-CoV-2 PLpro was used for blind docking analysis of some known drugs and bioactive substances. Fig. 1 A shows the overall workflow of Discovery Studio (DS) LibDock algorithm. Then SARS-CoV-2 PLpro structure was submitted to the SARS-CoV-2 PLpro structure hotspots methods to predict the protein-ligand binding sites.
Fig. 1.
Workflow of molecular docking, architecture of SARS-CoV-2 PLpro, and sequences alignment of PLprofrom coronaviruses. (A) A schematic workflow of protein-ligand complex formation by Discovery Studio. (B) Solvent-accessible surface representation of SARS-CoV PLpro is shown in blue. The two ubiquitin, ubiquitin-like binding subsites of SARS-CoV-2 PLpro and catalytic domain are shown in the solvent accessible surface representation with SUb1 shaded cyans, SUb2 shaded golden and red circle, respectively. (C) The PLpro from SARS-CoV-2 (7JIW), SARS (4OW0), MERS (4RF1), Rhinolophus affinis (KF569996.1) and MHV (5WFI). The secondary structure shown is the predicted by DSSP for SARS-CoV PLpro (5E6J). Similarity and alignment calculations were performed using ClustalW. Residue positions form the catalytic triads are marked with red triangle, while key residues forming ubiquitination and ISGylation binding sites are marked with blue stars. T, turn. The blocking loop2 (BL2) is boxed in gold. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In addition to the recommend drugs for COVID-19, we have also conducted molecular docking studies with SARS-CoV-2 PLpro for various natural compounds, antiviral drugs, antifungal drugs and anticancer drug (Table 1). Among SARS-CoV-2 PLpro, SARS PLpro, MERS PLpro, Rhinolophus affinis PLpro, and MHV PLpro, classic catalytic triad, two different ubiquitin/ubiquitin-like conjugates and ISG15 modified protein binding sites are quite conservative (Fig. 1B). And some of them may be related to protein stability, kinetics, ligand binding and catalytic performance [9,[22], [23], [24]]. Amino acid sequence alignment showed that SARS-CoV-2 and SARS PLpro (82.86%) as well as Rhinolophus affinis PLpro (83.49%) were highly similar, and there were significant differences between SARS-CoV-2 and MERS (29.84%) and MHV PLpro (29.52%) (Fig. 1C). Therefore, SARS-CoV-2 PLpro is more closely related to SARS and Rhinolophus affinis than MERS and MHV. Examples include the active conservative catalytic trinity of Cys111, His272 and Asp286. The catalytic Trinity accepts Cys111 in the form of –SH as nucleophilic agent, His272 as general acid and alkali, and Asp286 promotes the deprotonation of Cys111 [25,26].
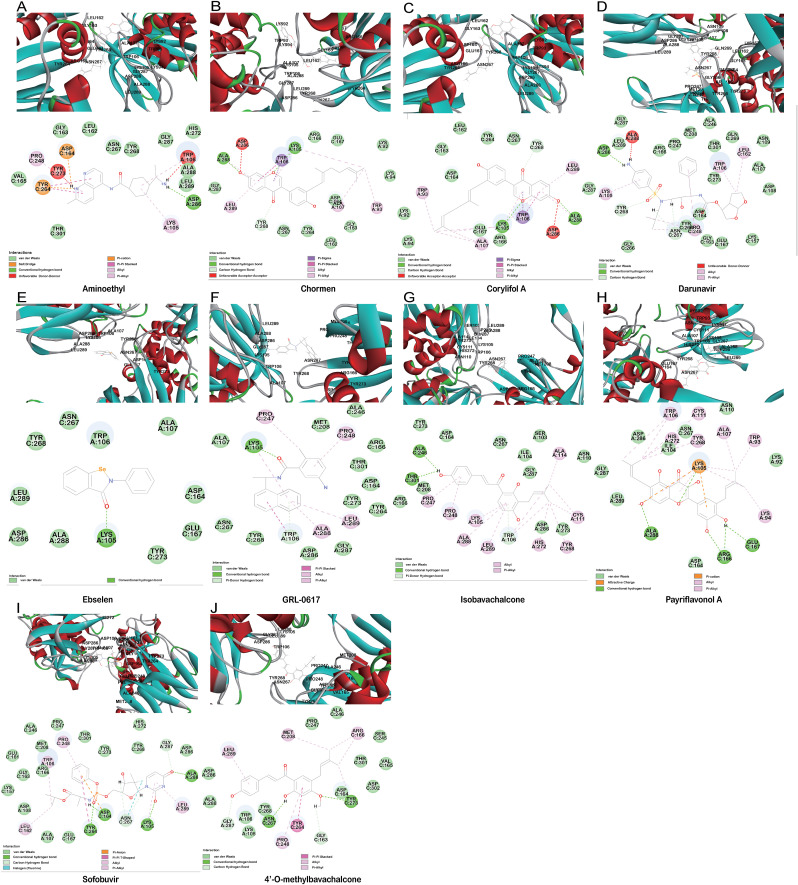
Aminoethyl, corylifol A, chromen, darunavir, ebselen, GRL-0617, isobavachalcone, papyriflavonol A, sofosbuvir and 4′-O-methylbavachalcone shows the formed interactions among the drug ligands near the catalytic triad. Aminoethyl and darunavir form conventional hydrogen bonds with Asp286 (Fig. 2 A and B). In addition, aminoethyl constructs ten van der waals with Leu162, Gly163, Val165, Asn267, Tyr268, His272, Gly287, Ala288, Leu289 and Thr301, two π-alkyl interactions with Lys105 and Pro248, two π-sigma with Asp164 and Tyr264, and two unfavorable donor-donor interactions of Trp106 and Tyr273. Darunavir constructs two carbon hydrogen bond with Asn267 and Tyr268, eighteen van der waals with Ala107, Asp108, Asn109, Lys157, Gly163, Asp164, Arg166, Glu167, Met208, Ala246, Pro247, Tyr264, Gly266, Gln269, Tyr273, Gly287, Leu289 and Thr301, four π-alkyl interactions with Lys105, Trp106, Leu162 and Pro248. However, the donor-donor interaction between Darunavir and Ala288 is unfavorable. Darunavir may block the binding of PLpro with target protein and exert its enzymatic activity. Chromen and corylifol A has almost the same site binding pattern, forming two conventional hydrogen bonds with Lys105 and Ala288 (Fig. 2C and D), a carbon hydrogen bond with Tyr268, ten van der waals with Lys92, Lys94, Leu162, Gly163, Asp164, Arg166, Glu167, Tyr264, Asn267 and Gly287, three π-alkyl interactions with Trp93, Ala107 and Leu289, a π-sigma with Trp106, and a unfavorable acceptor-acceptor sites with Asp286. Ebselen is an organic selenium compound that mimics the activity of glutathione. This compound forms a covalent bond between the selenium atom and thiols in the cysteine residue by forming a selenylsulfide (-Se-S-) linkage to inhibit HIV-1 capsid dimerization [27]. In the paper, we found that ebselen constructed a conventional hydrogen bonds with Lys105 (Fig. 2E), ten van der waals with Trp106, Ala107, Asp164, Glu167, Asn267, Tyr268, Tyr273, Asp286, Ala288 and Leu289. Although ebselen could not form -Se-S- with Cys111, it interacts with the PLpro active site. It GRL-0617 interacts with the residues in the active site through hydrogen bonding with Lys105 and Trp106, π-alkyl interaction with Pro247, Pro248, Ala288 and Leu289, van der waals forces of attraction with other residues (Fig. 2F). GRL-0617 is contained in the substrate cleft formed between the β11-β12 strands and the loop connecting α3 and α4 (α3-to-α4 loop), which occupies the S3–S4 pockets [4,6]. It leads to the loss of PLpro activity. Isobavachalcone and papyriflavonol A form van der waals forces with Asp286, π-alkyl hydrophobic interaction with Cys111, His272 and other sites (Fig. 2G and H). Sofosbuvir and 4′-O-methylbavachalcone construct four and two conventional hydrogen bonds, respectively (Fig. 2I and J). They also form van der waals with Asp286 and with other sites.
Fig. 2.
The minimum docked poses of the compounds along with their corresponding catalytic triad interaction plots within the active site of SARS-CoV-2 PLpro.
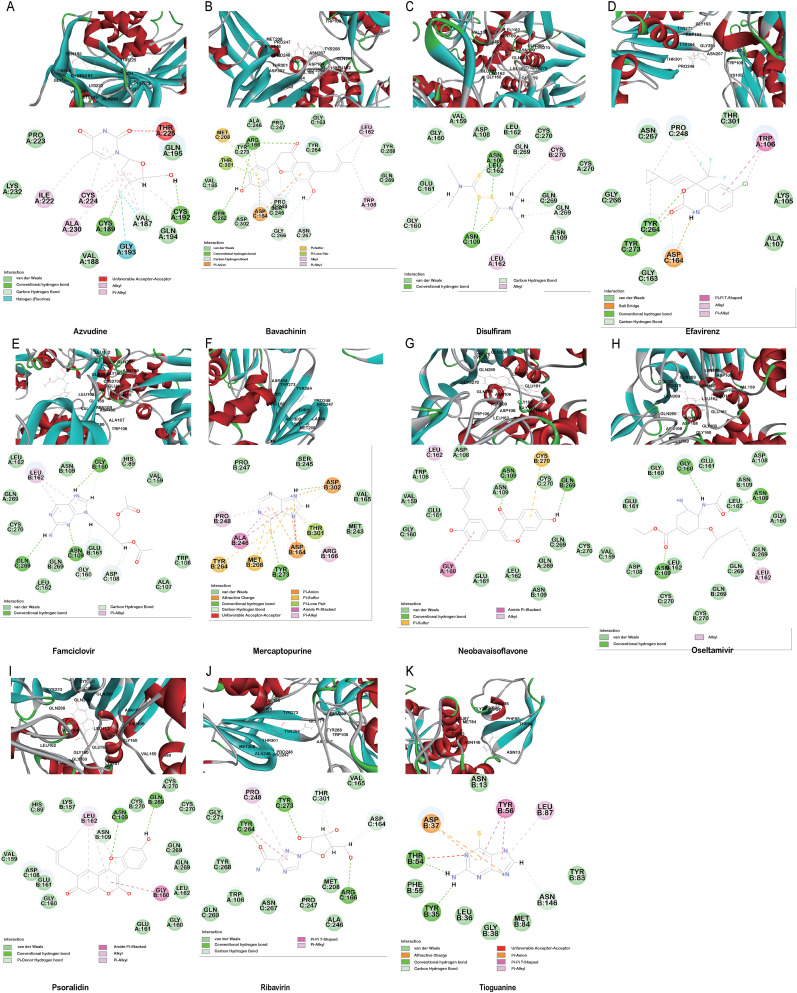
The PLpro can also inhibit host innate immune response by reversing host ubiquitination and ISGylation events [8]. In addition to blocking catalytic sites, PLpro binding, processing active sites of ubiquitin and ISG15 protein sites are highly desirable [22]. Azvudine, bavachinin, disulfiram, efavirenz, famciclovir, mercaptopurine, neobavaisoflavone, oseltamivir, psoralidin and ribavirin show interactions proximity to the sites, locating around Trp106, Asn109, Met208, Lys232, Pro247, Tyr268, Gln269, Thr301 (Fig. 3 A–J). Tioguanine (6-thioguanine, 6 TG) has been proven to be effective in inhibition the enzyme activity of SARS PLpro by forming of a hydrogen bond with Cys112 [19,28]. Here, it is found that 6 TG forms two conventional hydrogen bonds with Tyr35 and Thr54, a carbon hydrogen bond with Asn146, six van der waals with Asn13, Leu36, Gly38, Phe55, Tyr83 and Met84, π-alkyl with Leu87, π-π T-shaped interaction forces with Tyr56 and other forces as depicted in the Fig. 3K. It may affect the host ubiquitination and ISGylation events through the bond interaction with the N-terminal UBL domain.
Fig. 3.
The minimum docked poses of the compounds along with their corresponding ubiquitination and ISGylation interaction plots within the active site of SARS-CoV-2 PLpro.
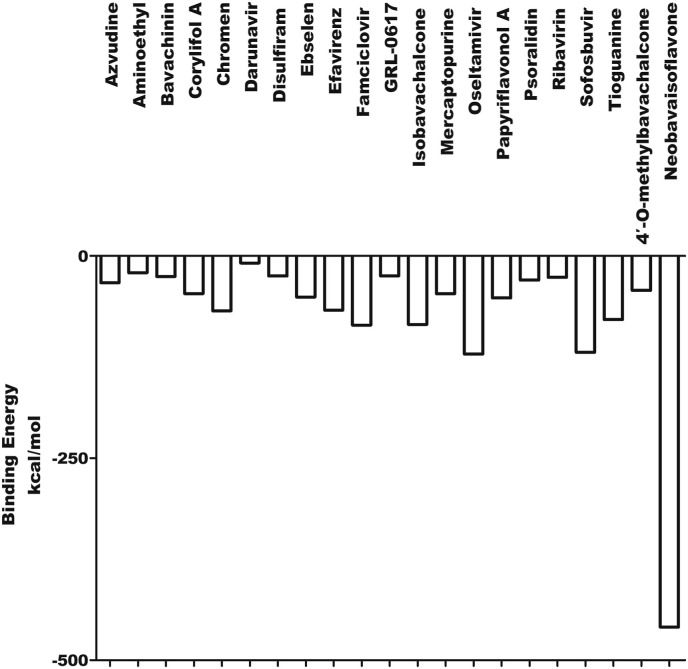
The results of blind molecular docking of 21 different classes of compounds showed that they might be potential anti-SARS-CoV-2 drugs because they bind to the active site of the papain-like protease of SARS-CoV-2 PLpro. Inhibition of this enzyme/protease is the key to blocking virus replication and reproduction. The inhibitor binding sites in SARS-CoV-2 protease are composed of key residues Cys111, His272 and Asn286 as evident from the recent study on the GRL-0617 and ebselen inhibitors for SARS-CoV-2 PLpro [4,29]. Through the blind docking study of all 21 molecules with SARS-CoV-2 protease, we found that these molecules are usually surrounded by the above mentioned residues, ubiquitination and ISGylation sites, which clearly indicates that those molecules can prevent the replication of SARS-CoV-2 virus and evade the host immune response. The changes areas of the important interacting residues in the ligands and protease active sites were listed in Supplementary Table 1. Although blind docking studies have shown that all molecules can be used as potential drugs for the treatment of COVID-19, based on the estimated binding energy (ΔG) value, we can infer that among all the compounds studied, neobavaisoflavone (with the highest negative minimum ΔG value (−459 kcal/mol) may be the best potential inhibitor of SARS-CoV-2, followed by oseltamivir (−121.55 kcal/mol) ˃ sofosbuvir (−119.44 kcal/mol) ˃ famciclovir (−85.61 kcal/mol) ˃ isobavachalcone (−84.75 kcal/mol) ˃tioguanine (−78.64 kcal/mol) ˃ chromen (−67.92 kcal/mol) ˃ efavirenz (−66.98 kcal/mol) ˃ papyriflavonol A (−51.99 kcal/mol) ˃ ebselen (−50.99 kcal/mol) > corylifol A (−46.78 kcal/mol) ˃ mercaptopurine (−46.61 kcal/mol) ˃ 4′-O-methylbavachalcone (−42.64 kcal/mol) ˃ azvudine (−33.1 kcal/mol) ˃ psoralidin (−29.89 kcal/mol) ˃ ribavirin (−26.49 kcal/mol) ˃ bavachinin (−25.59 kcal/mol) ˃ disulfiram (−24.84 kcal/mol) ˃ GRL-0617 (−24.62 kcal/mol) ˃ aminoethyl (−20.87 kcal/mol) ˃ darunavir (−8.74 kcal/mol) (Fig. 4 ).
Fig. 4.
The binding energy of the interaction between SARS-CoV-2 PLpro and 21 compounds.
In this study, the free energy of binding energy of compounds and PLpro was ranged from −459 kcal/mol to −8.74 kcal/mol, indicating that those compounds could interact with the SARS-CoV-2 PLpro. Our finding also suggests the potential binding modes with known therapeutic drugs for COVID-19. Since this study was performed using computational methods, special attention should be paid to these results and further experiments in vivo and in vitro are needed.
Funding
This work was supported by grants from National Key Plan for Research and Development of China [2016YFD0500300], National Natural Science Foundation of China [82072270 and 81871663], National Major S & T Project for the Prevention and Treatment of Major Infectious Diseases in China [2017ZX10004206-007], the Innovation Project of Shandong Academy of Medical Sciences, and Academic Promotion Programme of Shandong First Medical University [2019LJ001].
Declaration of competing interest
We declare no competing interests.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2020.11.083.
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2020.11.083
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Transparency document
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y., Zhang L. Social media WeChat infers the development trend of COVID-19. J. Infect. 2020;81:e82–e83. doi: 10.1016/j.jinf.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghahremanpour M.M., Tirado-Rives J., Deshmukh M., Ippolito J.A., Zhang C.H., de Vaca I.C., Liosi M.E., Anderson K.S., Jorgensen W.L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.28.271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosken Y.K., Cholko T., Lou Y.C., Wu K.P., Chang C.A. Insights into dynamics of inhibitor and ubiquitin-like protein binding in SARS-CoV-2 papain-like protease. Front. Mol. Biosci. 2020;7:174. doi: 10.3389/fmolb.2020.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden van Noort G.J., Geurink P.P., Ovaa H., Newman J., Riboldi-Tunnicliffe A., Czabotar P.E., Mitchell J.P., Feltham R., Lechtenberg B.C., Lowes K.N., Dewson G., Pellegrini M., Lessene G., Komander D. EMBO J; 2020. Mechanism and Inhibition of the Papain-like Protease, PLpro, of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Muller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E., Baker S.C., Ghosh A.K., Mesecar A.D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu N., Yang J., Zheng B., Zhang Y., Cao Y., Huan C., Wang S., Chang J., Zhang W. The pyrimidine analog FNC potently inhibits the replication of multiple enteroviruses. J. Virol. 2020;94 doi: 10.1128/JVI.00204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin M.H., Moses D.C., Hsieh C.H., Cheng S.C., Chen Y.H., Sun C.Y., Chou C.Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antivir. Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32:504–515. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seniya C., Yadav A., Khan G.J., Sah N.K. In-silico studies show potent inhibition of HIV-1 reverse transcriptase activity by a herbal drug. IEEE ACM Trans. Comput. Biol. Bioinf. 2015;12:1355–1364. doi: 10.1109/TCBB.2015.2415771. [DOI] [PubMed] [Google Scholar]
- 14.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2014;29:59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 15.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 16.Yuan S., Chu H., Zhang K., Ye J., Singh K., Kao R.Y., Chow B.K., Zhou J., Zheng B.J. A novel small-molecule compound disrupts influenza A virus PB2 cap-binding and inhibits viral replication. J. Antimicrob. Chemother. 2016;71:2489–2497. doi: 10.1093/jac/dkw194. [DOI] [PubMed] [Google Scholar]
- 17.Williams P.L., Yildirim C., Chadwick E.G., Van Dyke R.B., Smith R., Correia K.F., DiPerna A., Seage G.R., Hazra R., 3rd, Crowell C.S., A.R.T.T.s.o.t.P.H.I.V.A.C. Surveillance Monitoring for, Association of maternal antiretroviral use with microcephaly in children who are HIV-exposed but uninfected (SMARTT): a prospective cohort study. Lancet HIV. 2020;7:e49–e58. doi: 10.1016/S2352-3018(19)30340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.J.R. Semaan, M. Parmar, Famciclovir, StatPearls, Treasure Island (FL), 2020.
- 19.Chou C.Y., Chien C.H., Han Y.S., Prebanda M.T., Hsieh H.P., Turk B., Chang G.G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisman A.N., Valuev-Elliston V.T., Ozerov A.A., Khandazhinskaya A.L., Chizhov A.O., Kochetkov S.N., Pannecouque C., Naesens L., Seley-Radtke K.L., Novikov M.S. 1,6-Bis[(benzyloxy)methyl]uracil derivatives-Novel antivirals with activity against HIV-1 and influenza H1N1 virus. Bioorg. Med. Chem. 2016;24:2476–2485. doi: 10.1016/j.bmc.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Cazares-Cortazar A., Uribe-Noguez L.A., Mata-Marin J.A., Gaytan-Martinez J., de la Luz Martinez-Rodriguez M., Villavicencio-Ferrel P.E., Chapararro-Sanchez A., Mauss S., Ocana-Mondragon A. A decrease in hepatitis C virus RNA to undetectable levels in chronic hepatitis C patients after PegIFNalpha + RVB or sofosbuvir + NS5A inhibitor treatment is associated with decreased insulin resistance and persistent oxidative stress. Arch. Virol. 2020;165:2759–2766. doi: 10.1007/s00705-020-04797-y. [DOI] [PubMed] [Google Scholar]
- 22.Kandeel M., Kitade Y., Fayez M., Venugopala K.N., Ibrahim A. The emerging SARS-CoV-2 papain-like protease: its relationship with recent coronavirus epidemics. J. Med. Virol. 2020 doi: 10.1002/jmv.26497. [DOI] [PubMed] [Google Scholar]
- 23.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 24.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niemeyer D., Mosbauer K., Klein E.M., Sieberg A., Mettelman R.C., Mielech A.M., Dijkman R., Baker S.C., Drosten C., Muller M.A. The papain-like protease determines a virulence trait that varies among members of the SARS-coronavirus species. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su H., Su H., Zhou F., Huang Z., Ma X., Natarajan K., Zhang M., Huang Y. Molecular insights into small molecule drug discovery for SARS-CoV-2. Angew Chem. Int. Ed. Engl. 2020 doi: 10.1002/anie.202008835. [DOI] [PubMed] [Google Scholar]
- 27.Thenin-Houssier S., de Vera I.M., Pedro-Rosa L., Brady A., Richard A., Konnick B., Opp S., Buffone C., Fuhrmann J., Kota S., Billack B., Pietka-Ottlik M., Tellinghuisen T., Choe H., Spicer T., Scampavia L., Diaz-Griffero F., Kojetin D.J., Valente S.T. Ebselen, a small-molecule capsid inhibitor of HIV-1 replication. Antimicrob. Agents Chemother. 2016;60:2195–2208. doi: 10.1128/AAC.02574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., Chou C.Y., Chang G.G. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antivir. Chem. Chemother. 2009;19:151–156. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- 29.Mikolaj Zmudzinski W.R., Olech Kamila, Granda Jarosław, Giurg Mirosław, Burda-Grabowska Małgorzata, Zhang Linlin, Sun Xinyuanyuan, Lv Zongyang, Nayak Digant, Kesik-Brodacka Malgorzata, Olsen Shaun K., Hilgenfeld Rolf, Drag Marcin. 2020. Ebselen Derivatives Are Very Potent Dual Inhibitors of SARS-CoV-2 Proteases - PLpro and Mpro in in Vitro Studies, bioRxiv : the Preprint Server for Biology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.