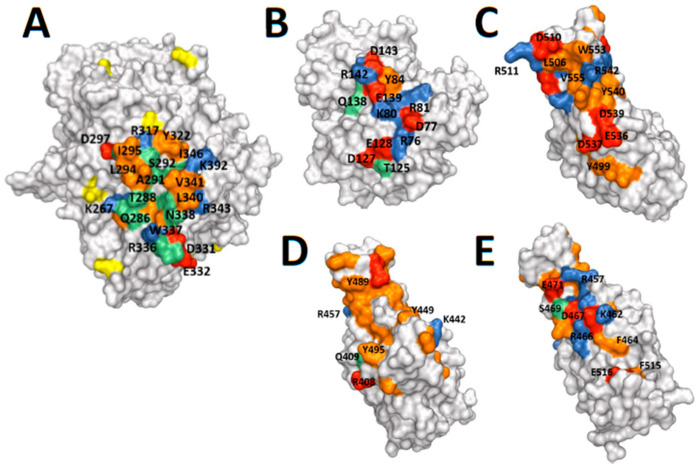
Figure 5.
Protein structures. (A) DPP4 monomer (PDB ID 1W1I) [46]. (B) Adenosine Deaminase (ADA) (PDB ID 1W1I) [46]. (C) MERS-CoV spike RBD (PDB ID 4L72) [45], and (D,E) SARS-CoV-2 spike RBD (PDB ID 6M0J) [48] shown with space-filling surfaces with similar orientation and scale. The observed and predicted interfacial binding sites are highlighted in green, with the charged residues highlighted in red (negative) and blue (positive), and the hydrophobic residues are highlighted in orange. In particular, compared to the other molecules SARS-CoV-2 spike RBD has an extensive hydrophobic surface, and fewer charged residues at the predicted DPP4 binding site that is in a similar location to MERS-CoV spike RBD (D) [28], but has more charges surfaces on the opposite side of SARS-CoV-2 spike RBD, which is a potential alternative binding site (E) [28]. The N-glycosylated residues in DPP4 are highlighted in yellow, and are sufficiently distant from the binding interface to expect that they do not bind to ADA [20].