Abstract
The diagnosis and prognosis of traumatic brain injury (TBI) is complicated by variability in the type and severity of injuries and the multiple endophenotypes that describe each patient's response and recovery to the injury. It has been challenging to capture the multiple dimensions that describe an injury and its recovery to provide clinically useful information. To address this challenge, we have performed an open-ended search for panels of microRNA (miRNA) biomarkers, packaged inside of brain-derived extracellular vesicles (EVs), that can be combined algorithmically to accurately classify various states of injury. We mapped GluR2+ EV miRNA across a variety of injury types, injury intensities, history of injuries, and time elapsed after injury, and sham controls in a pre-clinical murine model (n = 116), as well as in clinical samples (n = 36). We combined next-generation sequencing with a technology recently developed by our lab, Track Etched Magnetic Nanopore (TENPO) sorting, to enrich for GluR2+ EVs and profile their miRNA. By mapping and comparing brain-derived EV miRNA between various injuries, we have identified signaling pathways in the packaged miRNA that connect these biomarkers to underlying mechanisms of TBI. Many of these pathways are shared between the pre-clinical model and the clinical samples, and present distinct signatures across different injury models and times elapsed after injury. Using this map of EV miRNA, we applied machine learning to define a panel of biomarkers to successfully classify specific states of injury, paving the way for a prognostic blood test for TBI. We generated a panel of eight miRNAs (miR-150-5p, miR-669c-5p, miR-488-3p, miR-22-5p, miR-9-5p, miR-6236, miR-219a.2-3p, miR-351-3p) for injured mice versus sham mice and four miRNAs (miR-203b-5p, miR-203a-3p, miR-206, miR-185-5p) for TBI patients versus healthy controls.
Keywords: extracellular vesicles, miRNA, statistical learning, traumatic brain injury
Introduction
A key hurdle to improving the treatment of traumatic brain injury (TBI) is the current lack of molecular biomarkers that can be used to accurately diagnose a patient's injury, track their recovery, and guide their treatment.1–4 While there has been great interest in the discovery of TBI biomarkers and enormous progress made in the development of ultrasensitive platforms to detect sparse molecular markers in blood and cerebral spinal fluid,4–12 the development of clinically useful blood test has been confounded by the variability in both injuries and in individual patients’ response to injury.7
Much work has been done to discover biomarkers inspired by existing knowledge of the neuropathology of TBI. For example, degenerating axons and neurons are known to release neurofilaments and other proteins such as ubiquitin C-terminal hydrolase L1 (UCH-L1) into the cerebrospinal fluid and blood, and reactive astrocytes release calcium-binding protein B (S100B) and glial fibrillary acidic protein (GFAP) into the same fluid compartments.4,6,10,11,13,14 Additionally, various microRNA (miRNA) biomarkers have been clinically evaluated, including miR-21, miR-93, and miR-191.5,7–9 Currently discovered biomarkers have demonstrated positive predictive values for identifying injured patients versus healthy controls4–7,10,11,13,14 and limited effectiveness at predicting states of injury5–7 and recovery.5,7–9 Several groups have measured multiple markers and combined these multiple measures algorithmically to diagnose TBI.4,15 Recently, protein biomarkers (UCH-L1, GFAP) were approved as a diagnostic by the U.S. Food and Drug Administration (FDA) that can identify patients with intracranial lesions following a head injury without the need for a computed tomography (CT) x-ray scan (FDA, 2018).
However, despite these recent successes, because of the wide variety of injury types and severities endemic to TBI, and the unique and dynamically changing combination of multiple endophenotypes of an injured and recovering brain—including axonal shearing, inflammation, glial cell damage, brain edema, and vascular injury16—it has remained challenging to adequately map any biomarker or sets of biomarkers to an individual's unique state of injury and recovery to improve clinical outcomes.
To address this challenge, we present an open-ended approach for the development of a highly multiplexed TBI biomarker panel. Rather than relying solely on previous knowledge of the degenerative changes in the brain following a TBI, we have instead combined three technologies to identify combinations of markers to classify specific states of injury. We use Track Etched Magnetic Nanopore (TENPO) sorting17–20 to isolate brain-derived GluR2+ extracellular vesicles (EVs) directly from plasma, next-generation sequencing to map the miRNA biomarkers packaged inside these EV biomarkers, and statistical machine learning algorithms to algorithmically combine these biomarkers to classify the specific injury state. Here, our goal is to enrich for a subset of brain-derived EVs that express GluR2 protein that can provide a signature for various states of brain injury, rather than to isolate a pure brain-derived EV population. Our previous work in this area demonstrated the value of using the RNA cargo of EVs to diagnose specific injury states, but the discovery of these RNA marker was limited to only injured versus sham subjects using a murine blast model.17
In this work, we sequenced the brain-derived EV miRNA cargo in murine TBI models (n = 116) with a variety of injury types (controlled cortical impact [CCI] and Blast), multiple injury intensities, history of injuries, and time elapsed since the injury (1 h to 14 days). We also sequenced the brain-derived EV miRNA cargo in clinical samples (n = 36), to accomplish the following goals:
-
1.
We generated a map of biomarkers that follow injury, both in the acute phase and days to weeks after the injury, which we used to classify specific states of injury.
-
2.
We identified candidate brain-derived EV activated signaling pathways present in the GluR2+ EV miRNA, shared between the murine model and the clinical cohort, to connect the miRNA biomarkers to underlying mechanisms of TBI.
-
3.
We evaluated and validated strategies to apply statistical machine learning on the EV RNA biomarkers to classify injury types, intensities, history of prior injury, and time elapsed since injury.
-
4.
We validated that this machine learning–based biomarker panel approach has the potential for translation to clinical use.
Methods
Mouse plasma collection
All mouse work was approved by Institutional Animal Care and Use Committee (IACUC). Blood was collected from mice by cardiac puncture and collected in sodium citrate BD Vacutainer blood collection tubes (BD Biosciences). Blood was centrifuged at 1500 g for 10 min and plasma was recovered, followed by a second spin at 3000 g for 10 min to minimize cellular contamination.
Human sample collection
TBI subjects and healthy control subjects were enrolled in a prospective biorepository study at the University of Pennsylvania. The study was approved by the Institutional Review Board of the University of Pennsylvania (IRB Protocol # 825783; initial approval 10/16/2016; most recent approval 10/7/2018). All TBI subjects required admission to Penn Presbyterian Medical Center, the Level 1 trauma center of the University of Pennsylvania, for their traumatic injuries. All TBI subjects included in this analysis had: 1) high clinical suspicion of non-penetrating acute TBI, determined by the treating physicians for which a head CT scan was performed; 2) age ≥18 years; 3) interval between time of injury and enrollment <24 h; and 4) ability to obtain informed consent from the subject or a legally authorized representative. The majority of the TBI subjects sustained mild TBIs, as defined by a post-resuscitation Glasgow Coma Scale (GCS) score of 13–15. For this study, we analyzed plasma samples that were collected <24 h from injury and were immediately aliquoted and frozen at −80°C until further analyses.
CCI
CCI experiment was performed on adult male (12–14 weeks old) C57BL/6J mice (Charles River, MA). All CCI injuries were performed as previously described.71 Anesthetized mice (isoflurane 3.0% induction, 1.5–2.0% maintenance in medical grade air: 21% oxygen, 78% nitrogen) were placed in a stereotaxic frame. A manual craniotomy was performed on the right parietotemporal region of the skull midway between bregma and lambda using a 4 mm diameter trephine (CMA7431058, Harvard Apparatus, Holliston, MA). The skull flap was removed and the animal was loaded into the CCI machine. Isoflurane was stopped 30 sec before CCI. A moderate injury was produced at an average impact velocity of 2.4 m/sec with an impact depth of 1.0 mm, centering the impact at approximately −2.5 mm bregma. Sham control animals underwent craniotomy, placed in the stereotaxic holder, but no CCI was delivered. Following the CCI, the exposed area was sutured and mice were allowed to emerge from anesthesia in a heated cage.
Blast injury
Adult male (12–14 weeks old) C57BL/6J mice (Charles River, MA) were exposed to a Blast overpressure insult that mimicked Blast-induced traumatic brain injury.23 Anesthetized mice (isoflurane 3.0% induction, 1.5–2.0% maintenance) were used as sham controls. Anesthetized mice were prepared for injury by placing sound insulating foam into each ear canal. Mice were loaded into a holder positioned 1 cm away from the end of the shock tube, positioned with their snouts facing the shock tube. Head motion was limited with a metal rod encircling the snout and placing a cervical collar between the occiput and shoulders. A single overpressure of either 215 ± 18kPa or 415 ± 41 kPa was delivered and the animal was immediately removed from the holder assembly. We strictly followed the guidelines from the University of Pennsylvania office of University Laboratory Animal Resources using approved protocols, and animal care and use was in accordance with the guidelines specified by the IACUC of the University of Pennsylvania.
TENPO device
Using thermal evaporation, 200 nm layer of permalloy (Ni80Fe20) and 30 nm layer of gold were sequentially deposited (Kurt Lesker PVD-75; Singh Nanofabrication Facility, University of Pennsylvania) on a track-etched polycarbonate membrane with 600 nm pores (Whatman). Moisture-resistant polyester film (McMaster-Carr, 0.004″ thick) and solvent-resistant tape (McMaster-Carr) were cut using laser micromachining (Universal Laser VLS 3.50) and used to make layers of TENPO device. A reservoir was made using an optically clear cast acrylic sheet (McMaster-Carr) and an output was made using a polydimethylsiloxane piece. A tygon tubing was connected to the output and a negative pressure supply (Programmable Syringe Pump; Braintree Scientific).
EV capture using TENPO device
Biotin anti-human, mouse GluR1/GluR2 antibody (Bioss) was incubated with the sample for 20 min at room temperature with shaking and anti-biotin ultrapure microbeads (Miltenyi Biotec) was incubated with the sample for 20 min for room temperature subsequently. The sample was loaded to the reservoir of the TENPO device and a programmable syringe pump (Braintree) was used to apply negative pressure.
RNA isolation
Second part of the ExoRNeasy serum/plasma kit (Qiagen) was used for RNA extraction. We first added 700 μL of QIAzol lysis reagent (Qiagen) on chip and then collected the lysate using a negative pressure and used the second part of the ExoRNeasy serum/plasma kit. The exosomal RNA was stored at −80C or processed immediately for further analysis.
RNA sequencing
QIAseq miRNA library kit (Qiagen) was used to make a library of exosomal RNA. BioAnalyzer was used to quantify RNA. The library was sequenced using a HiSeq 2500 kit (Illumina, Next-Generation Sequencing Core; University of Pennsylvania).
Sequencing data and Kyoto Encyclopedia of Genes and Genomes pathway analysis
Expression was quantified using a modified version of the UPenn SCAP-T RNA-Seq expression pipeline (Fisher, S A., “Safisher/Ngs.” GitHub, 2017), aligning to the hg38 and mm10 genomes for human and mouse, respectively. The minimum fragment length allowed past the TRIM module was reduced to 16 bases. STAR72 version STAR_2.4.0h1 was used, and was allowed to map as few as 16 bases in a read by setting “—outFilterMatchNmin 16” and “—outFilterMatchNminOverLread 0.” The number of allowed mismatches was capped at one using “—outFilterMismatchNmax 1,” and “—alignIntronMax 1” was set to prohibit calling unannotated splices. Expression counts were normalized by DESeq273 and quantified using VERSE74, using Gencode 25 and UCSD mm10 gene annotations, combined with MirBase v21 annotations for 3p and 5p microRNA. For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, DIANA mirPath v. 3.0 was used to identify the pathways of target genes regulated by exosomal miRNAs.
Machine learning data analysis
Using Matlab (R2016b), we performed an averaging of scores generated by five different built-in statistical learning algorithms (KNN, SVM, LDA, Logistic regression, Naive Bayes). To prevent overfitting, we performed 5-fold cross-validation and averaged the performance from 500 repeated runs for the classification with small sample sizes. For the classification with enough sample sizes, we created a separate training set and an independent blinded test set to better prevent overfitting.
Results
RNA sequencing of the EV RNA cargo from TBI mouse models and a clinical cohort
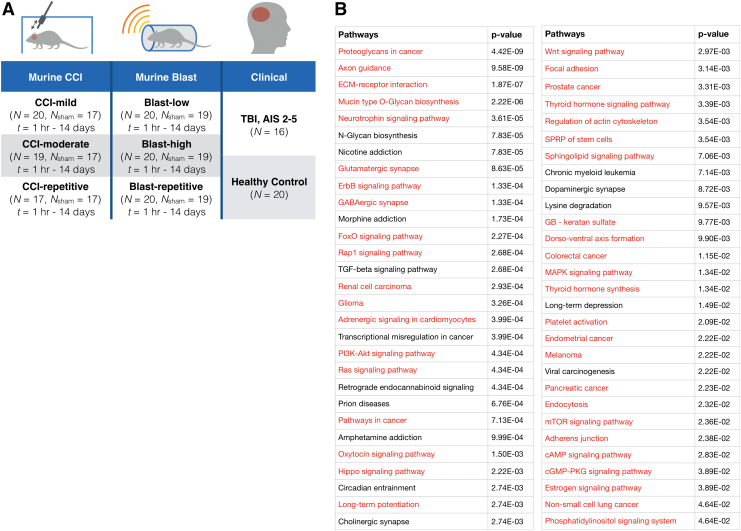
We used two different murine models of traumatic brain injury in this study, CCI and Blast, allowing us to control the injury and the time-point elapsed between injury and sample collection with a precision that is not possible in clinical samples.21 Our murine study was designed to include mice with a diverse set of injuries and recovery times. The study included 116 mice, of which 56 were given a CCI, 60 given a Blast injury,22,23 and 36 given sham condition (Fig. 1A). For each of these injury types, we used two different injury intensities. For the CCI injuries, we performed both mild (n = 20) and moderate injuries (n = 19).24 For the Blast injuries, we performed both low (n = 20, 215 kPa) and high (n = 20, 415 kPa) Blast pressure injuries. For repetitive injuries, we performed the second injury 24 h after the first injury, including mild CCI followed by mild CCI and low Blast followed by low Blast.
FIG. 1.
RNA sequencing to map the molecular changes in brain-derived extracellular vesicles (EVs). (A) A schematic of various injury states designed for this study. The injury states for mice include different types (CCI, Blast), different intensities within each type (mild CCI, moderate CCI, low Blast, high Blast), different histories of injury (single, double), and various time-points post-injury (1 h, 1 day, 4 days, 14 days) for all the states including sham controls. The injury states for human include traumatic brain injury (TBI) patients with Abbreviated Injury Score 2–5 and healthy controls. (B) We compared signaling pathways that were activated in brain-derived EVs of TBI mice to those of TBI human patients. A total of 56 pathways were found to be significantly activated (p < 0.05) in TBI patients, and of these 56 pathways, 41 pathways were detected to be significant (p < 0.05) in TBI mouse models. SPRP, signaling pathways regulating pluripotency; GB, glycosaminoglycan biosynthesis. Color image is available online.
For each of these injury conditions, we collected samples at t = 1 h, 1 day, 4 days, and 14 days after injury. Due to the limited volume of mouse plasma, we performed terminal blood collection for each combination of injury condition and time-point after injury, rather than perform longitudinal sample collection from the same animals. For each condition and time-point after injury, four to five biological replicates per condition were analyzed. For each condition and time-point after injury, we measured sham controls (n = 36) that were processed identically to the injured mice, with the exception of the injury itself. Additionally, we sequenced 16 TBI patients with an Abbreviated Injury Score (AIS) = 1–5 and 20 healthy controls. Healthy controls were used rather than orthopedic controls, as has been the standard in previous studies.5,25,26 For each sample, we used our recently developed magnetic nanopore device (TENPO) to specifically enrich for GluR2+ EVs from 1 mL of plasma from the human samples and 0.5 mL of plasma for the mice samples.17
Comparison of the EV RNA profile in mice with clinical samples
To validate that our animal models present EV RNA biomarkers representative of what is found in clinical samples, we first compared signaling pathways activated in brain-derived EVs of TBI mouse models (CCI, Blast) with those of TBI patients with various injury intensities (AIS = 1–5). We identified 56 significantly activated pathways in brain-derived EVs for TBI patients compared with healthy controls. Of these 56 pathways, 41 homologous pathways were co-identified in our TBI mouse models (Fig. 1B). The high percentage of co-identified pathways between mouse and human indicates the potential that the biomarkers that we find in our mouse model can be translated to clinical study, which encouraged us to use our mouse models to study the capability of our platform to identify specific injury states. In the sequencing data for the samples from the murine models and the clinical samples, we were able to measure a total of 451 miRNA and 109 miRNA (average normalized counts >10), respectively.
Dynamics of brain-derived EV miRNA cargo following injury
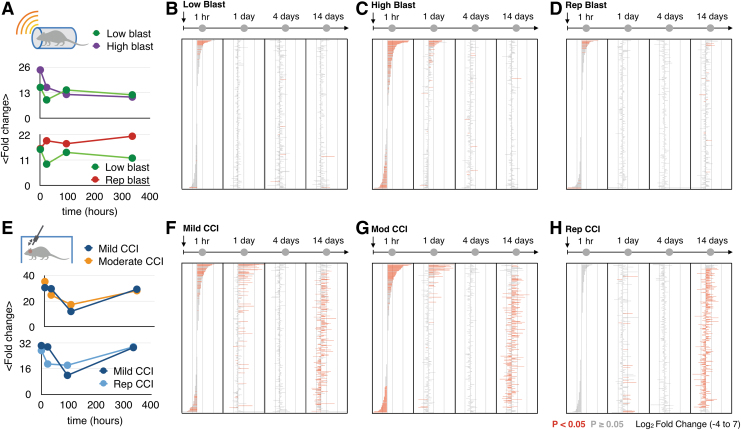
To track the dynamic changes in EV miRNA biomarkers at time-points following injury, we calculated the average fold changes of EV miRNA between injury states and their corresponding sham controls generated for each time-point. In these analyses, we included all EV miRNA biomarkers (n = 396) that were significantly differentially expressed in at least one injury state (adjusted p < 0.05), compared with its appropriate sham control (Fig. 2A, 2E). By profiling EV miRNAs from different injury states, we first identified four major trends:
FIG. 2.
Significantly different microRNAs (miRNAs) from different injury states. (A) The mean fold changes calculated using miRNA expression level changes of two different intensities of Blast injuries (low-215 kPa, high-415 kPa) and two different histories of Blast injuries (single, double) to their own sham controls at four time-points post-injury (1 h, 1 day, 4 days, 14 days). Error bars represent standard error from 396 extracellular vesicle (EV) miRNA biomarkers. (B) Differentially expressed miRNAs over time (1 h, 1 day, 4 days, 14 days) for low Blast injury. (C) Differentially expressed miRNAs over time (1 h, 1 day, 4 days, 14 days) for high Blast injury. (D) Differentially expressed miRNAs over time (1 h, 1 day, 4 days, 14 days) for repetitive Blast injury with two injuries. (E) The fold changes calculated using miRNA expression level changes of two different intensities of CCI (mild, moderate) and two different histories of CCIs (single, double) to their own sham controls at four time-points post-injury (1 h, 1 day, 4 days, 14 days). Error bars represent standard error from 396 EV miRNA biomarkers. Color image is available online.
-
1.
Higher injury intensities (Fig. 2C, 2G) generate more differentially expressed miRNAs than lower injury intensities (Fig. 2B, 2F) for both Blast injury and CCI.
-
2.
There are time-points after the injury with no significantly different miRNAs compared with sham controls.
-
3.
The molecular signatures of both Blast injury intensities and repetitive Blast become similar over time to those of sham controls (Fig. 2B-D), whereas the molecular signatures of both CCI intensities (Fig. 2F, 2G) indicate two different stages—acute response and a longer-term response to the injury.
-
4.
Repeat injuries appear to suppress the acute response (< 4 days) for both Blast (Fig. 2E) and CCI injuries (Fig. 2H), and more significantly for CCI.
Mice with repetitive mild CCI injuries had a pattern distinct from that of a single injury. For mice with a previous CCI, the EV miRNAs biomarkers in the acute phase were notably suppressed compared with mice with a single CCI (Fig. 2H). Notably, at 1 h, there was not a single EV miRNA that had a significant fold change, in contrast to the mice with only a single injury where there were 72 miRNA markers that had a significant fold change. At 14 days, the EV miRNA profile of the mice with repeat CCI was no longer suppressed and resembled that of mice with a single CCI at the same time-point.
Correlation analysis of brain-derived EV miRNA cargo following injury
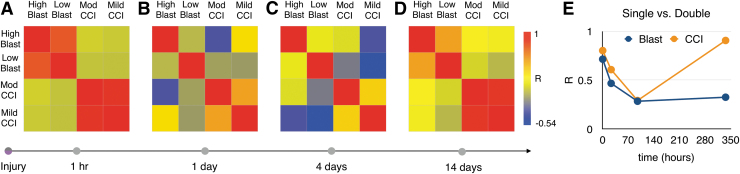
To study the dynamics of the biomarkers associated with different injury states, we characterized the similarity of EV RNA cargo between mice with different injury types and intensities over time. We found that the GluR2+ EV miRNA profiles for CCIs and Blast injuries were more different from one another than were the differences between subjects with different injury intensities or times elapsed after injury (Fig. 3A-D). In particular, in the acute phase for all four time-points post-injury, the Blast and CCI injuries had no or poor correlation with one another (R values from −0.5 to 0.5; Fig. 3A). Different injury intensities of the same injury type were similar to one another at short times after the injury (1 h; Fig. 3A) and at long times after the injury (14 days; Fig. 3B; R values from 0.5 to 1) but not at intermediate times (1 day, 4 days; Fig. 3C, 3D; R values from −0.25 to 0.75). For each of these analyses, we calculated the Pearson correlation coefficients (R) for pair-wise comparison of the EV RNA profile from each different injury types and intensities at different time-points post-injury.
FIG. 3.
Correlation of molecular changes of different injury states. The Pearson correlation coefficients calculated using microRNA expression level changes from different types (Blast vs. CCI) of injuries and different intensities (mild/low, moderate/high) of injuries within the types at (A) 1 h, (B) 1 day, (C) 4 days, (D) 14 days. (E) The Pearson correlation coefficients between single injury and repetitive injury (double) over time for Blast and CCI. Color image is available online.
For repetitive injuries, each injury type (CCI, Blast) showed a distinct trajectory of their correlation with a single injury over time. A single mild CCI and the repetitive CCI were highly correlated with one another at the acute phase (1 h), but then became less correlated over time at the 1-day and 4-day time periods. However, at 14 days we observed the most correlation between single and repeat injury (R = 0.91; Fig. 3E). A single low Blast and our repetitive Blast were the most correlated at the acute phase (1 h) and the correlation decayed gradually over time.
Enriched signaling pathways in brain-derived EV RNA cargo following TBI
To evaluate whether the identified molecular signatures in brain-derived EVs relate to underlying mechanisms of TBI, we performed KEGG pathway analysis using significantly differentially expressed miRNAs for each injury state. We identified 50 pathways that were universally significantly activated (adjusted p < 0.05) across all injury states compared with controls, which include glutamatergic synapse, axon guidance, PI3K-Akt signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway. Notably, proteoglycans in cancer pathway was the most activated pathway for both low and high intensities of Blast injury and moderate CCI (Supplementary Fig. S1).
Several interesting patterns emerged from the KEGG pathway analysis. Blast and CCI share more pathways in their acute response (1 h) than they do for their persistent response (14 days; Supplementary Fig. S2A, S2B). Mild and moderate CCI (Supplementary Fig. S2A) shares more pathways between their acute and persistent responses than either Low or High Blast does (Supplementary Fig. S2B). Mild and moderate CCI shares more pathways between their acute and persistent responses (Supplementary Fig. S2C) than Low or High Blast does (Supplementary Fig. S2D). Additionally, the pathways activated by Blast decays more over time while CCI has more pathways that persist out to the 14-day time period.
Classification of TBI using statistical learning–based classification on the brain-derived EV miRNA cargo
To identify combinations of miRNA biomarkers that can predict specific states of injury in individual patients, we apply statistical (i.e., machine) learning algorithms. These machine learning–defined biomarkers algorithmically combine individual EV miRNA into a composite marker that can be more accurate than its constituent features. To diminish the effects of overfitting, which can plague the application of machine learning to relatively small datasets (M < 100),27 we used an ensemble of machine learning algorithms (linear discriminate analysis, logistic regression, Naive Bayes, SVM, K-nearest neighbor). By averaging the prediction of multiple models, a more accurate prediction can be achieved than with any single model, because the fitting of each model to noise is averaged out across the multiple models.
The work flow of our application of machine learning to our EV miRNA data is as follows:
-
1.
On a training set of data, we use a least absolute shrinkage and selection operator (LASSO) to reduce the number of features from the entire sequencing dataset to a number of markers appropriate for a portable assay (k < 10). Performance is benchmarked by receiving operating characteristic analysis to map the tradeoff between sensitivity and specificity, wherein we calculate the area under the curve (AUC). We also report model accuracy (True Positives/Total Subjects). The input into our feature reduction algorithms were the normalized counts of 451 miRNAs (average raw counts >10) from the sequencing data (Fig. 1A).
-
2.
We then train a machine learning model using this discovered panel of biomarkers to classify the intended states of injury using a training set of data. To evaluate the models at this stage, we use k-fold cross-validation in which a single set of data is partitioned many times into a training and a test set and the accuracy averaged over these many combinations to guard against the effects of overfitting.
-
3.
We finally evaluate the models that we have created by testing them on a user blinded test set.
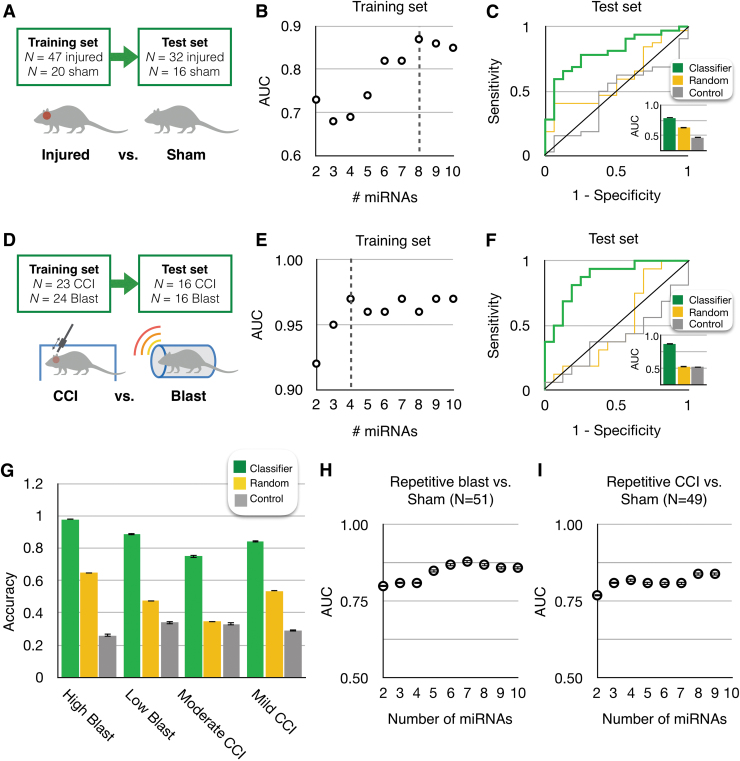
We first applied this workflow to the classification of mice with heterogeneous injury conditions (different time-points, intensities, types) from sham-injured mice. We included subjects with a diversity in injury severity, type, and time elapsed since injury to model the injuries observed in a clinical setting. We selected features and trained our machine learning model using a training set (n = 20 sham, n = 47 injured) and evaluated the classification using a separate user-blinded test set (n = 16 sham, n = 32 injured; Fig. 4A) Here, we included all the injuries except for the repetitive injuries. Using LASSO, we selected a panel of eight miRNAs (miR-150-5p, miR-669c-5p, miR-488-3p, miR-22-5p, miR-9-5p, miR-6236, miR-219a.2-3p, miR-351-3p; Supplementary Fig. S3A) and achieved an AUC = 0.87 on the training set (Fig. 4B). When we applied this model to classify the injury state of the mice in the independent blinded test set, we achieved AUC = 0.79 (Fig. 4C). To confirm that the performance is specific to the set of miRNA that we had selected, we compared this result to a control wherein we evaluated n = 5 sets of eight randomly selected biomarkers (AUC = 0.63). To confirm that the performance is specific to the miRNA signature identified by training our machine learning algorithm on our training set, we performed a negative control experiment in which we randomly shuffled class labels to create a random classifier. This control experiment resulted in an AUC = 0.47, equivalent to random guessing. The performance of our classifier was significantly better than using randomly selected features (p < 0.01) or using a randomly shuffled labels (p < 0.01).
FIG. 4.
Machine learning detected microRNA (miRNA) signatures for the diagnosis of traumatic brain injury (TBI) states. (A) Using machine learning, we classified injured mice versus uninjured mice (sham) in a heterogeneous set of mice with different injury intensities, post-injury times, and injury types, and sham mice with different post-anesthesia times. (B) We screened for miRNA panels using least absolute shrinkage and selection operator on the Training Set. The best performing panel based on its area under the curve (AUC) using cross-validation, was selected. (C) We used this panel of eight miRNA biomarkers on the user-blinded test set to classify injured versus uninjured. A receiver operating characteristic (ROC) curve is shown for this classifier, which achieved an AUC = 0.79. We compared this result to a control wherein we used five sets of randomly selected sets of eight randomly selected biomarkers (AUC = 0.63) and a control wherein we randomly shuffled the class labels (n = 5) to confirm the specificity of the TBI classifier (AUC = 0.47). (D) We classified CCI mice from Blast mice using the same workflow as above. (E) A panel of four miRNA biomarkers were selected. (F) An AUC = 0.87 was achieved, compared with AUC = 0.53 using randomly selected (n = 5) sets of four miRNA features, and AUC = 0.52 using (n = 5) randomly shuffled class labels. (G) We performed a four-way comparison to classify different elapsed time post-injury (1 h, 1 day, 4 days, 14 days) for high Blast, low Blast, mild CCI, and moderate CCI. (H) We classified repetitive Blast injury from sham controls with 5-fold cross-validation and screened different number of miRNA panels with their corresponding AUCs. (I) We classified repetitive CCI from sham controls with 5-fold cross-validation and screened different number of miRNA panels with their corresponding AUCs. Error bars in all plots represent Standard Error. In all ROC plots, for control analyses where multiple classifiers are considered, one example ROC curve is shown. Color image is available online.
To consider the effect of the heterogeneity of the underlying data on the performance of our classifications, we additionally evaluated the performance of classifying injured versus uninjured using the same panel of miRNA, but training and evaluating the machine learning to classify Blast versus Blast sham and CCI versus CCI sham separately. We found that when we used a separate training for Blast versus Blast sham (AUC = 0.81) and CCI versus CCI sham (AUC = 0.89), versus training them all together, we achieved a significantly improved performance (p < 0.01). When we additionally identified sets of features for Blast versus Blast sham and CCI versus CCI sham, we achieved AUC = 0.88 for Blast versus Blast sham using five miRNAs (miR-669c-5p, miR = 708-5p, miR-141-3p, miR-6538, miR-8112; Supplementary Fig. S3C) and AUC = 0.94 for CCI versus CCI sham using three miRNAs (miR-351-5p, miR-219a.2-3p, miR-409-3p; Supplementary Fig. S3D). We found that when we identified features and trained our model for Blast versus Blast sham and CCI versus CCI sham separately, versus using a single panel and training them all together, we achieved a significantly improved performance (p < 0.01).
We next applied our machine learning workflow, using the same dataset, to classify the type of injury that the mice were subject to (i.e., CCI vs. Blast; Fig. 4D). To validate our machine learning approach, the mice with CCI and Blast had purposefully heterogeneous injury conditions (different time-points, intensities). We created a training set (n = 24 Blast, n = 23 CCI) and validated the classifier using an independent blinded test set (n = 16 Blast, n = 16 CCI). Using LASSO, we selected a panel of four miRNAs (miR-143-3p, miR-219a.2-3p, miR-1191a, and miR-708-5p; Supplementary Fig. S3B) and achieved AUC = 0.97 on the training set (Fig. 4E). When we applied this model to classify the type of injury in the independent blinded test set, we achieved AUC = 0.87 (Fig. 4F). We compared the performance of this classifier to classification using n = 5 sets of four randomly selected miRNA (AUC = 0.53) and to classification using randomly shuffled class labels (AUC = 0.52). Our classifier achieved significantly better performance than these controls (p < 0.001 and p < 0.001, respectively).
We next separately attempted to classify the time elapsed after injury for animals that were injured with high Blast (415 kPa), low Blast (215 kPa), moderate CCI, and mild CCI injuries. We achieved accuracies of 98%, 89%, 75%, and 84%, respectively (Fig. 4G). In this analysis, because there were not enough subjects for an independent test set, we instead used 5-fold cross-validation. First, we evaluated the value of selecting features by comparing the performance of the features identified using LASSO to n = 5 sets of randomly selected features (random miRNA). Second, we included a negative control experiment wherein we randomly shuffled the labels on the training data (control). The accuracies of the TBI classifiers of all four groups were significantly different from their control tests (high Blast: p = 0.003 for random miRNA, p < 0.001 for control; low Blast: p < 0.001 for random miRNA, p < 0.001 for control; moderate CCI: p = 0.001 for random miRNA, p < 0.001 for control; mild CCI: p = 0.001 for random miRNA, p < 0.001 for control).
Additionally, we attempted to classify repetitive injuries from sham controls. For repetitive Blast injury, we achieved a classification with an AUC = 0.88 using seven miRNAs (miR-10a-5p, miR-154-5p, miR-669c-5p, miR-30c.1-3p, miR-199a-5p, miR-27a-3p, let-7e-3p; Fig. 4H). For repetitive CCI, we achieved an AUC = 0.84 using eight miRNAs (miR-1843a-5p, miR-219a.2-3p, miR-467c-5p, miR-181c-3p, miR-669c-5p, miR-1968-5p, miR-378b, miR-203-5p; Fig. 4I).
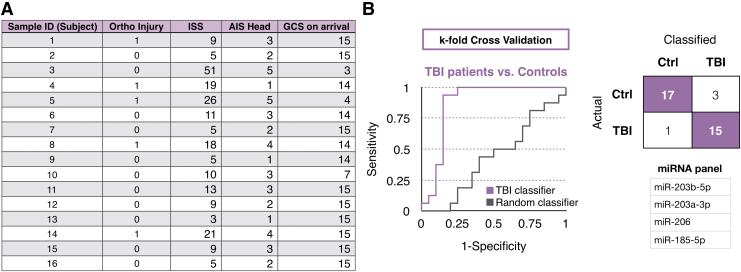
To demonstrate the translatability of the biomarker discovery methodology of our work using TBI mouse models to clinical diagnosis, we applied the same approach to human patient samples. We sequenced miRNAs packaged in GluR2+ EVs from n = 16 TBI patient samples and n = 20 control samples (Fig. 5A). With 109 detected miRNAs (average normalized counts >10), we used machine learning to classify injure patients versus controls (n = 20 controls, n = 16 TBI patients). We used k-fold (k = 5) cross-validation to guard against the effects of overfitting. We achieved an AUC = 0.84 classifying injured versus healthy (Fig. 5B). The panel of biomarkers to achieve this classification comprised of GluR2+ enriched EV miRNA, including miR-203b-5p, miR-203a-3p, miR-206, miR-185-5p (Supplementary Fig. S4). To confirm that the performance arises from a signature of injury and not to an artifact in the data, we performed a negative control experiment where we random shuffled the labels on the training data, which resulted in an AUC = 0.43, equivalent to random guessing.
FIG. 5.
Clinical diagnosis of traumatic brain injury (TBI) using microRNA (miRNA) signatures from brain-derived extracellular vesicles. (A) TBI patient data is included that specifies orthopedic injury score, injury severity score (ISS), Abbreviated Injury Score (AIS), and Glasgow Coma Scale (GCS). Sample IDs are blinded and labeled as numbers. (B) Classification of TBI patients and healthy controls using a machine learning algorithm. Randomly shuffled labels were created for random classifier. The receiver operating characteristic curve, the truth table, and the miRNA panel are included.’ Color image is available online.
Discussion
We developed an open-ended approach to identify multiplexed biomarkers for TBI that can classify specific injury states for individual subjects. To this end, we performed a multidimensional mapping of GluR2+ enriched EV miRNA using a recently developed magnetic nanopore based TENPO EV isolation technology and next-generation RNA sequencing. We sequenced the miRNA cargo of EVs from mice subjected to a variety of established models of TBI, with samples collected at multiple time-points elapsed after the injury. The profiles of EV miRNA for each injury state and their distinct dynamic change in the time after injury allowed us to classify the specific injury status of individuals using supervised machine learning. We have tested the potential translatability of our approach by applying it to a small cohort of TBI patients with different injury levels, as well as healthy controls, and demonstrated the ability to discriminate injured patients from healthy controls. Due to the differences between the rodent and human central nervous system and mechanisms of traumatic brain injury,28 we focused on testing the translatability of our methodology rather than discovering biomarkers from animal models and applying the same markers to TBI patients.
As has been done in previous studies,17,18 the biomarkers identified in this study can be implemented as a quantitative polymerase chain reaction (qPCR′A)-based test. In previous work by our lab, we showed that cancer could be diagnosed using QPCR measured RNA signatures present in EpCAM+ extracellular vesicles, which could not be resolved in the total extracellular vesicle population.18 To classify specific states of injury, we isolated a subset of the miRNA that has been sufficiently enriched to be brain-derived by targeting GluR2+ EVs. For future studies, it will be informative to compare the miRNA content derived from EVs to miRNA from cell-free plasma or serum.
The pathways that we identified in the GluR2+ EV miRNA, isolated from both clinical and pre-clinical samples, overlapped with several commonly explored and well documented changes in the brain after traumatic injury. Certainly, alterations in synaptic plasticity and maintenance of synaptic strength underlie the recovery response29–41 after injury, and our evidence shows that these pathways are likely present across both human and mouse sequencing data. Likewise, the putative involvement of dopaminergic, cholinergic, and glutamatergic synaptic signaling pathways are also consistent with past studies in pre-clinical42–45 and, in some cases, clinical studies.46,47 The involvement of these synapse subtypes also explains why several addiction and reward pathways appeared in our pathway analysis, as some of these synapse subtypes play key roles in different types of addiction. A number of intracellular signaling pathways consistent with past TBI studies also appeared in our profiling of brain derived exosomes, including those related to maintaining cytoskeletal integrity, key second messenger systems that include MAPK,48–50 PI3K-Akt,51–56 and cAMP,57,58 and both transcriptional and translational regulation (mTOR,59–64 FoxO65).
There are several aspects of the TENPO that can be further developed to expand the system's functionality to better improve diagnostics for TBI and to better study TBI's underlying mechanisms. In this study, we isolated brain-derived EVs using GluR2 expression on their surface, enriching for EVs shed from neurons.66 As the brain consists of multiple cell types, expanding the surface markers to separately target astrocytes and microglia, for example, can provide an increasingly comprehensive view of how these various cell types respond to injury.67–69 Additionally, the activated signaling pathways measured in this study can be further related to existing and emerging treatments to provide additional information on drug targeting and treatment guidance. Moreover, by collecting a larger and more diverse set of clinical samples from patients with TBI, we will be able to apply this technique to answer practical questions in clinical diagnostics. For example, we can profile the EV miRNA of TBI patients that develop secondary brain injury and those that do not, to better understand the multidimensional cascade of secondary brain injury and guide multiple therapeutic options for personalized and targeted therapies.70
Supplementary Material
Acknowledgments
We thank Tawny Meredith-Duliba and Erika Silverman from the Diaz-Arrastia lab at Penn for collecting and processing clinical samples that were used for this study. Funding was provided by the New Jersey Commission on Brain Injury Research (CSCR14IRG005) to DI and DM, National Institutes of Health NS 088276 to DM, Allen Foundation to DM and DI, U01 NS086090 to RDA, Health Research Formula Fund 4100077073 from Commonwealth of Pennsylvania to JK, the Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program, the National Institute of Health: 1R21CA182336-01A1 to DI and JK, and DI was supported by an American Cancer Society-CEOs Against Cancer-CA Division Research Scholar Grant, (RSG-15-227-01-CSM), the National Science Foundation's CAREER Award (#1554200), and The Hartwell Individual Research Award.
Author Disclosure Statement
DI is a co-founder of Chip Diagnostics. For the other authors, no competing financial interests exist.
Supplementary Material
Reference
- 1. Vos P. and Diaz-Arrastia R. (2014). Traumatic Brain Injury. John Wiley and Sons: Hoboken, NJ [Google Scholar]
- 2. Menon D.K., and Maas A.I.R. (2015). Traumatic brain injury in 2014: progress, failures and new approaches for TBI research. Nat. Rev. Neurol. 11, 71–72 [DOI] [PubMed] [Google Scholar]
- 3. Saatman K.E., Duhaime A.C., Bullock R., Maas A.I.R., Valadka A., and Manley G.T. (2008). Classification of traumatic brain injury for targeted therapies. J. Neurotrauma 25, 719–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diaz-Arrastia R., Wang K.K.W., Papa L., Sorani M.D., Yue J.K., Puccio A.M., McMahon P.J., Inoue T., Yuh E.L., and Lingsma H.F. (2014). Acute biomarkers of traumatic brain injury: relationship between plasma levels of ubiquitin C-terminal hydrolase-L1 and glial fibrillary acidic protein. J. Neurotrauma 31, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korley F.K., Diaz-Arrastia R., Wu A.H.B., Yue J.K., Manley G.T., Sair H.I., Van E., Jennifer, Everett A.D., TRACK-TBI investigators, Okonkwo D.O., Valadka A.B., Gordon W.A., Maas A.I., Mukherjee P., Yuh E.L., Lingsma H.F., Puccio A.M., and Schnyer D.M. (2016). Circulating brain-derived neurotrophic factor has diagnostic and prognostic value in traumatic brain injury. J. Neurotrauma 33, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Papa L., Lewis L.M., Silvestri S., Falk J.L., Giordano P., Brophy G.M., Demery J.A., Liu M.C., Mo J., Akinyi L., Mondello S., Schmid K., Robertson CS., Tortella F.C., Hayes R.L., and Wang K.K. (2012). Serum levels of ubiquitin C-terminal hydrolase distinguish mild traumatic brain injury from trauma controls and are elevated in mild and moderate traumatic brain injury patients with intracranial lesions and neurosurgical intervention. J. Trauma Acute Care Surg. 72, 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeter C.B., Hergenroeder G.W., Hylin M.J., Redell J.B., Moore A.N., and Dash P.K. (2013). Biomarkers for the diagnosis and prognosis of mild traumatic brain injury/concussion. J. Neurotrauma 30, 657–670 [DOI] [PubMed] [Google Scholar]
- 8. Staalnacke B., Bjornstig U., Karlsson K., and Sojka P. (2005). One-year follow-up of patients with mild traumatic brain injury: post-concussion symptoms, disabilities and life satisfaction at follow-up in relation to serum levels of S-100B and neuron-specific enolase in acute phase. J. Rehabil. Med. 37, 300–305 [DOI] [PubMed] [Google Scholar]
- 9. Gahlot G., Soni Y., Joshi G., and Rachit S. (2017). Clinical significance of serum biomarker S100B to predict outcome sfter traumatic brain injury. Indian J. Mednodent Allied Sci. 5, 24–29 [Google Scholar]
- 10. Shahim P., Zetterberg H., Tegner Y., and Blennow K. (2017). Serum neurofilament light as a biomarker for mild traumatic brain injury in contact sports. Neurology 88, 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shan R., Szmydynger-Chodobska J., Warren O.U., Mohammad F., Zink B.J., and Chodobski A. (2016). A new panel of blood biomarkers for the diagnosis of mild traumatic brain injury/concussion in adults. J. Neurotrauma 33, 49–57 [DOI] [PubMed] [Google Scholar]
- 12. Bogoslovsky T., Wilson D., Chen Y., Hanlon D., Gill J., Jeromin A., Song L., Moore C., Gong Y., and Kenney K., (2017). Increases of plasma levels of glial fibrillary acidic protein, tau, and amyloid beta up to 90 days after traumatic brain injury. J. Neurotrauma 34, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papa L., Brophy G.M., Welch R.D., Lewis L.M., Braga C.F., Tan C.N., Ameli N.J., Lopez M.A., Haeussler C.A., Giordano D.I.M., Silvestri S., Giordano P., Weber K.D., Hill-Pryor C., and Hack D.C. (2016). Time course and diagnostic accuracy of glial and neuronal blood biomarkers GFAP and UCH-L1 in a large cohort of trauma patients with and without mild traumatic brain injury. JAMA Neurol. 73, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pasinetti G.M., Ho L., Dooley C., Abbi B., and Lange G. (2012). Select non-coding RNA in blood components provide novel clinically accessible biological surrogates for improved identification of traumatic brain injury in OEF/OIF geterans. Am. J. Neurodegener. Dis. 1, 88. [PMC free article] [PubMed] [Google Scholar]
- 15. Di B., Alex P., Buonora J.E., Rhind S.G., Hutchison M.G., Baker A.J., Rizoli S.B., and Diaz-Arrastia R., and Mueller G.P. (2015). Blood biomarkers in moderate-to-severe traumatic brain injury: potential utility of a multi-marker approach in characterizing outcome. Front. Neurol. 6, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Golding E.M. (2002). Sequelae following traumatic brain injury: the cerebrovascular perspective. Brain Res. Rev. 38, 377–388 [DOI] [PubMed] [Google Scholar]
- 17. Ko J., Hemphill M., Yang Z., Sewell E., Na Y.J., Haber M., Sandsmark D., Fisher S.A., Torre D., Svane K., Omelchenko A., Firestein B., Diaz-Arrastia R., Kim J., Meaney D.F., and Issadore D. (2018). Diagnosis of traumatic brain injury using miRNA signatures in nanomagnetically isolated brain-derived extracellular vesicles. Lab Chip 18, 3617–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ko J., Bhagwat N., Yee S.S., Ortiz N., Sahmoud A., Black T., Aiello N.M., McKenzie L., O'Hara M., Redlinger C., Romeo J., Carpenter E.L., Stanger B.Z., and Issadore D. (2017). Combining machine learning and nanofluidic technology to diagnose pancreatic cancer using exosomes. ACS Nano 11, 11182–11193 [DOI] [PubMed] [Google Scholar]
- 19. Ko J., Bhagwat N., Yee S.S., Black T., Redlinger C., Romeo J., O'Hara M., Raj A., Carpenter EL., Stanger B.Z., and Issadore D. (2017). A magnetic micropore chip for rapid (< 1 hour) unbiased circulating tumor cell isolation and in situ RNA analysis. Lab Chip 17, 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muluneh M., Shang W., and Issadore D. (2014). Track-etched magnetic micropores for immunomagnetic isolation of pathogens. Adv. Healthc. Mater. 3.7, 1078–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiong Y., Mahmood A., and Chopp M. (2013). Animal models of traumatic brain injury. Nat. Rev. Neurosci. 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panzer M.B., Matthews K.A., Yu A.W., Morrison B., Meaney D.F., and Bass C.R. (2012). A multiscale approach to Blast neurotrauma modeling: part I—development of novel test devices for in vivo and in vitro blast injury models. Front. Neurol. 3, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beamer M., Tummala S.R., Gullotti D., Kopil C., Gorka S., Abel T., Cameron R., Morrison III B., Cohen A.S., and Meaney D.F. (2016). Primary Blast injury causes cognitive impairments and hippocampal circuit alterations. Exper. Neurol. 283, 16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Y., Mao H., Yang KH., Abel T., and Meaney D.F. (2014). A modified controlled cortical impact technique to model mild traumatic brain injury mechanics in mice. Front. Neurol. 5, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathias J.L., Dennington V., Bowden S.C., and Bigler E.D. (2013). Community versus orthopaedic controls in traumatic brain injury research: how comparable are they? Brain Inj. 27, 887–895 [DOI] [PubMed] [Google Scholar]
- 26. Rubenstein R., Chang B., Yue J.K., Chiu A., Winkler E.A., Puccio A.M., Diaz-Arrastia R., Yuh E.L., Mukherjee P., Valadka A.B., Gordon W.A., Okonkwo D.O., Davies P., Agarwal S., Lin F., Sarkis G., Yadikar H., Yang Z., Manley G.T., Wang K.K.W., the TRACK-TBI Investigators, Cooper S.R., Dams-O'Connor K., Borrasso AJ., Inoue T., Maas A.I.R., Menon D.K., Schnyer D.M., and Vassar M.J. (2017). Comparing plasma phospho tau, total tau, and phospho tau—total tau ratio as acute and chronic traumatic brain injury biomarkers. JAMA Neurol. 74, 1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ko J., Baldassano S.N., Loh P.L., Kording K., Litt B., and Issadore D. (2018). Machine learning to detect signatures of disease in liquid biopsies—a user's guide. Lab Chip 18.3, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petraglia A. L., Dashnaw M.L., Turner R.C., and Bailes J.E. (2014). Models of mild traumatic brain injury: translation of physiological and anatomic injury. Neurosurgery 75 Suppl 4, S34–S49 [DOI] [PubMed] [Google Scholar]
- 29. Carlson S.W., Henchir J., and Dixon C.E. (2017). Lateral fluid percussion injury impairs hippocampal synaptic soluble N-ethylmaleimide sensitive factor attachment protein receptor complex formation. Front. Neurol. 8, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mulherkar S., Firozi K., Huang W., Uddin M.D., Grill R.J., Costa-Mattioli M., Robertson C., and Tolias K.F. (2017). RhoA-ROCK inhibition reverses synaptic remodeling and motor and cognitive deficits caused by traumatic brain injury. Sci. Rep. 7, 10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen T., Wu Y., Wang Y., Zhu J., Chu H., Kong L., Yin L., and Ma H. (2017). Brain-derived neurotrophic factor increases synaptic protein levels via the MAPK/Erk signaling pathway and Nrf2/Trx axis following the transplantation of neural stem cells in a rat model of traumatic brain injury. Neurochem. Res. 42, 3073–3083 [DOI] [PubMed] [Google Scholar]
- 32. Perez E.J., Tapanes S.A., Loris Z.B., Balu D.T., Sick T.J., Coyle J.T., and Liebl D.J. (2017). Enhanced astrocytic d-serine underlies synaptic damage after traumatic brain injury. J. Clin. Invest. 127, 3114–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vascak M., Sun J., Baer M., Jacobs K.M., and Povlishock J.T. (2017). Mild traumatic brain injury evokes pyramidal neuron axon initial segment plasticity and diffuse presynaptic inhibitory terminal loss. Front. Cell. Neurosci. 11, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vogel E.W., Rwema S.H., Meaney D.F., Bass C.R., and Morrison B. (2017). Primary blast injury depressed hippocampal long-term potentiation through disruption of synaptic proteins. J. Neurotrauma 34, 1063–1073 [DOI] [PubMed] [Google Scholar]
- 35. Mayeux J., Katz P., Edwards S., Middleton J.W., and Molina P.E. (2017). Inhibition of endocannabinoid degradation improves outcomes from mild traumatic brain injury: a mechanistic role for synaptic hyperexcitability. J. Neurotrauma 34, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winston C.N., Noël A., Neustadtl A., Parsadanian M., Barton D.J., Chellappa D., Wilkins T.E., Alikhani A.D., Zapple D.N., Villapol S., Planel E., and Burns M.P. (2016). Dendritic spine loss and chronic white matter inflammation in a mouse model of highly repetitive head trauma. Am. J. Pathol. 186, 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Logue O.C., Cramer N.P., Xu X., Perl D.P., and Galdzicki Z. (2016). Alterations of functional properties of hippocampal networks following repetitive closed-head injury. Exp. Neurol. 277, 227–243 [DOI] [PubMed] [Google Scholar]
- 38. Chan J.L., Reeves T.M., and Phillips L.L. (2014). Osteopontin expression in acute immune response mediates hippocampal synaptogenesis and adaptive outcome following cortical brain injury. Exp. Neurol. 261, 757–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winston C.N., Chellappa D., Wilkins T., Barton D.J., Washington P.M., Loane D.J., Zapple D.N., and Burns M.P. (2013). Controlled cortical impact results in an extensive loss of dendritic spines that is not mediated by injury-induced amyloid-beta accumulation. J. Neurotrauma 30, 1966–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Almonte A.G., Qadri L.H., Sultan F.A., Watson J.A., Mount D.J., Rumbaugh G., and Sweatt J.D. (2013). Protease-activated receptor-1 modulates hippocampal memory formation and synaptic plasticity. J. Neurochem. 124, 109–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hunt R.F., Scheff S.W., and Smith B.N. (2011). Synaptic reorganization of inhibitory hilar interneuron circuitry after traumatic brain injury in mice. J. Neurosci. 31(18):6880–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shimada R., Abe K., Furutani R., and Kibayashi K. (2014). Changes in dopamine transporter expression in the midbrain following traumatic brain injury: an immunohistochemical and in situ hybridization study in a mouse model. Neurol. Res. 36, 239–46 [DOI] [PubMed] [Google Scholar]
- 43. Wagner A.K., Drewencki L.L., Chen X., Santos F.R., Khan A.S., Harun R., Torres G.E., Michael A.C., and Dixon C.E. (2009). Chronic methylphenidate treatment enhances striatal dopamine neurotransmission after experimental traumatic brain injury. J. Neurochem. 108, 986–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen Y.H., Huang E.Y., Kuo T.T., Miller J., Chiang Y.H., and Hoffer B.J. (2017). Impact of traumatic brain injury on dopaminergic transmission. Cell Transplant. 26, 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin S.S. and Dixon C.E. (2015). Alterations in cholinergic pathways and therapeutic strategies targeting cholinergic system after traumatic brain injury. J. Neurotrauma 32, 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arenth P.M., Russell K.C., Ricker J.H., and Zafonte R.D. (2011). CDP-choline as a biological supplement during neurorecovery: a focused review. PM R 3(6 Suppl 1), S123–S31 [DOI] [PubMed] [Google Scholar]
- 47. Frenette A.J., Kanji S., Rees L., Williamson D.R., Perreault M.M., Turgeon A.F., Bernard F., and Fergusson D.A. (2012). Efficacy and safety of dopamine agonists in traumatic brain injury: a systematic review of randomized controlled trials. J. Neurotrauma 29, 1–18 [DOI] [PubMed] [Google Scholar]
- 48. Chen T., Wu Y., Wang Y., Zhu J., Chu H., Kong L., Yin L., and Ma H. (2017). Brain-derived neurotrophic factor increases synaptic protein levels via the MAPK/Erk signaling pathway and Nrf2/Trx axis following the transplantation of neural stem cells in a rat model of traumatic brain injury. Neurochem. Res. 42, 3073–3083 [DOI] [PubMed] [Google Scholar]
- 49. Yang J., Wu Z., Renier N., Simon D.J., Uryu K., Park D.S., Greer P.A., Tournier C., Davis R.J., and Tessier-Lavigne M. (2015). Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell 160, 161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuo J.R., Cheng Y.H., Chen Y.S., Chio C.C., and Gean P.W. (2013). Involvement of extracellular signal regulated kinases in traumatic brain injury-induced depression in rodents. J. Neurotrauma 30, 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao Y.Y., Zhang Z.H., Zhuang Z., Lu Y., Wu L.Y., Ye Z.N., Zhang X.S., Chen C.L., Li W., and Hang C.H. (2018). Recombinant milk fat globule-EGF factor-8 reduces apoptosis via integrin β3/FAK/PI3K/AKT signaling pathway in rats after traumatic brain injury. Cell. Death Dis. 9, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He H., Liu W., Zhou Y., Liu Y., Weng P., Li Y., and Fu H. (2018). Sevoflurane post-conditioning attenuates traumatic brain injury-induced neuronal apoptosis by promoting autophagy via the PI3K/AKT signaling pathway. Drug Des. Devel. Ther. 12, 629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao Y., Li J., Wu L., Zhou C., Wang Q., Li X., Zhou M., and Wang H. (2016). Tetrahydrocurcumin provides neuroprotection in rats after traumatic brain injury: autophagy and the PI3K/AKT pathways as a potential mechanism. J. Surg. Res. 206, 67–76 [DOI] [PubMed] [Google Scholar]
- 54. Wang Z.F., Pan Z.Y., Xu C.S., and Li Z.Q. (2017). Activation of G-protein coupled estrogen receptor 1 improves early-onset cognitive impairment via PI3K/Akt pathway in rats with traumatic brain injury. Biochem. Biophys. Res. Commun. 82, 948–953 [DOI] [PubMed] [Google Scholar]
- 55. Zhang L., Ding K., Wang H., Wu Y., and Xu J. (2016). Traumatic brain injury-induced neuronal apoptosis is reduced through modulation of PI3K and autophagy pathways in mouse by FTY720. Cell. Mol. Neurobiol. 36, 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu C.H., Hung T.H., Chen C.C., Ke C.H., Lee C.Y., Wang P.Y., and Chen S.F. (2014). Post-injury treatment with 7,8-dihydroxyflavone, a TrkB receptor agonist, protects against experimental traumatic brain injury via PI3K/Akt signaling. PLoS One 9, e113397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Titus D.J., Oliva A.A., Wilson N.M., and Atkins C.M. (2015). Phosphodiesterase inhibitors as therapeutics for traumatic brain injury. Curr. Pharm. Des. 21, 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shen M., Wang S., Wen X., Han X.R., Wang Y.J., Zhou X.M., Zhang M.H., Wu D.M., Lu J., and Zheng Y.L. (2017). Dexmedetomidine exerts neuroprotective effect via the activation of the PI3K/Akt/mTOR signaling pathway in rats with traumatic brain injury. Biomed. Pharmacother. 95, 885–893 [DOI] [PubMed] [Google Scholar]
- 59. Don A.S., Tsang C.K., Kazdoba T.M., D'arcangelo G., Young W., and Zheng X.F. (2012). Targeting mTOR as a novel therapeutic strategy for traumatic CNS injuries. Drug Discov. Today 17, 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garling R.J., Watts L.T., Sprague S., and Digicaylioglu M. (2018). Progesterone modulates mTOR in the hippocampus of mice after traumatic brain injury. Neural Regen. Res. 13, 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Butler C.R., Boychuk J.A., and Smith B.N. (2017). Brain Injury-Induced Synaptic Reorganization in Hilar Inhibitory Neurons Is Differentially Suppressed by Rapamycin. eNeuro, ENEURO-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Butler C.R., Boychuk J.A., and Smith B.N. (2016). Differential effects of rapamycin treatment on tonic and phasic GABAergic inhibition in dentate granule cells after focal brain injury in mice. Exp. Neurol. 280, 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nikolaeva I., Crowell B., Valenziano J., Meaney D., and D'arcangelo G. (2016). Beneficial effects of early mTORC1 inhibition after traumatic brain injury. J. Neurotrauma 33, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rozas N.S., Redell J.B., Hill J.L., McKenna J. 3rd, Moore A.N., Gambello M.J., and Dash P.K. (2015). Genetic activation of mTORC1 signaling worsens neurocognitive outcome after traumatic brain injury. J. Neurotrauma 32, 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun L., Zhao M., Liu M., Su P., Zhang J., Li Y., Yang X., and Wu Z. (2018). Suppression of FoxO3a attenuates neurobehavioral deficits after traumatic brain injury through inhibiting neuronal autophagy. Behav. Brain Res. 337, 271–279 [DOI] [PubMed] [Google Scholar]
- 66. Isaac J.T.R., Ashby M.C., and McBain C.J. (2007). The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron 54, 859–871 [DOI] [PubMed] [Google Scholar]
- 67. Feldmann M., Pathipati P., Sheldon R.A., Jiang X., and Ferriero D.M. (2014). Isolating astrocytes and neurons sequentially from postnatal murine brains with a magnetic cell separation technique. J. Biol.l Methods 1, e11 [Google Scholar]
- 68. Korzhevskii D.E. and Kirik O.V. (2016). Brain microglia and microglial markers. Neurosci. Behav. Physiol. 46, 284–290 [Google Scholar]
- 69. Sharma K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U.K., Philips M.A., Rossner M.J., Mann M., and Simons M. (2015). Cell type—and brain region—resolved mouse brain proteome. Nat. Neurosci. 18, 1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xiong Y., Mahmood A., and Chopp M. (2009). Emerging treatments for traumatic brain injury. Expert Opin. Emerging drugs 14, 67–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith D., Soares H., Pierce J., Perlman K., Saatman K., Meaney D.F., Dixon C., and McIntosh T. (1995). A model of parasagittal controlled cortical impact in the mouse: cognitive and histopathologic effects. J. Neurotrauma 12, 169–178 [DOI] [PubMed] [Google Scholar]
- 72. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Love M.I, Huber W, and Anders S. (2014). Genome Biol. 15, 550, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu Q., Fisher S.A., Shallcross J., and Kim J. (2016). VERSE: a versatile and efficient RNA-Seq read counting tool. bioRxiv 053306 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.