Abstract
Simple Summary
Recently, there has been increased recognition of an association between the bacterial microbiome and cancer. In this study, we characterized the non-HPV head and neck squamous cell carcinoma (HNSCC) microbiome. We found a significant enrichment of Fusobacterium, depletion of Streptococcus, and the microbial signatures of twelve bacterial genera distinguishing HNSCC. With increased Fusobacterium—in particular, F. nucleatum—in our HNSCC cohort and its known association with prognosis in colorectal cancers (CRC), we sought to further characterize the association between clinical outcomes and F. nucleatum, and the host interaction with F. nucleatum. We identified a higher abundance of F. nucleatum in non-smokers and an improved survival, in contrast to CRC. An integrative analysis also identified that the enrichment of F. nucleatum was associated with host gene promoter methylation, suggesting that the bacterial mircobiome status may have a potential role as a prognostic biomarker and be involved in the pathogenesis of HNSCC.
Abstract
The role of oral microbiota in head and neck squamous cell carcinoma (HNSCC) is poorly understood. Here we sought to evaluate the association of the bacterial microbiome with host gene methylation and patient outcomes, and to explore its potential as a biomarker for early detection or intervention. Here we performed 16S rRNA gene amplicon sequencing in sixty-eight HNSCC patients across both tissue and oral rinse samples to identify oral bacteria with differential abundance between HNSCC and controls. A subset of thirty-one pairs of HNSCC tumor tissues and the adjacent normal tissues were characterized for host gene methylation profile using bisulfite capture sequencing. We observed significant enrichments of Fusobacterium and Peptostreptococcus in HNSCC tumor tissues when compared to the adjacent normal tissues, and in HNSCC oral rinses when compared to healthy subjects, while ten other bacterial genera were largely depleted. These HNSCC-related bacteria were discriminative for HNSCC and controls with area under the receiver operating curves (AUCs) of 0.84 and 0.86 in tissue and oral rinse samples, respectively. Moreover, Fusobacterium nucleatum abundance in HNSCC cases was strongly associated with non-smokers, lower tumor stage, lower rate of recurrence, and improved disease-specific survival. An integrative analysis identified that enrichment of F. nucleatum was associated with host gene promoter methylation, including hypermethylation of tumor suppressor genes LXN and SMARCA2, for which gene expressions were downregulated in the HNSCC cohort from The Cancer Genome Atlas. In conclusion, we identified a taxonomically defined microbial consortium associated with HNSCC that may have clinical potential regarding biomarkers for early detection or intervention. Host–microbe interactions between F. nucleatum enrichment and clinical outcomes or host gene methylation imply a potential role of F. nucleatum as a pro-inflammatory driver in initiating HNSCC without traditional risk factors, which warrants further investigation for the underlying mechanisms.
Keywords: HNSCC, microbiome, Fusobacterium, methylation, host-microbiome interaction
1. Background
Known etiological factors involved in the development of head and neck squamous cell carcinoma (HNSCC) include smoking, alcohol abuse, and areca nut chewing [1,2]. However, the prevalence of HNSCC without traditional risk factors has been increasing [3]. Human papillomavirus (HPV) plays an etiological role in a proportion of oropharyngeal squamous cell carcinoma but has no clear association with SCC of other sites in the head and neck region [3]. Emerging data implicate the involvement of the human bacterial microbiome in a variety of cancers, most notably Fusobacterium in colorectal cancer [4,5] and Helicobacter in gastric cancer [6,7]. The bacterial microbiome has also been proposed to interact with the host and play a role in carcinogenesis through host epigenetic alterations in the gut mucosa that are seen in ulcerative colitis and colorectal cancer [8,9,10].
In the head and neck region, the oral bacterial microbiome has been implicated in the development of dental caries with acidophilic bacteria that result in the demineralization of enamel and resultant caries [11]. Other poor oral health conditions, including chronic periodontitis and gingivitis [12], have also been implicated in HNSCC, as it may result in a change in the oral microbiome leading to chronic inflammation and cancer progression [13,14]. Recently, there have been studies reporting alterations in the microbiome of HNSCC [15,16,17,18]. A significant loss in microbial diversity, and changes in the relative abundance of some oral bacteria, such as Fusobacteria and Streptococcus, have been observed in HNSCC patients. However, many of these studies had limitations in sampling methods, did not control for HPV status and did not evaluate the host–microbiome interactions [17,18,19,20].
Here we sought to characterize the non-HPV HNSCC microbiome using both tissues and oral saliva rinses, in order to delineate the microbial consortium associated with HNSCC. Microbiota of HNSCC tumor tissues and the adjacent normal tissues were compared, and oral rinse samples from the same HNSCC patients were collected to compare them with those from matched healthy subjects. Finally, with these data we correlated the bacterial microbiome with host genome methylation pattern and patient outcomes.
2. Methods
2.1. Ethical Approval
This study has been approved by the approved by The Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee (CREC reference number 2015.396, 2017.143). Patients with head and neck squamous cell carcinoma who were admitted into the Prince of Wales Hospital and United Christian Hospital in Hong Kong and agreed with written informed consent were recruited between October 2015 and April 2018. All cases were reviewed by a pathologist. Separately, age, gender, and smoking-matched healthy individuals above 18 years of age with no history of malignancies were recruited from Prince of Wales Hospital.
2.2. HNSCC Tissue Collection and Processing
Tumor tissues and the adjacent normal (AN) tissues were collected at the time of surgery. Briefly, around 5 mm3 tumor samples were excised from the tumor mass without involving the margin. Paired normal tissues ≥ 5 cm away from the margin of the tumor were excised. These specimens were copiously irrigated with sterile saline to wash away surface contamination prior to collection. Tissue specimens were stored at −80 °C until further use.
2.3. Collection of Oral Rinse Samples
Oral rinse samples of HNSCC patients were collected in the hospital prior to surgery before any intraoperative antibiotics. In brief, pre-treatment 30 mL oral rinses were obtained with normal saline gargled twice, for 20 and 10 s respectively, as previously described [21,22]. Oral rinse samples from healthy subjects were collected using the same protocol. Oral rinses were sent to the laboratory within one hour, and were centrifuged at 1600× g for 10 min in 4 °C immediately. Supernatants were discarded and the cellular pellet was stored at −80°C until further use.
2.4. DNA Extraction of Tissues and Oral Rinses
Fresh frozen tissue samples of around 10–20 mg were manually homogenized into small pieces and treated with 20 μL proteinase K at 55 °C overnight to lyse cell membranes. Disaggregated samples were extracted for total DNA using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. Purified DNA was eluted into 100 μL elution buffer and stored in −20 °C. For oral rinsing, total DNA was extracted from pellets centrifuged from 1–2 mL of oral rinse solution using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), and eluted into 100 μL elution buffer for further use.
2.5. HPV Genotyping
HPV genotyping was performed using two PCR-based amplicon sequencing assays targeting the conserved L1 open reading frame (ORF) of HPV [23]. A pair of dual 12 bp barcodes were introduced to the PCR amplicons using forward and reverse primers; PCR mixtures were sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) at the Weill Cornell Medicine Genomics Resources Core Facility, New York, USA, using paired-end 150 bp reads. Following demultiplexing, short reads passing the quality filter were blastn-searched against a PV reference database using UPARSE software [24]. An operational taxonomic unit (OTU) count table was created using a 90% identity threshold assigning each OTU with a PV type.
2.6. 16S rRNA Gene PCR Amplification and Sequencing
Extracted DNA was used for oral microbiota profiling by sequencing the bacterial 16S rRNA gene V3-V4 region, with a primer set of 341F (5′−CCT ACG GGN GGC WGC AG−3′) and 806R (5′−GGA CTA CNV GGG TWT CTA AT−3′) plus minor modifications as previously described [25]. A pair of dual 12 bp barcodes was indexed to each amplicon set through the forward and reverse primers modified from the Earth Microbiome Project protocol [26]. Successful amplicons were equally pooled and sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) at the Weill Cornell Medicine Genomics Resources Core Facility, New York, USA, using paired-end 300 bp reads. For quality control, each sequencing batch included a mock community, DNA negative controls, and technical replicate samples.
2.7. 16S Sequence Data Processing and Community Composition Statistical Analyses
Following short reads demultiplexing, the QIIME2 (v2019.7) package [27], including pipelines for quality control, dada2 denoising, and sequence clustering, was applied to process 16S rRNA short reads into an amplicon sequence variant (ASV) count table. The SILVA v132 99% 16S rRNA gene reference database was used to assign taxonomic identities to ASVs at genus and phylum levels, respectively. All reads assigned to mitochondria and chloroplasts were excluded. The Fusobacterium ASVs were further assigned at the species level using pplacer [28] by placing short reads on a phylogenetic tree inferred from the 16S rRNA gene complete sequences of Fusobacterium to maximize phylogenetic likelihood.
In order to retain all samples for diversity analysis, reads from each sample were rarefied to a depth of 2000 mean reads, after repeating for 100 times, to normalize the data for differences in sequencing depth among samples. Bacterial taxa with ≥1% relative abundance in at least one sample were retained. The diversity of observed bacterial genus taking into consideration species richness, and the effective numbers of Shannon and Simpson indexes, were calculated. Pairwise Bray–Curtis dissimilarities between samples were calculated using scripts in R v3.4.0 package. Differences in community composition were assessed using permutational multivariate analysis of variance (PERMANOVA) in the Vegan R package. Principal coordinate analysis was performed to visualize associations between community composition. Comparisons of the relative abundances of ASVs or bacterial taxa between defined groups were performed using nonparametric Mann–Whitney Wilcoxon rank sum test (MWU), Wilcoxon signed rank test (WSR), Kruskal–Wallis test (KW), or Tukey’s honest significant difference (Tukey HSD) post hoc test, where appropriate. Receiver operating characteristic (ROC) curve analyses, including calculation of area under the ROC curve (AUC), were used to evaluate the ability of bacterial genera to distinguish cases from controls. A two-sided p value of ≤0.05 and/or a false discovery rate (FDR)-adjusted p value (q value) of ≤0.05 was used as the threshold for significance.
2.8. Microbial Rank Correlation and Identifying Indicator Markers
Networks between bacterial genera were determined by calculating the pairwise SparCC correlation coefficient using microbial reads counts [29]. Two bacteria genera were considered statistically correlated if the absolute value was ≥0.3 and the p value was ≤0.05. Cytoscape v3.7.1 was used to visualize the network [30].
2.9. Functional Prediction Based on 16S rRNA Gene Community Composition
Functional profiles of microbial communities were predicted using PICRUSt2 (https://github.com/picrust/picrust2/wiki) based on 16S rRNA gene sequences represented as Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) and metabolic MetaCyc pathway counts [31]. Nine samples with nearest sequenced taxon index (NSTI) values ≥0.15 were excluded due to low accuracy of the functional prediction. Differential abundances of KO pathways for the comparison between conditions were analyzed using the DESeq2 R package [32].
2.10. Fusobacterium nucleatum in Situ Hybridization (ISH) Analysis
Six primary HNSCC cases with F. nucleatum on qPCR in fresh tissue had archival formalin-fixed paraffin-embedded (FFPE) samples retrieved to determine the spatial distribution of Fusobacterium in HNSCC tissues. Briefly, 4 um-thick FFPE sections were subjected to RNA in situ hybridization with B-FNucleatum-fadA RNA probe and RNAscope® 2.5 HD Reagent Kit-BROWN according to the protocol suggested by the manufacturer (ACD). Negative control probe—DapB (catalogue number 310043), and positive control probe—Hs-PPIB (catalogue number 313901), were used for negative and RNA quality controls respectively. The slides were evaluated at high magnification (40×) fields per sample by a dedicated head and neck pathologist. The digital microscope camera Nikon DS-Ri2 digital microscope camera and software NIS Elements Imaging Software Version 4.60 were used to capture the images.
2.11. Quantitative PCR (qPCR) for Fusobacterium nucleatum
A TaqMan primer/probe set targeting the nusG gene was used to amplify Fusobacterium nucleatum DNA as previously described [33]. The fold difference (2-ΔΔCt) in F. nucleatum abundance in tumor versus AN tissue was calculated by subtracting ΔCttumor from ΔCtAN, where ΔCt is the difference in threshold cycle number for the nusG and PGT assay. PCR amplification was performed using Taqman Universal Master Mix, with primer and probe sequences as follows: Fusobacterium nugG forward primer, 5′ AAC CAT TAC TTT AAC TCT ACC ATG TTC A 3′; nugG reverse primer, 5′ TTG ACT TTA CAG AAG GAG ATT ATG TAA AAA TC 3′; nugG FAM probe, 5′ TCA GCA ACT TGT CCT TCT TGA TCT TTA AAT GAA CC 3′; PGT forward primer, 5′ ATC CCC AAA GCA CCT GGT TT 3′; PGT reverse primer, 5′ AGA GGC CAA GAT AGT CCT GGT AA 3′; PGT FAM probe, 5′ CCA TCC ATG TCC TCA TCT C 3′.
2.12. Human DNA CpG Methylation from HNSCC Tissue
Total DNA from HNSCC tumor and AN tissue was pre-prepared for illumine library using KAPA HTP Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) and then exposed to sodium bisulfite at 60 °C in the dark to convert unmethylated cytosine nucleotides into uracil nucleotides using EpiTect DNA Bisulfite Kit (Qiagen, Valencia, CA, USA). The SeqCap Epi CpGiant System interrogating more than 5.5 million CpG sites (Roche, USA) was used to probe bisulfite-converted human DNA libraries for Illumina HiSeq sequencing (Illumina, San Diego, CA, USA) at NovoGene, China, using paired-end 150 bp reads.
Short reads were quality trimmed using Trimmomatic [34] and mapped to the human genome (hg38) using Bismark [35]. Mapped bam files were sorted, and duplicated reads were removed. Methylation calls from bam files were further imported into methylKit [36] to call differentially methylated regions (DMRs) of 1000 bp size genome-wide. Promoters were defined by taking 3000 bp up- and down-stream of the transcription start site. Age, gender, smoking status, drinking status, and Fusobacterium abundance (Fuso-high vs. Fuso-low) for each individual were included in the model as confounding factors. Five groups of comparison were conducted, including (1) tumor vs. adjacent normal tissues, (2) Fusobacterium-high vs. Fusobacterium-low in tumor tissues, (3) Fusobacterium-high vs. Fusobacterium-low in adjacent normal tissues, (4) tumor vs. adjacent normal tissues in Fusobacterium-high group, and (5) tumor vs. adjacent normal tissues in Fusobacterium-low group. The methylation status was determined in percentage based on the actual C bases in the reads aligning to a given cytosine location in DMR. Hyper- and hypo-methylated regions were identified if the methylation difference were ≥15% (adjusted p ≤ 0.01), as measured by calculateDiffMeth in methylKit. The association between Fusobacterium enrichment and gene promoter methylation was regarded as significant if the tests of comparisons 1, 2, and 4 but not 3 and 5 met criteria, which were further repeated by a negative binomial generalized linear regression model test and a Spearman’s rank-order correlation test, complied with glm.nb and cor.test, respectively (p ≤ 0.05). A volcano plot was used to visualize the DMRs.
To explore the association of DNA methylation in the promoter region with gene transcription, The Cancer Genome Atlas (TCGA) HNSCC cohort RNA-seq raw counts datasets composed of 500 tumor and 38 control tissue samples were retrieved using R package curatedTCGAData. Differential gene expression profiling was measured using DESeq2 using a cutoff of log2 fold change (log2FC) ≥1 and adjusted p ≤ 0.01. The KEGG 2019 human database at Enrichr (https://amp.pharm.mssm.edu/Enrichr/) was accessed to predict gene pathways.
3. Results
3.1. Study Subjects
Sixty-eight HNSCC patients, after excluding 13 high-risk (HR) HPV-positive cases, were included in this study, from which samples including oral rinses, tumor tissues, and the adjacent normal (AN) tissue were collected. Oral rinses from sixty-eight age and gender matched non-HNSCC subjects were collected as controls. There were no significant differences between HNSCC patients and control subjects in age, gender, smoking, and alcohol history (Table S1). All samples were negative for high-risk HPV DNA by PCR.
3.2. Differential Oral Microbial Community in HNSCCs Compared to Controls
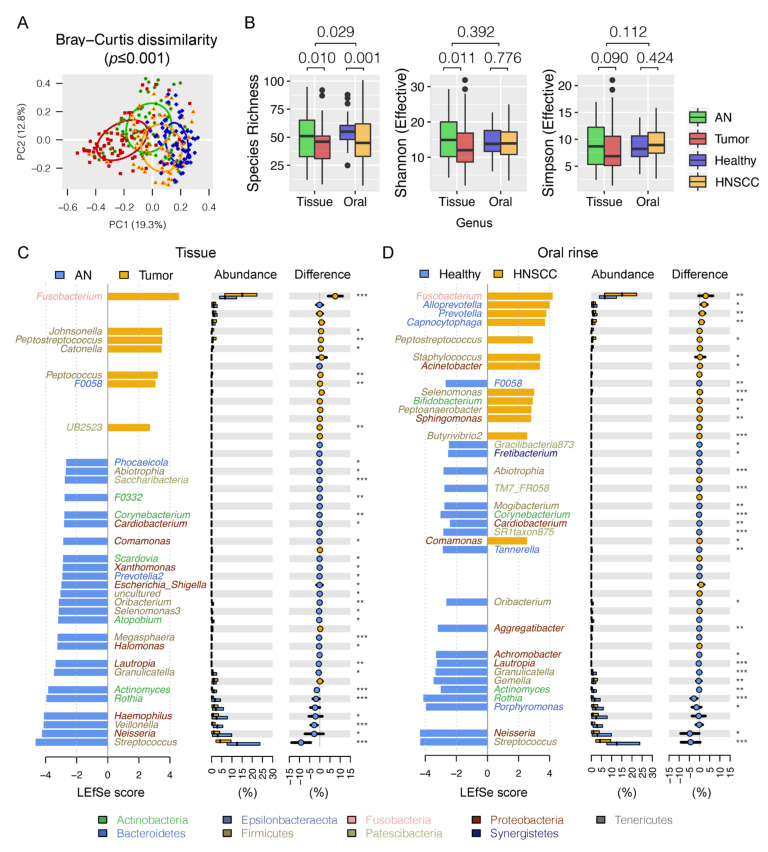
Overall, Firmicutes (relative abundance of 30.0 ± 1.0%), Proteobacteria (24.4 ± 1.2%), and Bacteroidetes (21.5 ± 0.7%) were the three most predominant bacterial phyla in the surveyed samples. When bacterial taxa were summarized at genus level, a permutational multivariate analysis of variance (PERMANOVA) using Bray–Curtis distances indicated that approximately 12.9% of variation in microbial composition could be attributed to the oral habitats (tissue vs. oral rinse) (df = 1, R2 = 0.086, pseudo-F = 26.483, p ≤ 0.001), disease status (HNSCCs vs. controls) (df = 1, R2 = 0.037, pseudo-F = 11.290, p ≤ 0.001), and the combination (df = 1, R2 = 0.006, pseudo-F = 1.887, p = 0.029), wherein oral habitat was the main source of variability, as supported by a principal coordinate analysis (PCoA) (Figure 1A). Decreased alpha diversity of oral microbiota was observed in tumor tissues when compared to the adjacent normal tissues, as measured by taxa richness and the effective numbers of Shannon index (Figure 1B). However, no significant differences in the effective numbers of Shannon or Simpson diversities were observed between oral rinses collected from HNSCC and healthy subjects.
Figure 1.
Oral microbiota dysbiosis associated with head and neck squamous cell carcinomas (HNSCCs). (A) Principal coordinates plot using Bray–Curtis dissimilarity. (B) Comparison of oral microbiota diversity summarized at the genus level. (C) Comparison of oral microbiota between HNSCC tumor (tumor) and adjacent normal (AN) tissues, with bacterial genera that best discriminated HNSCCs and controls by a linear discriminant analysis (LDA) for effect size (LEfSe). The bar length represents log10 LDA score. Difference in the relative abundance tested by pairwise Tukey HSD post hoc is shown on the right panel. (D) Comparison of oral microbiota between HNSCC oral rinses and healthy subjects. Adjusted Mann–Whitney U-test p value is marked with * if ≤ 0.05, ** ≤ 0.01, or *** ≤ 0.001.
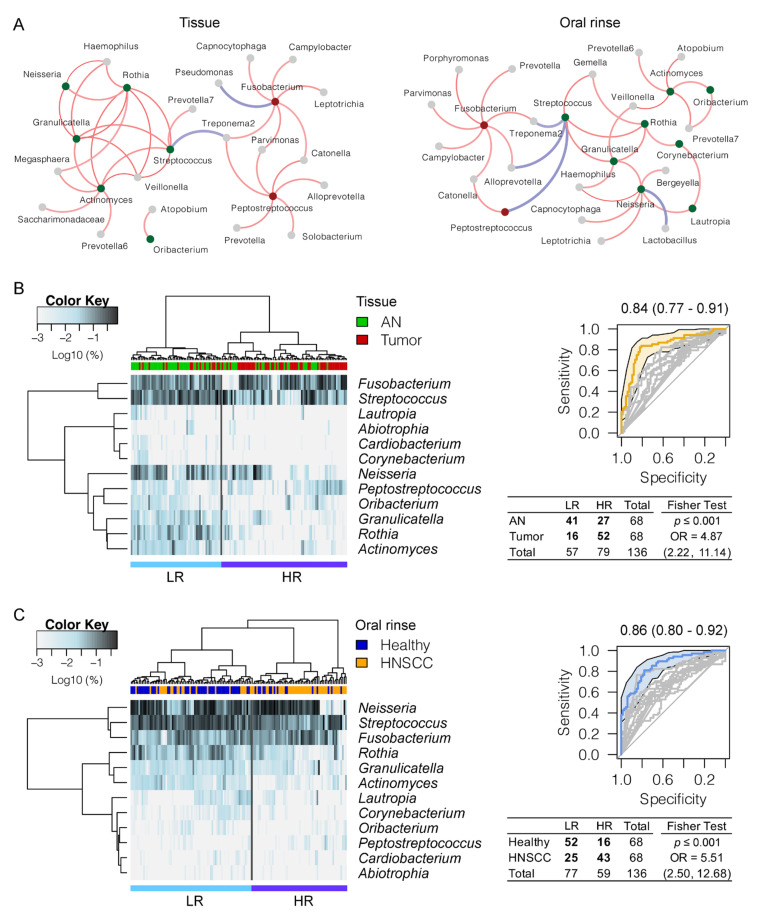
Using the linear discriminant analysis effect size (LEfSe) algorithm, we were able to distinguish oral bacteria between HNSCCs and controls. In total, there were seven significantly enriched and 25 depleted bacterial genera in HNSCC tumor tissues when compared to the adjacent normal tissues (Figure 1C, Table S2). Similarly, we found 13 bacterial genera largely dominated in abundance in HNSCC oral rinses, and 21 in healthy subjects (Figure 1D). These bacteria may represent pathologic or commensal microbiota in the upper aerodigestive tract. For example, Fusobacterium, the most predominant periodontal pathogen observed in 90% of the surveyed samples (≥1% relative abundance), was nearly double in relative abundance in HNSCC when compared to controls (tissue: 16.3% vs. 8.6%, p ≤ 0.001; oral rinse: 7.7% vs. 4.6%, p ≤ 0.001). In contrast, a dramatic reduction in relative abundance of the commensal Streptococcus in HNSCC was observed (tissue: 8.0% vs. 17.2%, p ≤ 0.001; oral rinse: 10.6% vs. 15.0%, p ≤ 0.001). In particular, we found two bacterial genera (Fusobacterium and Peptostreptococcus) to be consistently increased in relative abundances both in tumor tissues and HNSCC oral rinses, whereas ten taxa (Streptococcus, Neisseria, Rothia, Actinomyces, Granulicatella, Oribacterium, Lautropia, Corynebacterium, Abiotrophia, and Cardiobacterium) were significantly decreased, with area under the receiver operating curve (AUC) values ranging between 0.58 and 0.80 (Figure S1, Table 1), although bacteria may manifest as different colonies in different ecological habitats (tissues vs. oral rinses) (Figure S2). Using SparCC correlation analysis (r ≥ 0.3, p ≤ 0.01), we explored the relationships among bacterial genera with consistent changes in abundance both in tissue and oral rinse samples, and found two main network clusters represented by periodontal pathogens (e.g., Fusobacterium, Peptostreptococcus) and commensal bacteria (e.g., Streptococcus, Rothia, Granulicatella) (Figure 2A). The HNSCC-related pathogenic genera (red and green dots) further formed pathogen–pathogen or nonpathogen–nonpathogen collaborative connections (red lines) with other non-pathogenic bacteria (grey dots), such as Fusobacterium–Treponema2, Peptostreptococcus–Catonella, Neisseria–Haemophilus, and Granulicatella–Veillonella. An inhibitive network of bacteria (blue lines) was not common within the surveyed samples and was mainly observed between a pathogen and a nonpathogen (e.g., Streptococcus–Treponema2 and Streptococcus–Alloprevetolla).
Table 1.
Bacterial genera with coherent changes of the relative abundance between HNSCCs and controls across both tissues and oral rinses.
| AN | Tumor | Healthy | HNSCC | Tumor Tissue Compared to AN | HNSCC Oral Rinse Compared to Healthy Subjects | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taxonomy (Phylum; Genus) | Abundance (Mean ± smd) | Abundance (Mean ± smd) | Abundance (Mean ± smd) | Abundance (Mean ± smd) | Difference in Abundance | p Value (mwu Test) | LEfSe assign | LEfSe p adj | AUC Value | Difference in Abundance | p value (mwu test) | LEfSe Assign | LEfSe p adj | AUC Value |
| Fusobacteria; Fusobacterium | 8.60 ± 0.87 | 16.25 ± 1.51 | 4.60 ± 0.36 | 7.69 ± 0.72 | 7.65 (4.14,11.15) | 0.0000 | Tumor | 0.0002 | 0.69 (0.60, 0.78) | 3.09 (−0.42, 6.60) | 0.0027 | HNSCC | 0.0025 | 0.65 (0.55, 0.74) |
| Firmicutes; Peptostreptococcus | 0.79 ± 0.19 | 1.33 ± 0.19 | 0.06 ± 0.01 | 0.22 ± 0.06 | 0.54 (0.03, 1.05) | 0.0042 | Tumor | 0.0095 | 0.63 (0.53, 0.72) | 0.16 (−0.35, 0.67) | 0.0132 | HNSCC | 0.0130 | 0.62 (0.52, 0.72) |
| Firmicutes; Streptococcus | 17.16 ± 1.71 | 8.00 ± 1.33 | 15.00 ± 0.92 | 10.64 ± 1.02 | −9.17 (−13.86, −4.48) | 0.0000 | AN | 0.0000 | 0.75 (0.67, 0.83) | −4.36 (−9.05, 0.33) | 0.0001 | Healthy | 0.0001 | 0.70 (0.61, 0.79) |
| Proteobacteria; Neisseria | 6.80 ± 1.12 | 4.07 ± 0.96 | 18.82 ± 1.51 | 14.10 ± 1.28 | −2.73 (−7.25, 1.79) | 0.0076 | AN | 0.0400 | 0.60 (0.51, 0.70) | −4.72 (−9.24, −0.21) | 0.0222 | Healthy | 0.0228 | 0.61 (0.52, 0.71) |
| Actinobacteria; Rothia | 2.99 ± 0.48 | 1.21 ± 0.37 | 4.47 ± 0.57 | 1.77 ± 0.28 | −1.78 (−3.39, −0.18) | 0.0000 | AN | 0.0003 | 0.68 (0.59, 0.77) | −2.70 (−4.31, −1.10) | 0.0000 | Healthy | 0.0000 | 0.75 (0.67, 0.83) |
| Actinobacteria; Actinomyces | 1.99 ± 0.39 | 0.57 ± 0.16 | 0.87 ± 0.11 | 0.69 ± 0.13 | −1.42 (−2.24, −0.60) | 0.0000 | AN | 0.0000 | 0.73 (0.65, 0.82) | −0.18 (−1.00, 0.64) | 0.0100 | Healthy | 0.0097 | 0.63 (0.53, 0.72) |
| Firmicutes; Granulicatella | 1.42 ± 0.20 | 0.91 ± 0.25 | 1.20 ± 0.14 | 1.10 ± 0.40 | −0.52 (−1.48, 0.45) | 0.0126 | AN | 0.0127 | 0.62 (0.53, 0.72) | −0.10 (−1.07, 0.87) | 0.0002 | Healthy | 0.0002 | 0.68 (0.59, 0.77) |
| Proteobacteria; Lautropia | 0.47 ± 0.14 | 0.05 ± 0.02 | 0.64 ± 0.14 | 0.27 ± 0.07 | −0.42 (−0.80, −0.04) | 0.0008 | AN | 0.0096 | 0.60 (0.53, 0.67) | −0.37 (−0.75, 0.00) | 0.0000 | Healthy | 0.0000 | 0.71 (0.62, 0.80) |
| Proteobacteria; Cardiobacterium | 0.13 ± 0.04 | 0.05 ± 0.02 | 0.09 ± 0.01 | 0.09 ± 0.03 | −0.09 ( −0.18, 0.01) | 0.0192 | AN | 0.0410 | 0.58 (0.50, 0.65) | −0.00 (−0.10, 0.10) | 0.0020 | Healthy | 0.0020 | 0.65 (0.56, 0.73) |
| Actinobacteria; Corynebacterium | 0.15 ± 0.04 | 0.08 ± 0.05 | 0.27 ± 0.03 | 0.09 ± 0.03 | −0.07 (−0.21, 0.08) | 0.0008 | AN | 0.0091 | 0.60 (0.53, 0.68) | −0.18 (−0.32, −0.04) | 0.0000 | Healthy | 0.0000 | 0.80 (0.72, 0.87) |
| Firmicutes; Abiotrophia | 0.14 ± 0.08 | 0.08 ± 0.04 | 0.08 ± 0.02 | 0.02 ± 0.01 | −0.06 (−0.22, 0.11) | 0.1402 | AN | 0.0106 | 0.58 (0.52, 0.65) | −0.06 (−0.23, 0.10) | 0.0001 | Healthy | 0.0001 | 0.66 (0.59, 0.73) |
| Firmicutes; Oribacterium | 0.77 ± 0.16 | 0.72 ± 0.25 | 0.15 ± 0.02 | 0.11 ± 0.02 | −0.05 (−0.59, 0.49) | 0.0383 | AN | 0.0050 | 0.64 (0.54, 0.73) | −0.04 (−0.58, 0.49) | 0.0123 | Healthy | 0.0121 | 0.62 (0.53, 0.71) |
Figure 2.
A refinement of twelve “core” bacterial genera discriminating HNSCCs from controls. (A) A SparCC correlation analysis (r ≥ 0.3, p≤ 0.01) showing connections between pathogen–pathogen, nonpathogen–nonpathogen, and pathogen–nonpathogen bacteria. Grey dots indicate bacterial genera with no significant differences in abundance between HNSCC and controls. Red and green dots indicate bacterial genera enriched in HNSCCs and controls, respectively. Connections with red and blue lines indicate collaborative and inhibitive relationships, respectively. Hierarchical cluster analyses of tissues (B) and oral rinses (C) using distance matrix of 12 discriminative bacterial genera classified oral microbial communities into two clades, namely, high-risk and low-risk, based on the dendrogram topologies. The receiver operating characteristic (ROC) analysis of combined twelve genera achieved AUCs of 0.84 and 0.86 in tissues and oral rinses, respectively. Clustering between the two groups was evaluated using a two-tailed Fisher exact test.
3.3. Panel of the Bacterial Microbiome Useful in Differentiating HNSCC from Controls
To evaluate the potential use of oral microbiota for biomarkers in HNSCC, we performed hierarchical clustering to assign the surveyed samples into two groups, namely, high risk (HR) and low risk (LR), based on the relative abundance of twelve HNSCC-related bacterial genera with consistent changes both in tissue and oral rinse samples (Figure 2B,C). Sample clustering into the HR group comprised the majority of HNSCCs (76%, 52/68 of tumor tissues; 63%, 43/68 of HNSCC oral rinses), and the LR group was primarily controls (60%, 41/68 of adjacent normal tissues; 76%, 52/68 of healthy oral rinses), with odds ratios of 4.87 (p ≤ 0.001) and 5.51 (p ≤ 0.001) in tissues and oral rinses, respectively. A combination of these 12 bacterial genera achieved an AUC of 0.84 (95% CI 0.77–0.91) in tissues and 0.86 (95% CI 0.80–0.92) in oral rinses for distinguishing HNSCCs from controls, suggesting the potential utility of bacterial discrimination for biomarkers in HNSCC clinical management.
3.4. Pathways Related to Bacterial Adaptation and Infectious Disease Enriched in HNSCC
Tumor microenvironment may enrich for functionally similar microbial communities. We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) to predict Kyoto Encyclopaedia of Genes and Genomes (KEGG) Ortholog (KO) abundance profiles based on the composition of microbial communities. Differentially abundant and depleted pathways were observed between HNSCCs and controls (Table S3), including twenty functions with consistent changes both in tissue and oral rinse habitats (Figure S3). The most significant enrichments (p ≤ 0.05) involved the plant–pathogen interaction (ko04626) and epithelial cell signaling (ko05120), which were mainly contributed by Fusobacterium and Leptotrichia (Table S4). In contrast, depletions in abundance of Rothia, Lautropia, Neisseria, and several commensal bacteria were largely responsible for the suppression of pathways related to proteasome (ko03050), p53 signaling pathway (ko04115), and ribosome biogenesis in eukaryotes (ko03008). We also observed enriched pathways related to bacterial cell motility (ko02030: bacterial chemotaxis, ko02040: flagellar assembly) in tumor tissues, implying a favorable microenvironment of niche adaptation for periodontal pathogens such as Treponema2, Fusobacterium, and Catonella.
3.5. Fusobacterium nucleatum is Associated with HNSCC Patient Outcomes
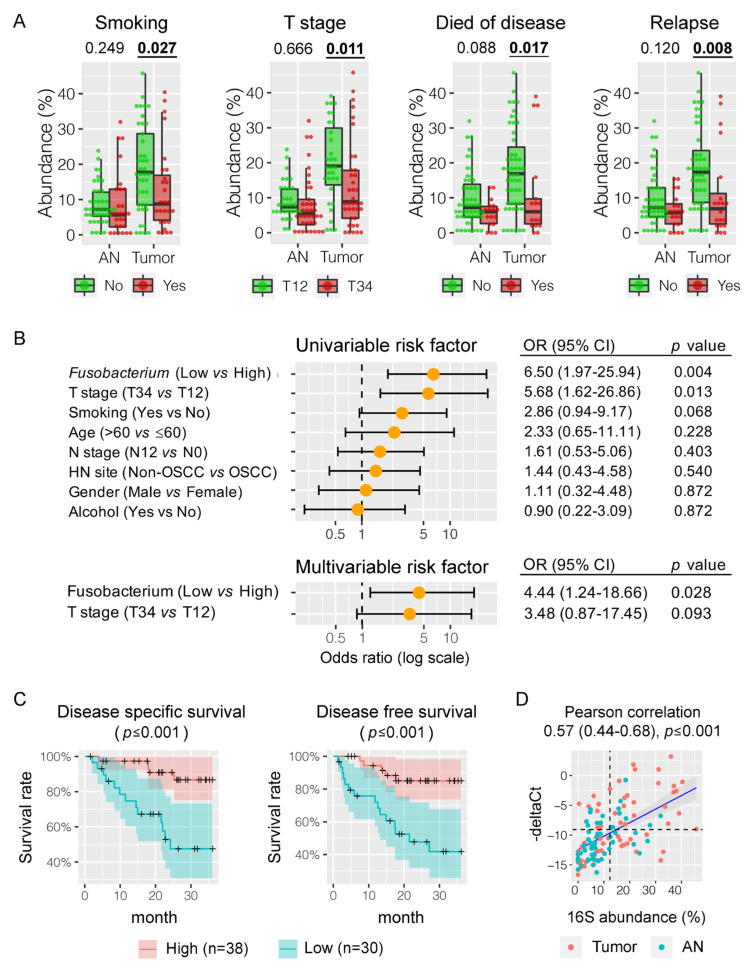
In order to explore the association of the bacterial microbiome with patient outcomes, we compared the relative abundances of bacterial genera with patient factors, including age, gender, smoking, alcohol, T stage, N stage, relapse, and survival. Interestingly, Fusobacterium enrichment in HNSCC tumor tissues was significantly associated with non-smokers (mean abundance of 18.7% vs. 12.5%, p = 0.027), lower tumor stage (19.9% vs. 13.2%, p = 0.011), better cancer-specific survival (18.0% vs. 11.0%, p = 0.017), and a lower rate of relapse (18.5% vs. 11.0%, p = 0.008) (Figure 3A and Figure S4A). The association was further confirmed by pairwise comparison between tumor and the adjacent normal tissues, for example, in non-smokers (18.7% vs. 8.6%, p ≤ 0.001) but not in smokers (12.5% vs. 8.6%, p = 0.132). Using the third quartile of Fusobacterium relative abundance in the adjacent normal tissues as a cutoff point (12.3%), we divided the surveyed HNSCC patients into Fusobacterium-high (n = 38) and -low (n = 30) groups based on the abundance values of Fusobacterium in tumor tissues. Both univariate and multivariate regression analyses demonstrated that the abundance of Fusobacterium was an independent predictor of HNSCC-specific survival (Figure 3B), with significantly better 3-year disease-specific survival (DSS) (86.7% vs. 47.6% at 36 months, p ≤ 0.001) and disease-free survival (DFS) (85.0% vs. 41.8% at 36 months, p ≤ 0.001) (Figure 3C). Evaluation of 16Sr RNA gene short reads using a phylogenetic topology algorithm and qPCR validation targeting on the conserved nusG gene confirmed that these clinical outcomes are related to F. nucleatum (Figure 3D and Figure S4B,C). The effect of F. nucleatum enrichment in HNSCC tumor tissues was not significantly influenced by age, gender, alcohol consumption, or N stage.
Figure 3.
Association of Fusobacterium abundance with HNSCC patient outcomes. (A) High abundance of Fusobacterium in HNSCC tumor tissues was positively associated with non-smokers, low T classification, better disease specific survival, and lower recurrence of disease. (B) Both univariate and multivariate regression analyses demonstrated that the high amount of Fusobacterium was an independent predictor of HNSCC disease specific survival. (C) A higher relative abundance of Fusobacterium in tumor tissue was correlated with improved 3-year disease-specific survival (DSS) and disease-free survival (DFS). (D) Quantitative PCR (qPCR) targeting the nusG gene confirmed the high abundance of Fusobacterium nucleatum in HNSCC tumor tissue.
We also observed several other bacteria associated with patient outcomes (Figure S5). For example, HNSCC tumor tissue-enriched Catonella and Johnsonella were usually observed in non-smokers or non-drinkers. The amount of Peptostreptococcus was positively related to more advanced tumor stage. Depletions of Neisseria, Haemophilus, and Rothia in HNSCC cases were associated with worse cancer-specific survival.
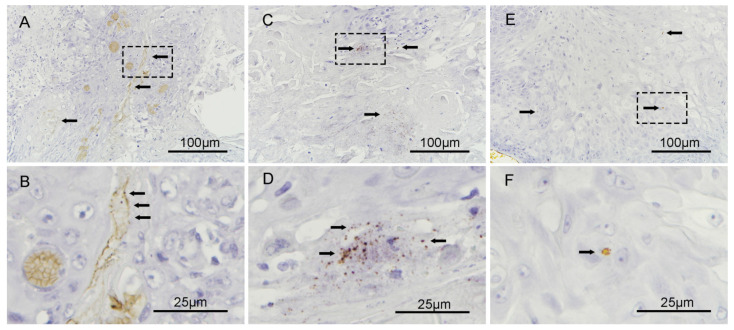
3.6. Fusobacterium nucleatum Localizes to the Intracellular Compartments of Tumor Cells
F. nucleatum has been reported as a highly invasive pathogen. To determine the spatial location of F. nucleatum in HNSCC tumors, RNA in situ hybridization (ISH) analysis for F. nucleatum was performed on six qPCR positive tumors from the surveyed cohort. Invasive F. nucleatum was noted focally in isolated cells with a morphology consistent with malignant cells and in the biofilm of mucosa adjacent to the tumor (Figure 4). There was no F. nucleatum noted in the stroma.
Figure 4.
RNA ISH analysis of F. nucleatum in HNSCC tumors. (A) and (B) demonstrate an F. nucleatum biofilm-like lining on the surfaces of the tumor cells at 200×; the black arrows point to the biofilm. (C) and (D) demonstrate the intracellular localization of F. nucleatum in tumor cells located adjacent to keratin whorls of squamous cell carcinoma at 200×. (E) and (F) further demonstrate the localization of F. nucleatum in the intracellular compartments of tumor cells in an oral cavity squamous cell carcinoma at 200×.
3.7. Enrichment of Fusobacterium is Associated with Host Gene Promoter Methylation in HNSCC
To evaluate the potential effects of HNSCC-related bacteria in host genetic and epigenetic alterations, we performed CpG DNA methylation using bisulfite capture sequencing in a random subset of paired tumor tissues and adjacent normal tissues, including 18 patients grouped as Fusobacterium-high and 13 as Fusobacterium-low. We mainly focused on CpG islands in the promoter region (up to 3kb) because of its association with gene expression. Overall, 2503 and 7432 differentially methylated regions (DMRs) were hyper- and hypo-methylated in HNSCC tumors when compared to adjacent normal tissues (≥15% methylation difference, adjusted p ≤ 0.01) and may regulate 1316 and 4515 genes, respectively (Figure S6A and Table S5). Genes with conflicting DMRs were removed (n = 366). By integrating HNSCC tumor tissue RNA-seq transcriptome profiles from The Cancer Genome Atlas (TCGA) and an enrichment analysis using KEGG human database, 156 genes with hyper-methylated promoters in our surveyed tumor tissues also had downregulated gene expression in the TCGA HNSCC cohort (log2FC ≤ −1, adjusted p ≤ 0.01) and involved multiple pathways, including the cholinergic synapse (CHRM2, GNG4, KCNJ12, ITPR1, PIK3R1), AMPK signaling pathway (CAB39L, LEPR, PIK3R1, CPT1B, CFTR), peroxisome (ACOX2, EPHX2, IDH2, ACOX3), and PI3K–Akt signaling pathway (CHRM2, RELN, GNG4, CHAD, FGF20, PIK3R1, EPHA2, FGF10) (p ≤ 0.01) (Figure S6B). Similarly, 494 highly expressed genes in TCGA cohort (log2FC ≥ 1, adjusted p ≤ 0.01) were found to be hypo-methylated in their promoter regions, with the top five most enriched KEGG pathways being related to cytokine–cytokine receptor interaction, cell adhesion, osteoclast differentiation, amoebiasis, and ECM–receptor interaction (Figure S6C).
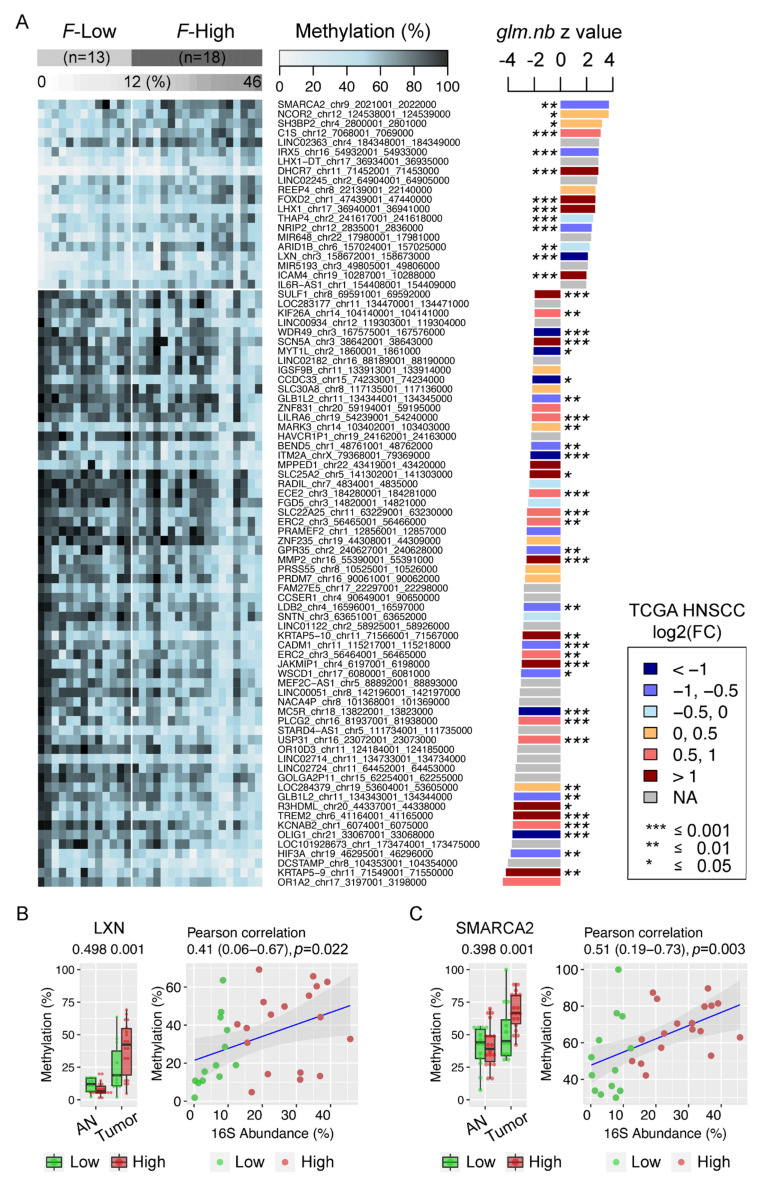
Next, we identified promotor regions that were differentially methylated between Fusobacterium-high and Fusobacterium-low tumor tissues to investigate whether HNSCC-enriched microbiota are associated with host gene methylation profiles. Interestingly, 20 (20) hyper- and 63 (61) hypo-methylated DMRs (genes) were positively and negatively related to Fusobacterium enrichment (≥15% methylation difference, adjusted p ≤ 0.01), respectively, which was further confirmed by a negative binomial generalized linear regression model test (glm.nb) and a Spearman’s rank-order correlation test (p ≤ 0.05) (Figure 5A and Figure S7, Table S5). We further compared gene expression profiles in the TCGA HNSCC cohort and found consistent associations between hypermethylation and downregulation in genes LXN, IRX5, NRIP2, and SMARCA2 (log2FC ≤ −0.5, adjusted p ≤ 0.01). Tumor suppressor gene (TSG) latexin (LXN) expression negatively correlates with the prognosis of several human cancers and plays significant roles in inflammatory response, tumor cell migration, invasion and apoptosis (Figure 5B); and TSG-SMARCA2 is involved in ATP-dependent chromatin remodeling related to DNA repair and replication (Figure 5C), implying a potential role of F. nucleatum infection in gene dysregulation related to the inflammatory response and cell proliferation through epigenetic silencing. Similarly, F. nucleatum may be also associated with promoted gene expression in HNSCC, including SULF1, TREM2, MMP2, KRTAP5-10, SCN5A, JAKMIP1, KRTAP5-9, ECE2, LILRA6, KCNAB2, ERC2, KIF26A, USP31, PLCG2, and SLC22A25 (log2FC ≥ 0.5, adjusted p ≤ 0.01), given the observed hypomethylation of gene promoters related to the enrichment of F. nucleatum.
Figure 5.
Bisulfite DNA capture sequencing demonstrating differentially methylated human gene promoters in Fusobacterium-enriched HNSCC tumor tissues. (A) A heat map of methylation profiles of host gene promoters between Fusobacterium-high (F-High) and Fusobacterium-low (F-Low) HNSCC tumor tissues. Negative binomial generalized linear regression model z values (glm.nb, p ≤ 0.05) are shown in the right panel of the heat map, with colors indicating gene expression changes (log2FC) in the TCGA HNSCC cohort. (B) Positive correlation between Fusobacterium enrichment and TSG-LXN promoter hypermethylation in HNSCC tumor tissues. (C) Positive correlation between Fusobacterium enrichment and TSG-SMARCA2 promoter hypermethylation in HNSCC tumor tissues. AN: adjacent normal tissues. Tumor: HNSCC tumor tissues.
4. Discussion
In this study, we sought to characterize oral microbiota dysbiosis in a subgroup of HPV-negative HNSCCs and their interactions with clinical outcomes and host epigenetic alteration. Our findings revealed specific oral microbial signatures in HNSCC with significant enrichment of Fusobacterium and depletion of Streptococcus, which is consistent with other studies [1,15,17,19]. We observed a consistent bacterial pattern in both tissue and oral rinse communities, including twelve bacterial genera that provided microbial signatures in HNSCC with high sensitivity and specificity, thereby offering reliable applications as novel biomarkers to complement cancer diagnostics [20,21,22,23]. Network analysis found collaborative relationships between periodontal pathogens or between commensal taxa, implying distinct roles of oral bacteria in forming community in disease progresses.
We identified HNSCC non-smokers as having a higher abundance of Fusobacterium nucleatum in tumor tissues, while the enrichment in smokers was not significant. Associations of lower tumor stage and improved cancer-specific survival with enrichment of F. nucleatum in HNSCC cases were also novel observations. The finding of better clinical outcomes associated with elevated levels of F. nucleatum in HNSCC when comparing tumor with adjacent normal tissue is, however, in contrast to poorer survival and worse prognosis of F. nucleatum infection in colorectal cancer (CRC) [37,38,39] and esophageal cancer [40]. Long-term smoking, alcohol consumption, and high-risk human papillomavirus (HPV) infection are established oncogenic drivers in HNSCC [41]. For HNSCC patients lacking these traditional risk factors, however, chronic infection and inflammation as factors in carcinogenic feedback loops incorporating the resident microbiota are increasingly recognized in the pathogenesis of cancer, highlighting a potential role of F. nucleatum acting as a “driver” in premalignant and/or early lesions of HNSCC that may initiate cancer development, which is subsequently outcompeted by opportunistic bacterial passengers, resulting in a lower abundance at later stages of disease [42,43]. This hypothesis is in conjunction with known F. nucleatum pro-carcinogenic features associated with bacterial invasion and inflammation (e.g., flagella assembly and bacterial chemotaxis) [24,25,27], metabolic pathways (e.g., homolactic fermentation) [26], production of DNA-damage compounds [44], and promoted cell proliferation (e.g., E-cadherin/β-catenin signaling via its FadA adhesin) [45], suggesting that enrichment of F. nucleatum in early stages of tumorigenesis is an independent risk factor in patients lacking major risk factors for HNSCC.
Interestingly, an integrative analysis identified that the enrichment of F. nucleatum was associated with host gene promoter methylation, which further implies its pathophysiologic role in regulating transcriptome level involvement in pro-inflammatory and cell proliferation processes [9,10]. The tumor suppressor gene (TSG) latexin (LXN) has been downregulated in a number of human cancers, including lymphoma, gastric carcinoma, and thyroid carcinoma [37,38,39], and the TCGA HNSCC cohort, in congruence with its promoter hypermethylation in Fusobcaterium-high tumor tissues. LXN has also been shown to be involved in the inflammatory response with inhibition of tumor growth and colony formation [40,42]. Overexpression of LXN inhibits the activity of the nuclear factor NF-kB pathway in inflammation by binding to a ribosomal protein subunit 3 (Rps3) [43], and also increased apoptosis and lowered proliferative activity [46]. This is in broad agreement with our observation of loss of LXN expression likely by F. nucleatum-associated epigenetic silencing in the HNSCC tumor microenvironment, where bacterial invasion may interfere with the pro-inflammatory immunomodulatory gene signature of LXN in blocking tumor immune cell evasion. We also observed a similar association between F. nucleatum enrichment and TSG-SMARCA2 promoter hypermethylation. The genes encoding the ATPase of the chromatin remodeling SW1/SNF complexes SMARCA4 and the alternative SMARCA2 are usually mutated or silenced in tumors, suggesting a role as tumor suppressor [47]. The SWI/SNF complex has roles in regulating gene expression and cancer development. Mutations of SMARCA2 are rarely observed but gene silencing related to epigenetic regulation in tumor has been reported [48,49], which is in line with strong hypermethylation observed in our Fusobacterium-enriched HNSCC tumor tissues when compared to Fusobacterium-low tumor tissues. In contrast to hypermethylation, hypomethylation of sulfatase 1 (SULF1) and matrix metalloproteinase 2 (MMP2) promoters was observed to be associated with the enrichment of F. nucleatum in our HNSCC tumor tissues. Both genes were overexpressed in human cancers, such as gastric, lung, or colorectal cancer [50,51], and were reported to remodel or breakdown extracellular matrix (ECM) [44], resulting in dysregulated cell signaling pathways (e.g., Wnt signaling pathway) [41], or release of growth factors such as TGF-β [45] that affect cell immune tolerance, adhesion, proliferation, and migration [52]. Whether F. nucleatum directly up-regulates SULF1 and/or MMP2 through epigenetic mechanisms or ECM remodeling by gene degradation plays a role in initial HNSCC tumor growth remains an interesting question warranting further investigations.
In this study, notable similarities in the relative abundances of bacteria were observed across both tissues and oral rinses, suggesting the potential to use oral rinses as non-invasive samples to reflect tissue microbial composition. There can be certain differences if difference sample courses are used, highlighting the importance of assessing the microbiota across both tissue and oral rinses, strengthening our findings. Given the prominent role of somatic mutations in TP53 in smoking-related HNSCCs, for example, dramatic enrichment of F. nucleatum in nonsmokers, may explain it in part as a pathogen causing DNA damage in patients without traditional high-risk factors [47,48,49]. However further prospective evaluations of patient factors including comorbidities will be needed to validate the clinical outcomes in F nucleatum-enriched HNSCC tumors. Furthermore, F. nucleatum may regulate host gene transcriptome profiles through epigenetic mechanisms and act as a pro-inflammatory driver involving multiple cell signaling pathways.
5. Conclusions
In conclusion, this study improves our understanding of microbial communities associated with HNSCC. Enriched F. nucleatum abundance in HNSCC non-smokers and an improved survival suggest possible pathologic roles of bacterial genera in patients that may serve as targets for HNSCC intervention through host epigenetic modifications. Future studies investigating the functional roles of HNSCC-related bacterial taxa in the host genomic and epigenetic changes will be essential to elucidating driver or passenger roles in HNSCC. Finally, a unique HNSCC microbial signature may facilitate the use of oral bacteria as biomarkers for disease screening and prevention.
Acknowledgments
We thank the anonymous participants who provided samples for this study.
Abbreviations
| HNSCC | Head and neck squamous cell carcinoma |
| AUC | Area under the receiver operating curve |
| HPV | Human papillomavirus |
| AN | Adjacent normal |
| ORF | Open reading frame |
| OUT | Operational taxonomic unit |
| ASV | amplicon sequence variant |
| PERMANOVA | Permutational multivariate analysis of variance |
| MWU | Mann–Whitney Wilcoxon rank sum test |
| WSR | Wilcoxon signed rank test |
| KW | Kruskal–Wallis test |
| Tukey HSD | Tukey’s honest significant difference |
| ROC | Receiver operating characteristic |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | Kyoto Encyclopedia of Genes and Genomes Orthology |
| FFPE | Formalin-fixed paraffin-embedded |
| DMRs | Differentially methylated regions |
| TCGA | The cancer genome atlas |
| HR | High risk |
| PCoA | Principal coordinate analysis |
| LefSe | Linear discriminant analysis effect size |
| LDA | Linear discriminant analysis |
| PICRUSt2 | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 |
| DSS | Disease specific survival |
| DFS | Disease free survival |
| ISH | In situ hybridization |
| Glm nb | generalized linear regression model test |
| TSG | tumor suppressor gene |
| LXN | Latexin |
| qPCR | Quantitative PCR |
| F-Low | Fusobacterium-low |
| F-High | Fusobacterium-high |
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/11/3425/s1. Figure S1: A refinement of twelve HNSCC-related bacterial genera discriminating HNSCCs from controls both in tissues (tumor vs adjacent normal) and oral rinses (HNSCC vs healthy). Figure S2: Bacterial genera showing different preference of colonization between tissue and oral rinse communities. Figure S3: Microbial Functional prediction based on 16S rRNA gene community composition using PICRUSt2. Figure S4: Association of Fusobacterium abundance with HNSCC patient outcomes. Figure S5: Association of other bacterial genera abundance with HNSCC patient outcomes. Figure S6: Bisulfite DNA capture sequencing of paired tumor and AN tissue samples from thirty-one HNSCC patients. Figure S7: Heat map of methylation profiles of host gene promoters showing differentially methylated status between Fusobacterium-high and Fusobacterium-low HNSCC tumor tissues. Table S1: Demographic information of patients with HNSCC compared to the healthy subjects. Table S2: Relative abundance of bacterial genera in designated groups. Pairwise Tukey HSD post-hoc test and non-parametric Wilcoxon Signed Rank test were used to detect statistical significance. Table S3: Discriminant analysis of predicted KEGG KO pathways between cases and controls across tissues and oral rinses using DESeq2. Table S4: Contribution in relative abundance of oral bacteria in predicted KEFF KO pathways. Table S5: Association of Fusobacterium enrichment with host gene promoter methylation.
Author Contributions
Study design, Z.C., J.Y.K.C. and P.K.S.C.; support with materials, K.C.A.C., S.F., C.W.K.N., L.L., E.H.L.L., Z.Y., E.W.Y.W. and J.W.L.; experiments, P.Y.W., S.F., P.L. and C.C.; data interpretation and analysis, Z.C., J.Y.K.C., P.K.S.C., P.Y.W., L.C., P.L., Q.M., J.Y., S.H.W., W.K.K.W., R.J.L., S.S.B., K.M., C.C., K.W.L., V.W.Y.L., A.S.L.C. and A.B.W.C.; manuscript writing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
The work was partially supported by funding agents from the Stanley Ho Medical Foundation (JYKC), the Research Grants Council of the Hong Kong Special Administrative Region, China (project numbers CUHK 14109716 and 14108818—J.Y.K.C.; and project number CUHK 14161017—Z.C.), the Direct grant (project number CUHK 4054350—Z.C.), and the Seed Fund for Gut Microbiota Research (P.K.S.C.) from the Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong Special Administrative Region, China. The funders had no role in study design, data collection, analysis, interpretation, or writing of the report.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hsiao J.-R., Chang C.-C., Lee W.-T., Huang C.-C., Ou C.-Y., Tsai S.-T., Chen K.-C., Huang J.-S., Wong T.-Y., Lai Y.-H., et al. The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogens. 2018;39:778–787. doi: 10.1093/carcin/bgy053. [DOI] [PubMed] [Google Scholar]
- 2.Lin A. Radiation therapy for oral cavity and oropharyngeal cancers. Dent. Clin. North. Am. 2018;62:99–109. doi: 10.1016/j.cden.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Isayeva T., Li Y., Maswahu D., Brandwein-Gensler M. Human Papillomavirus in non-oropharyngeal head and neck cancers: A systematic literature review. Head Neck Pathol. 2012;6:104–120. doi: 10.1007/s12105-012-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn J., Sinha R., Pei Z., Dominianni C., Wu J., Shi J., Goedert J.J., Hayes R.B., Yang L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guevarra L.A., Jr., Afable A.C.F., Belza P.J.O., Dy K.J.S., Lee S.J.Q., Sy-Ortin T.T., Albano P.M.S.P. Immunogenicity of a Fap2 peptide mimotope of Fusobacterium nucleatum and its potential use in the diagnosis of colorectal cancer. Infect. Agents Cancer. 2018;13:1–6. doi: 10.1186/s13027-018-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forman D., Newell D.G., Fullerton F., Yarnell J.W., Stacey A.R., Wald N., Sitas F. Association between infection with Helicobacter pylori and risk of gastric cancer: Evidence from a prospective investigation. BMJ. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansson L.R., Engstrand L., Nyren O., Lindgren A. Prevalence of helicobacter pylori infection in subtypes of gastric cancer. Gastroenterology. 1995;109:885–888. doi: 10.1016/0016-5085(95)90398-4. [DOI] [PubMed] [Google Scholar]
- 8.Mima K., Nishihara R., Qian Z.R., Cao Y., Sukawa Y., Nowak J.A., Yang J., Dou R., Masugi Y., Song M., et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahara T., Hirata I., Nakano N., Tahara S., Horiguchi N., Kawamura T., Okubo M., Ishizuka T., Yamada H., Yoshida D., et al. Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget. 2017;8:61917–61926. doi: 10.18632/oncotarget.18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Ye J., Fang D., Lv L., Wu W., Shi D., Li Y., Yang L., Bian X., Wu J., et al. Multi-omic profiling reveals associations between the gut mucosal microbiome, the metabolome, and host DNA methylation associated gene expression in patients with colorectal cancer. BMC Microbiol. 2020;20:1–13. doi: 10.1186/s12866-020-01762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson S.N., Snesrud E., Liu J., Ong A.C., Kilian M., Schork N.J., Bretz W. The dental plaque microbiome in health and disease. PLoS ONE. 2013;8:e58487. doi: 10.1371/journal.pone.0058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tezal M., Sullivan M.A., Hyland A., Marshall J.R., Stoler D., Reid M.E., Loree T.R., Rigual N.R., Merzianu M., Hauck L., et al. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2009;18:2406–2412. doi: 10.1158/1055-9965.EPI-09-0334. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Jia J.W.H. Metagenome-wide association studies: Fine-mining the microbiome. Nat. Rev. Genet. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Ganly I. The oral microbiome and oral cancer. Clin. Lab. Med. 2014;34:711–719. doi: 10.1016/j.cll.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Yang J., Feng Q., Chen B., Li M., Liang C., Li M., Li Z., Xu Q., Zhang L., et al. Compositional and functional analysis of the microbiome in tissue and saliva of oral squamous cell carcinoma. Front. Microbiol. 2019;10:1439. doi: 10.3389/fmicb.2019.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C.-Y., Yeh Y.-M., Yu H.-Y., Chin C.-Y., Hsu C.-W., Liu H., Huang P.-J., Hu S.-N., Liao C.-T., Chang K.-P., et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front. Microbiol. 2018;9:862. doi: 10.3389/fmicb.2018.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganly I., Yang L., Giese R.A., Hao Y., Nossa C.W., Morris L.G.T., Rosenthal M., Migliacci J., Kelly D., Tseng W., et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int. J. Cancer. 2019;145:775–784. doi: 10.1002/ijc.32152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero-Preston R., White J.R., Godoy-Vitorino F., Rodríguez-Hilario A., Navarro K., González H., Michailidi C., Jedlicka A., Canapp S., Bondy J., et al. High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget. 2017;8:110931–110948. doi: 10.18632/oncotarget.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Funchain P., Bebek G., Altemus J., Zhang H., Niazi F., Peterson C., Lee W.T., Burkey B.B., Eng C. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017;9:1–10. doi: 10.1186/s13073-017-0405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrero-Preston R., Godoy-Vitorino F., Jedlicka A., Rodríguez-Hilario A., González H., Bondy J., Lawson F., Folawiyo O., Michailidi C., Dziedzic A., et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7:51320–51334. doi: 10.18632/oncotarget.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho A.L., Jerónimo C., Kim M.M., Henrique R., Zhang Z., Hoque M.O., Chang S., Brait M., Nayak C.S., Jiang W.-W., et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin. Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 22.Rosas S.L., Koch W., Carvalho M.G.D.C., Wu L., Califano J., Westra W., Jen J., Sidransky D. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–942. [PubMed] [Google Scholar]
- 23.Wong M.C.S., Vlantis A.C., Liang M., Wong P.Y., Ho W.C.S., Boon S.S., Sze R.K.H., Leung C., Chan P.K.S., Chen Z. Prevalence and epidemiologic profile of oral infection with alpha, beta, and gamma Papillomaviruses in an Asian Chinese population. J. Infect. Dis. 2018;218:388–397. doi: 10.1093/infdis/jiy160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z., Hui P.C., Hui M., Yeoh Y.K., Wong P.Y., Chan M.C.W., Wong M.C.S., Ng S.C., Chan F.K.L., Chan P.K.S. Impact of preservation method and 16S rRNA hypervariable region on gut microbiota profiling. mSystems. 2019;4:e00271-18. doi: 10.1128/mSystems.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert J.A., Jansson J.K., Knight R. The Earth microbiome project: Successes and aspirations. BMC Biol. 2014;12:1–4. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsen F.A., Kodner R.B., Armbrust E.V. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohart F., Eslami A., Matigian N., Bougeard S., Cao K.-A.L. MINT: A multivariate integrative method to identify reproducible molecular signatures across independent experiments and platforms. BMC Bioinform. 2017;18:128. doi: 10.1186/s12859-017-1553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:002832. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A., et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krueger F., Andrews S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E.M., Melnick A., Mason C.E. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seed R.I., Taurozzi A.J., Wilcock D.J., Nappo G., Erb H.H.H., Read M.L., Gurney M., Archer L.K., Ito S., Rumsby M.G., et al. The putative tumour suppressor protein Latexin is secreted by prostate luminal cells and is downregulated in malignancy. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-41379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M., Osisami M., Dai J., Keller J.M., Escara-Wilke J., Mizokami A., Keller E.T. Bone microenvironment changes in latexin expression promote chemoresistance. Mol. Cancer Res. 2017;15:457–466. doi: 10.1158/1541-7786.MCR-16-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Sun C., Tan Y., Li L., Zhang H., Liang Y., Zeng J., Zou H. Transcription levels and prognostic significance of the NFI family members in human cancers. PeerJ. 2020;8:e8816. doi: 10.7717/peerj.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Zhang C., Li Z., Wang C., Jia J., Gao T., Hildebrandt G., Zhou D., Bondada S., Ji P., et al. Latexin inactivation enhances survival and long-term engraftment of hematopoietic stem cells and expands the entire hematopoietic system in mice. Stem Cell Rep. 2017;8:991–1004. doi: 10.1016/j.stemcr.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw R., Beasley N. Aetiology and risk factors for head and neck cancer: United Kingdom National multidisciplinary guidelines. J. Laryngol. Otol. 2016;130:S9–S12. doi: 10.1017/S0022215116000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsunaga K., Kikuchi J., Wada T., Furukawa Y. Latexin regulates the abundance of multiple cellular proteins in hematopoietic stem cells. J. Cell. Physiol. 2011;227:1138–1147. doi: 10.1002/jcp.22834. [DOI] [PubMed] [Google Scholar]
- 43.You Y., Wen R., Pathak R.D., Li A., Li W., Clair D.S., Hauerjensen M., Zhou D., Liang Y. Latexin sensitizes leukemogenic cells to gamma-irradiation-induced cell-cycle arrest and cell death through Rps3 pathway. Cell Death Dis. 2014;5:e1493. doi: 10.1038/cddis.2014.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng F., Zhang Y., Lu Z., Zhang S., Pan Y. Fusobacterium nucleatum Caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol. 2020;39:144–151. doi: 10.1089/dna.2019.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue Z., Zhou Y., Wang C., Zheng J., Zhang P., Zhou L., Wu L., Shan Y., Ye M., He Y., et al. Latexin exhibits tumor-suppressor potential in pancreatic ductal adenocarcinoma. Oncol. Rep. 2015;35:50–58. doi: 10.3892/or.2015.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ehrenhöfer-Wölfer K., Puchner T., Schwarz C., Rippka J., Blaha-Ostermann S., Strobl U., Hörmann A., Bader G., Kornigg S., Zahn S., et al. SMARCA2-deficiency confers sensitivity to targeted inhibition of SMARCA4 in esophageal squamous cell carcinoma cell lines. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-48152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J., He K., Zhang Y., Song J., Shi Z., Chen W., Shao Y. Inactivation of SMARCA2 by promoter hypermethylation drives lung cancer development. Gene. 2019;687:193–199. doi: 10.1016/j.gene.2018.11.032. [DOI] [PubMed] [Google Scholar]
- 49.Glaros S., Cirrincione G.M., Muchardt C., Kleer C.G., Michael C.W., Reisman D. The reversible epigenetic silencing of BRM: Implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- 50.Salem N., Kamal I., Al-Maghrabi J., Abuzenadah A.M., Peer-Zada A.A., Qari Y., Al-Ahwal M.S., Al-Qahtani M., Buhmeida A. High expression of matrix metalloproteinases: MMP-2 and MMP-9 predicts poor survival outcome in colorectal carcinoma. Futur. Oncol. 2016;12:323–331. doi: 10.2217/fon.15.325. [DOI] [PubMed] [Google Scholar]
- 51.Lee H.-Y., Yeh B.-W., Chan T.-C., Yang K.-F., Li W.-M., Huang C.-N., Ke H.-L., Li C.-C., Yeh H.-C., Liang P.-I., et al. Sulfatase-1 overexpression indicates poor prognosis in urothelial carcinoma of the urinary bladder and upper tract. Oncotarget. 2017;8:47216–47229. doi: 10.18632/oncotarget.17590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker C., Mojares E., Hernández A.D.R. Role of extracellular matrix in development and cancer progression. Int. J. Mol. Sci. 2018;19:3028. doi: 10.3390/ijms19103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.