Abstract
Translation of traumatic brain injury (TBI) research findings from bench to bedside involves aligning multi-species data across diverse data types including imaging and molecular biomarkers, histopathology, behavior, and functional outcomes. In this review we argue that TBI translation should be acknowledged for what it is: a problem of big data that can be addressed using modern data science approaches. We review the history of the term big data, tracing its origins in Internet technology as data that are “big” according to the “4Vs” of volume, velocity, variety, veracity and discuss how the term has transitioned into the mainstream of biomedical research. We argue that the problem of TBI translation fundamentally centers around data variety and that solutions to this problem can be found in modern machine learning and other cutting-edge analytical approaches. Throughout our discussion we highlight the need to pull data from diverse sources including unpublished data (“dark data”) and “long-tail data” (small, specialty TBI datasets undergirding the published literature). We review a few early examples of published articles in both the pre-clinical and clinical TBI research literature to demonstrate how data reuse can drive new discoveries leading into translational therapies. Making TBI data resources more Findable, Accessible, Interoperable, and Reusable (FAIR) through better data stewardship has great potential to accelerate discovery and translation for the silent epidemic of TBI.
Keywords: analytics, big data, data sharing, FAIR principles, traumatic brain injury
Introduction
Traumatic brain injury (TBI) is a prevalent disorder impacting millions of individuals without a widely accepted therapeutic approach. TBI impacts 69 million individuals worldwide.1,2 The estimated economic burden of TBI is more than $60 billion annually in the United States alone3 and some estimates suggest it costs the global economy $400 billion worldwide.4 Paucity of therapeutic options results in a lack of clinical consensus and poor follow-up for TBI patients, which is especially detrimental for individuals with persistent post-injury symptoms.5 This stands in sharp contrast to the large number of potential therapeutics discovered in basic and pre-clinical models of TBI.6–8
Altogether, this suggests that translation of TBI research from basic animal models into human therapeutics lacks a well-defined pipeline.9 This special issue of the Journal of Neurotrauma highlights barriers to translation and recent exciting achievements that help to overcome these barriers, from novel approaches of animal models to biomarkers and regulatory innovations. In the present article we focus on the role of data science in accelerating translation. We frame our discussion around the concept that raw research data and unpublished “dark data” are under-utilized resources for driving discovery.10 We argue that better data stewardship has great potential to advance translation from basic research to therapy. In particular, we focus on the principle that organizing and federating numerous small datasets can produce big data that open new opportunities to apply modern machine learning tools for data-driven discovery. We contrast the treatment of pre-clinical research data with clinical data and point to recent advances in data-driven discovery in clinical TBI that are rapidly advancing precision medicine. Our review is intended to cover translation between pre-clinical and clinical data science without delving deeply into either side of this translational divide. Where possible, we refer interested readers to other reviews focused in depth on issues specific to clinical or pre-clinical domains.
Big data/small data—what's the difference?
The term big data was first coined in early 2000s in the Internet technology field to describe data that are difficult to work with because of being “big” according to at least one of the “3Vs”: volume, velocity, and variety.11 In recent years, veracity has been added as a fourth V as it became apparent that the accuracy of data is an increasing challenge as diverse aspects of society, including social media transition to the digital world. There have been various proposals to add more Vs,12 but we will limit our discussion here to these “classic” 4Vs as we feel they are most relevant to neurotrauma big data. The 4Vs challenge the limits of traditional database infrastructures, analytical approaches, and human accessibility, making knowledge extraction difficult. These challenges have led to innovations in data management approaches in the last decade. Examples include cloud storage—enormous enterprise data centers—to manage data volume; parallel on-chip processing (as opposed to hard-disk processing) of data streams to manage high-velocity data such as social media threads and multi-sensor data from mobile phones; and machine learning algorithms to manage data variety. Veracity, the uncertainty of data, may be remedied by provenance tracking and quality controls taken during collection and aggregation steps.
To find examples of high-volume, high-velocity neurotrauma data one only has to enter a busy intensive care unit (ICU) or neuroradiology suite. High-velocity physiological data collection (blood pressure, heart rate, oxygen saturation levels, etc.) is now possible using integrated digital systems, as equipment can be set to record continuously for days, producing very large data file sizes.13,14 Neuroimaging using modern 3 Tesla magnetic resonance imaging (MRI) scanners produce many terabytes of data in the process of detecting microlesions that predict TBI outcome.15
However, when considering the alignment of pre-clinical to clinical TBI data to promote translation, it becomes evident that translation does not involve particularly high data volume or velocity. This does not mean that translational TBI data are a “small data” problem. By its very nature, TBI involves heterogeneous injuries that impact the complex architecture of the brain and trillions of synapses in highly unpredictable ways. This constitutes the ultimate example of data variety. To grapple with heterogeneity, researchers typically collect various types of data including high-resolution imaging, physiology, molecular biology, behavior, and cognition. Often, multiple types of data are collected from a single subject, and within a single study or article different subjects are represented by different subsets of variables split across different figures. In this sense we would argue that TBI translation is indeed a big-data problem, and specifically a problem of variety and veracity.
For example, consider our own experience with the Moody Project for Translational TBI Research (Moody Project), an effort aimed at: 1) characterizing acute and chronic TBI using small and large animal models and 2) repurposing U.S. Food and Drug Administration (FDA)-approved drugs and testing novel drugs, devices, and adult stem cell-based therapies for treatment of TBI.16–20 This large-scale project was designed to bring together domain expertise in genomics, proteomics, histopathology, and behavioral outcome measures to explore the multi-modal effects of TBI over time in a pre-clinical model. Tens of thousands of genes were probed, hundreds of protein targets were assayed, and behavioral tests of motor function, memory, and cognitive function were collected. Harmonizing these disparate datasets presents a number of challenges from a logistical data perspective and provides an example of the considerations that must be acknowledged when dealing with this unique form of big data. With a large project where data collection spans many disciplines, there is the inherent variability in data structure that must be reconciled.
Each domain tends to collect and organize data in a way that is most practical or amenable to its field. When tasked with harmonizing this dataset, the first task was to curate and translate the unique vernacular and domain-specific shorthand into a clear and concise data dictionary, so that all data fields could be quickly and easily understood between researchers. Where possible, terminology was standardized across domains to keep shared traits consistent and readily identifiable. Aspects as simple as how different labs may refer to a time-point (e.g., “3m” vs. “3 months” vs. “3 mon post-injury,” etc.), or animal identification (“S34” vs. “Subject 34” vs. “34,” etc.) are essential for data harmonization.
After assessing the datasets and noting fields where missing data were present, we quickly came to realize that we needed a project-level set of common data elements (CDE) that spanned across domains before any analysis could begin. Further, we found harmonizing end-point data, such as genomic/proteomic/histological data, with longitudinal data such as behavioral measures required flexibility in our data structure (e.g., restructuring between “long form” and “wide form” view in a spreadsheet of repeated measures) and clarity in our variable naming conventions. We also found that, in the case of genomic datasets, a certain amount of first-order dimension reduction made data harmonization more manageable. For example, to explore how gene expression and behavioral recovery interact after TBI, we first used a data-driven approach to pare down the 45,610 genes probed using a topological data analysis tool, followed by factor analysis, which identified a subset of 79 genes that appeared to have strong correlation with injury conditions, brain regions, and time-points at which data were collected.21 This stepwise dimension reduction approach allowed for better manageable data integration with the behavioral dataset.22
This type of multi-dimensional analytic workflow has been termed syndromics or syndromic analysis and involves applying a data-driven or machine learning approach to heterogeneous neurotrauma outcome measures. The goal of this analysis is to visualize the neurotrauma “syndromic space” across the full landscape of end-points (for examples, see14, 23–26) and then use this visualization to help manage data variety and to determine the robustness and veracity of outcome patterns. Syndromic analysis can also be used to generate additional hypotheses and identify new therapeutic targets that we can test using pre-clinical models and clinical discovery studies.24,27,28 Together, this illustrates one potential set of solutions for big-data problems routinely encountered in translational TBI research.
What is the “long tail”? What are “dark data”?
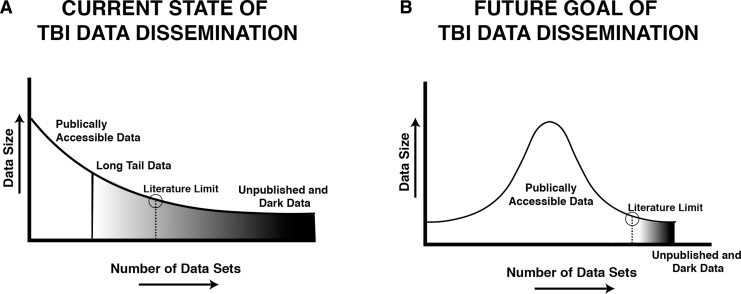
The problem of data variety leads to a curiously skewed distribution of TBI data in published literature29 that parallels a phenomenon observed in online marketing and in public health, the so called long tail of product (e.g., data) dissemination. Specifically, plotting the volume of each dataset (y-axis) as a function of the number of datasets (x-axis) produces a highly non-normal distribution, with relatively few datasets representing a bulk of the high-volume “big” data that is publicly available (e.g., published) (Fig. 1). The vast majority of the datasets collected extend to the right of the distribution into the long tail of data distribution, reflecting datasets of relatively modest size and high variety (Fig. 1A). Dissemination of these work products in the form of digital data is a rare phenomenon, even though long-tail data collectively comprise the majority of data collected in neurotrauma.
FIG. 1.
Shortening the long tail of dark data for traumatic brain injury (TBI) research. (A) The current state of TBI data consists of a relatively small number of large, publicly accessible datasets reflected schematically as a right-skewed distribution. The majority of data collected by the field exists in the long tail of the distribution, with most datasets consisting of relatively modest data sizes as either gray data that are difficult to access beyond summaries reported in publications; or dark data that are inaccessible, locked in non-digital formats. (B) The goal of digital data stewardship is to make TBI data Findable, Accessible, Interoperable, and Reusable (FAIR),56 thereby shortening the long tail of dark data, and making a greater proportion of the data in the TBI literature publicly accessible to drive new discoveries and accelerate translation.
It has been suggested that the long-tail phenomenon reflects the concentration of centralized data release by the traditional peer-reviewed publication system,30 where page limits and other costs prohibit publishing data in their full form. The modern scientific peer review system is centered around a 17th century data dissemination model, essentially unchanged since the first scientific journal,31 where data are reported in a highly refined form with accompanying narratives, summary figures, and tables. Indeed, given the high costs of traditional publishing, the literature typically only contains data in the form of summaries, graphs, or tables with very few examples of high-volume raw data being released as independent publications that can be directly accessed (e.g., gene accession numbers). These artifacts of the traditional publishing system result in the long-tail data that contain large quantities of semi-accessible data (“gray data”32) as well as inaccessible and unpublished data (dark data) (Fig. 1A).29
Recent estimates suggest that the long tail of dark data comprises approximately 85% of the data collected in the biomedical research enterprise worldwide.33,34 For this reason, it has been argued that published literature represents a small, highly selected subset of findings that reflect 15% of the data that happen to conform to expectations (i.e., hypotheses) of the article authors and fit into a tidy narrative “story.”29 Based on Bayesian statistical arguments, some prominent epidemiologists have suggested that the majority of published articles contain “false-positive” findings that contribute to irreproducibility in the biomedical research literature.35,36 A recent umbrella meta-analysis indicates that pressure to publish high-impact research results in suppression of dark data and “gray” literature (dissertations, abstracts, personal communications, and non-published works), resulting in systematic over-estimation of effect sizes in the published literature, contributing to systematic patterns of scientific irreproducibility.37
The central question of the current review then becomes: how do we shorten the long tail of dark data to produce a more comprehensive data dissemination model than traditional scientific publishing? (Fig. 1B). We believe the answer lies in new data dissemination tools that enable “data publication” and other forms of public release of long-tail and dark data.
Why publish long-tail or dark data?
The problems of bias and research inefficiency introduced by long-tail and dark data have been reported in several central nervous system (CNS) injury models.29,34,38–40 For example, systematic reviews and meta-analyses within the field of pre-clinical stroke have revealed a substantial overstatement of effect size in studies with poor reporting on key features such as blinding/randomization and subject attrition.39,41,42 In addition, meta-analysis tools that estimate selective reporting and publication bias suggest that around 30% of completed studies in pre-clinical stroke are not reported in published literature, likely because these results detracted from the authors' hypotheses, resulting in a major overstatement of effect sizes in the literature.39 In the field of spinal cord injury (SCI), a similar impact of dark data has been reported in meta-analyses of rho/rock inhibitors and cell-based therapies.40,43 This overstatement of effect sizes is a critical problem that has been shown to directly contribute to irreproducibility and failures in translation, as clinical trials are often based on highly selected and low-quality pre-clinical evidence of efficacy.36 Indeed, it has been demonstrated using objective bibliometric methods that article quality metrics are inversely correlated to effect size; that is, low-quality articles report the highest effect sizes, independent of citations or the impact factor of the journal.36,38,39
To date, there have been relatively few meta-analyses examining publication bias in TBI44; however, such efforts are underway.45 It is noteworthy that meta-analysis methods estimate dark data based on effect size in reported articles and impute completed but unreported studies.46 They do not speak to long-tail data, smaller packets of information such as partially completed studies that were halted early due to perceived futility, adverse health events (drug side effects, husbandry issues, etc.), data about non-primary outcomes, and meta-data about collected experiments. These examples of long-tail data are estimated to comprise the majority of data collected in biomedicine with “file-drawer” dark data representing an estimated $200+ billion in annual research investment worldwide.34,47 This suggests that failing to publish long-tail and dark data contributes to systematic biases, irreproducibility, misinformation, and fiscal waste in the system of biomedical research.35,48 A number of countermeasures have been presented to overcome publication bias.49
In addition to avoiding the negative impact of publication bias, mining long-tail and dark data can yield direct positive benefits. Our own experience demonstrates opportunities for novel discoveries, high-impact publications, and perhaps even accelerated translation through reuse and publication of long-tail and dark data. Within the SCI research community, grassroots efforts have begun to yield a culture of data sharing and pooled analysis that has launched new areas of inquiry.50–52 For example, our team developed the VISION-SCI data repository, pooling retrospective data from 13 SCI research laboratories; at the time of writing it contains data on more than 4000 animals and more than 2700 variables.52 Re-analysis of these legacy data using modern machine learning approaches has yielded novel insights with high potential for translation, even in very old data.
For example, re-analysis of data collected from 1994–96 as part of the Multicenter Animal Spinal Cord Injury Study (MASCIS)53 in combination with cervical SCI model development studies from the early 2000s54 revealed a previously unreported null effect of the steroid methylprednisolone on motor and histological outcomes.24 However, a recent machine intelligence tool known as topological data analysis (TDA) revealed that random variability in mean arterial blood pressure at the time of injury was a major predictor of long-term motor outcome, eclipsing the effect size of any drug-based therapeutic effects. In addition, this finding developed an unexpected direction: high blood pressure specifically predicted worse outcomes than low blood pressure. This was surprising because high blood pressure was previously unrecognized as a clinical predictor of outcome; indeed, most clinical focus has been on avoiding low blood pressure with vasopressors with little attention paid to the impact of high blood pressure.55 Yet, hypertension is a very robust predictor in multiple models of SCI58 and is now being examined in clinical studies. If these findings are confirmed in ongoing clinical studies, this will have immediate translational implications, because a number of anti-hypertensive drugs can offer new opportunities for precision medicine in SCI. This provides a strong argument for publishing long-tail data for reuse by outside researchers and data scientists to drive new discoveries.57
Other examples arguing for publishing long-tail data come from a similar retrospective effort in TBI and poly-neurotrauma models. Nielson and colleagues24 applied TDA machine intelligence to simultaneously re-assess the full set of multi-dimensional end-points in a prior study of controlled cortical impact (CCI) TBI versus contusive SCI versus polytrauma with both TBI and SCI.58 Machine learning revealed unexpected motor improvement when TBIs were ipsilateral to SCI, despite human intuition that this should cause bilateral impairment by impacting the corticospinal tract bilaterally at two different levels: damaging the right corticospinal tract at the level of the motor cortex and left corticospinal tract at the level of the spinal cord below the decussation of the pyramids.24 The functional improvement with bilateral injuries was shown to be of a very large effect and consistent when all end-points are considered in ensemble by the machine learning tool, although effects were subtle at individual end-points tested using older, less-sensitive statistical methods.58 Similar workflows have been extended to pooled clinical TBI data from the TRACK-TBI pilot and TBI Endpoints Development (TED) datasets.25,27
In a second example, Haefeli and associates59 pooled long-tail data from three separate pre-clinical trials of combinatorial therapeutics for TBI involving the anti-inflammatory drug minocycline, a neurotrophic drug acting on the p75 NTR system, and various types of rehabilitation therapy. Given the design complexity, a complete statistical analysis would have involved more than 300 analyses of variance for the 202 animals in the pooled dataset. For illustration purposes, Haefeli and associates ran all of these analyses and demonstrated that only 10% of the tested comparisons yielded statistical significance at a level that would survive statistical correction for multiple comparisons, suggesting that detection of significance is an improbable event considering all possible versions of the “truth” about therapeutic effects. Yet the unsupervised machine learning approach of non-linear principal component analysis (NLPCA) demonstrated a more nuanced “precision medicine” finding that the neurotrophic drug improved outcome but was undermined by certain forms of rehabilitation.
On the other hand, minocycline amplified the efficacy of the neurotrophin drug. Once the machine learning tool identified these effects, hypotheses could be generated and directly interrogated using hypothesis testing approaches. Haefeli and associates assessed scientific reproducibility empirically using a non-parametric cross-validation approach: external cross-validation across distinct experiments and internal cross-validation through 2000 iterations of balanced bootstrapping. The bootstrapping approach is a tool that depends on modern computers to assess reproducibility: the pooled population of subjects is randomly subsampled many different times with statistical analysis performed separately on each subsample.60 In the study by Haefeli and associates, these analyses revealed precise confidence intervals and effect sizes for therapeutic effects using the full set of long-tail data including both previously published61 and unpublished data, and demonstrated a reliable effect of neurotrophic agent therapy under certain rehabilitation conditions.
Together these early examples from SCI and TBI demonstrate the potential scientific value of long-tail and dark data and provide a rationale for publishing these data. In addition, they provide examples of general machine learning analytic pipelines (which should also be published online in programming development platforms such as GitHub) that can be used in both pre-clinical discovery and clinical data.25,27,62 Examples of such cross-species precision translation are beginning to be seen in the field of neurotrauma,63 demonstrating new opportunities for seamless integration of data across species within a single framework.
Incentives for publishing long-tail/dark data
The examples highlighted above involve data harmonization and curation efforts by dedicated data scientists working closely with the original data collectors to iteratively refine data curation and analysis. However, it is possible for such efforts to be less labor intensive if data stewardship for future dissemination is considered at the time of data collection. This, of course, requires that such stewardship be incentivized. Incentives for dissemination of long-tail and dark data include policy guidance, as well as a system of “carrots” and “sticks.”
The dominant, emerging policy for data stewardship was presented in a highly cited article by Wilkinson and co-workers64 that suggested all biomedical research data should be made Findable, Accessible, Interoperable, and Reusable (FAIR) (see box).
The FAIR data principles have been endorsed by major funders including the U.S. National Institutes of Health (NIH),65 the U.S. Veteran's Affairs Health System,66 non-profit groups such as the International Neuroinformatics Coordinating Facility (INCF67), and major journals.68 At the time of writing, these endorsements are framed in terms of encouraging researchers to adhere to FAIR stewardship. However, it is not a stretch to imagine that these will become mandates in coming years as the role of long-tail and dark data becomes better appreciated as major work products of the publicly funded biomedical research enterprise worldwide. Some funders, such as the Bill and Melinda Gates Foundation,69 are already enforcing data sharing under certain circumstances and have the ability to withhold funds unless data are made FAIR. This provides a clear example of sticks that are designed to incentivize sharing long-tail and dark data.
What about carrots? There are clear benefits of FAIR data sharing to donators, researchers, other investigators in the field, and the community at large (e.g., taxpayers). Other fields have issued a number of challenge initiatives to incentivize data sharing and collaboration, including the NIH Precision Medicine Initiative (now All of Us70), the Cancer Moonshot,71 the Sudden Unexpected Death in Epilepsy (SUDEP) Grand Challenge,72 among many others.
Data sharing increases transparency and reproducibility by allowing outside groups to corroborate findings using the same data with different analytic techniques. Data sharing also enables larger return on investment as reuse of the same dataset can leverage prior investments in research dollars and researcher data collection time. For example, the VISION-SCI database was developed using funding from a single NIH R01 grant ($1 million) and contains data from 26 prior grants including 16 from NIH. An NIH reporter query suggests that data collected from NIH alone involved a prior investment of more than $60 million in long-tail and dark data that were stored in inaccessible formats such as paper records and non-standardized spreadsheets.52 In other words, simply by making data FAIR, this work generated a 60-fold return on investment. In a similar manner, researchers can get a career boost simply by making their data FAIR and gaining citations for their data if a digital object identifier (DOI) is assigned.73 Established community repositories can serve as the issuers of such DOIs using international data citation standards,74 and can cross-index these DOIs with electronic libraries such as the California Digital Library75 and the Internet Archive.76 Future users of data will be able to give credit directly to data donors though DOI citation much like the current system of article citation, and data citations may benefit academic promotion in tenure decisions.
FAIR data sharing may also prevent researchers from wasting time on futile experiments by granting access to prior negative studies (that are “published” as a dataset), thus focusing taxpayer dollars more effectively. The wait to publicly release data from repositories following publication is lessened by automated search tools such as Wide-Open77 that recently triggered the public release of 400 overdue datasets, and emerging tools such as Google Dataset Search, among others. Finally, a major incentive with FAIR data sharing is that interoperable datasets can be pooled together to gain much higher sample sizes than can be achieved in a single laboratory, providing sensitivity to outcome patterns in larger datasets that may not appear in smaller, individual lab datasets. In addition, through the process of allowing their data to be pooled, individual laboratories may gain access to a wide community of data scientists who can help annotate their data, and add meta-data and new analysis pathways. These derivative work-products may then be added back to the original data as a form of enrichment, enabling new uses for data. This “crowd-sourcing” process has potential to create a “virtuous cycle” of open data sharing and analysis that leads to ever-increasing quality improvement and data value.78
The FAIR Data Principles As Applied to TBI Long-tail and Dark-Data
Findable: Long-tail and dark data should have a unique and consistent identifier such as a digital object identifier (DOI), similar to that of published papers.
Accessible: Once TBI data have been found, they can be accessed by both human scientists and machines such as computers running analytics, visualization, and indexing engines.
Interoperable: TBI data should contain well-defined formal annotations that enable data to be automatically harmonized with multiple software tools using widely understood language(s) and knowledge representations.
Reusable: TBI data should have well-developed user licensing rules and provide sufficient information to track data back to its source (provenance).
Disincentives for publishing long-tail/dark data and how do we overcome them?
Although the benefits and potential incentives for sharing long-tail and dark data are clear, it remains difficult to do so in the current scientific career ecosystem. It is worth examining some of the disincentives and barriers to disseminating data in an attempt to overcome them. First, data sharing is currently time-consuming, especially for older datasets that did not have data sharing in mind at the time of collection. Our own experiences with building the VISION-SCI repository from paper records suggest that this task is not insurmountable; however, managing legacy TBI data requires a unique combination of deep domain knowledge in both neurotrauma and data science. Currently, this is a rare combination of skills, limiting the potential workforce that can help with data “wrangling” from legacy data. As the science workforce becomes more populated with dedicated data science/biomedical science cross-training programs, such projects will become less cumbersome. Examples of these programs include the NIH Big-Data to Knowledge (BD2K) initiative, which has specialized award programs such as the BD2K RoAD-Trip (Data Science Rotations for Advancing Discovery), dedicated to data science bootcamp training for established biomedical researchers.79
A related disincentive is that data sharing can be costly, and may be considered an unfunded mandate, especially for traditional NIH grants that are dedicated to testing targeted hypotheses using collected data rather than curating and sharing data. Making data FAIR may take time and effort from new projects to devote to data curation of older projects. In some cases, this may not be technically legal to do in terms of effort reporting. The NIH rules do not explicitly prohibit designating the amount of effort in National Institute of Neurological Diseases and Stroke (NINDS) grants for data curation, but scientific reviewers (i.e., the neurotrauma community) would need to accept this practice during grant review process. As such, accepting data stewardship costs as part of grant review and funding decision may require explicit guidance for peer reviewers, and perhaps a change in culture about the importance of this funding designation.
Other cost-related issues include: Who pays for data storage? Who pays for database maintenance? These issues are commonly considered as operating costs in for-profit businesses; however, they are difficult to justify in grant reviews. Once a grant ends, it may become impossible to continue to fund data hosting and maintenance without a sustainable business model. This remains a largely unsolved problem in neurotrauma; however, business models for scientific journals may be repurposed to help support ongoing costs of making data FAIR. In the cancer field, federally funded clinical trial data are maintained in databases developed by consortia. Large multi-site neurotrauma groups such as TRACK-TBI (clinical TBI), Operation Brain Trauma Therapy (OBTT), the Moody Project (pre-clinical TBI), or the emerging Open Data Commons for SCI and Open Data Commons for TBI initiatives might be viable resources to support a sustainable data repository.50
For any business model to be feasible, data ownership and stewardship issues, as well as licensing agreements need to be solved. One very important question is who actually owns the data? In the United States, the Bayh-Dole Act automatically assigns intellectual property to universities, and faculty data collectors are considered stewards of the intellectual property.80 On the other hand, federal funders such as the NIH have mandated that NIH-funded data be released to the public after an embargo period, and this policy has been realized in the form of PubMed Central (PMC). A similar model could be extended to long-tail and dark data once data are made citable and FAIR. Such data release would be then covered under an open access publishing license such as the creative commons BY (CC-BY) licensing agreement. A related concern raised by some investigators is that an open access model does not allow researchers to approve data access, and some researchers have stated their fear of public misinterpretation of data or misuse by special interest groups.29,50,81 However, it is our opinion that these same issues exist in the current dissemination model for open access publications. It is less clear how making long-tail and dark data underlying these publications more accessible fundamentally increases risks beyond the existing system of publication followed by public scrutiny. It would seem that making source data more citable would only improve the self-correcting nature of scientific and public discourse.
A final set of disincentives relate to reputational concerns that competing researchers or malicious actors from the public will “weaponize” raw data to attack individuals who share their data. In some of the FAIR data workgroups,50,82,83 researchers have expressed their personal fears of backlash from competing researchers, special interests groups, and even anti-research terrorist groups84 using these raw data against them. This concern seems centered around the notion that long-tail and dark data may contain embarrassing secrets that would call into question the validity of the conclusions in associated published articles. Given the current reproducibility crisis, it is worth considering how the culture of data sharing can evolve such that researchers are rewarded for sharing data independent of the conclusions made from these data. Such credit attribution models currently exist in digital e-commerce market place (e.g., clicks, mouse-overs, and views result in ad revenues going to content providers) and e-commerce transactional tools provide examples of encryption-based digital security. Such models may be repurposed to credit attribution in academic data dissemination as well. The rise of individual citation metrics such as the h-index provides a glimpse into this type of attribution system.85
Big data options for TBI studies
At the time of writing, there are relatively limited big-data tools available to academic researchers and there is a strong need for plug-and-play tools that are easy to use and adaptable for a wide variety of research datasets. The NIH and U.S. Department of Defense (DoD) have jointly invested in the Federal Interagency Traumatic Brain Injury Research (FITBIR) informatics system, which provides secure access to clinical TBI datasets.3,86 Variability in data collection (and labeling of data fields) among investigators, labs, and TBI research subdomains in FITBIR are partly ameliorated by application of the TBI CDE project of the NIH's NINDS.87 The NINDS CDE workgroups defined a common vocabulary and set of protocols for clinical CDE data collection that should make data harmonization easier in the future. Use of the clinical CDEs is now a mandate for NIH- and DoD-funded clinical TBI studies and their use has enabled the development of harmonized multi-study datasets such as the TED meta-dataset88 and has helped facilitate regulatory pathway development of the first FDA-endorsed biomarkers for TBI.89,90 The CDE effort has been extended to pre-clinical TBI common data elements efforts that are currently underway.91 In theory, the FITBIR system has potential to create opportunities for FAIR data reuse.
However, whether there will be widespread reuse of these data by third-party researchers remains an open question. Adoption of such systems involves incorporating user-centered design principles that consider the workstyles of neurotrauma researchers instead of solely those of computer scientists and informaticians. To date, this has been hard to achieve using centralized development teams that are not integrated with the research community.
In contrast, there are some research community-driven efforts that provide alternative models for FAIR data sharing of long-tail and dark data. One model is OBTT, a collaborative research group whose goal is to screen and validate previously tested therapies in three animal models of TBI. To accomplish this, the members of OBTT created a scoring matrix to evaluate all tested therapies across the three testing sites. The scores allocated to the motor, cognitive, neuropathology, and serum biomarker categories were 4, 10, 4, and 4, respectively. However, the tasks and category “subscores” differed between the sites. For example, the Miami site subdivided the Morris water maze results into five different subscores, whereas the Pittsburgh site used two subscores.92
OBTT is composed of six sites: 1) The Safar Center for Resuscitation Research, University of Pittsburgh School of Medicine; 2) The Miami Project to Cure Paralysis, University of Miami School of Medicine; 3) The Neuroprotection Program at Walter Reed Army Institute of Research; 4) Virginia Commonwealth University; 5) Banyan Biomarkers, Inc.; and 6) The Center for Neuroproteomics and Biomarkers Research, University of Florida. Their data were sent to a central data store, masked, and discussed at monthly conference calls. Despite having “negative” findings for their first 4 out of 5 drugs tested, the OBTT group published articles for each drug as well as a synthesis article explaining the details of the design and workflow used. These works were published in a special issue of the Journal of Neurotrauma.93 The rationale for the study design and workflow choices of OBTT and OBTT-Extended Studies were a topic of discussion at the 2016 Moody Project TBI Symposium, held in Galveston, Texas. Discussion evolved into guidelines for pre-clinical therapy testing for TBI and were recently published,9 to share lessons learned with the neurotrauma community.
In addition to OBTT, the Moody Project group (at The University of Texas Medical Branch at Galveston, with collaborations at the University of California San Francisco, the University of Minnesota, and the University of Pennsylvania) also maintains a central database containing gene expression, proteomics, histopathology, behavior, surgical, and physiological outcome data before, during, and up to 1 year post-TBI (with and without drug, device, or stem-cell-based therapies; in three species of animal and using five different experimental models of TBI).
To complement these community-rooted efforts, several groups are focused on building scalable FAIR data-sharing infrastructure for neurotrauma long-tail and dark data. One example is our experience in building the VISION-SCI repository.52 We have partnered with the Neuroscience Information Framework (NIF)/SciCrunch group to develop an open data commons for SCI94 (http://odc-sci.org) and are developing similar infrastructure for TBI that enables community-driven data management, uploading, hosting, and citation as well as an application programming interface (API) that promotes interoperability. It is possible that this system architecture can be extended to include TBI, with proper support. The hope is that such systems will apply agile, user-centered design to help support sharing of long-tail and dark data from diverse research groups within the field.
Concluding Remarks and Overall Benefits of Data Sharing
Large, shared individual TBI datasets lend themselves to precision and targeted personalized medicine and can uncover previously unseen findings (such as the bimodal deleterious mean arterial pressure ranges) that could change clinical practices and improve the lives of TBI survivors.24,25,27,59,95 Additionally, once a populated centralized public database exists with pre-clinical and clinical TBI data, these results can be combined with outcome data from other neuro- and non-neuro-related databases to determine if TBI impacts other comorbidities, chronic diseases, aging, and immune responses to allergens and infectious diseases. Data from repositories have been used to create models and simulations in the fields of Alzheimer's disease,96 cardiovascular health,97 and to predict new drug targets and drug response biomarkers.98 In general, public databases can stimulate research by generating new hypotheses/areas of research,99 reducing the number of unnecessary repeated experiments, and lead to novel findings due to a larger sample size and access to more powerful statistical analyses that uncover previously unnoticed patterns. Barriers to easy and FAIR data sharing still exist,100–104 but with continued support for data repositories and increased interest in recognition for publication of all data (even long-tail, dark data, and “negative findings”), we are confident that the neurotrauma community can overcome these challenges.
Acknowledgments
We thank the Moody Project for Translational Traumatic Brain Injury Research Team for thoughtful discussions and sharing data. Supported in part by the Darrell K. Royal Research Fund for Alzheimer's Disease (BEH); Mission Connect, a program of the TIRR Foundation (BEH); Moody Project for Translational TBI Research (BEH); NIH BD2K TCC Data Science RoAD-Trip Fellowship (BEH); NIH/NINDS: UG3NS106899 (ARF), R01NS088475 (ARF); Department of Veterans Affairs: 1I01RX002245 (ARF), I01RX002787 (ARF); Wings for Life Foundation (ARF); and Craig H. Neilsen Foundation (ARF).
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Dewan M.C., Rattani A., Gupta S., Baticulon R.E., Hung Y.-C., Punchak M., Agrawal A., Adeleye A.O., Shrime M.G., Rubiano A.M., Rosenfeld J.V., and Park K.B. (2018). Estimating the global incidence of traumatic brain injury. J. Neurosurg. 84, 1–18 [DOI] [PubMed] [Google Scholar]
- 2. Roozenbeek B., Maas A.I.R., and Menon D.K. (2013). Changing patterns in the epidemiology of traumatic brain injury. Nat. Rev. Neurol. 9, 231–236 [DOI] [PubMed] [Google Scholar]
- 3. Yue J.K., Vassar M.J., Lingsma H.F., Cooper S.R., Okonkwo D.O., Valadka A.B., Gordon W.A., Maas A.I.R., Mukherjee P., Yuh E.L., Puccio A.M., Schnyer D.M., Manley G.T.; and TRACK-TBI Investigators. (2013). transforming research and clinical knowledge in traumatic brain injury pilot: multicenter implementation of the common data elements for traumatic brain injury. J. Neurotrauma 30, 1831–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maas A.I.R., Menon D.K., Adelson P.D., Andelic N., Bell M.J., Belli A., Bragge P., Brazinova A., Buki A., Chesnut R.M., Citerio G., Coburn M., Cooper D.J., Crowder A.T., Czeiter E., Czosnyka M., Diaz-Arrastia R., Dreier J.P., Duhaime A.-C., Ercole A., van Essen T.A., Feigin V.L., Gao G., Giacino J., Gonzalez-Lara L.E., Gruen R.L., Gupta D., Hartings J.A., Hill S., Jiang J.-Y., Ketharanathan N., Kompanje E.J.O., Lanyon L., Laureys S., Lecky F., Levin H., Lingsma H.F., Maegele M., Majdan M., Manley G., Marsteller J., Mascia L., McFadyen C., Mondello S., Newcombe V., Palotie A., Parizel P.M., Peul W., Piercy J., Polinder S., Puybasset L., Rasmussen T.E., Rossaint R., Smielewski P., Söderberg J., Stanworth S.J., Stein M.B., Steinbüchel, von N., Stewart W., Steyerberg E.W., Stocchetti N., Synnot A., Ao Te B., Tenovuo O., Theadom A., Tibboel D., Videtta W., Wang K.K.W., Williams W.H., Wilson L., Yaffe K.; and InTBIR Participants and Investigators. (2017). Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 16, 987–1048 [DOI] [PubMed] [Google Scholar]
- 5. Seabury S.A., Gaudette, É., Goldman D.P., Markowitz A.J., Brooks J., McCrea M.A., Okonkwo D.O., Manley G.T., and the TRACK-TBI Investigators, Adeoye O., Badjatia N., Boase K., Bodien Y., Bullock M.R., Chesnut R., Corrigan J.D., Crawford K., Diaz-Arrastia R., Dikmen S., Duhaime A.-C., Ellenbogen R., Feeser V.R., Ferguson A., Foreman B., Gardner R., Giacino J., Gonzalez L., Gopinath S., Gullapalli R., Hemphill J.C., Hotz G., Jain S., Korley F., Kramer J., Kreitzer N., Levin H., Lindsell C., Machamer J., Madden C., Martin A., McAllister T., Merchant R., Mukherjee P., Nelson L., Noel F., Palacios E., Perl D., Puccio A., Rabinowitz M., Robertson C., Rosand J., Sander A., Satris G., Schnyer D., Sherer M., Stein M., Taylor S., Temkin N., Toga A., Valadka A., Vassar M., Vespa P., Wang K., Yue J., Yuh E., and Zafonte R. (2018). Assessment of follow-up care after emergency department presentation for mild traumatic brain injury and concussion. JAMA Netw. Open 1, e180210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kochanek P.M., Dixon C.E., Mondello S., Wang K.K.K., Lafrenaye A., Bramlett H.M., Dietrich W.D., Hayes R.L., Shear D.A., Gilsdorf J.S., Catania M., Poloyac S.M., Empey P.E., Jackson T.C., and Povlishock J.T. (2018). Multi-center pre-clinical consortia to enhance translation of therapies and biomarkers for traumatic brain injury: Operation Brain Trauma Therapy and beyond. Front. Neurol. 9, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hawryluk G.W.J., and Bullock M.R. (2016). Past, present, and future of traumatic brain injury research. Neurosurg. Clin. North Am. 27, 375–396 [DOI] [PubMed] [Google Scholar]
- 8. Kochanek P.M., and Clark R.S.B. (2016). Traumatic brain injury research highlights in 2015. Lancet Neurol. 15, 13–15 [DOI] [PubMed] [Google Scholar]
- 9. DeWitt D.S., Hawkins B.E., Dixon C.E., Kochanek P.M., Armstead W., Bass C.R., Bramlett H.M., Buki A., Dietrich W.D., Ferguson A.R., Hall E.D., Hayes R.L., Hinds S.R., LaPlaca M.C., Long J.B., Meaney D.F., Mondello S., Noble-Haeusslein L.J., Poloyac S.M., Prough D.S., Robertson C.S., Saatman K.E., Shultz S.R., Shear D.A., Smith D.H., Valadka A.B., VandeVord P., and Zhang L. (2018). Pre-clinical testing of therapies for traumatic brain injury. J. Neurotrauma 35, 2737–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huie J.R., Almeida C.A., and Ferguson A.R. (2018). Neurotrauma as a big-data problem. Curr. Opin. Neurol. 31, 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laney D. 3D data management: controlling data volume, velocity and variety. 2001. https://blogs.gartner.com/doug-laney/files/2012/01/ad949-3D-Data-Management-Controlling-Data-Volume-Velocity-and-Variety.pdf (Last accessed March20, 2019)
- 12. [no date]. httpswww.elderresearch.comblog-v-of-big-data (Last accessed January, 23, 2019)
- 13. Sorani M.D., Hemphill J.C., Morabito D., Rosenthal G., and Manley G.T. (2007). New approaches to physiological informatics in neurocritical care. Neurocrit. Care 7, 45–52 [DOI] [PubMed] [Google Scholar]
- 14. Hemphill J.C., Andrews P., and De Georgia M. (2011). Multimodal monitoring and neurocritical care bioinformatics. Nat. Rev. Neurol. 7, 451–460 [DOI] [PubMed] [Google Scholar]
- 15. Yuh E.L., Mukherjee P., Lingsma H.F., Yue J.K., Ferguson A.R., Gordon W.A., Valadka A.B., Schnyer D.M., Okonkwo D.O., Maas A.I.R., Manley G.T.; and TRACK-TBI Investigators. (2013). Magnetic resonance imaging improves 3-month outcome prediction in mild traumatic brain injury. Ann. Neurol. 73, 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerson J., Castillo-Carranza D.L., Sengupta U., Bodani R., Prough D.S., DeWitt D.S., Hawkins B.E., and Kayed R. (2016). Tau oligomers derived from traumatic brain injury cause cognitive impairment and accelerate onset of pathology in Htau mice. J. Neurotrauma 33, 2034–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez U.A., Zeng Y., Deyo D., Parsley M.A., Hawkins B.E., Prough D.S., and DeWitt D.S. (2018). Effects of mild blast traumatic brain injury on cerebral vascular, histopathological, and behavioral outcomes in rats. J. Neurotrauma 35, 375–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boone D.K., Weisz H.A., Bi M., Falduto M.T., Torres K.E.O., Willey H.E., Volsko C.M., Kumar A.M., Micci M.-A., DeWitt D.S., Prough D.S., and Hellmich H.L. (2017). Evidence linking microRNA suppression of essential prosurvival genes with hippocampal cell death after traumatic brain injury. Sci. Rep. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sell S.L., Johnson K., DeWitt D.S., and Prough D.S. (2017). Persistent Behavioral Deficits in Rats after Parasagittal Fluid Percussion Injury. J. Neurotrauma 34, 1086–1096 [DOI] [PubMed] [Google Scholar]
- 20. Esenaliev R.O., Petrov I.Y., Petrov Y., Guptarak J., Boone D.R., Mocciaro E., Weisz H., Parsley M.A., Sell S.L., Hellmich H., Ford J.M., Pogue C., DeWitt D., Prough D.S., and Micci M.-A. (2018). Nano-pulsed laser therapy is neuroprotective in a rat model of blast-induced neurotrauma. J. Neurotrauma 35, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. [No authors cited]. (2017). Abstracts from the 35th Annual National Neurotrauma Symposium July 7–12, 2017 Snowbird, Utah. J. Neurotrauma 34, A1–A163 [DOI] [PubMed] [Google Scholar]
- 22. [no authors cited]. (2018). The 3rd Joint Symposium of the International and National Neurotrauma Societies and AANS/CNS Section on Neurotrauma and Critical Care August 11–16, 2018 Toronto, Canada. J. Neurotrauma 35, A2–A285 [Google Scholar]
- 23. Zakrasek E.C., Nielson J.L., Kosarchuk J.J., Crew J.D., Ferguson A.R., and McKenna S.L. (2017). Pulmonary outcomes following specialized respiratory management for acute cervical spinal cord injury: a retrospective analysis. Spinal Cord 55, 559–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nielson J.L., Paquette J., Liu A.W., Guandique C.F., Tovar C.A., Inoue T., Irvine K.-A., Gensel J.C., Kloke J., Petrossian T.C., Lum P.Y., Carlsson G.E., Manley G.T., Young W., Beattie M.S., Bresnahan J.C., and Ferguson A.R. (2015). Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury. Nat. Commun. 6, 8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nielson J.L., Cooper S.R., Yue J.K., Sorani M.D., Inoue T., Yuh E.L., Mukherjee P., Petrossian T.C., Paquette J., Lum P.Y., Carlsson G.E., Vassar M.J., Lingsma H.F., Gordon W.A., Valadka A.B., Okonkwo D.O., Manley G.T., Ferguson A.R.; and TRACK-TBI Investigators. (2017). Uncovering precision phenotype-biomarker associations in traumatic brain injury using topological data analysis. PLoS One 12, e0169490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beauparlant J., van den Brand R., Barraud Q., Friedli L., Musienko P., Dietz V., and Courtine G. (2013). Undirected compensatory plasticity contributes to neuronal dysfunction after severe spinal cord injury. Brain 136, 3347–3361 [DOI] [PubMed] [Google Scholar]
- 27. Huie J.R., Diaz-Arrastia R., Yue J.K., Sorani M.D., Puccio A.M., Okonkwo D.O., Manley G.T., Ferguson A.R.; and TRACK-TBI Investigators. (2019). testing a multivariate proteomic panel for traumatic brain injury biomarker discovery: a TRACK-TBI pilot study. J. Neurotrauma 36, 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haines C.J., and Read M.D. (1983). Characteristic fetal heart rate changes in severe rhesus isoimmunization. Aust. N. Z. J. Obstet. Gynaecol. 23, 114–116 [DOI] [PubMed] [Google Scholar]
- 29. Ferguson A.R., Nielson J.L., Cragin M.H., Bandrowski A.E., and Martone M.E. (2014). Big data from small data: data-sharing in the “long tail” of neuroscience. Nat. Neurosci. 17, 1142–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The Economist. (2017). The findings of medical research are disseminated too slowly. March 25, 2017, 1–6 [Google Scholar]
- 31. Oldenburg H. (1665). An introduction to this tract. Philosophical Transactions of the Royal Society of London 1, 1–2 [Google Scholar]
- 32. Hopewell S., McDonald S., Clarke M., and Egger M. (2007). Grey literature in meta-analyses of randomized trials of health care interventions. Cochrane Database Syst. Rev. 19, MR000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Macleod M.R., Michie S., Roberts I., Dirnagl U., Chalmers I., Ioannidis J.P.A., Salman R.A.-S., Chan A.-W., and Glasziou P. (2014). Biomedical research: increasing value, reducing waste. Lancet 383, 101–104 [DOI] [PubMed] [Google Scholar]
- 34. Chan A.-W., Song F., Vickers A., Jefferson T., Dickersin K., Gøtzsche P.C., Krumholz H.M., Ghersi D., and van der Worp H.B. (2014). Increasing value and reducing waste: addressing inaccessible research. Lancet 383, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ioannidis J.P.A. (2005). Why most published research findings are false. PLoS Med. 2, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ioannidis J.P.A. (2017). Acknowledging and overcoming nonreproducibility in basic and preclinical research. JAMA 317, 1019–1020 [DOI] [PubMed] [Google Scholar]
- 37. Fanelli D. (2010). Do pressures to publish increase scientists' bias? An empirical support from US States Data. PLoS One 5, e10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsilidis K.K., Panagiotou O.A., Sena E.S., Aretouli E., Evangelou E., Howells D.W., Salman R.A.-S., Macleod M.R., and Ioannidis J.P.A. (2013). Evaluation of excess significance bias in animal studies of neurological diseases. PLoS Biol. 11, e1001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sena E.S., van der Worp H.B., Bath P.M.W., Howells D.W., and Macleod M.R. (2010). Publication bias in reports of animal stroke studies leads to major overstatement of efficacy. PLoS Biol. 8, e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watzlawick R., Sena E.S., Dirnagl U., Brommer B., Kopp M.A., Macleod M.R., Howells D.W., and Schwab J.M. (2014). Effect and reporting bias of RhoA/ROCK-blockade intervention on locomotor recovery after spinal cord injury. JAMA Neurol. 71, 91. [DOI] [PubMed] [Google Scholar]
- 41. Holman C., Piper S.K., Grittner U., Diamantaras A.A., Kimmelman J., Siegerink B., and Dirnagl U. (2016). Where have all the rodents gone? The effects of attrition in experimental research on cancer and stroke. PLoS Biol. 14, e1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van der Worp H.B., Howells D.W., Sena E.S., Porritt M.J., Rewell S., O'Collins V., and Macleod M.R. (2010). Can animal models of disease reliably inform human studies? PLoS Med. 7, e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watzlawick R., Rind J., Sena E.S., Brommer B., Zhang T., Kopp M.A., Dirnagl U., Macleod M.R., Howells D.W., and Schwab J.M. (2016). Olfactory ensheathing cell transplantation in experimental spinal cord injury: effect size and reporting bias of 62 experimental treatments: a systematic review and meta-analysis. PLoS Biol. 14, e1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jackson M., Srivastava A., and Cox C. (2017). Preclinical progenitor cell therapy in traumatic brain injury: a meta-analysis. J. Surg. Res. 214, 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirst T.C., Watzlawick R., Rhodes J.K., Macleod M.R., and Andrews P.J.D. (2016). Study protocol: a systematic review and meta-analysis of hypothermia in experimental traumatic brain injury: why have promising animal studies not been replicated in pragmatic clinical trials? Evid. Based Preclin. Med. 3, e00020–e00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zwetsloot P.-P., Van Der Naald M., Sena E.S., Howells D.W., IntHout J., De Groot J.A., Chamuleau S.A., Macleod M.R., and Wever K.E. (2017). Standardized mean differences cause funnel plot distortion in publication bias assessments. eLife 6, e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Glasziou P., Altman D.G., Bossuyt P., Boutron I., Clarke M., Julious S., Michie S., Moher D., and Wager E. (2014). Reducing waste from incomplete or unusable reports of biomedical research. Lancet 383, 267–276 [DOI] [PubMed] [Google Scholar]
- 48. Mlinarić A., Horvat M., and Šupak Smolčić V. (2017). Dealing with the positive publication bias: why you should really publish your negative results. Biochemia Medica 27, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carroll H.A., Toumpakari Z., Johnson L., and Betts J.A. (2017). The perceived feasibility of methods to reduce publication bias. PLoS One 12, e0186472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Callahan A., Anderson K.D., Beattie M.S., Bixby J.L., Ferguson A.R., Fouad K., Jakeman L.B., Nielson J.L., Popovich P.G., Schwab J.M., Lemmon V.P.; and FAIR Share Workshop participants. (2017). Developing a data sharing community for spinal cord injury research. Exp. Neurol. 295, 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Callahan A., Abeyruwan S.W., Al-Ali H., Sakurai K., Ferguson A.R., Popovich P.G., Shah N.H., Visser U., Bixby J.L., and Lemmon V.P. (2016). RegenBase: a knowledge base of spinal cord injury biology for translational research. Database 2016, baw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nielson J.L., Guandique C.F., Liu A.W., Burke D.A., Lash A.T., Moseanko R., Hawbecker S., Strand S.C., Zdunowski S., Irvine K.-A., Brock J.H., Nout-Lomas Y.S., Gensel J.C., Anderson K.D., Segal M.R., Rosenzweig E.S., Magnuson D.S.K., Whittemore S.R., McTigue D.M., Popovich P.G., Rabchevsky A.G., Scheff S.W., Steward O., Courtine G., Edgerton V.R., Tuszynski M.H., Beattie M.S., Bresnahan J.C., and Ferguson A.R. (2014). Development of a database for translational spinal cord injury research. J. Neurotrauma 31, 1789–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Basso D.M., Beattie M.S., Bresnahan J.C., Anderson D.K., Faden A.I., Gruner J.A., Holford T.R., Hsu C.Y., Noble L.J., Nockels R., Perot P.L., Salzman S.K., and Young W. (1996). MASCIS Evaluation of Open Field Locomotor Scores: Effects of Experience and Teamwork on Reliability. Multicenter Animal Spinal Cord Injury Study. J. Neurotrauma 13, 343–359 [DOI] [PubMed] [Google Scholar]
- 54. Gensel J.C., Tovar C.A., Hamers F.P.T., Deibert R.J., Beattie M.S., and Bresnahan J.C. (2006). Behavioral and Histological Characterization of Unilateral Cervical Spinal Cord Contusion Injury in Rats. J. Neurotrauma 23, 36–54 [DOI] [PubMed] [Google Scholar]
- 55. Hawryluk G., Whetstone W., Saigal R., Ferguson A., Talbott J., Bresnahan J., Dhall S., Pan J., Beattie M., and Manley G. (2015). Mean Arterial Blood Pressure Correlates with Neurological Recovery after Human Spinal Cord Injury: Analysis of High Frequency Physiologic Data. J. Neurotrauma 32, 1958–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kepler C.K., Schroeder G.D., Martin N.D., Vaccaro A.R., Cohen M., and Weinstein M.S. (2015). The effect of preexisting hypertension on early neurologic results of patients with an acute spinal cord injury. Spinal Cord 53, 763–766 [DOI] [PubMed] [Google Scholar]
- 57. Neff E.P. (2018). Dark data see the light. Lab Animal 47, 45–48 [DOI] [PubMed] [Google Scholar]
- 58. Inoue T., Lin A., Ma X., McKenna S.L., Creasey G.H., Manley G.T., Ferguson A.R., Bresnahan J.C., and Beattie M.S. (2013). Combined SCI and TBI: recovery of forelimb function after unilateral cervical spinal cord injury (SCI) is retarded by contralateral traumatic brain injury (TBI), and ipsilateral TBI balances the effects of SCI on paw placement. Exp. Neurol. 248, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haefeli J., Ferguson A.R., Bingham D., Orr A., Won S.J., Lam T.I., Shi J., Hawley S., Liu J., Swanson R.A., and Massa S.M. (2017). A data-driven approach for evaluating multi-modal therapy in traumatic brain injury. Sci. Rep. 7, 42474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Efron B., and Gong G. (1983). A Leisurely Look at the Bootstrap, the Jackknife, and Cross-Validation. Am. Statistician 37, 36–48 [Google Scholar]
- 61. Lam T.I., Bingham D., Chang T.J., Lee C.C., Shi J., Wang D., Massa S., Swanson R.A., and Liu J. (2013). beneficial effects of minocycline and botulinum toxin-induced constraint physical therapy following experimental traumatic brain injury. Neurorehabil. Neural Repair 27, 889–899 [DOI] [PubMed] [Google Scholar]
- 62. Mabray M.C., Talbott J.F., Whetstone W.D., Dhall S.S., Phillips D.B., Pan J.Z., Manley G.T., Bresnahan J.C., Beattie M.S., Haefeli J., and Ferguson A.R. (2016). multidimensional analysis of magnetic resonance imaging predicts early impairment in thoracic and thoracolumbar spinal cord injury. J. Neurotrauma 33, 4093–40962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Friedli L., Rosenzweig E.S., Barraud Q., Schubert M., Dominici N., Awai L., Nielson J.L., Musienko P., Nout-Lomas Y., Zhong H., Zdunowski S., Roy R.R., Strand S.C., van den Brand R., Havton L.A., Beattie M.S., Bresnahan J.C., Bézard E., Bloch J., Edgerton V.R., Ferguson A.R., Curt A., Tuszynski M.H., and Courtine G. (2015). Pronounced species divergence in corticospinal tract reorganization and functional recovery after lateralized spinal cord injury favors primates. Sci. Transl. Med. 7, 302ra134–302ra134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., Bouwman J., Brookes A.J., Clark T., Crosas M., Dillo I., Dumon O., Edmunds S., Evelo C.T., Finkers R., Gonzalez-Beltran A., Gray A.J.G., Groth P., Goble C., Grethe J.S., Heringa J., 't Hoen P.A.C., Hooft R., Kuhn T., Kok R., Kok J., Lusher S.J., Martone M.E., Mons A., Packer A.L., Persson B., Rocca-Serra P., Roos M., van Schaik R., Sansone S.-A., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M.A., Thompson M., van der Lei J., van Mulligen E., Velterop J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., and Mons B. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. https://commonfund.nih.gov/sites/default/files/NIHStrategicPlanforDataScienceFinal.pdf (Last accessed January23, 2019)
- 66. https://sparcopen.org/news/2015/department-of-veterans-affairs-va-public-access-plan-to-use-pmc-platform-for-articles/ (Last accessed January23, 2019)
- 67. https://www.incf.org/activities/standards-and-best-practices/what-is-fair (Last accessed January23, 2019)
- 68. Announcement: FAIR data in Earth science. (2019). Nature 565, 134–134 [DOI] [PubMed]
- 69. https://www.gatesfoundation.org/How-We-Work/General-Information/Information-Sharing-Approach (Last accessed January24, 2019)
- 70. https://allofus.nih.gov (Last accessed January23, 2019) 2019
- 71. https://www.cancer.gov/researchkey-initiatives/moonshot-cancer-initiative (Last accessed January23, 2019)
- 72. https://www.epilepsy.com/living-epilepsy/our-programs/sudep-institute/sudep-challenge-initiative (Last accessed January23, 2019)
- 73. Bierer B.E., Crosas M., and Pierce H.H. (2017). Data authorship as an incentive to data sharing. N. Engl. J. Med. 376, 1684–1687 [DOI] [PubMed] [Google Scholar]
- 74. http://www.datacite.org (Last accessed January23, 2019)
- 75. http://www.cdlib.org (Last accessed January23, 2019)
- 76. http://archive.org (Last accessed January23, 2019)
- 77. Grechkin M., Poon H., and Howe B. (2017). Wide-Open: accelerating public data release by automating detection of overdue datasets. PLoS Biol. 15, e2002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ferguson A.R., Stück E.D., and Nielson J.L. (2011). Syndromics: a bioinformatics approach for neurotrauma research. Transl. Stroke Res. 2, 438–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jagodnik K.M., Koplev S., Jenkins S.L., Ohno-Machado L., Paten B., Schurer S.C., Dumontier M., Verborgh R., Bui A., Ping P., McKenna N.J., Madduri R., Pillai A., and Ma'ayan A. (2017). Developing a framework for digital objects in the Big Data to Knowledge (BD2K) commons: report from the Commons Framework Pilots Workshop. J. Biomed. Informatics 71, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Markel H. (2013). Patents, profits, and the American people: the Bayh-Dole Act of 1980. N. Engl. J. Med. 369, 794–796 [DOI] [PubMed] [Google Scholar]
- 81. Johnson G. (2009). A vet's view of animal research. httpspenntoday.upenn.edu--researchvets-view-animal-research (Last accessed January24, 2019)
- 82. (2016). Spinal Cord Injury Preclinical Data Workshop: developing a FAIR share community. httpsmeetings.ninds.nih.govHomeIndex (Last accessed January23, 2019)
- 83. FAIR data, metadata, and data sharing in neurotrauma. httpsneuronline.sfn.orgArticlesProfessional-DevelopmentFAIR-Data-Metadata-and-Data-Sharing-in-Neurotrauma (Last accessed January23, 2019)
- 84. Kordower J.H. (2009). Animal rights terrorists: what every neuroscientist should know. J. Neurosci. 29, 11419–11420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hirsch J.E. (2007). Does the h index have predictive power? Proc. Natl. Acad. Sci. 104, 19193–19198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thompson H.J., Vavilala M.S., and Rivara F.P. (2015). Common data elements and federal interagency traumatic brain injury research informatics system for TBI research. Annu. Rev. Nurs. Res. 33, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Saatman K.E., Duhaime A.-C., Bullock R., Maas A.I.R., Valadka A., Manley G.T.; and Workshop Scientific Team and Advisory Panel Members. (2008). Classification of traumatic brain injury for targeted therapies. J. Neurotrauma 25, 719–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Manley G.T., Mac Donald C.L., Markowitz A., Stephenson D., Robbins A., Gardner R.C., Winkler E.A., Bodien Y., Taylor S.., Yu K., Kannan L., Kumar A., MaCrea M., and Wang K.K.W. (2017). The Traumatic Brain Injury Endpoints Development (TED) Initiative: Progress on a public-private regulatory collaboration to accelerate diagnosis and treatment of traumatic brain injury. J. Neurotrauma 34, 2721–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. https://www.fda.gov/downloads/Drugs/DevelopmentApprovalProcess/UCM550255.pdf (Last cited January 23, 2019)
- 90. https://www.fda.gov/downloads/Drugs/DevelopmentApprovalProcess/DrugDevelopmentToolsQualificationProgram/BiomarkerQualificationProgram/UCM605365.pdf (Last cited January 23, 2019)
- 91. Smith D.H., Hicks R.R., Johnson V.E., Bergstrom D.A., Cummings D.M., Noble L.J., Hovda D., Whalen M., Ahlers S.T., LaPlaca M., Tortella F.C., Duhaime A.-C., and Dixon C.E. (2015). Pre-clinical traumatic brain injury common data elements: toward a common language across laboratories. J. Neurotrauma 32, 1725–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kochanek P.M., Bramlett H.M., Dixon C.E., Shear D.A., Dietrich W.D., Schmid K.E., Mondello S., Wang K.K.W., Hayes R.L., Povlishock J.T., and Tortella F.C. (2016). approach to modeling, therapy evaluation, drug selection, and biomarker assessments for a multicenter pre-clinical drug screening consortium for acute therapies in severe traumatic brain injury: operation brain trauma therapy. J. Neurotrauma 33, 513–522 [DOI] [PubMed] [Google Scholar]
- 93. Rasmussen T.E., and Crowder A.T. (2016). Synergy in science and resources. J. Neurotrauma 33, 511–512 [DOI] [PubMed] [Google Scholar]
- 94. http://odc-sci.org (Last accessed January23, 2019)
- 95. Butte A.J. (2017). Big data opens a window onto wellness. Nat. Biotechnol. 35, 720–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Haas M., Stephenson D., Romero K., Gordon M.F., Zach N., and Geerts H. (2016). Big data to smart data in Alzheimer's disease: Real-world examples of advanced modeling and simulation. Alzheimer's Dement. 12, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 97. Hemingway H., Asselbergs F.W., Danesh J., Dobson R., Maniadakis N., Maggioni A., van Thiel G.J.M., Cronin M., Brobert G., Vardas P., Anker S.D., Grobbee D.E., Denaxas S.; and Innovative Medicines Initiative 2nd programme, Big Data for Better Outcomes, BigData@Heart Consortium of 20 academic and industry partners including ESC. (2018). Big data from electronic health records for early and late translational cardiovascular research: challenges and potential. Eur. Heart J. 39, 1481–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen B., and Butte A.J. (2016). Leveraging big data to transform target selection and drug discovery. Clin. Pharmacol. Ther. 99, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schönberger V.M., and Ingelsson E. (2018). Big Data and medicine: a big deal? J. Intern. Med. 283, 418–429 [DOI] [PubMed] [Google Scholar]
- 100. Figueiredo A.S. (2017). Data sharing: convert challenges into opportunities. Front. Public Health 5, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Longo D.L., and Drazen J.M. (2016). Data sharing. N. Engl. J. Med. 374, 276–277 [DOI] [PubMed] [Google Scholar]
- 102. Adam N.R., Wieder R., and Ghosh D. (2017). Data science, learning, and applications to biomedical and health sciences. Ann. N. Y. Acad. Sci. 1387, 5–11 [DOI] [PubMed] [Google Scholar]
- 103. Khachaturian A.S., Meranus D.H., Kukull W.A., and Khachaturian Z.S. (2013). Big data, aging, and dementia: pathways for international harmonization on data sharing. Alzheimer's Dement. 9, S61–S62 [DOI] [PubMed] [Google Scholar]
- 104. Payakachat N., Tilford J.M., and Ungar W.J. (2015). National Database for Autism Research (NDAR): big data opportunities for health services research and health technology assessment. Pharmacoeconomics 34, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]