Abstract
Objective: It has been demonstrated that the transcription factors TAZ (transcriptional coactivator with PDZ-binding motif), paired box gene 8 (PAX8), and NK2 homeobox 1 (NKX2-1) are coexpressed in the nucleus of thyroid cells. Furthermore, TAZ is known to enhance the transcriptional activity of PAX8 and NKX2-1 as well as the key thyroid-specific gene, thyroglobulin (TG), suggesting a critical role for TAZ in the control of thyroid cell speciation. We previously reported that the small molecule ethacridine, identified as a TAZ activator, was able to induce thyroid-specific transcription in endodermal cells differentiated from human embryonic stem (hES) cells using activin A. Since transcription factors are epigenetically regulated in cell differentiation, we investigated the epigenetic changes in the promoter regions of these key transcription factors during in vitro differentiation of hES cells into thyrocytes.
Methods: We initially profiled chromatin accessibility using the technique of Assay for Transposase Accessible Chromatin sequencing (ATAC-seq), and then examined DNA methylation and histone acetylation in the promoter regions of the three selected thyroid transcription factors and the thyroid-specific genes during hES cell differentiation.
Results: ATAC-seq analysis showed enriched chromatin accessibility of TAZ, NKX2-1, and PAX8 after exposure to activin A and ethacridine. There were no methylation changes found in the NKX2-1, PAX8, and TAZ promoters by bisulfite sequencing. In contrast, acetylation of histone H4, specifically acetylation of lysine 16, was observed in each of the promoters when measured by chromatin immunoprecipitation polymerase chain reaction assays, which correlated with the activity and expression of NKX2-1 and PAX8 as well as sodium/iodide symporter, thyroid stimulating hormone receptor, and TG genes.
Conclusions: These results indicate that ethacridine treatment of activin A-derived endodermal hES cells leads to enhanced chromatin accessibility, which, in turn, allows histone H4 acetylation in the regulation of active genes for speciation of thyroid follicular cells from hES cells.
Keywords: epigenetic modification, TAZ, NKX2-1, PAX8, thyroid, human embryonic stem cells, acetyl histone H4, acetylation of histone H4 at lysine 16, ATAC-seq
Introduction
With the advent of pluripotent stem cells, including embryonic and induced pluripotent stem cells, we and others have aimed to recapitulate thyroid cell differentiation (1–4). Direct thyroid reprogramming of pluripotent stem cells into thyroid epithelial cells has been successfully achieved using transfection and overexpression of key transcription factors, or by the direct molecular induction of the same transcription factors (1–4). The most studied transcription factors include TAZ (transcriptional coactivator with PDZ-binding motif), which itself is a coactivator of the well-known transcription factors called paired box gene 8 (PAX8) and NK2 homeobox 1 (NKX2-1), and which play a central role in thyroid-specific gene transcription. These three proteins, when coexpressed in the nucleus of differentiated thyroid cells, lead to the enhancement of thyroid gene expression (5). We found that ethacridine, identified as a TAZ activator, was able to induce thyroid-specific transcription in endodermal cells differentiated from human embryonic stem (hES) cells (2,6). However, even with these manipulations, the conversion rate has remained relatively low (<50%). One factor for this is that the reprogrammed cells appear at various stages of speciation, thus suggesting the existence of a series of molecular barriers, which are spatiotemporally regulated. One such well-recognized barrier is the accessibility of the specific chromatin regions for transcription factors to enable them to bind to and maintain stable cell fate changes during the period of differentiation.
Cell reprogramming is mostly an epigenetic process leading to modifications to cytosine bases in the DNA and the histones that package the genome (7,8). Thus, the epigenetic status of a cell implies its chromatin structure and DNA accessibility, which in turn influences gene expression. Cell fate changes such as those that occur during normal development involve dynamic repatterning of the epigenetic landscape. The process of reprogramming somatic cells to pluripotency and the epigenetic changes during that process have been extensively characterized (9). Hence, elucidating the epigenetic dynamics during thyroid cell differentiation in vitro is critical to improve the efficiency and quality of thyroid cell speciation and plasticity during thyroid cell fate determination.
One of the ways epigenetic modifications, such as acetylation and DNA methylation, work is by restricting or promoting the accessibility of DNA transcription factors to the gene and thus regulating their expression. Chromatin accessibility can be studied using the Assay for Transposase Accessible Chromatin sequencing (ATAC-seq), which is a sensitive tool to detect accessible DNA regions over the entire genome (10). Hence, analysis of transcription factor DNA sequence motifs in regions that display altered accessibility will indicate the transcription factor families involved in the regulation of gene expression. We now know that many cell types harbor an epigenetic signature that defines its identity (11), and recent advances in profiling epigenome modifications have resulted in the identification of many such modifications during development. Interestingly, after each cell division, cells may or may not retain the epigenetic pattern similar to the parental cell (12) and upon differentiation, the epigenetic signatures of daughter cells may be altered according to the new cell identity and the environment in which they are cultured and propagated.
In this study, we have begun to examine some of the epigenetic changes in thyroid cells by looking at the chromatin changes in the promoter regions of important transcription factors during thyroid differentiation of hES cells. We focused on the specific regulatory changes on the thyroid transcription factors TAZ, NKX2-1, and PAX8 by examining their DNA methylation status and acetylation of histone H4 (AcH4), especially acetylation of lysine 16 (H4K16ac), in the minimal promoter regions of these key transcription factors.
Materials and Methods
Cell culture and treatment
The hES cells (H9) were maintained in feeder-free culture conditions with mTeSR medium (Stemcell Technologies, Vancouver, BC) on six-well plates coated with Matrigel (BD Biosciences, Franklin Lakes, NJ). The culture medium was changed daily, and the cells were passaged every 4–5 days at ratios of 1:3 to 1:6. Cells were cultured in a humidified chamber in a 5% CO2-air mixture at 37°C. After hES cells were passaged into culture dishes for two days, hES cells were washed once with phosphate-buffered saline (PBS) and cultured in RPMI 1640 medium (Life Technologies Corporation, Grand Island, NY) containing 1% B27 (Life Technologies) supplemented with 100 ng/mL of activin A (R&D Systems, Minneapolis, MN) for four days. Subsequently, the supplements were changed to 5 μM ethacridine (Sigma, St. Louis, MO) for one day to enhance the transcriptional activity of PAX8 and NKX2-1. After validating the enhancement of gene expression by real-time polymerase chain reaction (PCR), the cells were further exposed to thyrotropin (1 mIU/mL) for the induction of differentiation up to 21 days. Cells were harvested for analysis at different time points.
ATAC-sequencing
ATAC-seq was performed according to Activemotif Inc. (10). A total of 5000 cells were resuspended in cold PBS according to the ATAC-seq protocol. Chromatin was extracted and processed for Tn5-mediated tagmentation and adapter incorporation, according to the manufacturer's protocol (Nextera DNA Sample Preparation Kit; Illumina®, San Diego, CA) at 37°C for 30 minutes. Reduced-cycle amplification was carried out in the presence of compatible indexed sequencing adapters. The quality of the libraries was assessed by a DNA-based fluorometric assay (Thermo Fisher Scientific, Waltham, MA) and automated capillary electrophoresis (Agilent Technologies, Inc., Santa Clara, CA). Up to three samples per lane were pooled and run on a HiSeq2500 Illumina sequencer with a paired-end read of 50 bp. The following steps were followed in analyzing the ATAC seq data: (i) The paired-end 42 bp sequencing reads generated by Illumina sequencing were used for “Peak Finding.” The peak caller protocol used was MACS2 as reported (13), which used both reads from each aligned fragment. The generic term “Interval” was used to describe genomic regions with local enrichments in tag numbers. (ii) To identify the density of transposition events along the genome, the genome was divided into 32 bp bins, and the number of fragments in each bin was determined. “Interval” or “Merged Region” intensity values were calculated based on the number of fragments within the 32 bp bins. For normalization of the data, the tag number of all samples was reduced (by random sampling) to the number of tags present in the smallest sample. (iii) Then “Merged Region” analysis was carried out by comparing peak metrics between two or more samples, overlapping intervals were grouped into “Merged Regions,” which are defined by the start coordinate of the most upstream interval and the end coordinate of the most downstream interval. In locations where only one sample had an Interval, this Interval defined the Merged Region. (iv) After defining the Intervals and Merged Regions, their genomic locations along with their proximities to gene annotations and other genomic features were determined. In addition, average and peak fragment densities within intervals and merged regions were compiled. Further, DESseq2 analysis of these raw data was carried out to report fold changes or log 2 ratios with adjusted p-values.
Quantification of TAZ protein by immunofluorescence
TAZ protein in ethacridine-treated cells was quantitated by measuring fluorescence intensity (FI) in the nuclear and cytoplasmic compartments of stained cells (2,14). Acquired immunofluorescent images of TAZ were loaded into Adobe Photoshop for analysis in which green intensity stained using Alexa 488 was quantified for FI. FI was calculated using corrected total cell fluorescence (CCTF) as described here: CCTF = Integrated intensity − (Area of selected cell × Mean fluorescence of background readings). At least three positive images were counted in each group, and mean and standard deviation (SD) were plotted as indicated in the graph.
RNA isolation and reverse transcription PCR
Total RNA was extracted from cultured cells using the RNeasy Mini Kit Isolation System (Qiagen Ltd., Manchester, UK), which includes a digestion step with DNase I. RNA quantity and quality were assessed by ultraviolet spectrophotometry. Complementary DNA synthesis was performed using the SuperScript III First-Strand Synthesis System (Invitrogen Corp., Carlsbad, CA). The real-time quantitative reverse transcription PCR was carried out using predesigned quantitative PCR (qPCR) assays (intercalating dye-based assays; Integrated DNA Technologies) and employing the QuantStudio 3 real-time PCR system (Applied Biosystems, Foster City, CA). Relative expression levels of each gene in real time were analyzed using the 2−ΔΔCT method and normalized to the expression of the housekeeping gene GAPDH. Data presented (M – standard error of the mean [SEM]) are obtained from three independent experiments in which all sample sets were analyzed in triplicate.
Promoter regions
In this study, promoters of NKX2-1, PAX8, and TAZ were defined to be regions around the annotated transcription start site ±1 kbp available at www.genecopoeia.com/product/search/view_seq_promoter.php?cid
Bisulfite sequencing PCR
For CpG island analysis, genomic DNA was extracted from the treated cells at different stages of differentiation using DNeasy Blood and Tissue Kit from QIAGEN. The DNA was then bisulfite converted (∼500 ng/reaction) using the EpiJET Bisulfite Conversion Kit from Thermo Scientific. Two to 4 μL of the 10 μL bisulfite-converted DNA was PCR amplified for the promoter regions of indicated genes using primers designed by online software MethPrimer. The PCR cleaned up and TA-cloned into pGEMT-easy vector (Promega, Madison, WI). Twelve positive clones in each sample were randomly picked, and their DNA sequenced using T7/M13 universal primers. Finally, the sequences were analyzed using open access software QUMA for CpG islands.
Chromatin immunoprecipitation and qPCR
Chromatin immunoprecipitation (ChIP) assays were performed (using the Acetyl-Histone H4 Immunoprecipitation [CHIP] Assay Kit and ChIPAb+ Acetyl-Histone H3 [Lys27]—ChIP Validated Antibody and Primer Set from Millipore Corp) according to the manufacturer's instructions. In brief, a total of 1 × 106 cells were crosslinked with 1% formaldehyde in culture medium for 10 minutes at 37°C. After terminating the crosslinking reaction, the cells were washed with ice-cold PBS containing 0.1 mmol/L phenylmethylsulfonyl fluoride and 1 μg/μL Leupeptin and resuspended in 200 μL sodium dodecyl sulfate (SDS) lysis buffer. The cells were sonicated (Diagenode Bio-ruptor, Denville, NJ) to shear DNA into 500 bp fragments, and the sonicated samples were separated by centrifugation at 13,000g for 10 minutes at 4°C. Supernatant fractions were diluted 10 × with ChIP dilution buffer. A portion of diluted cell supernatant (1%, or ∼20 μL) was kept for use as a control “input DNA” in the subsequent qPCR analysis. The remaining portion was precipitated by incubation with antibodies against acetyl-histone H4 or Acetyl-Histone H4 (Lys16), or Acetyl-Histone H3 (Lys27), or immunoglobulin G (IgG) control at 4°C overnight. Protein A/G–agarose beads were added, followed by incubation at 4°C for 2 hours. The mixture was washed once with low-salt immune complex wash buffer, once with high-salt immune complex wash buffer, once with LiCl immune complex wash buffer, and twice with Tris-EDTA buffer, and subsequently eluted in elution buffer (1% SDS, 0.1 mol/L NaHCO3). The crosslinking of the DNA complexes was reversed by the addition of 200 mmol/L NaCl and incubation at 65°C for 4 hours. DNA was subsequently treated with 2 μL proteinase K (10 μg/μL) at 45°C for 1 hours, collected using the PCR Purification Kit (QIAGEN) and subjected to real-time PCR. This technique employed a preclearing step of sonicated crosslinked DNA, which reduces nonspecific binding before the immunoprecipitation by addition of anti-H4IgG. In addition, the protocol used a no-antibody immunoprecipitation control, and each experiment included inactivated DNA samples.
The qPCR primers of promoter regions of interested genes were designed using an algorithm from Integrated DNA technologies, and three pairs of primers covering the promoter regions were selected as shown in Supplementary Table S1. qPCR was performed for the target region of the indicated gene promoter with SYBR® Green PCR Master Mix (Applied Biosystems) on QuantStudio 3 real-time PCR system (Applied Biosystems) per manufacturer protocols. The ΔΔCT method was then used for the data analysis, in which the value of target DNA fragments that were enriched in each sample was normalized to the value of the 5% input DNA of each sample. The qPCR data were expressed as the means ± SDs. The ChIP assays combined with subsequent quantitative PCR (ChIP-qPCR) assay were repeated twice to confirm the reproducibility of the results.
Data analysis
All values are expressed as mean ± SEM. All samples were analyzed by Student's t-test or analysis of variance as described. A p-value of <0.05 was considered significant.
Results
Effect of ethacridine on TAZ translocation and gene expression
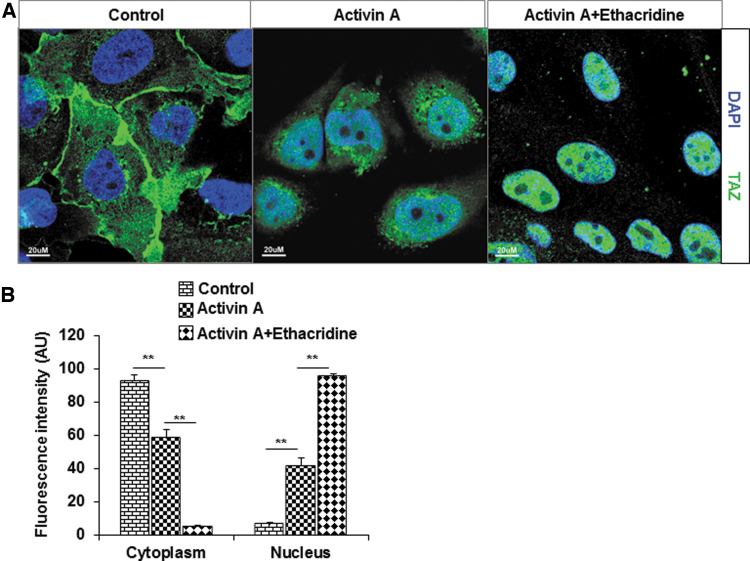
As previously published (2), we observed enhanced expression of NKX2-1, PAX8, and TAZ after ethacridine treatment of activin A-derived hES cells (data not shown). Since the effect of TAZ on the activity of transcription factors is thought to be exerted by its translocation into the nucleus, we studied the effect of ethacridine on TAZ translocation in hES cells by immunofluorescence imaging (Fig. 1A). Quantitative analysis of immunofluorescent images showed that in control cells, the TAZ was largely localized in the cytoplasm. Activin A treatment showed minimal translocation of TAZ into the nucleus but on treatment with 5 μM of ethacridine with activin A, a large degree of TAZ translocation into the nucleus was observed (Fig. 1A, B). These observations showed that ethacridine has the ability to enhance translocation of TAZ into the nucleus.
FIG. 1.
Translocation of TAZ in hES cells. (A) The nuclear translocation of TAZ on treatment with 5 μM of ethacridine was detected by immunostaining using a specific anti-TAZ antibody. Left panel: In hES cells, TAZ is predominantly localized in the cytoplasm. Middle panel: In activin A-treated cells, TAZ translocated minimally to the nucleus. Right panel: In activin A + ethacridine-treated cells, most TAZ is translocated to the nucleus. (B) The relative abundances of TAZ in cells in cytoplasm and nucleus were quantified from set of 10 cells per image and expressed as arbitrary units of FI. Data are means ± SEM of three independent experiments (**p < 0.01). AU, authorized unite; DAPI, 4′, 6-diamidino-2-phenylindole; FI, fluorescence intensity; hES, human embryonic stem; SEM, standard error of the mean; TAZ, transcriptional coactivator with PDZ-binding motif.
Chromatin modifications during differentiation of hES cells into thyroid cells
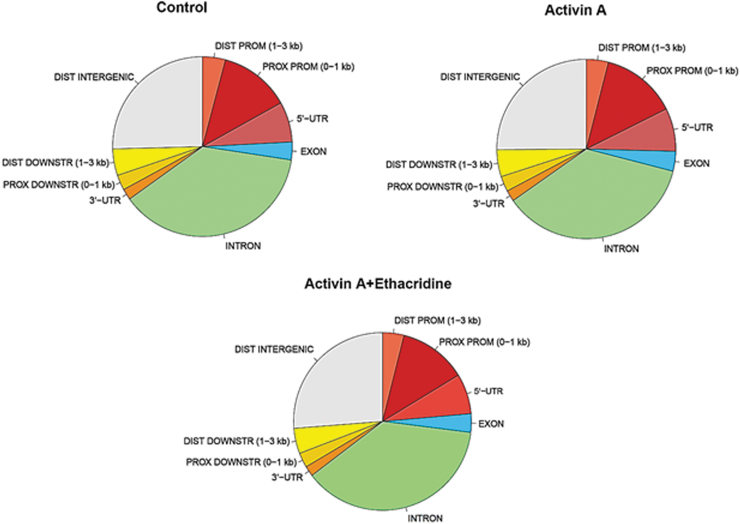
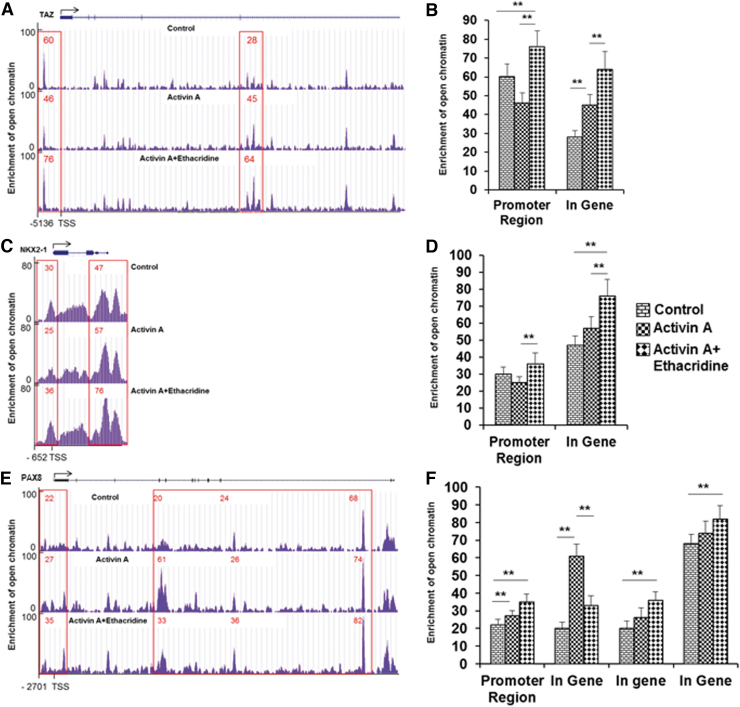
The epigenetic landscape in thyroid cell differentiation from hES cells was determined by ATAC-seq. To examine these changes, the open chromatin regions (or peaks) were identified across all tested samples and performed in duplicate. Comparison of total chromatin changes by this method in untreated, and activin A- and ethacridine-treated hES cells showed broadly similar profiles of chromatin accessibility (Fig. 2; Supplementary Table S2), suggesting that the influence of these treatments on chromatin accessibility may be subtle and likely to be specific to particular promoter regions rather than throughout the genome. However, we did find categories of peaks that were specifically related to activin A or ethacridine exposure while directing thyroid differentiation from hES cells. By differential analysis, there was a total of 505 open regions, of which 461 were more open in the activin A group and 44 more open in the ethacridine-treated group. Some of the enriched chromatin accessibility regions were either highly specific to one of the treated samples or shared among the different groups. Figure 3 illustrates some of these changes in the TAZ, NKX2-1, and PAX8 gene promoters and, arbitrarily selected, intragene exonic regions. The open chromatin peaks in the TAZ (WWTR1) promoter and gene regions were more enriched in ethacridine-treated hES cells in comparison with activin A-treated and untreated hES cells, respectively (Fig. 3A, 3B). The same pattern was seen in the NKX2-1 regions (Fig. 3C, D) and PAX8 regions (Fig. 3E, F). There were clear intragene changes induced by activin A and by ethacridine in all three genes examined. The activin A-induced changes were less significant than those seen with ethacridine, which suggested that activin A regulated gene expression during thyroid cell differentiation by involving additional factors rather than just chromatin accessibility. However, the changes at the promoter regions were significant, suggesting physiologic consequences for gene activity of these transcription factors (see summary in Table 1).
FIG. 2.
The landscape of open chromatin regions (or peaks) across all tested samples. The total open chromatin regions (or peaks) were identified across all tested samples and performed in duplicate. Comparison of three samples of control, activin A, or activin A + ethacridine-treated hES cells showed broadly similar profiles of chromatin accessibility. DIST DOWNSTR, distal downstream; DIST PROM, distal promoter; PROX DOWNSTR, proximal downstream; PROX PROM, proximal promoter; UTR, untranslated region.
FIG. 3.
Changes of chromatin accessibility in regulatory regions of TAZ (A, B), NKX2-1 (C, D), and PAX8 (E, F). (A, C, E) The average ATAC-seq signals of TAZ, NKX2-1, and PAX8 for each category. (B, D, F). The graph transformed ATAC-seq signal for all samples. (A, B) The open chromatin peaks in the TAZ (WWTR1) promoter and gene regions were enriched in ethacridine-treated hES cells in comparison with activin A-treated and untreated control hES cells, respectively. (C, D) The open chromatin peaks in the NKX2-1 promoter and gene regions were enriched in ethacridine-treated hES cells in comparison with activin A-treated and untreated control hES cells, respectively. (E, F) The open chromatin peaks in the PAX8 promoter and gene regions were enriched in ethacridine-treated hES cells in comparison with activin A-treated and untreated control hES cells, respectively. **p < 0.01. ATAC-seq, Assay for Transposase Accessible Chromatin sequencing; NKX2-1, NK2 homeobox 1; PAX8, paired box gene 8; TSS, transcriptional start site.
Table 1.
Summary of Epigenetic Changes of Genes
| ATAC-seq |
H4K16 |
|||||
|---|---|---|---|---|---|---|
| Activin A |
Activin A + ethacridine |
Activin A | Activin A + ethacridine | |||
| Promoter | In-gene | Promoter | In-gene | |||
| TAZ | ± | ++ | ++ | ++++ | ++ | ++ |
| NKX2-1 | ± | + | ++ | +++ | +++ | ++++ |
| PAX8 | + | ++ | + | ++ | ++ | ++++ |
| NIS | ± | + | ± | ++ | ++ | +++ |
| TSHR | ± | + | ± | ++ | ++ | +++ |
| TG | ± | + | ± | ++ | ++ | +++ |
Notes: Compared with untreated control.
ATAC-seq, Assay for Transposase Accessible Chromatin sequencing; H4K16, acetylation of lysine 16; NIS, sodium/iodide symporter; NKX2-1, NK2 homeobox 1; PAX8, paired box gene 8; TAZ, transcriptional coactivator with PDZ-binding motif; TG, thyroglobulin; TSHR, thyroid stimulating hormone receptor.
Examining the thyroid-specific data, we found few peaks that were specifically related to activin A or ethacridine exposure. Sodium/iodide symporter (NIS), thyroid stimulating hormone receptor (TSHR), and thyroglobulin (TG) showed only intragene changes (Supplementary Fig. S1A–C). This similarly indicated that additional factors must also be at work inducing thyroid gene expression and not simply chromatin accessibility.
DNA methylation changes during differentiation of hES cells into thyroid cells
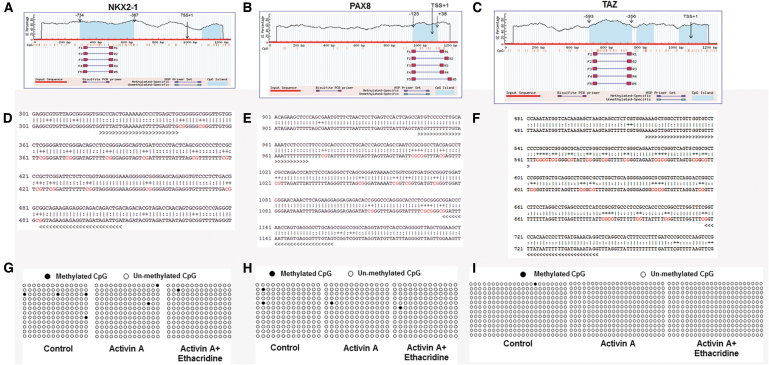
To determine if the changes in gene expression were also influenced by changes in DNA methylation, we examined methylation of the promoter regions of the two key transcription factors, NKX2-1 and PAX8, and coactivator TAZ, by examining the CpG islands within the 1Kb region of the promoter. We used the online software MethPrimer to predict CpG islands located in the core promoters of the NKX2-1 and PAX8 genes (Fig. 4A–C). DNA methylation patterns of the CpG sites at the core promoter of NKX2-1 and PAX8 were determined using bisulfite-assisted sequencing of the different treatment groups. In total, 14 CpG sites for NKX2-1, 13 CpG sites for PAX8, and 23 CpG sites for TAZ (Fig. 4D–F) were predicted in the promoter regions. The DNA methylation percentages of the 14 CpG sites in NKX2-1, 13 CpG sites in PAX8, and 23 CpG sites in TAZ were then assessed using QUMA software. As shown in Figure 4G–I, the DNA methylation level changes were insignificant between the different treatments (untreated hES cells, activin A-treated and activin A plus ethacridine-treated hES cells) resulting in an average 2.3%, −1.2%, and −0.59%, respectively, for NKX2-1; 1.2%, −0.6%, and −0.6%, respectively, for PAX8; and no methylation of the TAZ promoter. Thus, the DNA methylation status of the core promoters of NKX2-1, PAX8, and TAZ did not appear to play a major role in thyroid cell differentiation from hES cells. We were not able to find CpG islands in the TG promoter region and very few in NIS and TSHR promoters.
FIG. 4.
Methylation analysis of NKX2-1, PAX8, and TAZ (A–C). Blue background indicates the GC percentage, and dashed lines show the core promoter region. The x-axis denotes the bp position in the 5′-UTR relative to TSS. MSP denotes the methylated-specific primers for bisulfite-assisted sequencing (D, E, and F); the sequence of the core promoter of the gene. The methylation loci are marked in red letters (G, H, and I). The percentages of the CpG sites were analyzed using QUMA software. Each line represents one individual bacterial clone, and each circle one single CpG dinucleotide. Open circles and black circles show unmethylated and methylated CpGs, respectively. MSP, methylated-specific primer.
AcH4 around the NKX2-1 and PAX8 promoters
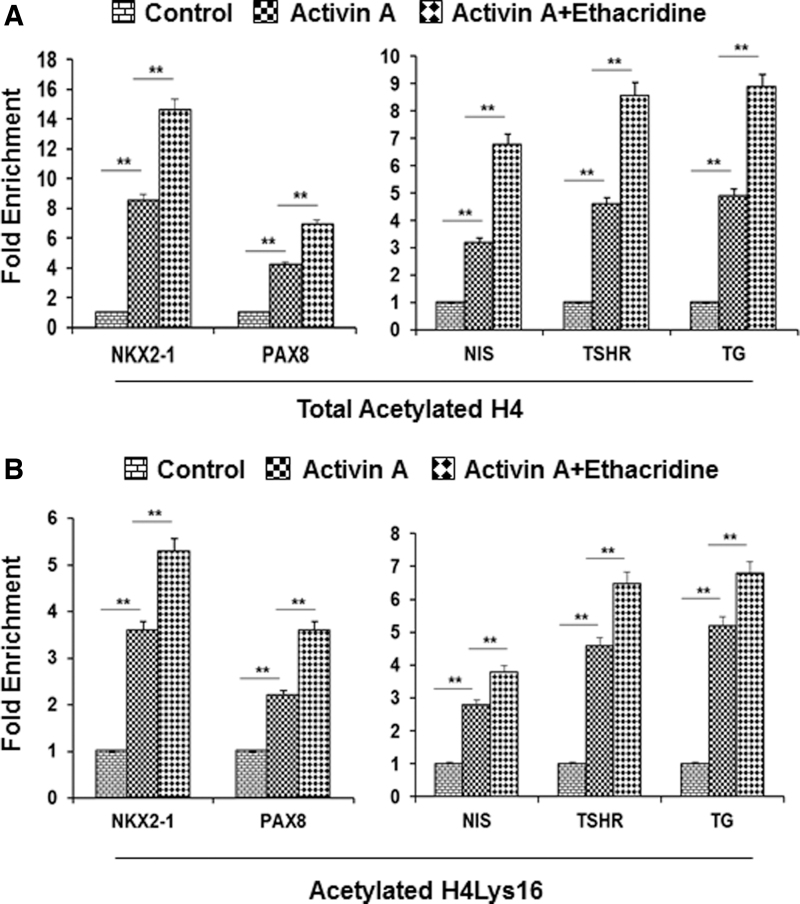
To correlate promoter accessibility with total histone H4 acetylation related to TAZ, NKX2-1, and PAX8 expression, ChIP-qPCR in their promoter regions were carried out in the three different groups of cells. Primers were chosen in the promoter regions of TAZ (Supplementary Fig. S2), NKX2-1 (Supplementary Fig. S3, upper panel), and PAX8 (Supplementary Fig. S3, lower panel). We found that in comparison with the untreated control hES cells, the expression of acetylated histone H4 was increased by ∼7-fold for NKX2-1 and 4-fold for PAX8 in activin A-treated cells, and increased by ∼16-fold for NKX2-1 and 5-fold for PAX8 in activin A plus ethacridine-treated cells (Fig. 5A, left panel). The acetylation changes in these genes' regions correlated with the enriched chromatin accessibility in ATAC-seq (Supplementary Fig. S4), suggesting acetylation as a possible cause of increased chromatin accessibility (see summary in Table 1). In contrast, activin A with ethacridine translocated TAZ to the nucleus without increasing H4 acetylation (Supplementary Fig. S2).
FIG. 5.
Acetylation of histones in the promoter region of thyroid transcription factors and thyroid-specific genes after activin A and ethacridine enrichment: (A) Acetylation of histone H4 in the promoter region of the thyroid transcription factors: NKX2-1 and PAX8 (left panel) and the thyroid-specific genes: NIS, TSHR, and TG (right panel) (B). Acetylation of histone H4 Lys16 (H4K1Ac) in the promoter region of thyroid transcription factors: NKX2-1 and PAX8 (left panel) and the thyroid-specific genes: NIS, TSHR, and TG (right panel). After cell fixation and harvesting, chromatin DNA was isolated, sheared by sonication, and immunoprecipitated with monoclonal antibody against acetylated histone H4. The resulting enriched genomic DNA was purified and used in ChIP-qPCR assays. PCR primers were designed to cover the promoter region of thyroid transcription factors: NKX2-1 and PAX8 and the thyroid-specific gene: NIS, TSHR, and TG. Data are expressed as mean ± SEM and represent one of three separate experiments. **p < 0.01 by analysis of variance two-way analysis. ChIP-qPCR, ChIP assays combined with subsequent quantitative PCR; NIS, sodium/iodide symporter; PCR, polymerase chain reaction; TG, thyroglobulin; TSHR, thyroid stimulating hormone receptor.
Surprisingly, despite the lack of changes seen in the chromatin studies, enrichment of acetylated histone H4 for thyroid-specific genes TG, TSHR, and NIS was observed in both the activin A and activin A plus ethacridine groups (Fig. 5A, right).
AcH4 on lysine 16 and H3K27 around the NKX2-1 and PAX8 promoters
H4K16Ac is particularly interesting because this is the most acetylatable site on the H4 N-terminal tail and is said to influence the formation of a compact higher order chromatin structure. H4K16 is catalyzed by specific histone lysine acetyltransferases (15) and has been demonstrated to have a critical role in regulating transcription (16). Since H4K16ac-dependent transcriptional regulation is likely to be a cell-type-specific process, we used ChIP-qPCR in the promoters of the genes of interest in our three different groups of cells. We found that in comparison with the untreated control hES cells, the enrichment of H4K16Ac was increased in the NKX2-1 and PAX8 promoters in activin A and ethacridine-treated cells (Fig. 5B, left panel). Consistent with the H4 enrichment in NIS, TG, and TSHR promoter regions, we also found that H4K16Ac enriched these gene promoters (Fig. 5B, right panel). These results again suggested that H4K16Ac regulation of chromatin accessibility is induced by activin A and ethacridine while directing hES cell differentiation into thyroid cells. However, there was no influence on the TAZ gene, which appears to be mobilized by nuclear translocation as described earlier (Supplementary Fig. S2). However, histone H3K27 acetylation on the three thyroid transcription factors in the different groups of human ES cells showed no changes (Supplement Fig. S5).
Discussion
Since epigenetics is a critical and reversible change in the chromatin state, we examined its role in thyroid cell differentiation by assessing two key epigenetic changes involved in the regulation of thyroid gene expression in human thyroid cells differentiated from hES cells. We characterized epigenetic modifications specifically in the thyroid transcriptional factors: TAZ, NKX2-1, and PAX8 and the thyroid-specific genes: TG, TSHR, and NIS during thyroid cell differentiation by first examining chromatin accessibility using ATAC-Seq followed by examination of the methylation and histone acetylation status of their promoter regions.
We first used ATAC-seq with our in vitro model of thyroid cell differentiation to define the activin A and/or ethacridine regulatory changes on the transcription factors that are most involved in thyroid cell speciation. We identified regions in the promoters and intragene regions of TAZ, NKX2-1, and PAX8 that were more accessible after exposure to activin A and/or ethacridine, thus providing a contributing mechanism for control of hES cell fate into thyrocytes. However, it is important to note that chromatin accessibility does not always correlate with transcriptional status, because in addition to merely gaining access to the DNA there remains histone modifications and methylation status of DNA that profoundly influence transcription initiation and output. Thus, it is essential that chromatin accessibility be integrated with other epigenetic datasets particularly at promoters and with transcriptional activity of a given gene.
Hence, the integration of changes in acetylated histone H4 with the chromatin accessibility data provided an even more complete picture of epigenetic regulation of thyroid cell differentiation. For example, the enrichment of acetylated histone H4 in NKX2-1 and PAX8 showed a positive correlation with promoter accessibility shown by ATAC-seq. There were also various degrees of enrichment of acetylated histone H4 on TG, TSHR, and NIS. Four lysine residues in the NH2-terminal tail domain of histone H4 are subject to reversible acetylation (positions 5, 8, 12, and 16). H4 Lys16 acetylation is unique since it plays a vital role in the maintenance of chromatin structure, and their hyperacetylation would lead to the unfolding of the nucleosomal fiber (17) and contribute to transcriptional upregulation (18). Here, we found that in comparison with untreated hES cells, the enrichment of H4K16Ac was increased in NKX2-1 and PAX8 by activin A, and further enhanced by ethacridine and consistent with the H4K16Ac enrichment seen in the NIS, TG, and TSHR promoter regions. These data indicated that H4K16Ac may play a major role during the early stages of thyroid cell differentiation. In contrast to the marked changes in acetylated H4, we did not observe any significant change in DNA methylation occurring in the treated cells during thyroid cell differentiation.
Although we did not find any significant changes in the methylation status in promoter regions, the lack of methylation changes does not necessarily suggest that these regions could not be influenced by methylation changes in upstream and downstream regions from the promoters. However, these were not examined in this study because of the focus on the promoter regions and the importance of PAX8 and NKX2-1 binding to the promoter sites. In contrast, we found that H4K16 acetylation levels correlated with gene expression and with chromatin accessibility in a subtle but highly specific manner. Therefore, in conclusion, we can say that both activin A and ethacridine can induce subtle changes in chromatin accessibility, especially in the promoter region of key transcriptional factors NKX2-1 and PAX8, and that TAZ, by its ability to translocate to the nucleus, has a major influence on such changes. Furthermore, promoter region acetylation changes in H4K16 appear to be a marker in the transcriptional control of thyroid speciation rather than methylation changes.
Supplementary Material
Authors' Contributions
R.M. designed and performed the experiments, analyzed the data, and wrote the article; S.M. designed experiments; R.L. designed experiments, data analysis, and article writing; and T.F.D. designed experiments, performed data analysis, and article editing.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported in part by NIH grant DK069713, a VA Merit Award BX000800 (to T.F.D.) and the Segal Family Endowment.
Supplementary Material
References
- 1. Kurmann AA, Serra M, Hawkins F, Rankin SA, Mori M, Astapova I, Ullas S, Lin S, Bilodeau M, Rossant J, Jean JC, Ikonomou L, Deterding RR, Shannon JM, Zorn AM, Hollenberg AN, Kotton DN. 2015. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell 17:527–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma R, Morshed SA, Latif R, Davies TF. 2017. TAZ induction directs differentiation of thyroid follicular cells from human embryonic stem cells. Thyroid 27:292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antonica F, Kasprzyk DF, Opitz R, Iacovino M, Liao XH, Dumitrescu AM, Refetoff S, Peremans K, Manto M, Kyba M, Costagliola S. 2012. Generation of functional thyroid from embryonic stem cells. Nature 491:66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma R, Morshed SA, Latif R, Davies TF. 2015. Thyroid cell differentiation from murine induced pluripotent stem cells. Front Endocrinol (Lausanne) 6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Di Palma T, D'Andrea B, Liguori GL, Liguoro A, de Cristofaro T, Del Prete D, Pappalardo A, Mascia A, Zannini M. 2009. TAZ is a coactivator for Pax8 and TTF-1, two transcription factors involved in thyroid differentiation. Exp Cell Res 315:162–175 [DOI] [PubMed] [Google Scholar]
- 6. Kawano S, Maruyama J, Nagashima S, Inami K, Qiu W, Iwasa H, Nakagawa K, Ishigami-Yuasa M, Kagechika H, Nishina H, Hata Y. 2015. A cell-based screening for TAZ activators identifies ethacridine, a widely used antiseptic and abortifacient, as a compound that promotes dephosphorylation of TAZ and inhibits adipogenesis in C3H10T1/2 cells. J Biochem 158:413–423 [DOI] [PubMed] [Google Scholar]
- 7. Theunissen TW, Jaenisch R. 2014. Molecular control of induced pluripotency. Cell Stem Cell 14:720–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernstein BE, Meissner A, Lander ES. 2007. The mammalian epigenome. Cell 128:669–681 [DOI] [PubMed] [Google Scholar]
- 9. Hernandez C, Wang Z, Ramazanov B, Tang Y, Mehta S, Dambrot C, Lee YW, Tessema K, Kumar I, Astudillo M, Neubert TA, Guo S, Ivanova NB. 2018. Dppa2/4 facilitate epigenetic remodeling during reprogramming to pluripotency. Cell Stem Cell 23:396–411 e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Surani MA, Hayashi K, Hajkova P. 2007. Genetic and epigenetic regulators of pluripotency. Cell 128:747–762 [DOI] [PubMed] [Google Scholar]
- 12. Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12:7–18 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morshed A, Dutta P. 2018. Mathematical model for tissue-level hypoxic response in microfluidic environment. J Biomech Eng 140:011009-1-011009-10. [DOI] [PubMed] [Google Scholar]
- 15. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 16. Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. 2012. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 11:163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shia WJ, Pattenden SG, Workman JL. 2006. Histone H4 lysine 16 acetylation breaks the genome's silence. Genome Biol 7:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. 2009. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 16:825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.