Abstract
The recognition of ACE2 by the receptor-binding domain (RBD) of spike protein mediates host cell entry. The objective of the work is to identify SARS-CoV2 spike variants that emerged during the pandemic and evaluate their binding affinity with ACE2. Evolutionary analysis of 2178 SARS-CoV2 genomes identifies RBD variants that are under selection bias. The binding efficacy of these RBD variants to the ACE2 has been analyzed by using protein-protein docking and binding free energy calculations. Pan-proteomic analysis reveals 113 mutations among them 33 are parsimonious. Evolutionary analysis reveals five RBD variants A348T, V367F, G476S, V483A, and S494P are under strong positive selection bias. Variations at these sites alter the ACE2 binding affinity. A348T, G476S, and V483A variants display reduced affinity to ACE2 in comparison to the Wuhan SARS-CoV2 spike protein. While the V367F and S494P population variants display a higher binding affinity towards human ACE2. Reorientation of several crucial residues at the RBD-ACE2 interface facilitates additional hydrogen bond formation for the V367F variant which enhances the binding energy during ACE2 recognition. On the other hand, the enhanced binding affinity of S494P is attributed to strong interfacial complementarity between the RBD and ACE2.
Keywords: Spike glycoprotein, ACE2, Population variants, Selection, Binding free energy
1. Introduction
The recent emergence of a novel coronavirus imposes a gigantic impact on our society, the public health care system, and the economy. Among several viral proteins, the spike glycoprotein (S) enables virus entry into the host cells [1,2]. Receptor recognition in coronaviruses is complex and yet to be resolved completely [3]. The spike protein sequences among coronaviruses belong to different genera are diverse. However, SARS-CoV2 (betacoronavirus) and HCoV-NL63 (alphacoronavirus) both recognize human angiotensin-converting enzyme 2 (ACE2) [4]. On the other hand, MERS-CoV recognizes dipeptidyl peptidase 4 (DPP4) [5], although it belongs to betacoronavirus. Murine hepatitis virus (MHV), another betacoronavirus mediates cell entry using the carcinoembryonic antigen cell adhesion molecules 1 (CEACAM 1) [6]. Gammacoronaviruse IBV recognizes the sialic acid receptor [7]. Coronaviruses achieve this host receptor recognition diversity through genetic mutation and recombination events in their S gene. This diversity in receptor recognition also results in tissue and cellular tropism. The presence of a particular receptor and its expression are critical determinants of cellular tropism in CoVs. While accessory genes of SARS-CoV2 play a crucial role in host tropism, spike glycoprotein appears to be the critical determinant.
Spike glycoprotein protrudes from the viral envelop as a clove-shaped homo-trimer [8]. Each monomer consists of two segments, S1 and S2. The S1 domains from the three trimers entangle to form the ectodomain of viral spike while the S2 domains of the trimer associate to form the spike stalk, transmembrane, and small intracellular domains [9]. The subdomain that recognizes the host cell surface receptor, the receptor-binding domain (RBD), exists within the S1 [10]. During virus entry, the RBD binds to the peptidase domain (PD) of the ACE2 receptor which opens another cleavage site on S2. The cleavage by host proteases mediates the fusion of the viral membrane to the host membrane. The RBD comprises a core domain and an extension known as the receptor-binding motif (RBM). The RBM directly binds to the viral binding motif (VBM) in the PD of ACE2 [11]. Structural comparisons of the RBD-ACE2 complex of SARS-CoV and SARS-CoV2 reveal a highly similar binding mechanism. However, there are few variations at the RBM that drastically alter the binding affinity of RBD to the ACE2 in the case of SARS-CoV and SARS-CoV2. Real-time surface plasmon resonance (SPR) assays revealed that the equilibrium dissociation constants (KD) of SARS-CoV2 S1 domain binding to human ACE2 is 94.6 ± 6.5 nM, while that for SARS-RBD is 408.7 ± 11.1 nM [12]. Alterations of few residues at the RBD lead to a ∼4 fold increase in binding affinity in SARS-CoV2 which might account for its high rate of infectivity due to effective receptor recognition mechanism. RNA viruses are highly mutation prone and during the progression of the pandemic, the virus encountered diverse host genetic diversity and different cellular microenvironments which may lead to diverse selection pressure in different viral strains. Thus it is very crucial to explore the genetic diversity of the S gene across the world from the large genomic pool to identify the population variants of the SARS-CoV2 and evaluates its effect on host receptor binding. Here, we adopt a pan-genomic approach to analyze SARS-CoV2 genomes around the world to identify population variants of spike protein and evaluates their binding efficiency with the human ACE2 receptor using protein-protein docking, free energy calculations, and molecular dynamics simulation.
2. Materials and method
Full-length SARS-CoV2 spike protein sequences were obtained from the ViPR database. Sequences with missing regions and unresolved amino acids were discarded. Final 2178 manually curated spike protein sequences were then clustered using the CD-HIT program [13]. The clustering resulted in 114 spike protein clusters. Sequences were aligned using the MAFFT program and analyzed for several composition analysis using MAGA-X 10.1 [14]. The best-suited amino acid substitution model identification was carried out by performing the Maximum Likelihood fits on 56 different models and the Jones-Taylor-Thornton (JTT) model with discrete Gamma distributed (+G) rate (shape parameter = 0.47) was found to be the best substitution model, judged by the lowest Bayesian Information Criterion and Akaike Information Criterion score. The conservedness of the SARS-CoV2 spike protein was evaluated by using the CONSURF web-server [15]. Evolutionary distances between sequences and substitution rate analysis were carried out using the MEGA-X [14]. Corresponding gene sequences of 114 spike variants were obtained from the NCBI nucleotide database and aligned by using the MUSCLE alignment tool. Site-wise selection pressure analysis was performed with the SLAC (Single Likelihood Ancestor Counting) method using the Datamonkey web-server (https://www.datamonkey.org/).
The crystal structure of the SARS-CoV2 RBD bound to the PD domain of the human ACE2 complex was considered (PDB ID: 6M0J) [16]. Five RBD mutants were considered in the study (S494P, V483A, G476S, A348T, and V367F). Both wild-type and mutant complexes were refined using the GalaxyWeb [17]. The binding free energies of wild-type RBD and its five population variants to the human ACE2 receptor were calculated by using the MM/GBSA method implemented in the HawkDock server [18]. The RBD-ACE2 complexes for wild-type, V367F, and S494P, obtained from the protein-protein docking refinement, were then studied by molecular dynamics simulation using the AMBER-ILDN force field [19] with the aid of GROMACS 2018.1 package [20]. All the complexes were solvated in a triclinic box containing TIP3P water. The dimension was chosen in such a way that all the protein atoms were at least 10 Å apart from the box edges. An appropriate number of counter ions were added to neutralize the charge of the system. Then, 500 steps of energy minimization were performed using the steepest descent algorithm, followed by 10 ns of position restrained dynamics and 2 ns NVT simulation. Finally, 100 ns of NPT simulation were carried out for each system. The temperature was maintained by coupling to a Nosé–Hoover thermostat with a time constant for coupling set to 0.1 ps. The constant pressure of 1 bar was maintained by using the isotropic Parrinello–Rahman barostat with the time constant for coupling set to 2 ps.
3. Result and discussion
3.1. Analysis of spike variants
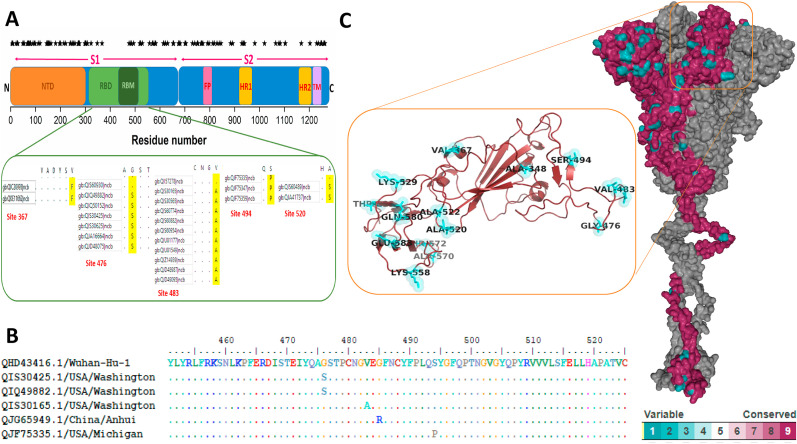
The alignment of 2178 spike protein sequences identifies 113 variable sites among the total 1273 residues. Among them, 33 are parsimonious sites, i. e, at least two sequence variations have been observed in two different strains. All the variations are probed on the entire spike sequence and shown in Fig. 1 A. The density of variations is high on the N-terminal domain and around the S1/S2 cleavage site. In comparison, the receptor-binding domain (RBD) is more conserved and the numbers of variations are less. We have explored the presence of parsimonious sites in the RBD since these sites represent positively selected non-synonymous mutations. Fig. 1A reveals five parsimonious sites in the RBD. V367F variation is observed in two viral strains obtained from Hong Kong and the USA. G476S variants are widespread and found in seven viral strains reported from the Washington State, USA. The most common RBD variant is V483A which was reported in different states in the USA. Another variant, S494P, is observed in several SARS-CoV2 strains from Michigan, USA. The A520S variant was reported in two viral strains from Washington, USA.
Fig. 1.
(A) Distribution of mutational sites over the entire spike protein. (B) RBM alignment of SARS-CoV2 variants. (C) Conserved and variable regions on the monomeric spike protein in the prefusion trimeric complex are shown. Detailed identification of variable residues within the RBD domain is highlighted within the square.
RBD consists of a core region and a flexible extension, known as RBM which directly interacts with the peptidase domain of ACE2. The RBM alignment of spike RBD variants is shown in Fig. 1B. We have used CONSURF analysis to probe the conserved and variable regions on the monomeric spike protein in the prefusion trimeric complex based on the SARS-CoV2 spike protein sequence alignment and the result is shown in Fig. 1C. Evident from the figure, the N-terminal domain of the S1 region contains more variable sites. However, there are significant discrete patches of the variable regions on the RBD which are directly involved in host ACE2 protein recognition (Shown in the inset). Detailed identification of variable residues within the RBD domain is highlighted within the square (Fig. 1C).
Comparison of selection sites and alignment reveals three RBM population variants under selection bias, G476S, V483A, and S494P that are in close contact with the ACE2 binding surface. Besides, we have considered two other sites that are under positive selection bias, 348th, and 367th site. Although these two sites are not at the RBM, the sites are positioned within the loop region that connects the core region of the RBD to the RBM region. Sequence alignment reveals the population variants for those sites are A348T and V367F.
3.2. Analysis of evolutionary rate and selection bias
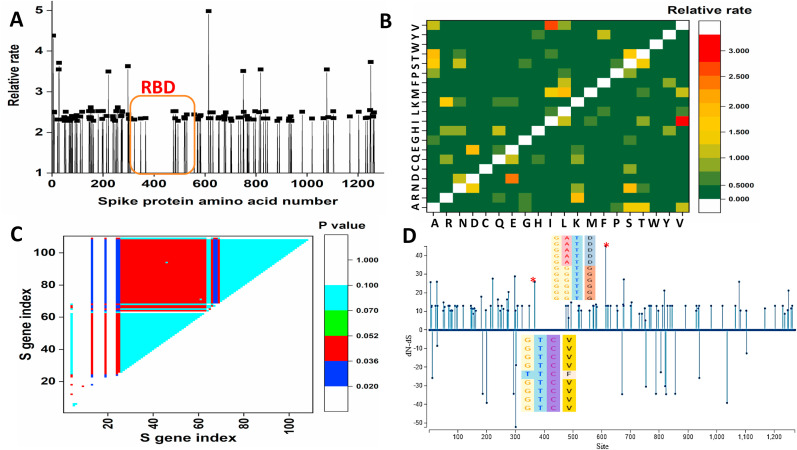
Representative spike protein sequences from each of the 114 clusters have been aligned by using the program MAFFT. The Jones-Taylor-Thornton (JTT) model with non-uniformity of evolutionary rates among sites modeled by using a discrete Gamma distribution with a shape parameter of 0.47 which is the most suitable substitution model, based on maximum parsimony fits, is used to estimate the rate at each site and the result is shown in Fig. 2 A. The relative rate of amino acid substitution is shown in Fig. 2B. The NTD of the S1 domain of the spike displays more sites with a higher relative rate. Leu5 displays a high relative rate and is often substituted by phenylalanine in many variants. Ala26 also displays a high mutational probability and often changes to either valine or serine at the site. Notably, alanine displays a very high mutational probability among all the 20 amino acids in spike proteins of SARS-CoV2 which is possibly dictated by the compositional preferences of spike proteins. Ser221 is mutated to tryptophan and leucine in two spike variants. Glu298 at the interface of the NTD and RBD displays a higher rate of mutation and substituted by lysine and glycine, two very different amino acids. RBD, on the other hand, is very less prone to mutation. Only residue 483 displays a marginally high evolutionary rate. The highest rate has been observed at Asp614 where it is frequently mutated to glycine. In the S2 domain, Ser750 shows a high evolutionary rate and is substituted to arginine and isoleucine in two strains isolated from Anhui province, China. Ile818 is also highly mutation prone and substituted to valine and serine in two strains isolated from the USA and China, respectively. Notably, in the entire dataset, isoleucine to valine substitution is most frequent, displays the highest rate of substitution (Fig. 2B). Ala1078 also displays a high rate of substitution and is often replaced with serine and valine. Close to the C-terminal end, Cys1250 is substituted with tyrosine and phenylalanine in two viral sequences. However, this unusual substitution is rare in the entire dataset (Fig. 2B).
Fig. 2.
(A) Distribution of relative rate on each residue of SARS-CoV2 spike protein (B) The relative rate value for the substitution of an amino acid to another one calculated from the maximum likelihood analysis. (C) Identification of the SARS-CoV2 S gene under positive selection bias by using the codon-based test. The probability of rejecting the null hypothesis of strict-neutrality in favor of the alternative hypothesis for positive selection is shown in different colors. (D) Selection site analysis of SARS-CoV2 spike protein under the positive selection bias. Corresponding alignment at the particular codon position (indicated by ∗) under high positive selection bias is shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
A codon-based test has been used to infer selection pressure in the S gene using the Nei-Gojobori method. Spike variants that display significant positive selection bias are shown in Fig. 2C. Evident from the figure, the spike gene of five viral strains (Three from India and two from the USA) are under strong positive selection pressure. Further, SLAC (Single Likelihood Ancestor Counting) method has been used to infer the ancestral state using the maximum likelihood method and also elucidates the selection pressure, purifying/positive, at each site by measuring dS, dN, and difference of the values at each site.
The number of sites under selection bias is shown in Fig. 2D. Numbers of positively selected sites are distributed throughout the gene. The highest selection pressure has been observed at the 614th site where aspartic acid to glycine variations are commonly observed in the population. This nonsynonymous substitution is due to a change in the 2nd codon from A to G. Another significant non-synonymous mutation has been observed in the RBD region at 367th position due to the substitution from G to T at the 1st codon position. This substitution leads to an amino acid change from valine to phenylalanine.
3.3. Effect of RBD mutations on ACE2 recognition: structural and binding free energy analysis
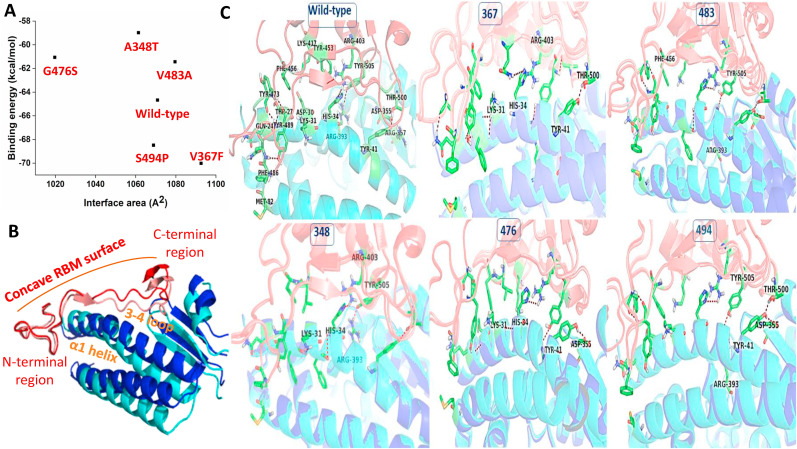
The RBD-ACE2 crystal complex has been refined extensively using a series of Monte Carlo (MC) simulations and short molecular dynamics simulations, followed by energy minimization that allows the repacking of the protein-protein binding interface. This rigorous refinement of the complex leads to tighter packing at the protein-protein interface. The interface area between the SARS-CoV2 RBD and the peptidase domain of ACE2 increases by 225 Å2 in comparison to the crystallographic form. The calculated binding free energy is −64.67 kcal/mol (Fig. 3 A). Fig. 3B reveals a comparison between the crystal structure of the RBD-ACE2 complex (blue) and the refined complex (red). Overall the structure remains highly similar and the calculated RMSD is 2.6 Å. The peptidase domain undergoes more structural changes compared to the RBD. Overall, both the RBD and PD regions move towards each other closely. The loopy overhang of the RBM that is in close contact with the N-terminal domain of the α1 helix of the PD maintains the crystallographic conformation in the refined complex. However, the opposite region of the concave surface of the RBM undergoes conformational changes to allow optimum interaction with the loop between the β3 and β4 antiparallel strands, called the 3–4 loop (Fig. 3B). Free energy decomposition analysis using MM/GBSA method reveals Phe486 of RBD is the highest energetic contributor (−4.9 kcal/mol). The residue is involved in hydrogen bonding interaction with Tyr489, another important residue that largely contributes to the total binding free energy (−4.6 kcal/mol). Interestingly, another significant energetic contribution comes from the van der Waals interaction between phe486 of RBD and Met82 of ACE2 at the N-terminal region of the α1 helix that sterically locks the loopy N-terminal region of the concave RBM surface. The 2nd highest energetic contribution comes from the His34 of ACE2 (−4.68 kcal/mol) which forms a direct hydrogen bond with Arg403 in the middle of the concave RBM of Spike protein. Close to this interaction site, another hydrogen bonding interaction between Tyr505 of RBM and Arg393 of ACE2 contributes highly to the total binding free energy. At the C-terminal end of the concave surface, the primary binding energy contribution comes Asp335 of the 3–4 loop of the ACE2 (−4.14 kcal/mol) which is involved in a salt-bridge interaction with Lys337. The interaction is a part of an interaction network that tethered the C-terminal loop of the RBM to the ACE2. Two important residues, Thr500 of spike RBM and Tyr41 of α1 helix of ACE2, are involved in hydrogen bonding interactions also strengthen the binding ability of SARS-CoV2 Spike protein to human ACE2 (Fig. 3C).
Fig. 3.
(A) Scatter plot of the calculated RBD-ACE2 interfacial area and the binding free energy for wild-type and spike variants are shown. (B) The structural alignment of the crystal structure of the RBD-ACE2 complex (blue and red) with the refined complex (light blue and light red) is shown. (C) The binding regions of the RBD-ACE2 complex for wild-type and different spike variants are shown. RBD and ACE2 are shown as transparent cartoons with pink and cyan color, respectively. Residues that contribute significantly to the binding free energy are shown as green sticks. Residues that contribute differently for each spike variant in comparison to Wuhan SARS-CoV2 spikes are labeled. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Effects of RBM mutations on the binding affinity to ACE2
The binding ability of five spike population variants, A348T, V367F, G476S, V483A, and S494P with ACE2 has been explored using the protein-protein docking and binding free energy calculations. Comparisons of calculated binding free energies of spike RBD and its several population variants with human ACE2 along with the interface area of all RBD-ACE2 complexes are shown in Fig. 3A.
The A348T, G476S, and V483A variants display reduced affinity to ACE2 in comparison to the Wuhan SARS-CoV2 spike protein (Wild-type). While the V367F and S494P population variants display a higher binding affinity towards human ACE2, compared to the Wild-type spike protein. The binding affinity does not correlate well with the protein-protein interface area between the RBD and ACE2 in all the complexes. Therefore, the altered binding affinity is primarily dictated by the altered specific interactions between RBD and ACE2 in each mutant (Fig. 3A). Changes in interaction patterns that are associated with altered binding affinity are highlighted in Fig. 3C for each mutant.
In V367F, the enhanced binding energy is primarily contributed by an altered orientation of Lys31 which enhances its contribution to the binding free energy (−4.59 kcal/mol). It involves two additional hydrogen bonds formation. Another major enhanced energetic contribution comes from Arg303 which reorients to form strong hydrogen bonds with nearby Gln493. This reorientation leads to the loosening of the hydrogen bonding interaction with His34 by ∼1 kcal/mol. Another important observation for this spike variant is that both Tyr41 and Thr500 reorient in a way that both are involved in a strong hydrogen bond formation which gains energetic contribution by more than ∼1 kcal/mol. S494P RBD variant almost very similarly binds with ACE2 like the Wuhan SARS-CoV2 RBD. The enhanced binding energy is primarily contributed by Tyr505 (−4.98 kcal/mol) which is now involved in a strong hydrogen bonding network with Arg393. Another important energetic contribution comes from the altered orientation of Tyr41 which is now involved with two hydrogen-bonding interactions with Asp355 and Thr500. As a result its binding energy contribution increase by ∼1 kcal/mol. The reduction of binding affinity of the V483A mutant in comparison to Wuhan SARS-CoV2 is primarily due to the altered orientation of Tyr505 which weakens the hydrogen bonding interaction with Arg393 by more than 1 kcal/mol. A348T RBD variant shows the least binding affinity to ACE2 among all the population variants. In this RBD-ACE2 complex, the Tyr505-Arg393, and Arg403-His34 hydrogen bonding interactions strengthen between the middle of the concave RBM of spike variant and α1 of ACE2. However, the rest of the hydrogen bonds on both the side of the concave interfaces weakens, some of them are lost. Particularly, the Lys31 lost all the hydrogen-bonding interactions which significantly lowers the binding affinity. For the G476S variant, the overall interfacial contacts between the RBD and ACE2 reduces significantly, evident from a reduction of RBD and ACE2 interfacial area by ∼50 Å2 in comparison to the Wuhan RBD-ACE2 complex. Tyr41 reorients significantly to form a hydrogen bonding interaction with Asp355. As a result of its side-chain reorientation, Asp355 loses its contribution from the salt-bridge interaction with Arg357. The involvement of His34 in the hydrogen bonding interaction also reduces by ∼0.5 kcal/mol. Likewise, Lys31 also loses its hydrogen bonding interaction which reduces the binding energy contribution by 0.5 kcal/mol.
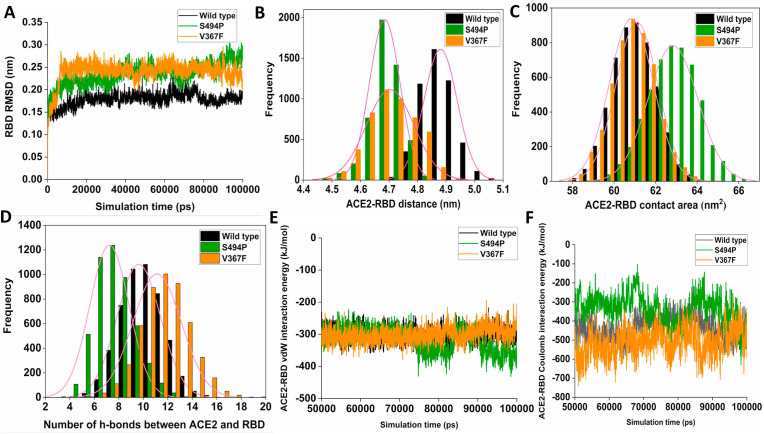
Furthermore, molecular dynamics simulations have been used to explore the driving forces behind the enhanced binding affinity of the two spike variants. Root mean square deviation analysis over the entire trajectory reveals that the V367F and S494P RBD variants remain stable throughout the simulation when complexed with ACE2 (Fig. 4 A). During the simulation. Both the variants approach closely to the ACE2 in comparison to the wild-type (Fig. 4B). However, the driving forces behind the improved receptor recognition for the two variants are distinctively different. The buried RBM-ACE2 interfacial surface area increases by 2 nm2 for the S494P variant in comparison to the wild-type and V367F variant during the simulation (Fig. 4C). On the other hand, the V367F variant forms a higher number of hydrogen bonds with the ACE2 in comparison to the wild-type. However, the number of hydrogen bonds between ACE2 and RBD decreases significantly for the S494P variants (Fig. 4D) during the simulation. This indicates the enhanced binding affinity of S494P is attributed to strong interfacial complementarity during ACE2 recognition, while the V367F variant interacts with the ACE2 mediated by a higher number of hydrogen bonds. Interaction energy decomposition also shows that the van der Waals energy increases between the RBD and ACE2 for the S494P variant during the later simulation timescale (Fig. 4E), while the interaction between the RBD and ACE2 is driven by electrostatic interactions for the V367F variant.
Fig. 4.
Variations of root mean square deviation (RMSD) (A), Distribution of RBD-ACE2 distances (B), Distribution of RBD-ACE2 interfacial area (C), number of hydrogen bonds (D), van der Waals interaction energy (E), and electrostatic interactions energy (F) between RBD and ACE2 obtained from the molecular dynamics simulations for wild-type, V367F and S494P variant of RBD are shown.
4. Conclusion
Here, the genetic diversity of the S gene of 2178 SARS-CoV2 genomes across the world reveals 113 mutations, among them 33 are parsimonious. Comparisons of selection sites reveal three RBM population variants G476S, V483A, and S494P that are in close contact with the ACE2 binding region. Another two sites, 348th and 367th, close to the RBM are also under strong positive selection bias. Interestingly, the V367F and S494P population variants display a higher binding affinity towards human ACE2. The enhanced binding affinity of S494P is attributed to strong interfacial complementarity during ACE2 recognition, while the V367F variant interacts with the ACE2 mediated by a higher number of hydrogen bonds. In V367F, the reorientation of Lys31 facilitates two additional hydrogen bonds formation which enhances its binding free energy contribution. Besides, a significant energetic contribution comes from the formation of strong hydrogen bonds by the reorientation of Arg303 and Thr500 at the interface. The result shows the emergence of two spike RBD variants with enhanced ACE2 binding affinity. Thus it is important to monitor the emergence of these strains by analyzing more viral genomes with the progression of the pandemic.
Declaration of competing interest
Authors declare no conflict of interest.
Acknowledgment
The author gratefully acknowledges Microsoft AI for Health program for providing Azure cloud capabilities to carry out the research (Microsoft AI for Health COVID-19 grant ID: 00011000243).
References
- 1.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan, Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astuti I. Ysrafil, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86:2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devaux C.A., Rolain J.-M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song W., Wang Y., Wang N., Wang D., Guo J., Fu L., Shi X. Identification of residues on human receptor DPP4 critical for MERS-CoV binding and entry. Virology. 2014;471–473:49–53. doi: 10.1016/j.virol.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirai A., Ohtsuka N., Ikeda T., Taniguchi R., Blau D., Nakagaki K., Miura H.S., Ami Y., Yamada Y.K., Itohara S., V Holmes K., Taguchi F. Role of mouse hepatitis virus (MHV) receptor murine CEACAM1 in the resistance of mice to MHV infection: studies of mice with chimeric mCEACAM1a and mCEACAM1b. J. Virol. 2010;84:6654–6666. doi: 10.1128/JVI.02680-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambepitiya Wickramasinghe I.N., de Vries R.P., Weerts E.A.W.S., van Beurden S.J., Peng W., McBride R., Ducatez M., Guy J., Brown P., Eterradossi N., Gröne A., Paulson J.C., Verheije M.H. Novel receptor specificity of avian gammacoronaviruses that cause enteritis. J. Virol. 2015;89:8783–8792. doi: 10.1128/JVI.00745-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;80–:1444–1448. doi: 10.1126/science.abb2762. 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y., Niu B., Gao Y., Fu L., Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Mega X. Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N., ConSurf An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Ko J., Park H., Heo L., Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40:W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinforma. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]