Abstract
Aim
We aimed to systematically examine the effects of enzymatic activity of 38 human CYP2C9 alleles and 21 human CYP3A4 alleles, including wild-type CYP2C9.1 and CYP3A4.1, which contain the 24 CYP2C9 novel alleles (*36–*60) and 6 CYP3A4 novel alleles (*28–*34) newly found in the Chinese population, on sildenafil metabolism through in vitro experiment.
Methods
The recombinant cytochrome P450 alleles protein of CYP2C9 and CYP3A4 expressed in insect baculovirus expression system were reacted with 10–500 µM sildenafil for 30 minutes at 37°C, and the reaction was terminated by cooling to −80°C immediately. Next, we used ultra-performance liquid chromatography tandem mass spectrometry (UHPLC-MS/MS) detection system to detect sildenafil and its active metabolite N-desmethyl sildenafil.
Results
The intrinsic clearance (Vmax/Km) values of most CYP2C9 variants were significantly altered when compared with the wild-type CYP2C9*1, with most of these variants exhibiting either reduced Vmax and/or increased Km values. Four alleles (CYP2C9*11, *14, *31, *49) exhibited no markedly decreased relative clearance (1-fold). The relative clearance of the remaining thirty-three variants exhibited decrease in different levels, ranging from 1.81% to 88.42%. For the CYP3A4 metabolic pathway, when compared with the wild-type CYP3A4*1, the relative clearance values of four variants (CYP3A4*3, *10, *14 and *I335T) showed significantly higher relative clearance (130.7–134.9%), while five variants (CYP3A4*2, *5, *24, *L22V and *F113I) exhibited sharply reduced relative clearance values (1.80–74.25%), and the remaining nine allelic variants showed no statistical difference. In addition, the kinetic parameters of two CYP3A4 variants (CYP3A4*17 and CYP3A4*30) could not be detected, due to the defect of the CYP3A4 gene.
Conclusion
These findings were the first evaluation of all these infrequent CYP2C9 and CYP3A4 alleles for sildenafil metabolism; when treating people who carry these CYP2C9 and CYP3A4 variants, there should be more focus on the relation of dose intensity, side effects and therapeutic efficacy when administering sildenafil. The study will provide fundamental data on effect of CYP2C9 and CYP3A4 allelic variation on sildenafil metabolism for further clinical research.
Keywords: allelic variants, CYP2C9 polymorphisms, CYP3A4 polymorphisms, enzymatic activity, sildenafil, individual treatment
Introduction
Sildenafil is a novel inhibitor of phosphodiesterase type 5 enzyme (PDE5). It can increase levels of cyclic guanosine monophosphate (cGMP) by inhibiting its breakdown via PDE5. Ultimately, intracellular calcium levels decrease by these pathways, thereby smooth muscle cells will be relaxed.1,2 PDE5 is mainly distributed in penile corpora cavernosa, which is the basis for the success of sildenafil in the treatment of erectile dysfunction (ED).3 Sildenafil was the first oral medicine to treat male erectile dysfunction approved by the FDA. Additionally, numerous research has shown that besides the penis, PDE5 is also distributed in cardiac, brain, vasculature, central nervous system (CNS) and so on, which implies that sildenafil is effective for pulmonary arterial hypertension (PAH), acute mountain sickness (AMS), Raynaud’s disease and stroke.4–6
Sildenafil is extensively metabolized to its active metabolite, namely, N-desmethyl sildenafil (Figure 1). Plasma concentrations of this metabolite are approximately 40% of those seen for sildenafil.7 In vitro experiments showed that the N-desmethyl sildenafil has a similar selective characteristic for PDE enzymes to sildenafil, but for PDE5, the activity of metabolite is 2.5-fold lower compared with sildenafil.8 Therefore, the metabolic rate of sildenafil affects the plasma drug concentrations, thus further leading to either reduced therapeutic efficiency or adverse drug reaction. Evidence from sildenafil metabolism in rat and human liver microsomes inferred that CYP3A4 and CYP2C9 are the major metabolic enzymes in the biotransformation.8
Figure 1.
Chemical structure of the analytes and metabolic pathway of sildenafil.
The cytochromes P450 (CYPs) are the most important drug metabolic enzymes in human liver, which are capable of catalyzing the oxidative biotransformation processes of ~90% current therapeutic drugs.9 CYP2C9 belongs to the CYP2C subfamily and constitutes ~20% of the CYPs protein content in human liver microsomes, and it is responsible for clearing ~15% of clinical drugs that undergo Phase I metabolic reaction.10,11 CYP2C9 plays a major role in the hydroxylation metabolism processes of acidic or neutral drug, mainly including antihypertensives losartan, anticoagulants warfarin, anticonvulsants phenytoin, hypoglycemic agents glimepiride and tolbutamide, anti-inflammatory drugs diclofenac and flurbiprofen.12 Like other CYP2C enzymes, CYP2C9 manifests prominent genetic polymorphisms, and it can lead to wide interindividual variations in drug metabolism.13 Up to now, a total of 60 allelic variants of CYP2C9 have been reported (http://www.cypalleles.ki.se/cyp2c9.htm). The most common allelic variants, CYP2C9*2 (Arg144Cys) and *3 (Ile359Leu), have been well studied a highly impact on its enzymatic activity both in clinical studies and in vitro.14,15 The CYP2C9 *2 variant has a C>T transition at position 432 of exon 3 encoding arginine, causing an exchange of cysteine at position 144 (Arg144Cys) of CYP2C9 protein, while the analysis of the variant CYP2C9 *3 demonstrated A>T1077 transversion in exon 7 which encodes an amino acid exchanging isoleucine to leucine at position 359 (Ile359Leu).16 The CYP2C9*2 and CYP2C9*3 allelic variants result in substantial decrease in CYP2C9 enzyme activity due to reduced interaction with NADPH-CYP-reductase or with CYP2C9 substrate, respectively.17–19 These loss-of-function mutations of CYP2C9 have been demonstrated to be less active in valproate metabolism than the wild type CYP2C9*1.20 CYP2C9*2 and CYP2C9*3 alleles thus lead to genetically determined poor metabolism of valproic acid. A separate study on individuals identified with variants CYP2C9 *2 and CYP2C9 *3 showed significantly reduced enzymatic activity for phenytoin metabolism, as individuals who presented these variants had a reduction of 27–54% of the phenytoin metabolism, exhibiting a higher predisposition to phenytoin toxicity at the usual dosages.21–23 The prevalence of these two alleles displays inter-ethnic differences. CYP2C9*2 is almost absent in Asian and African populations, whereas it is present in 10–19% of Europeans. The frequency of CYP2C9*3 is 3–6% in Asians, 1–5% in Africans and 6–9% in Europeans.9,24,25 In a recent study, Dai et al sequenced all nine exons of CYP2C9 in 2127 healthy Chinese people and discovered 37 sorts of new mutation sites, 21 of which could lead to amino acid coding change. Twenty-one sorts of new mutation sites have been nominated as new alleles CYP2C9*36-*56 by Human CYP450 Allele Nomenclature Committee.26 In subsequent studies, Luo et al further analyzed the CYP2C9 nine exons in previous 2127 healthy Chinese people and found three novel mutation positions that could result in amino acid variation, which have been named CYP2C9*58 (P337T), CYP2C9*59 (I434F) and CYP2C9*60 (L467P).27–29
CYP3A4 is another important and abundantly expressed CYP450 drug-metabolizing enzyme, which belongs to the human CYP3A subfamily and constitutes ~30% of the total CYPs in human liver.30 It is in charge of clearing approximately half of clinical drugs that experience phase I metabolic reaction, such as anticancer drugs (tamoxifen), Hypnotics (zolpidem), antibiotics (erythrocin), Hormone Drugs (prednisolone), anticonvulsants (carbamazepine), opioids (fentanyl), calcium channel blockers (nifedipine), antihistamines (loratadine) and so forth.31–33 Previous research suggested that the gene polymorphism of CYP3A4, leads to up to 60-fold different expression levels and enzyme activity of CYP3A4 between the individual human livers.34 Finally, the different enzyme activity and expression levels may further lead to interindividual variations of substrate metabolism, which would finally result in treatment failure or adverse medicine reactions. Therefore, the relation of individual CYP3A4 gene polymorphisms to metabolic effects of clinical drug will be meaningful for further studies.
Up to now, Human CYP Allele Nomenclature Committee has named 53 nonsynonymous mutations and 34 variants of CYP3A4 (https://www.pharmvar.org/gene/CYP3A4). In past research, Hu et al systematically analyzed all thirteen exons’ genetic sequence of CYP3A4 in 1114 healthy Chinese people and found five sorts of allelic variants (1G, 4, 5, 18B, 23), which have been reported in other populations.35 In addition, seven novel allele variants were also identified and designated CYP3A4*28−*34. 25 The frequency of these new allele variants was detected at only 0.04%. Whereas, based on nearly 14 hundred million Chinese population, it makes sense to study the relationship of new allele variants and drug metabolism in guiding clinical medication.
However, little attention has been paid to the effect of CYP3A4 and CYP2C9 allele variants on sildenafil metabolism. In the present study, we systematically assessed the enzymatic activities of 38 CYP2C9 and 20 CYP3A4 allele variants, including wild-type and their novel identified variants, toward the metabolism of sildenafil, to provide evidence for the individual therapy of this important vasodilator in clinical application.
Materials and Methods
Chemicals and Reagents
Sildenafil (purity 99.0%), its metabolite N-desmethyl sildenafil (purity 99.0%), and the Internal Standard (IS) Midazolam (over 99% purity) were purchased from J&K Scientific Ltd. (Hong Kong, China). The NADPH-Na4 was bought from Roche (Basel, Switzerland). Wild-type and recombinant human cytochrome P450 microsomes (CYP2C9 and CYP3A4) and CYP b5 expressed in insect baculovirus expression system were acquired from Beijing Hospital (Beijing, China). We got mass spectrometry analytical–grade methanol, acetonitrile and other organic solvents from Merck (Darmstadt, Germany). The ultrapure water was gained via Merck Millipore Milli-Q Advantage 10 system (Billerica, MA, USA). All other materials and reagents used were acquired from Sunflower and Technology Ltd. (Beijing, China). Spodoptera frugiperda (Sf)21 insect cells were purchased from Invitrogen (Carlsbad, CA, USA).
Microsomes’ Preparation
Wild-type and recombinant human CYP2C9 and CYP3A4 microsomes and purified cytochrome b5 were prepared according to previously reported method.26,36–38 In brief, cDNAs encoding CYP2C9 and CYP3A4 allelic variants were amplified by overlap extension PCR using wild-type CYP2C9 and CYP3A4 cDNAs as template, and ligated to the previously constructed dual expression baculovirus vector pFastBac-OR so as to obtain the ultimate expression vector pFastBac-OR-CYP2C9 and pFastBac-OR-CYP3A4. To ensure that no errors were introduced during PCR amplification, all of the cDNA regions were confirmed by gene sequencing. We transfected the resulting pFastBac-OR-CYP2C9 and CYP3A4 plasmid into DH10Bac to get the corresponding recombinant Bacmid. The recombinant Bacmid containing fused CYPOR and CYP2C9 or CYP3A4 gene was confirmed by PCR and then transfected into Sf21 cells. Three days after transfection, Sf21 cells were collected. Then the target proteins were released from the cells by sonication and recombinant microsomes were further collected by differential centrifugation. Finally, the microsomal preparations were quantified by measured difference spectra.
Instrumentation
We implemented the compound analysis by liquid chromatographic system of Waters ACQUITY UPLC I-Class Plus (Waters Technologies, Milford, MA, USA) coupled to a Waters Xevo TQD Mass Spectrometer (Waters Technologies), equipped with a vacuum degasser, an automatic injector, a column heating device and a binary gradient pump. Waters Masslynx 4.1 workstation (Waters Technologies) was used for data collection and instrument control.
UPLC-MS/MS
N-desmethyl sildenafil and Midazolam (IS) were separated by an XBridge BEH C18 column (2.1 mm*50 mm, 1.7 µm particle size, Waters Corp.) at 40°C constant column temperature. The temperature of the automatic injector was maintained at 4°C. The mobile phase of gradient elution was made up of acetonitrile (B) and 0.1% formic acid (A) at 0.3 mL/min solvent flow, and the injection sample size was 2 µL. The optimization gradient elution process was performed as follows: 35% B (0–0.4 minutes), solvent B increased linearly 35–90% (0.4–0.8 minutes), maintained at 90% B (0.8–1.5 minutes), then solvent B decreased linearly to 35% (1.5–1.8 minutes), and finally maintained at 35% B (1.8–2.5 minutes). The running time of the whole elution process was 2.5 min. After complete detection of each sample, autosampler and pipeline access road were washed (cleaning procedure: 50% methanol in water, 0.3 mL/min, 2.5 min). Under previously mentioned chromatography separating quantitative conditions, retention time of N-desmethyl sildenafil and Midazolam (IS) were 0.73 and 0.85 minutes, respectively. The N-desmethyl sildenafil was quantified by a seven point standard curve.
The multiple reaction monitoring mode and positive electrospray ionization (ESI) of XEVO TQD triple-quadrupole MS spectrometer (Waters Technologies) were used to detect target products, optimized instrument parameters were as follows: ionization source temperature 150°C, desolvation temperature 500°C, capillary voltage 3500 V, and 1000 liter/h nitrogen was applied to evaporate the solvent. The ions of target compound detected by multiple reaction monitoring were m/z 461.0→283.0, m/z 326.2→291.0 for N-desmethyl sildenafil and Midazolam (IS), respectively. In addition, the collision energy for N-desmethyl sildenafil and Midazolam (IS) were set to 30, 40 V, respectively.
Conditions and Methods for Incubation
The reaction system contained the following ingredients: 5 pmol wild-type CYP450 (CYP2C9*1 and CYP3A4*1) microsomes or 5 pmol other recombinant CYP450 (CYP2C9 and CYP3A4) microsomes, 5 pmol prepared CYP b5 (0.21 μg/μL, P450/b5 = 1:1) and 4.74 µL Sildenafil in 100mM Tris–HCl buffer (pH7.4). Sildenafil standard was dissolved in methanol, and working solutions of respective concentration levels prepared by methanol diluting. The final content of sildenafil in the reaction system was determined from 10–500 µM according to pretest study. First, the prepared incubation mixture was preincubated in a 37°C shaking incubator for five minutes. Then we added the NADPH regenerating system into the incubation mixture to initiate the reaction, and the final volume of 200 µL, that this reaction was last for 30 min at 37°C. Finally, the reaction was terminated by rapidly transferring to −80°C refrigerator. All experimental conditions about the sildenafil N-demethylation activity assay were based on linearity range with reaction time, substrate and microsome protein concentration respectively, incubations were repeated three times for each substrate concentration and the data are displayed as the mean ± S.D (standard deviation) from three experiments. 40 µL Midazolam (500 ng/mL) as the internal standard was added to the enzyme reaction mixture and then 400 µL acetonitrile was added for protein precipitation. Then the incubations were put under vortex movement for 2 min, and centrifuged to discard sediment at 14,000 rpm for 10 min at 4°C. We used ultrapure water 1:1 diluting the supernatant and then 2 µL of the processed mixture was injected into the UPLC-MS/MS instrument for following quantitative analysis of Sildenafil and its metabolite.
Statistical Analysis
Michaelis-Menten analysis and enzyme kinetic parameters were performed by program Prism 8.0 (GraphPad Software Inc., SanDiego, CA, USA). The kinetic data of each CYP mutant allele recombinant microsome on sildenafil substrates are shown as mean ± S.D of 3 parallel tests. Differences among wild type and allelic mutants were compared with one-way ANOVA . Comparison between Vmax, Km and Clint of each variant was done with Dunnett’s test. All experimental results were analyzed with statistical analysis software SPSS version 17.0 (SPSS Inc., Chicago, IL, USA), and a p value lower than 0.05 means statistical significance.
Results
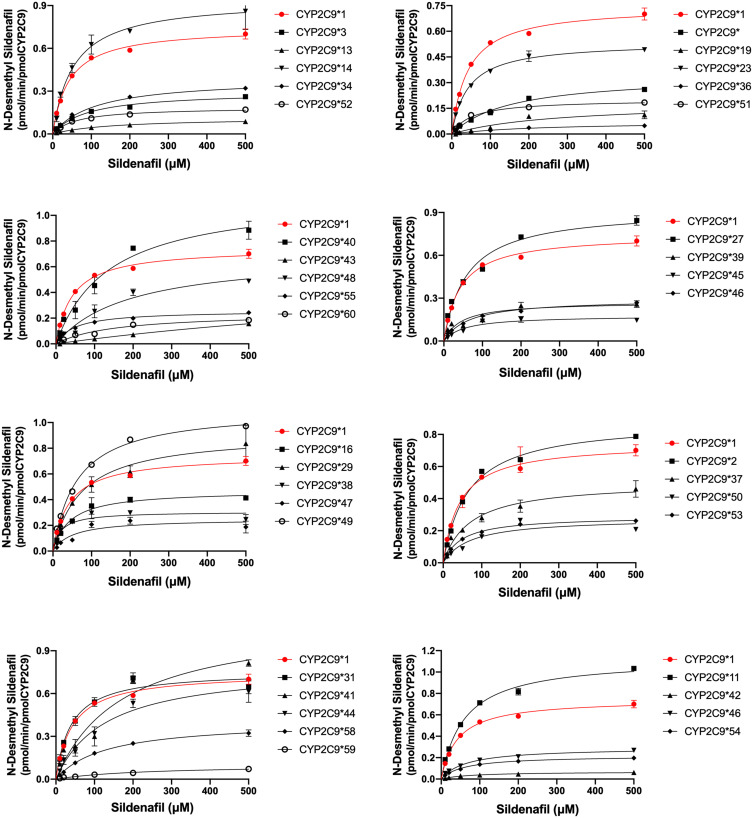
To acquire the accurate catalytic activities of different genotypes CYP2C9 on sildenafil metabolism, the catalytic activities of wild-type CYP2C9*1 and 37 allelic variants were assessed in this study. Michaelis-Menten equation plots of sildenafil for CYP2C9 wild-type and allelic variants are shown in Figure 2, and the details about the kinetic parameters of each variant are summarized in Table 1. As shown in Figure 2 and Table 1, the resulting enzyme kinetic parameters of Km, Vmax and CLint for sildenafil of wild type CYP2C9*1 were 42.75 µM, 0.75 pmol/min/pmol, 17.46 µL/min/µmol. Almost all of the tested variants showed altered values in Vmax or Km compared with CYP2C9*1, which resulted in change of the CLint (Vmax/Km) values of these variants for sildenafil metabolism. Thirty-seven allelic variants could be divided into two groups according to their relative clearance for sildenafil compared with wild type: four variants (CYP2C9*11, *14, *31, *49) exhibited no significant difference by statistic analysis in CLint value; the remaining thirty-three alleles (CYP2C9*2, *3, *8, *13,*16, *19, *23,*27, *29, *34, *36, *37, *38, *39, *40, *41, *42, *43, *44, *45, *46, *47, *48, *50, *51, *52, *53, *54, *55, *56, *58, *59, *60) exhibited significant reduction of CLint values (1.81–88.42%). Five of the thirty-three variants (CYP2C9*2,*23, *27, *29, *38) showed higher or flat CLint values (71.05–88.42% relative clearance) compared with the most common allelic variant CYP2C9*2 (78.10% relative clearance), which showed approximately 20% declined activity for sildenafil metabolism in comparison with wild type, and was considered as the slight-reduction group; sixteen variants (CYPC29*16, *34, *37, *39, *40, *41, *44, *46, *47, *48, *50, *51, *53, *54, *55, *56) exhibited lower CLint values (20.05–60.63% relative clearance) than CYP2C9*2, but higher than another common allelic variant CYP2C9*3 (18.89% relative clearance), and then, they could be considered as the moderate-reduction group; the remaining twelve variants (CYP2C9*3, *8, *13, *19, *36, *42, *43, *45**52, *58, *59, *60) exhibited lower CLint values (1.82–19.17% relative clearance) than CYP2C9*3 and could be considered as the significant-reduction group.
Figure 2.
Michaelis–Menten curves of the enzymatic activity of the recombinant wild-type CYP2C9*1 and 37 variants toward sildenafil N-demethylation (each point represents the mean ±SD of three parallel experiments).
Table 1.
Michaelis-Menten Kinetic Parameters for Demethylation Activities of Wild-Type CYP2C9*1 and 37 Mutant CYP2C9 Alleles Toward Sildenafil
| Variants | Vmax (pmol/min/pmol) | Km (µM) | CLint (µL/min/µmol) | Relative Clearance (% of Wild Type) |
|---|---|---|---|---|
| CYP2C9*1 | 0.75±0.02 | 42.75±2.45 | 17.46±0.45 | 100.00 |
| CYP2C9*2 | 0.88±0.05 | 64.75±6.99 | 13.71±0.79 | 78.10 |
| CYP2C9*3 | 0.30±0.01** | 89.98±3.22** | 3.30±0.07** | 18.89 |
| CYP2C9*8 | 0.35±0.00* | 153.73±0.06** | 2.25±0.01** | 12.87 |
| CYP2C9*11 | 1.13±0.01** | 60.71±0.60 | 18.55±0.23 | 106.20 |
| CYP2C9*13 | 0.11±0.01 | 155.23±12.01* | 0.74±0.02** | 4.21 |
| CYP2C9*14 | 0.96±0.13 | 57.48±22.69 | 17.72±3.96 | 100.72 |
| CYP2C9*16 | 0.47±0.00* | 45.96±8.98 | 10.58±0.12 | 60.64 |
| CYP2C9*19 | 0.19±0.06* | 280.63±114.29 | 0.70±0.08** | 3.96 |
| CYP2C9*23 | 0.54±0.01* | 42.57±1.45 | 12.60±0.13* | 72.09 |
| CYP2C9*27 | 0.92±0.02* | 59.77±1.89* | 15.44±0.29 | 88.42 |
| CYP2C9*29 | 0.96±0.20 | 85.60±48.25 | 12.49±3.53 | 71.05 |
| CYP2C9*31 | 0.76±0.03 | 39.48±3.94 | 19.30±1.18 | 110.66 |
| CYP2C9*34 | 0.39±0.03** | 111.70±8.74* | 3.51±0.16** | 20.05 |
| CYP2C9*36 | 0.07±0.00** | 224.97±12.59* | 0.32±0.00** | 1.82 |
| CYP2C9*37 | 0.51±0.09 | 73.53±33.54 | 7.69±2.54 | 41.00 |
| CYP2C9*38 | 0.31±0.03** | 20.47±5.50 | 15.59±3.01 | 87.04 |
| CYP2C9*39 | 0.28±0.03** | 46.46±14.72 | 6.24±1.44* | 34.91 |
| CYP2C9*40 | 1.17±0.12 | 140.93±26.79 | 8.36±0.69** | 47.83 |
| CYP2C9*41 | 1.13±0.12 | 178.47±52.69 | 6.63±1.40* | 37.11 |
| CYP2C9*42 | 0.07±0.00** | 83.50±31.53 | 0.89±0.25*** | 5.00 |
| CYP2C9*43 | 0.46±0.23 | 1,044.00±540.35 | 0.45±0.03** | 2.38 |
| CYP2C9*44 | 0.82±0.16 | 142.40±68.39 | 6.37±1.93 | 34.98 |
| CYP2C9*45 | 0.18±0.02*** | 54.15±13.77 | 3.39±0.54*** | 19.17 |
| CYP2C9*46 | 0.29±0.02** | 66.54±11.44 | 4.48±0.51*** | 25.62 |
| CYP2C9*47 | 0.25±0.04** | 52.47±15.16 | 4.91±0.69** | 27.95 |
| CYP2C9*48 | 0.70±0.06 | 193.07±33.91 | 3.68±0.29*** | 20.97 |
| CYP2C9*49 | 1.11±0.01** | 63.31±2.64 | 17.51±0.53 | 100.15 |
| CYP2C9*50 | 0.28±0.00** | 70.54±3.24* | 3.94±0.11** | 22.56 |
| CYP2C9*51 | 0.20±0.00** | 51.84±2.83 | 3.89±0.15** | 22.27 |
| CYP2C9*52 | 0.19±0.01** | 62.53±6.19 | 3.00±0.16** | 17.13 |
| CYP2C9*53 | 0.29±0.02** | 47.46±1.69 | 6.09±0.18** | 34.84 |
| CYP2C9*54 | 0.22±0.01** | 60.71±5.28 | 3.61±0.22** | 20.69 |
| CYP2C9*55 | 0.26±0.00** | 49.92±3.98 | 5.18±0.32*** | 29.64 |
| CYP2C9*56 | 0.37±0.02** | 76.22±7.41 | 4.83±0.20** | 27.64 |
| CYP2C9*58 | 0.41±0.02** | 129.13±12.93 | 3.18±0.15** | 18.24 |
| CYP2C9*59 | 0.10±0.00** | 231.80±33.88 | 0.45±0.04** | 2.54 |
| CYP2C9*60 | 0.25±0.04* | 156.97±58.23 | 1.64±0.27*** | 9.20 |
Notes: *Significantly different from wild-type CYP2C9, *p < 0.05, **p < 0.01, ***p < 0.001.
Abbreviation: N.D., not determined.
As Table 1 shows, fourteen allelic variants had a lower Vmax value than wild-type (CYP2C9*16, *23, *37, *39, *45, *46, *47, *50-*56), and the value of Km having no obvious variation (1-fold), therefor these variants showed lower CLint than wild-type. Meanwhile, the CLint values of six variants (CYP2C9*40, *41, *43, *44, *48, *58) were lower compared with the wild type CYP2C9*1 due to high level of Km values and Vmax values of these variants having no significant difference (1-fold). As a result, the Vmax values of ten variants (CYP2C9*2, *14, *29, *31, *37, *40, *41, *43, *44, *48) showed no significant difference compared with the CYP2C9*1, whereas the Km values of seven variants (CYP2C9*3, *8, *13, *27, *34, *36, *50) showed a significant difference compared with the CYP2C9*1 (P<0.05). The CLint values of twenty-six variants for sildenafil metabolism (CYP2C9*3, *8, *13, *19, *23, *34, *36, *39, *40, *41, *42, *43, *45, *46, *47, *48, *50-*56, *58, *59, *60) showed significant difference compared with the CYP2C9*1 (*P<0.05, **P<0.01, ***P<0.001).
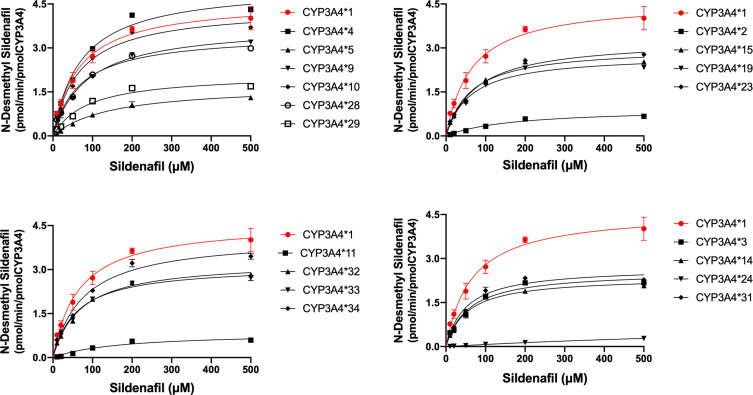
Michaelis-Menten equation plots of sildenafil for CYP3A4 wild-type and allelic variants are shown in Figure 3, and the details about the kinetic parameters of each variant are displayed in Table 2. The N-desmethyl sildenafil was not detected in CYP3A4*17, which indicated that the enzyme activity was too weak to result in any sildenafil metabolism. The resulting enzyme kinetic parameters of Km, Vmax and CLint for sildenafil of wild type CYP3A4*1 were 64.02 µM, 9.29 pmol/min/pmol, 0.15 µL/min/µmol. As shown in Table 2, except for the variant of CYP3A4*17, almost all of the allelic variants showed changed Vmax values compared with the wild-type, the five variants (CYP3A4*5, *24, *L22V, *H324Q and *F113I) exhibited decreased Vmax values (36.1–98.8%), the remaining thirteen variants displayed increased Vmax values compared to CYP3A4*1 (229.3–106.2%), CYP3A4*14 and *F113I are the only variants that had no statistical difference with the wild-type on Vmax values. Meanwhile, most CYP3A4 allelic variants presented significant alterations of values in Km: eight variants (CYP3A4*2, *5, *9, *11, *23, *24, *I427V and *F113I) showed considerably higher Km values (1.11–20.58 fold) compared with CYP3A4*1, while only two variants, CYP3A4*H32Q and *14, exhibited decreased Km values: 80.7% and 83.7%, and the remaining eight variants showed no statistical difference in Km value. Therefore, based on the CLint values of sildenafil metabolism, allelic variants were classified into three categories. Four variants (CYP3A4*3, *10, *14 and *I335T) showed significantly higher CLint values (130.7–134.9%) compared with the CYP3A4*1. Five variants (CYP3A4*2, *5, *24, *L22V and *F113I) exhibited sharply reduced CLint values (1.80–74.25%) compared with the CYP3A4*1, particularly, CYP3A4*24 whose CLint values were extremely decreased to 1.80% compared with the CYP3A4*1. The remaining nine allelic variants (CYP3A4*4, *9, *11, *15, *19, *23, *H324Q, *I427V and *A370S) exhibited no statistical difference in CLint values compared with the CYP3A4*1.
Figure 3.
Michaelis–Menten curves of the enzymatic activity of the recombinant wild-type CYP3A4*1 and 22 variants toward sildenafil N-demethylation (each point represents the mean ±SD of three parallel experiments).
Table 2.
Michaelis-Menten Kinetic Parameters for Demethylation Activities of Wild-Type CYP3A4*1 and 22 Mutant CYP3A4 Alleles Toward Sildenafil
| Variants | Vmax (pmol/min/pmol) | Km (µM) | CLint (µL/min/µmol) | Relative Clearance (% of Wild Type) |
|---|---|---|---|---|
| CYP3A4*1 | 9.29±0.45 | 64.02±5.07 | 0.15±0.02 | 100.03 |
| CYP3A4*2 | 13.39±0.43*** | 169.77±11.26*** | 0.08±0.00** | 55.54 |
| CYP3A4*3 | 10.68±0.17** | 56.99±3.79 | 0.19±0.01* | 132.00 |
| CYP3A4*4 | 10.91±0.10** | 71.13±1.20 | 0.15±0.00 | 107.79 |
| CYP3A4*5 | 6.01±0.43** | 147.87±26.30** | 0.04±0.00** | 28.91 |
| CYP3A4*9 | 10.82±0.14** | 85.07±6.63* | 0.13±0.01 | 89.53 |
| CYP3A4*10 | 12.88±0.15*** | 67.05±1.03 | 0.19±0.00* | 134.97 |
| CYP3A4*11 | 21.31±0.74*** | 167.87±16.17*** | 0.13±0.01 | 89.56 |
| CYP3A4*14 | 9.87±0.20 | 53.13±3.69* | 0.19±0.01* | 130.70 |
| CYP3A4*15 | 10.06±0.10* | 62.29±3.60 | 0.16±0.01 | 113.64 |
| CYP3A4*17 | ND | ND | ND | ND |
| CYP3A4*19 | 10.65±0.29* | 62.22±2.04 | 0.17±0.00 | 120.34 |
| CYP3A4*23 | 11.33±0.30** | 76.10±4.29* | 0.15±0.00 | 104.76 |
| CYP3A4*24 | 3.36±0.56*** | 1,317.67±298.59** | 0.00±0.00*** | 1.80 |
| CYP3A4*28 | 5.91±0.13*** | 68.89±5.69 | 0.09±0.01** | 60.47 |
| CYP3A4*29 | 9.18±0.03 | 86.90±2.89** | 0.11±0.00* | 74.25 |
| CYP3A4*30 | ND | ND | ND | ND |
| CYP3A4*31 | 7.49±0.02** | 51.67±3.65* | 0.15±0.01 | 102.20 |
| CYP3A4*32 | 13.20±0.31*** | 69.34±2.67 | 0.19±0.00* | 133.84 |
| CYP3A4*33 | 10.45±0.33* | 60.64±3.97 | 0.17±0.01 | 121.15 |
| CYP3A4*34 | 11.78±0.40** | 77.36±5.91* | 0.15±0.01 | 107.11 |
Notes: *Significantly different from wild-type CYP3A4, *p < 0.05, **p < 0.01, ***p < 0.001.
Abbreviation: ND, not determined.
Discussion
Owing to the fact that the carriers of these low-frequency CYP450 mutations are hard to find, it is difficult to carry out metabolism evaluations in vivo; therefore, targeted drug in vitro metabolic study is a great way to identifythe metabolic enzyme activities of variation individual. The evidence so far suggests that genetic polymorphisms in CYP450 is a main reason of individual variation in drug reactions, perhaps resulting in the problems of medications’ effectiveness and adverse reactions. So, our research is crucially important for reflecting complicated pharmacogenetics in clinical individualized medication. Sildenafil has robust efficacy in vasodilatation; patients with defective CYP450 on long-term treatment with sildenafil may have higher risks of ADRs (adverse drug reaction), such as renal toxicity and induce or worsen heart. Precision medicine can achieve clinical individualized medication based on enzyme metabolic abilities of each CYP450 genotype. Patients with increased CYP450 enzymatic activity should up their doses, while patients with decreased enzymatic activity should use smaller doses or extend dosing interval.
To further investigate the effect of the CYP2C9 allele on the metabolism of sildenafil, the recombinant insect microsomes were utilized to screen the 38 CYP2C9 variants. Among these variants, some exhibited lower enzyme activity than wild-type CYP2C9*1, in agreement with previous research findings, while some led to no change in enzyme activity, a finding that was not corroborated by previous studies.
CYP2C9*2 and CYP2C9*3, two widely investigated variants, are present in approximately 8–14% and 10–15% of Caucasian individuals, but are not as prevalent in Asian and black populations. The amino acid change Arg144Cys exists on the external surface of the protein CYP2C9*2. Using recombinant insect microsomes, Wang et al and Xia et al observed that CYP2C9*2(R144C) had lower CLint values for flurbiprofen and diclofenac, about 61.36% and 89.48% of wild type, respectively.39,40 Compared with CYP2C9*1, its enzyme activity was only slightly reduced. In this study, CYP2C9*2 resulted in a small increase in Vmax (1.2 times) and Km (1.5 times) for the catalysis of sildenafil, thereby leading to a small decrease in clearance rate. This finding was consistent with previous studies. Our research also adds to previous empirical evidence by showing that, compared to wild-type CYP2C9*1, CYP2C9*3 sildenafil Clint was significantly reduced. Blaisdell et al demonstrated that compared to wild-type CYP2C9*1, there was a marked decrease in enzyme activity in tolbutamide metabolism for CYP2C9*8 (R150H). However, Wang et al and Pan et al found that CYP2C9*8 was associated with lower CLint values for flurbiprofen and carvedilol.39,41,42 In our study, we found that CYP2C9*8 showed significant reduction in Vmax (46%) and 3.6-fold higher Km, which caused a significantly lower clearance rate for sildenafil. Apparent discrepancies of CYP2C9*8 might be explained by the R150H substitution altering an essential interaction between the CYPs enzyme and the NADPH-CYP450 reductase.
Some studies on tolbutamide and carvedilol metabolism have shown that compared with the wild-type CYP2C9*1, the CYP2C9*11 (R335W) allele is linked to a decrease in enzyme activity.28,29 However, in some other studies, it has been noted that CYP2C9*11 may have little to do with the dosage requirements of warfarin, and that it shows a significant increase in the enzyme activity of bosentan metabolism.43,44 In this study, the catalytic activity of the CYP2C9*11 allele on sildenafil was not significantly different from that of the wild type. First identified in a Chinese poor metabolizer of lornoxicam, CYP2C9*13 (Leu90Pro) is a variant that occurs among roughly 2% of the Chinese population.45,46 The catalytic activity of CYP2C9*13 on diclofenac and tolbutamide decreases in vitro.47 In our study, CYP2C9*13 had lower Vmax, higher Km, and lower CLint enzyme activity than the CYP2C9*1 allele. However, this non-synonymous mutation of CYP2C9 was located in the non-heme binding region and is far away from the substrate binding pocket. Therefore, it is unclear why there was CYP2C9*13-mediated reduction in catalytic activity.
CYP2C9*14, CYP2C9*16, and CYP2C9*19 were all first discovered in Southeast Asians,47 and the CLint of tolbutamide metabolism appears to be moderately or significantly reduced in vitro.48 However, in this study, there was no significant difference in the CLint value of CYP2C9*14 for sildenafil. Compared with the wild type, only the CLint value of sildenafil for CYP2C9*19 was significantly reduced (3.9%), which resulted from a 3.9-fold reduction in Vmax, a 6.5-fold increase in Km value, and a 25.3-fold reduction in CLint. These findings are consistent with those of previous studies.
In spite of the fact that the enzymatic function of CYP2C9*23 was not ascertained from Veenstra et al’s study,49 our study showed that allelic variation leads to a reduction in the clearance of sildenafil, while Km has no significant change. CYP2C9*27 (Arg150Leu) and CYP2C9*29 (Pro279Thr) were originally found in the Japanese population, but compared with the CYP2C9*1, these two alleles showed similar enzymatic activities against diclofenac.50 In our study, there was no significant change in the clearance rate of sildenafil by these two alleles, which is consistent with the results of previous studies. CYP2C9*31 was considered to be a functionally defective allelic variant,51 but our research shows that there was no significant change in the clearance rate of CYP2C9*31. CYP2C9*34 was first discovered in 724 Japanese subjects. Compared with wild type CYP2C9*1, the catalytic activity of CYP2C9*34 showed no significant change.52 Our results discovered that CYP2C9*34 significantly reduced the CLint value of sildenafil metabolism.
CYP2C9*36 has an M1V amino acid substitution that results from 1A>G nucleotide substitution. This alteration leads to a decrease in the expression level of CYP2C9 protein. In this study, CYP2C9*36 resulted in a significant decrease in Vmax of Sildenafil metabolism (9.3%) and a significant increase in Km value (5-fold), leading to a significant reduction in clearance rate. Therefore, we infer that CYP2C9*36 will have an effect on its enzymatic activity due to its low protein expression. Our results indicated that CYP2C9*42(R124Q) and CYP2C9*43(R124W) also significantly reduced the CLint of sildenafil in vitro, a finding consistent with the study by Dai et al.36 Our results demonstrated that compared with wild type CYP2C9*1, CYP2C9*37, *39, *41, *46, *47, *48, *50, *51, *52, *53, *55 and *56 showed lower CLint values for sildenafil in vitro. Similarly, these findings were also corroborated by Dai et al.36 However, when sildenafil was used as a substrate, the relative catalytic activity between these CYP2C9 allelic variants appeared to be completely different. For example, CYP2C9*40(F110S) and CYP2C9*54(S343R) are both predicted to be rapidly metabolizing allele variants,36 but our results showed that the activity of either of them was reduced. CYP2C9*38(G96A) and CYP2C9*49(I222V) showed impaired CYP2C9 activity in the study by Dai et al.53 However, we found that it had no effect on the enzyme activity of sildenafil metabolism compared with wild-type CYP2C9*1. The two variants CYP2C9*44 (T130M) and CYP2C9*45 (R132W) are similar new coding allelic variants, which were first discovered in Chinese populations and showed similar reduced activity in vitro.36 In contrast, our results showed that the CLint value was much lower for CYP2C9*45 than for CYP2C9*44.
For the newly discovered variants CYP2C9*58 (P337T), CYP2C9*59 (I434F), and CYP2C9*60 (L467P), all showed significantly lower CLint. We believe that the different characteristics of these substrates might be the main reason for this discrepancy of enzymatic activity, because previous research has indicated that decrease in the CLint of nine various substrates could change from 3-fold for diclofenac 4-hydroxylation to 27-fold for piroxicam 5-hydroxylation in vitro for the typical allele variant CYP2C9*3.36
We carefully analyzed 22 CYP3A4 variants and the wild-type CYP3A4*1 to display reliable and complete data with regard to the effects of CYP3A4 allele on the metabolism in vitro of sildenafil. The metabolic activity of wild-type CYP3A4*1 was chosen as the positive contrast. Dai et al constructed the CYP3A4 *17 and identified the enzymatic activity by metabolizing chlorpyrifos and testosterone in an Escherichia coli system, results showed that CYP3A4*17 had an F189S substitution in exon 7 that caused decreased metabolic activity of chlorpyrifos and testosterone compared with the wild type.54 Our results were consistent with past research, CYP3A4*17 exhibited remarkably low enzymatic activity toward sildenafil that no metabolites had been detected. Similar to CYP3A4 *17, the novel allelic variant CYP3A4*30 had premature termination codon mutation that produces truncated protein, resulting in the metabolites of CYP3A4*30 not being detectable and it means that the CYP3A4*30 had no metabolic activity on sildenafil. Therefore, people who carry these two CYP3A4 alleles can be categorized as PMs (poor metabolizers) for sildenafil and much more attention should be paid to sildenafil-caused adverse reactions.
Hsieh et al sequenced all thirteen exons of CYP3A4 in 102 Taiwanese Chinese people and discovered three sorts of new mutation sites including CYP3A4*4 and CYP3A4*5.55 By studying the ratio of free cortisol to 6beta-hydroxycortisol, it was confirmed that these mutations obviously decreased the activity of CYP3A4. But for the sildenafil metabolism, we discovered only CYP3A4*5 exhibited reduced activity, the CYP3A4*4 showed no statistical difference in CLint values compared with the wild type.
Eiselt et al indicated that compared with CYP3A4*1, CYP3A4*9 and CYP3A4*10 exhibited no obviously different catalytic activity in steroid hydroxylase, whereas the catalytic activity of CYP3A4*11 was decreased significantly.56 In our study, we found that CYP3A4*9 and CYP3A4*11 showed no statistical difference in CLint values, while CYP3A4*10 exhibited slight increment in Vmax (1.38-fold), which caused a higher clearance rate for sildenafil. For apparent CYP3A4*10 and *11 difference might be interpreted by the activity of substrate-dependent, D174H and T363M substitution potentially leading to a change in an essential interaction between NADPH and CYP3A4 in oxidative drug metabolism.
The CYP3A4*2 (S222P) and CYP3A4*3 (M445T) variant alleles make up approximately 2.7% and 4% of Caucasians, respectively.57 Miyazaki et al’s research showed reduced metabolic activity toward midazolam, testosterone and nifedipine in the Escherichia coli protein expression system.58 In the present study, in agreement with previous research, CYP3A4*2 exhibited a significantly higher Km and slightly higher Vmax, which induced lower CLint (55.54%) compared with wild-type. For CYP3A4*3 (M445T) carried genetic mutation of 1334T>C in exon twelve, which is located in heme binding section, and may promote the heme binding of CYP3A4.57 Consequently, compared with CYP3A4*1, CYP3A4*3 exhibited a slight increment in CLint value (1.32-fold of CYP3A4*1) with increased Vmax and reduced Km value.
Two variations, CYP3A4*14 and *15, were first detected in liver donors (fifty-three Caucasians and twenty-one African-Americans).59 The allelic frequency of CYP3A*14 (L15P) in different races was between 3% and 4.2%, the allelic frequency of CYP3A*15 (R162Q) was 2%, but only detected in Black populations.59 Lamba et al used midazolam as substrate to measure the enzyme activities of CYP3A4*14 and CYP3A4*15, and found metabolic activity of midazolam hydroxylation was not substantially altered compared with wild-type.59 Our study showed that CYP3A4*14 showed higher CLint values of sildenafil (130.7%) in vitro, whereas CYP3A4*15 exhibited no obvious changes.
Drögemöller et al sequenced all the coding exons, flanking intronic regions of CYP3A4 in 159 South Africans from 3 South African population groups, and detected 2 novel non-synonymous variants (R162W and Q200H), named CPY3A4*23 and CPY3A4*24.60 Our previous study also identified CYPY3A4*23, for the first time in Chinese populations.35 Although the allelic variant function of CYP2C9*23 was not revealed by Veenstra’s study (2005), our research showed that CPY3A4*24 caused a massive reduction in clearance value (close to 0% of wild type) with significantly decreased Vmax (36%) and sharply increased Km (20-fold), relative to wild type, whereas CYP2C9*23 showed no significant changes in enzymatic functions.
In the present study, CYP3A4*32 (I335T) exhibited a slightly increased Vmax value (1.42-fold of wild-type) and the km value was not increased significantly, which caused a higher CLint rate for sildenafil compared with wild-type in vitro. Our result showed that CYP3A4*28 and CYP3A4*29 exhibited various degrees of decreased relative clearance rate for sildenafil with a lower Vmax value but a higher Km value when compared with CYP3A4*1. The remaining three allelic variants (CYP3A4*31, CYP3A4*33 and CYP3A4*34) exhibited both Vmax and Km values pointing toward the same trend, as the metabolic activities of these variants for sildenafil showed no significant change compared with CYP3A4*1. These results are not consistent with our previous study for amiodarone, lidocaine and Ibrutinib.37,38,61 We thought the metabolic activities among CYP3A4 variants were influenced by affinity differences when using different substrates.
Conclusion
In summary, the present study provides another way to analyze drug metabolism in vitro, and functionally assessed the enzymatic activity of wild-type, 37 variants of CYP2C9 and 20 variants of CYP3A4 in the metabolism of sildenafil, including 30 recently detected new coding variants. For all we know, this is the first academic thesis to systematically study all these rare allele variants for sildenafil metabolism. Although gene frequency of these rare CYP genetic variations in the Chinese population are minimum 0.1% and maximum 1.8%, against the background of more than 1.4 billion people in China, getting accurate data for rare CYP alleles in the Chinese population and exploring effects of alleles on drug metabolism and activity, will be valuable and important in clinical treatment. These data provide new insights about CYP2C9 and CYP3A4 gene polymorphisms and their impact on enzymatic activity, offer better comprehension of pharmacogenomics that will help to understand diversity of sildenafil dose–response relationship, and may contribute to offer treatment in an effective and safe manner, and these results can be used as the theoretical basis for individualized medication in Chinese populations.
Acknowledgments
The authors thank the members of the Beijing Institute of Geriatrics of the Ministry of Health for their advice and assistance. This work was supported by National Natural Science Foundation of China (NSFC 81973397) and supported by grants from the Jinhua Science and Technology Bureau (No. 2019jzk014). In addition, Peng-fei Tang wants to thank Xue-meng Wu, in particular, for the care and support received during the process of writing and in life.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Eardley I, Ellis P, Boolell M, Wulff M. Onset and duration of action of sildenafil for the treatment of erectile dysfunction. Br J Clin Pharmacol. 2002;53(Suppl 1\(Suppl 1)):61s–65s. doi: 10.1046/j.0306-5251.2001.00034.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francis SH, Corbin JD. Sildenafil: efficacy, safety, tolerability and mechanism of action in treating erectile dysfunction. Expert Opin Drug Metab Toxicol. 2005;1(2):283–293. doi: 10.1517/17425255.1.2.283 [DOI] [PubMed] [Google Scholar]
- 3.Leiblum SR. After sildenafil: bridging the gap between pharmacologic treatment and satisfying sexual relationships. J Clin Psychiatry. 2002;63(Suppl 5):17–22; discussion 23–15. [PubMed] [Google Scholar]
- 4.Fazan R Jr, Huber DA, Silva CAA, Dias da Silva VJ, Salgado MCO, Salgado HC. Sildenafil acts on the central nervous system increasing sympathetic activity. J Appl Physiol. 2008;104(6):1683–1689. doi: 10.1152/japplphysiol.01142.2007 [DOI] [PubMed] [Google Scholar]
- 5.Bocchi EA, Guimarães G, Mocelin A, Bacal F, Bellotti G, Ramires JF. Sildenafil effects on exercise, neurohormonal activation, and erectile dysfunction in congestive heart failure: a double-blind, placebo-controlled, randomized study followed by a prospective treatment for erectile dysfunction. Circulation. 2002;106(9):1097–1103. doi: 10.1161/01.CIR.0000027149.83473.B6 [DOI] [PubMed] [Google Scholar]
- 6.Galiè N, Ghofrani HA, Torbicki A, et al. Sildenafil citrate therapy for pulmonary arterial hypertension. N Engl J Med. 2005;353(20):2148–2157. doi: 10.1056/NEJMoa050010 [DOI] [PubMed] [Google Scholar]
- 7.Walker DK, Ackland MJ, James GC, et al. Pharmacokinetics and metabolism of sildenafil in mouse, rat, rabbit, dog and man. Xenobiotica. 1999;29(3):297–310. doi: 10.1080/004982599238687 [DOI] [PubMed] [Google Scholar]
- 8.Hyland R, Roe EG, Jones BC, Smith DA. Identification of the cytochrome P450 enzymes involved in the N-demethylation of sildenafil. Br J Clin Pharmacol. 2001;51(3):239–248. doi: 10.1046/j.1365-2125.2001.00318.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 10.Van Booven D, Marsh S, McLeod H, et al. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics. 2010;20(4):277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz UI. Clinical relevance of genetic polymorphisms in the human CYP2C9 gene. Eur J Clin Invest. 2003;33(Suppl 2):23–30. doi: 10.1046/j.1365-2362.33.s2.6.x [DOI] [PubMed] [Google Scholar]
- 12.Zhou SF, Zhou ZW, Huang M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology. 2010;278(2):165–188. doi: 10.1016/j.tox.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Wang J, Huang SQ, Su HH, Zhou SF. Genetic polymorphism of the human cytochrome P450 2C9 gene and its clinical significance. Curr Drug Metab. 2009;10(7):781–834. doi: 10.2174/138920009789895480 [DOI] [PubMed] [Google Scholar]
- 14.Iida I, Miyata A, Arai M, et al. Catalytic roles of CYP2C9 and its variants (CYP2C9*2 and CYP2C9*3) in lornoxicam 5ʹ-hydroxylation. Drug Metab Dispos. 2004;32(1):7–9. doi: 10.1124/dmd.32.1.7 [DOI] [PubMed] [Google Scholar]
- 15.Lee CR. CYP2C9 genotype as a predictor of drug disposition in humans. Methods Find Exp Clin Pharmacol. 2004;26(6):463–472. [PubMed] [Google Scholar]
- 16.Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6(4):341–349. doi: 10.1097/00008571-199608000-00007 [DOI] [PubMed] [Google Scholar]
- 17.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267(1):83–90. [PubMed] [Google Scholar]
- 18.Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH: cytochromeP450 oxidoreductase. Pharmacogenetics. 1997;7(3):203–210. doi: 10.1097/00008571-199706000-00005 [DOI] [PubMed] [Google Scholar]
- 19.Wei L, Locuson CW, Tracy TS. Polymorphic variants of CYP2C9: mechanisms involved in reduced catalytic activity. Mol Pharmacol. 2007;72(5):1280–1288. doi: 10.1124/mol.107.036178 [DOI] [PubMed] [Google Scholar]
- 20.Ho PC, Abbott FS, Zanger UM, Chang TK. Influence of CYP2C9 genotypes on the formation of a hepatotoxic metabolite of valproic acid in human liver microsomes. Pharmacogenomics J. 2003;3(6):335–342. doi: 10.1038/sj.tpj.6500210 [DOI] [PubMed] [Google Scholar]
- 21.Hung CC, Lin CJ, Chen CC, Chang CJ, Liou HH. Dosage recommendation of phenytoin for patients with epilepsy with different CYP2C9/CYP2C19 polymorphisms. Ther Drug Monit. 2004;26(5):534–540. doi: 10.1097/00007691-200410000-00012 [DOI] [PubMed] [Google Scholar]
- 22.Dagenais R, Wilby KJ, Elewa H, Ensom MHH. Impact of genetic polymorphisms on phenytoin pharmacokinetics and clinical outcomes in the Middle East and North Africa Region. Drugs R D. 2017;17(3):341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tate SK, Depondt C, Sisodiya SM, et al. Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci U S A. 2005;102(15):5507–5512. doi: 10.1073/pnas.0407346102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurose K, Sugiyama E, Saito Y. Population differences in major functional polymorphisms of pharmacokinetics/pharmacodynamics-related genes in Eastern Asians and Europeans: implications in the clinical trials for novel drug development. Drug Metab Pharmacokinet. 2012;27(1):9–54. doi: 10.2133/dmpk.DMPK-11-RV-111 [DOI] [PubMed] [Google Scholar]
- 25.Hirota T, Eguchi S, Ieiri I. Impact of genetic polymorphisms in CYP2C9 and CYP2C19 on the pharmacokinetics of clinically used drugs. Drug Metab Pharmacokinet. 2013;28(1):28–37. doi: 10.2133/dmpk.DMPK-12-RV-085 [DOI] [PubMed] [Google Scholar]
- 26.Dai DP, Wang YH, Wang SH, et al. In vitro functional characterization of 37 CYP2C9 allelic isoforms found in Chinese Han population. Acta Pharmacol Sin. 2013;34(11):1449–1456. doi: 10.1038/aps.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo SB, Li CB, Dai DP, et al. Characterization of a novel CYP2C9 mutation (1009C>A) detected in a warfarin-sensitive patient. J Pharmacol Sci. 2014;125(2):150–156. doi: 10.1254/jphs.13189FP [DOI] [PubMed] [Google Scholar]
- 28.Dai DP, Li CB, Wang SH, et al. Identification and characterization of a novel CYP2C9 allelic variant in a warfarin-sensitive patient. Pharmacogenomics. 2015;16(13):1475–1486. doi: 10.2217/pgs.15.89 [DOI] [PubMed] [Google Scholar]
- 29.Dai D-P, Wang S-H, Li C-B, et al. Identification and functional assessment of a new CYP2C9 allelic variant CYP2C9*59. Drug Metab Dispos. 2015;43(8):1246–1249. doi: 10.1124/dmd.115.063412 [DOI] [PubMed] [Google Scholar]
- 30.Zanger UM, Turpeinen M, Klein K, Schwab M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal Bioanal Chem. 2008;392(6):1093–1108. [DOI] [PubMed] [Google Scholar]
- 31.Werk AN, Cascorbi I. Functional gene variants of CYP3A4. Clin Pharmacol Ther. 2014;96(3):340–348. doi: 10.1038/clpt.2014.129 [DOI] [PubMed] [Google Scholar]
- 32.Li AP, Kaminski DL, Rasmussen A. Substrates of human hepatic cytochrome P450 3A4. Toxicology. 1995;104(1–3):1–8. doi: 10.1016/0300-483X(95)03155-9 [DOI] [PubMed] [Google Scholar]
- 33.Zhou SF. Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab. 2008;9(4):310–322. doi: 10.2174/138920008784220664 [DOI] [PubMed] [Google Scholar]
- 34.Westlind A, Löfberg L, Tindberg N, Andersson TB, Ingelman-Sundberg M. Interindividual differences in hepatic expression of CYP3A4: relationship to genetic polymorphism in the 5ʹ-upstream regulatory region. Biochem Biophys Res Commun. 1999;259(1):201–205. doi: 10.1006/bbrc.1999.0752 [DOI] [PubMed] [Google Scholar]
- 35.Hu GX, Dai DP, Wang H, et al. Systematic screening for CYP3A4 genetic polymorphisms in a Han Chinese population. Pharmacogenomics. 2017;18(4):369–379. doi: 10.2217/pgs-2016-0179 [DOI] [PubMed] [Google Scholar]
- 36.Dai DP, Xu RA, Hu LM, et al. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. Pharmacogenomics J. 2014;14(1):85–92. doi: 10.1038/tpj.2013.2 [DOI] [PubMed] [Google Scholar]
- 37.Fang P, Tang PF, Xu RA, et al. Functional assessment of CYP3A4 allelic variants on lidocaine metabolism in vitro. Drug Des Devel Ther. 2017;11:3503–3510. doi: 10.2147/DDDT.S152366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang CC, Zheng X, Liu TH, et al. Functional characterization of 21 CYP3A4 variants on amiodarone metabolism in vitro. Xenobiotica. 2019;49(1):120–126. doi: 10.1080/00498254.2017.1414971 [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Bao SH, Pan PP, et al. Effect of CYP2C9 genetic polymorphism on the metabolism of flurbiprofen in vitro. Drug Dev Ind Pharm. 2015;41(8):1363–1367. doi: 10.3109/03639045.2014.950274 [DOI] [PubMed] [Google Scholar]
- 40.Xia MM, Wang L, Pan PP, et al. The role of CYP2C9 genetic polymorphisms in the oxidative metabolism of diclofenac in vitro. Pharmazie. 2014;69(12):898–903. [PubMed] [Google Scholar]
- 41.Blaisdell J, Jorge-Nebert LF, Coulter S, et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14(8):527–537. doi: 10.1097/01.fpc.0000114759.08559.51 [DOI] [PubMed] [Google Scholar]
- 42.Pan PP, Weng QH, Zhou CJ, et al. The role of CYP2C9 genetic polymorphism in carvedilol O-desmethylation in vitro. Eur J Drug Metab Pharmacokinet. 2016;41(1):79–86. doi: 10.1007/s13318-014-0245-2 [DOI] [PubMed] [Google Scholar]
- 43.King BP, Khan TI, Aithal GP, Kamali F, Daly AK. Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics. 2004;14(12):813–822. doi: 10.1097/00008571-200412000-00004 [DOI] [PubMed] [Google Scholar]
- 44.Chen M, Zhang Y, Pan P, et al. Effects of cytochrome P450 2C9 polymorphism on bosentan metabolism. Drug Metab Dispos. 2014;42(11):1820–1825. doi: 10.1124/dmd.114.060244 [DOI] [PubMed] [Google Scholar]
- 45.Si D, Guo Y, Zhang Y, Yang L, Zhou H, Zhong D. Identification of a novel variant CYP2C9 allele in Chinese. Pharmacogenetics. 2004;14(7):465–469. doi: 10.1097/01.fpc.0000114749.08559.e4 [DOI] [PubMed] [Google Scholar]
- 46.Guo Y, Zhang Y, Wang Y, et al. Role of CYP2C9 and its variants (CYP2C9*3 and CYP2C9*13) in the metabolism of lornoxicam in humans. Drug Metab Dispos. 2005;33(6):749–753. doi: 10.1124/dmd.105.003616 [DOI] [PubMed] [Google Scholar]
- 47.Guo Y, Wang Y, Si D, Fawcett PJ, Zhong D, Zhou H. Catalytic activities of human cytochrome P450 2C9*1, 2C9*3 and 2C9*13. Xenobiotica. 2005;35(9):853–861. doi: 10.1080/00498250500256367 [DOI] [PubMed] [Google Scholar]
- 48.DeLozier TC, Lee SC, Coulter SJ, Goh BC, Goldstein JA. Functional characterization of novel allelic variants of CYP2C9 recently discovered in southeast Asians. J Pharmacol Exp Ther. 2005;315(3):1085–1090. doi: 10.1124/jpet.105.091181 [DOI] [PubMed] [Google Scholar]
- 49.Veenstra DL, Blough DK, Higashi MK, et al. CYP2C9 haplotype structure in European American warfarin patients and association with clinical outcomes. Clin Pharmacol Ther. 2005;77(5):353–364. doi: 10.1016/j.clpt.2005.01.019 [DOI] [PubMed] [Google Scholar]
- 50.Maekawa K, Fukushima-Uesaka H, Tohkin M, et al. Four novel defective alleles and comprehensive haplotype analysis of CYP2C9 in Japanese. Pharmacogenet Genomics. 2006;16(7):497–514. doi: 10.1097/01.fpc.0000215069.14095.c6 [DOI] [PubMed] [Google Scholar]
- 51.Matimba A, Del-Favero J, Van Broeckhoven C, Masimirembwa C. Novel variants of major drug-metabolising enzyme genes in diverse African populations and their predicted functional effects. Hum Genomics. 2009;3(2):169–190. doi: 10.1186/1479-7364-3-2-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin T, Maekawa K, Kamide K, et al. Genetic variations of CYP2C9 in 724 Japanese individuals and their impact on the antihypertensive effects of losartan. Hypertens Res. 2008;31(8):1549–1557. doi: 10.1291/hypres.31.1549 [DOI] [PubMed] [Google Scholar]
- 53.Dai DP, Wang SH, Geng PW, Hu GX, Cai JP. In vitro assessment of 36 CYP2C9 allelic isoforms found in the Chinese population on the metabolism of glimepiride. Basic Clin Pharmacol Toxicol. 2014;114(4):305–310. doi: 10.1111/bcpt.12159 [DOI] [PubMed] [Google Scholar]
- 54.Dai D, Tang J, Rose R, et al. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001;299(3):825–831. [PubMed] [Google Scholar]
- 55.Hsieh KP, Lin YY, Cheng CL, et al. Novel mutations of CYP3A4 in Chinese. Drug Metab Dispos. 2001;29(3):268–273. [PubMed] [Google Scholar]
- 56.Eiselt R, Domanski TL, Zibat A, et al. Identification and functional characterization of eight CYP3A4 protein variants. Pharmacogenetics. 2001;11(5):447–458. doi: 10.1097/00008571-200107000-00008 [DOI] [PubMed] [Google Scholar]
- 57.Sata F, Sapone A, Elizondo G, et al. CYP3A4 allelic variants with amino acid substitutions in exons 7 and 12: evidence for an allelic variant with altered catalytic activity. Clin Pharmacol Ther. 2000;67(1):48–56. doi: 10.1067/mcp.2000.104391 [DOI] [PubMed] [Google Scholar]
- 58.Miyazaki M, Nakamura K, Fujita Y, Guengerich FP, Horiuchi R, Yamamoto K. Defective activity of recombinant cytochromes P450 3A4.2 and 3A4.16 in oxidation of midazolam, nifedipine, and testosterone. Drug Metab Dispos. 2008;36(11):2287–2291. doi: 10.1124/dmd.108.021816 [DOI] [PubMed] [Google Scholar]
- 59.Lamba JK, Lin YS, Thummel K, et al. Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics. 2002;12(2):121–132. doi: 10.1097/00008571-200203000-00006 [DOI] [PubMed] [Google Scholar]
- 60.Drögemöller B, Plummer M, Korkie L, et al. Characterization of the genetic variation present in CYP3A4 in three South African populations. Front Genet. 2013;4:17. doi: 10.3389/fgene.2013.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu RA, Wen J, Tang P, et al. Functional characterization of 22 CYP3A4 protein variants to metabolize ibrutinib in vitro. Basic Clin Pharmacol Toxicol. 2018;122(4):383–387. doi: 10.1111/bcpt.12934 [DOI] [PubMed] [Google Scholar]