Abstract
KRAS mutations are found in approximately one third of non-small cell lung cancer (NSCLC) patients. In this study, we aim to investigate whether KRAS G12/G13 mutant allele fraction (MAF) in cell-free DNA (cfDNA) can provide meaningful prognostic information in NSCLC. Multiplex droplet-digital PCR was used to quantitatively assess KRAS G12/G13 MAF in cfDNA from 114 pre-treated advanced disease NSCLC patients. In 14 patients, changes in KRAS G12/G13 MAF were longitudinally monitored during treatment. Plasma KRAS G12/G13 status was associated with poor patients’ outcome in terms of progression-free survival (PFS) (p < 0.001) and overall survival (OS) (p < 0.001). In multivariate analysis, the detection of plasma KRAS mutations was an independent predictor of adverse PFS (HR = 3.12; p < 0.001) and OS (HR = 2.53; p = 0.002). KRAS G12/G13 MAF at first treatment evaluation (T1) was higher (p = 0.013) among patients experiencing progressive disease compared to those with disease control, and increased KRAS MAF at T1 was associated (p = 0.005) with shorter PFS. On the contrary, no association was observed between tissue KRAS mutation status and patients’ prognosis. Our results show that ddPCR-based detection of KRAS G12/G13 mutations in plasma could serve as an independent biomarker of unfavorable prognosis in NSCLC patients. Changes in KRAS MAF can provide valuable information for monitoring patient outcome during treatment.
Keywords: ctDNA, NSCLC, ddPCR, liquid biopsy, prognostic biomarker, molecular testing
1. Introduction
In advanced NSCLC, treatment paradigms have now shifted from histology-based to genotype-based approaches [1]. Current guidelines for genetic testing include screening for the presence of activating alterations in epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1 (ROS1), or v-Raf murine sarcoma viral oncogene homolog B (BRAF) prior to the initiation of first-line treatment in NSCLC [2]. Beyond these biomarkers, Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations, and specifically the KRAS G12C point mutation, will likely soon be approved as biomarkers for the selection of patients eligible for treatment with direct inhibitors such as AMG510 [3].
KRAS mutations, and in particular single amino acid substitutions in codon 12 (e.g., G12C, G12V, and G12D) are the most prevalent gain-of-function alterations found in 20–40% of lung adenocarcinomas [4]. These oncogenic mutations, hold the KRAS oncoprotein in a constitutively active state and are associated with the development and progression of several cancers, including NSCLC [5]. Nevertheless, the prognostic and predictive value of KRAS mutations in NSCLC is still not well established [6,7].
To date, the majority of studies are based on tumor tissue genotyping [7]. Although tissue biopsy is considered as the “gold standard” source for the molecular profiling of cancer, in advanced NSCLC, access to tumor tissue is often limited [8,9]. Additionally, the procedure is invasive and cannot capture the temporal and spatial heterogeneity of the tumor [9]. To this end, circulating tumor DNA (ctDNA), the fraction of circulating cell-free DNA (cfDNA) that derives from cancer cells, represents a valuable analyte within “liquid biopsy” by providing an alternative, reliable and non-invasive source of clinically valuable information [10,11]. Furthermore, ctDNA levels have been correlated with tumor load in lung cancer patients and dynamic changes have the potential to monitor tumor burden and the biological alterations of the tumor under the pressure of anticancer treatment [12].
Detection and quantification of tumor-derived mutations in cfDNA is technically challenging, considering that cfDNA molecules are highly fragmented (134–144 bp) by nature and diluted by cfDNA of non-malignant origin [11,13]. The third generation PCR technology, droplet digital PCR (ddPCR), has the potential to fulfill the need for rapid, robust and highly sensitive detection of mutations of clinical importance in cfDNA, with mutant allele frequencies (MAF) as low as 0.01% [13]. The sensitivity of ddPCR is superior compared to standard quantitative PCR assays in detecting low target copies and has a simplified workflow compared to other digital PCR technologies [8,14].
Based on the above, we hypothesized that the concentration of KRAS mutations in plasma cfDNA is associated with outcomes in patients with advanced NSCLC treated with chemotherapy and that monitoring of ctDNA levels may provide indications of treatment efficacy. In this study, we applied a sensitive multiplex ddPCR-based assay to detect and quantify the seven most common hotspot mutations in codons 12 and 13 (G12A, G12C, G12D, G12V, G12R, G12S, and G13D) of KRAS oncogene in cfDNA isolated from plasma samples obtained from NSCLC patients prior to initiating first-line treatment for recurrent or metastatic disease. Our primary goal was to investigate whether plasma KRAS G12/G13 MAF, both as a categorical variable and as a continuous measure, has prognostic implications in patients treated with first-line systemic therapy. An additional aim of our study was to evaluate the variation of KRAS G12/G13 MAF in a subset of serially acquired plasma samples from patients with baseline mutant cfDNA, to explore potential associations with response to treatment.
2. Materials and Methods
2.1. Study Population
A total of 114 whole blood samples were obtained from patients with advanced NSCLC before starting first-line systemic therapy, at the Department of Medical Oncology, University General Hospital of Heraklion, Crete, Greece, between 2017 and 2020. All patients received standard first-line regimens according to current treatment guidelines. Patients’ baseline characteristics such as age, smoking history, the Eastern Cooperative Oncology Group Performance Status, histological subtype, number of metastatic organs, TNM stage and PD-L1 status were also collected for statistical analyses. Data concerning tissue genotyping were also collected, and patients carrying EGFR mutations or ALK translocations in tumor samples were excluded from the analysis.
Follow-up data included progression-free survival (PFS) and overall survival (OS). PFS was defined as the time elapsed between the initiation of first-line treatment and first documented disease progression or death. OS was determined as the time interval from the beginning of first-line treatment to the date of death from any cause (Table 1). Response to treatment was evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST) 1.1, and described as partial response (PR), stable disease (SD), and progressive disease (PD) [15].
Table 1.
Clinicopathological characteristics of NSCLC patients included in the study.
| Variable | No. of Patients (%) |
|---|---|
| TNM Stage a | |
| III | 24 (21.05) |
| IV | 88 (77.19) |
| Unknown | 2 (1.75) |
| Number of metastatic organs | |
| ≤2 | 88 (77.19) |
| >2 | 26 (22.81) |
| Smoking history | |
| Non-smokers | 7 (6.14) |
| Former smokers | 33 (28.95) |
| Smokers | 68 (59.65) |
| Unknown | 6 (5.26) |
| PD-L1 status b | |
| Negative | 37 (32.46) |
| Positive | 45 (39.47) |
| Unknown | 32 (28.07) |
| Gender | |
| Males | 92 (80.70) |
| Females | 22 (19.30) |
| ECOG Performance status c | |
| 0 | 86 (75.44) |
| 1 | 25 (21.93) |
| 2 | 2 (1.75) |
| Unknown | 1 (0.88) |
| Histological subtype | |
| Adenocarcinoma | 91 (79.82) |
| Squamous cell carcinoma | 15 (13.16) |
| Other | 8 (7.02) |
| First-Line Treatment | |
| Chemotherapy | 86 (75.44) |
| Combination chemo-immunotherapy | 20 (17.54) |
| Immunotherapy alone | 5 (4.39) |
| Unknown | 3 (2.63) |
| KRAS tissue status d | |
| Wild-Type | 61 (53.51) |
| G12C | 16 (14.04) |
| G12D | 10 (8.77) |
| G12V | 5 (4.39) |
| G13D | 2 (1.75) |
| G12S | 1 (0.88) |
| G12A | 1 (0.88) |
| Unknown | 18 (15.79) |
a Tumor Node Metastasis (TNM) staging system. b PD-L1 status was determined by immunohistochemistry. c Eastern Cooperative Oncology Group Performance Status. d Determined by Sanger sequencing.
This study complied with the ethical standards of the World Medical Association’s Declaration of Helsinki, as revised in 2013. The research protocol was approved by the institutional ethics committee of the University Hospital of Heraklion (20/12-07-17), and all participants provided written informed consent to participate in the study.
2.2. Plasma Processing and Cell-Free DNA (cfDNA) Extraction
Pre-treatment peripheral blood samples were collected from all NSCLC patients in K2 EDTA blood collection tubes. The plasma fraction was obtained by a two-step centrifugation protocol, within 2 h upon receipt. Firstly, blood was centrifuged at 1300× g for 15 min at 4 °C. Then, the supernatant was collected and centrifuged at 2000× g for 15 min at 4 °C. Isolated plasma samples were stored at −80 °C until further processing.
Total cfDNA was extracted from 2 mL of plasma from each sample, using the QIAmp circulating nucleic acid kit (Qiagen) according to the manufacturer’s protocol. cfDNA was eluted in 60 μL of nuclease-free water and quantified using the Qubit dsDNA HS Assay kit (Life Technologies) on Qubit fluorometer 2.0 (Life Technologies). The concentration of cfDNA was expressed in ng/μL (median cfDNA concentration = 1.45 ng/μL and range 0.25–800.0 ng/μL; Table 2).
Table 2.
Descriptive statistics for continuous variables of the study in NSCLC patients.
| Percentiles | ||||||
|---|---|---|---|---|---|---|
| Variable | No. of Patients | Mean ± S.E. a | Range | 25th | 50th (Median) | 75th |
| KRAS G12/G13 MAF b | ||||||
| MUTc plasma samples | 27 | 7.02 ± 1.49 | 0.2–28.27 | 1.53 | 3.95 | 10.04 |
| KRAS G12/G13 copies/mL plasma | ||||||
| MUT c plasma samples | 27 | 1553.6 ± 729.38 | 19.70–18,600.0 | 62.50 | 192.0 | 1080.0 |
| cfDNA concentration d (ng/μL) | 114 | 15.88 ± 8.86 | 0.25–800.0 | 0.70 | 1.45 | 3.19 |
| Patients’ Age (years) | 114 | 65.59 ± 0.91 | 37.0–84.0 | 60.00 | 67.00 | 72.25 |
| PFS (months) | 114 | 4.97 ± 0.52 | 0.13–25.58 | 1.60 | 2.79 | 5.53 |
| OS (months) | 114 | 9.05 ± 0.92 | 0.13– 44.68 | 1.95 | 5.08 | 13.07 |
a Standard error. b MAF, Mutant allele frequency. c MUT, KRAS G12/G13 mutant samples. d Concentration was determined using Qubit fluorometer 2.0 (Life Technologies).
2.3. Blood-Based KRAS G12/G13 Mutation Analysis via Droplet Digital PCR (ddPCR)
ddPCR was performed using the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) and the KRAS G12/G13 Screening Multiplex Kit (Bio-Rad). This multiplex assay can distinguish wild-type from mutant KRAS exon 2 sequence, but is not able to distinguish among individual mutations in codons 12/13. In particular, the seven most common mutations in KRAS exon 2 (G12A, G12C, G12D, G12R, G12S, G12V, and G13D) included in this multiplex assay were quantified in plasma by ddPCR.
Each ddPCR reaction mixture contained the 1X KRAS G12/G13 multiplex screening kit assay reagent (Bio-Rad) which includes primers and probes (wild-type probe labeled with HEX dye and mutant probe marked with FAM dye), 1X ddPCR Supermix for probes (no UTP; Bio-Rad), 5U restriction enzyme MseI (New England Biolabs), and 4 μL of cfDNA template, adjusted to a final volume of 20.0 μL with DEPC-treated water. Thereafter, samples were mixed with 70.0 μL of droplet generator oil for probes (Bio-Rad) and partitioned into up to 20,000 droplets using QX200 droplet generator (Bio-Rad). Emulsified samples were transferred on 96-well plates (Bio-Rad) and endpoint PCR was performed on a C1000 Touch thermal cycler (Bio-Rad). The following thermal cycling conditions were used, according to the manufacturer’s recommendations: 95 °C for 10 min, and 40 cycles of 94 °C for 30 s, 55 °C for 1 min, and 98 °C for 10 min. The plate was then transferred and read in the FAM and HEX channels using the QX200 droplet reader (BioRad). Each sample was tested in two technical replicates and every ddPCR run included negative template controls (NTCs) and positive template controls (PTCs) to calculate fluorescence thresholds. The PTCs used were synthetic DNA fragments for wild-type only and mutant sequence (gBlock gene fragments of ∼500 bp covering KRAS exon 2, Integrated DNA Technologies), and gDNA extracted from cancer cell lines with known KRAS G12/G13 mutation status (HT-29, wild-type; LS174T, heterozygous for G12D; A-549, homozygous for G12S).
Data analysis was achieved using the QuantaSoft Analysis Pro Software (Version 1.0.596) to assign positive and negative droplets and to provide an absolute quantification of target DNA (target copies/μL of reaction). Threshold was manually set based on positive control samples for each channel, and a cut-off of three positive droplets (Rule of three) and the limit of detection of the assay were used to call a sample KRAS G12/G13 mutant. Wells with less than 10,000 accepted droplets were excluded from further analyses.
KRAS G12/G13 mutant allele concentration (copies/µL reaction, CMUT) and wild-type (WT) allele concentration (copies/µL reaction, CWT) were used for the calculation of MAF using the equation: (CMUT/(CMUT + CWT)) × 100. Moreover, since it has been reported that MAF might be influenced by variable levels of non-tumor wild-type DNA present in each sample [13], we also calculated the number of KRAS G12/G13 mutant copies/mL of original plasma used from each patient.
2.4. Determination of the Detection Limit of the ddPCR-Based KRAS G12/G13 Multiplex Assay
The limit of detection (LoD) is an important performance parameter to be established for validation of ddPCR measurements, and is defined as the lowest mutant allele fraction (MAF) that can be reliably detected and is distinguishable from the background or negative control.
For the determination of the LoD of the KRAS G12/G13 multiplex assay, a positive control mutant template (gBlock carrying the KRAS c.34G > C/p.G12R mutation) was diluted in a background of wild-type DNA (gBlock, KRAS exon 2 wild type sequence) to obtain a series of standard samples with the desired MAF range (50%, 10%, 1%, 0.2%, 0.1%). Additionally, 2 mL of pooled plasma from three healthy donors was spiked with the same standard samples with MAFs of 50%, 10%, 1%, 0.2%, 0.1%, and cfDNA was extracted as described above.
Each dilution point from standard samples and cfDNA extracted from spiked plasma, were analyzed in triplicate using ddPCR and the resulting data were merged during the calculation of the MAF of mutant KRAS.
2.5. Serial Monitoring of KRAS G12/G13-Mutated cfDNA in NSCLC Patients
In a subset of 14 patients with detectable KRAS G12/G13 mutations at baseline (T0; prior to treatment initiation), longitudinal sampling was performed at the time of first treatment response evaluation (T1; at 6–9 weeks according to treatment schedule) and on disease progression or end of first-line treatment, whichever occurred first (T2). For 6 patients, plasma samples were available at all three time points and for the remaining 8, only samples obtained at T0 and T1 were available due to disease progression at the time of first evaluation of treatment response.
At each time-point, ddPCR was carried out as described above to determine KRAS G12/G13 MAF and mutant copies/mL of plasma, and to monitor changes between baseline and T1 or T2 time points. Patients were divided into disease control group if they exhibited PR or SD (non-PD) and disease progression group if they experienced disease progression (PD).
2.6. Tissue-Based KRAS Mutational Analysis via SANGER Sequencing
In 96 out of 114 cases, KRAS mutation status was determined in formalin fixed paraffin embedded (FFPE) tissues using standard testing via Sanger sequencing and genomic DNA extracted from manually microdissected tumor cells. For the remaining 18 cases, KRAS genotyping data were not available due to inadequate tissue sample for further molecular analyses.
For Sanger sequencing, specific primers were designed to flank the KRAS exon 2 region which includes the hotspot mutations in codons 12 and 13. The primer sequences were as follows: KRAS Forward: 5′-TAAGGCCTGCTGAAAATGAC-3′ and KRAS Reverse: 5′-GTCCTGCACCAGTAATATGC-3′, resulting in a PCR amplicon of 165 bp.
PCR products were purified with the Nucleospin PCR clean-up kit (Macherey-Nagel), according to the manufacturer’s protocol. Cycle sequencing analysis was done in a final volume of 10.0 μL using: 2.0 μL of purified PCR product, BigDye Terminator v3.1 Sequencing reagents (Applied Biosystems, Foster City, CA, USA), and 125 nM each of the forward and reverse primers. The cycling conditions were as follows: 96 °C for 2 min, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 2 min. The reaction products were resolved by capillary electrophoresis on an ABI 3130 Genetic Analyzer (Applied Biosystems). Electropherograms were screened for gene alterations using the Sequencing Analysis software v5.4 (Applied Biosystems) and aligned using NCBI’s BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.7. Statistical Analysis
KRAS G12/G13 mutation load in cfDNA (MAF and copies/mL plasma) as assessed by ddPCR and patients’ clinical features and follow-up data, were subjected to statistical analyses using the IBM SPSS Version 23 software. KRAS G12/G13 mutation load was analyzed as both a categorical (presence versus absence of KRAS G12/G13 mutation in cfDNA) and continuous measure (log-transformed values of MAF, and KRAS G12/G13 mutant copies/mL). The association between KRAS G12/G13 plasma mutation status and clinicopathological features was examined by the chi-squared test or Fisher’s exact test. The Mann–Whitney U, Jonckheere–Terpstra and Kruskal-Wallis tests were employed to scrutinize the differences of KRAS G12/G13 MAF between distinct groups of NSCLC samples, as appropriate.
Survival analysis was performed by constructing Kaplan–Meier PFS and OS curves and significance was evaluated using the log-rank test. Univariate and multivariable Cox proportional hazards models were constructed for the endpoint of interest, in order to determine the prognostic value of KRAS G12/G13 mutation load and other clinical parameters. The full multivariate regression model was adjusted for important clinical factors and currently used strong prognostic indicators for NSCLC, including TNM stage, number of metastatic organs at diagnosis and performance status. Hazard ratios (HR) and 95% confidence intervals (CI) were also calculated.
For longitudinal samples, additional analyses were performed to evaluate whether variation of KRAS G12/G13 MAF and mutant copies/mL in cfDNA is associated with disease progression during treatment in NSCLC patients. In more detail, we calculated the KRAS G12/G13 MAF in each time point, and thereafter the MAF ratio (T1/T0 and T2/T0) to investigate the fold change of mutational load from baseline. An arbitrary cut-off of ≥1.5-fold change from baseline in KRAS G12/G13 MAF was used to dichotomize our patient cohort into KRAS G12/G13 MAF-increase group and KRAS G12/G13 MAF-steady/decrease group. Similar analysis was also performed for KRAS G12/G13 mutant copies/mL and the same cut-off of ≥1.5-fold change from baseline was used to dichotomize our patient cohort. The impact of KRAS G12/G13 mutational load change from baseline was assessed using the Kaplan–Meier method. The Mann–Whitney U test was used to compare KRAS G12/G13 MAF in the disease control (non-PD) and disease progression (PD) groups.
The Cohen’s Kappa coefficient (k) was used to measure the level of inter-technique agreement of KRAS mutation detection between plasma and paired tissue samples.
For all statistical analyses, p values less than 0.05 were regarded as statistically significant.
3. Results
3.1. Patients’ Characteristics
A total of 114 cases with recurrent or metastatic NSCLC were included in this study. Most patients (75.44%) received first-line platinum-based chemotherapy. Among 96 patients with available KRAS tissue genotyping data: 35 (36.46%) carried a KRAS codon 12/13 mutation in tumor tissue and 61 (63.54%) were KRAS codon 12/13 wild-type. The frequency of detection of specific KRAS mutations among mutated samples was as follows: G12C, 45.71% (N = 16/35); G12D, 28.57% (N = 10/35), G12V, 14.29% (N = 5/35), G13D, 5.71% (N = 2/35), G12S, 2.86% (N = 1/35) and G12A, 2.86% (N = 1/35). These mutations are all included in the ddPCR KRAS G12/G13 multiplex kit used for plasma-based analyses. Patients’ characteristics are summarized in Table 1.
3.2. Determination of the Limit of Detection of the ddPCR KRAS G12/G13 Multiplex Assay
Before applying ddPCR to detect KRAS G12/G13 mutations in cfDNA of patient samples, we determined the LoD of the assay using standard DNA samples with a MAF range of 50%, 10%, 1%, 0.2% and 0.1% and cfDNA extracted from spiked plasma using the same standards with the desired MAFs. The observed MAFs using ddPCR were, for the standard samples, 53.10%, 10.67%, 1.06%, 0.34% and 0.0%, respectively, and for the spiked samples, 56.10%, 10.62%, 1.26%, 0.39%, and 0.16%, respectively. The linearity range of 0.2% to 50% (concordance correlation coefficient: R2 = 0.997 for standards and R2 = 0.991 for spiked standards) indicates that samples with MAF levels higher than 0.2% can be reliably quantified and steadily detected with the ddPCR KRAS G12/G13 multiplex assay. Mutant alleles at a frequency of 0.1% were not consistently detected, and therefore the LoD of this KRAS ddPCR kit was defined as 0.2%, equivalent to two mutant copies per 1000 copies of DNA input and in agreement with the reported assay sensitivity [8].
3.3. Detection of KRAS G12/G13 Mutations in Plasma Cell-Free DNA via ddPCR: Association with Patients’ Clinicopathological Features and Response to Treatment
Plasma KRAS G12/G13 mutational load was analyzed quantitively in 114 plasma samples using the multiplex ddPCR assay. Twenty-seven (23.68%) out of 114 patients were carriers of KRAS G12/G13 mutations in plasma cfDNA. In these patients, the median plasma KRAS G12/G13 MAF was 3.95% (range, 0.20–28.27%) and the median KRAS mutant copies/mL of plasma were 192.0 copies/mL (range, 19.70–18,600.0 copies/mL). The remaining 87 patients (76.32%) were plasma KRAS G12/G13 wild-type (Table 2).
We further investigated potential associations between KRAS G12/G13 mutation status in cfDNA with the clinicopathological parameters of patients. In our cohort, KRAS G12/G13 cfDNA status was negatively associated (p = 0.031) with disease control during treatment, since KRAS mutations were detected in 37.50% of patients experiencing stable disease (SD) or partial response (PR) as compared to 62.50% of patients presenting progressive disease (PD) at first disease evaluation. KRAS cfDNA mutation status was also associated with the presence of bone metastases (p = 0.022) but not with liver (p = 0.844) or brain metastases (p = 0.899). No other statistically significant associations were observed between KRAS G12/G13 MAF in cfDNA and TNM stage (p = 0.213), smoking status (p = 0.135), PD-L1 status (p = 0.239), histologic subtype (p = 0.183), performance status (p = 0.379), gender (p = 0.361), and number of metastatic organs at diagnosis (p = 0.064) (Table 3).
Table 3.
Associations of KRAS G12/G13 MAF in ctDNA with clinicopathological data of NSCLC patients.
| Variable | No. of Patients | Mean ± S.E. a | Range | p-Value |
|---|---|---|---|---|
| TNM Stage b | 0.213 c | |||
| III | 24 | 0.34 ± 0.17 | 0.0–3.89 | |
| IV | 88 | 2.06 ± 0.56 | 0.0–28.27 | |
| Smoking status | ||||
| Non-smokers | 7 | 0.05 ± 0.03 | 0.0–0.21 | 0.135 d |
| Former smokers | 33 | 3.07 ± 1.06 | 0.0–28.27 | |
| Smokers | 68 | 1.41 ± 0.53 | 0.0–26.14 | |
| PD-L1 status | ||||
| Negative | 37 | 1.01 ± 0.38 | 0.0–8.59 | 0.239 c |
| Positive | 45 | 2.23 ± 0.84 | 0.0–26.14 | |
| Histologic type | ||||
| Adenocarcinoma | 91 | 1.85 ± 0.46 | 0.0–26.14 | 0.183 d |
| Squamous cell carcinoma | 15 | 0.17 ± 0.06 | 0.0–0.67 | |
| Other | 8 | 3.54 ± 3.53 | 0.0–28.27 | |
| Performance status e | ||||
| 0 | 86 | 1.29 ± 0.37 | 0.0–19.62 | 0.379 c |
| 1 or 2 | 27 | 3.27 ± 1.43 | 0.0–28.27 | |
| Gender | ||||
| Males | 92 | 1.38 ± 0.41 | 0.0–28.27 | 0.361 c |
| Females | 22 | 3.27 ± 1.49 | 0.0–26.14 | |
| Number of metastatic organs | ||||
| ≤2 | 88 | 1.30 ± 0.39 | 0.0–28.27 | 0.064 c |
| >2 | 26 | 3.25 ± 1.38 | 0.0–26.14 |
a Standard error. b TNM staging system. c Calculated by the “Mann–Whitney U test”. d Calculated by the “Jonckheere-Terpstra Test”. e Eastern Cooperative Oncology Group Performance Status.
3.4. KRAS G12/G13 Mutation Status in Plasma is Associated with Poor PFS and OS in NSCLC Patients
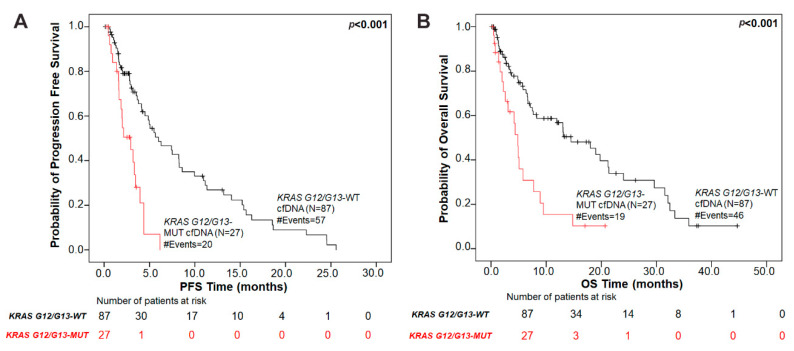
Next, we performed Kaplan–Meier survival analysis to investigate whether KRAS G12/G13 mutation status in cfDNA provides meaningful prognostic information in patients with NSCLC. Firstly, KRAS G12/G13 mutational load in plasma was used as a categorical variable (presence versus absence of KRAS G12/G13 mutations in cfDNA) to dichotomize our patient cohort into two groups. NSCLC patients in the KRAS G12/G13-mutant cfDNA group had significantly (p < 0.001) shorter PFS intervals compared to patients in the KRAS G12/G13-wild type cfDNA group (Figure 1). The median PFS of patients with KRAS G12/G13-mutant cfDNA was 2.93 months (95% CI = 1.61–6.15 months) as compared to 5.98 months (95% CI = 4.14–25.58 months) for patients in the KRAS G12/G13-wild type cfDNA group.
Figure 1.
Association of plasma KRAS G12/G13 mutation status with prognosis in NSCLC patients. Kaplan–Meier survival curves of (A) progression-free survival (PFS) and (B) overall survival (OS) based on KRAS G12/G13 mutation status in plasma. p values were calculated via the log-rank algorithm. The median PFS of patients with KRAS G12/G13-mutant cfDNA was 2.93 months as compared to 5.98 months for patients in the KRAS G12/G13-wild type cfDNA group (p < 0.001). The median OS duration for patients with KRAS G12/G13 mutations in plasma was 4.87 months versus 14.50 months for KRAS G12/G13-wild type plasma group (p < 0.001).
Regarding OS, NSCLC patients harboring KRAS G12/G13-mutant cfDNA exhibited a significantly (p < 0.001) inferior OS compared with patients in the KRAS G12/G13-wild type cfDNA group (Figure 1). The median OS duration was 4.87 months (95% CI = 2.24–7.73 months) and 14.50 months (95% CI = 7.50–21.37 months), respectively.
Using univariate Cox regression analysis, we further confirmed that KRAS G12/G13 mutation status in cfDNA is a prognostic indicator of unfavorable PFS and OS in NSCLC patients. In more details, NSCLC patients with KRAS G12/G13-mutant cfDNA had significantly higher risk of relapse ((HR) = 3.42, (95% CI) = 1.92–6.07, p < 0.001) and significantly increased risk of death ((HR) = 2.72, (95% CI) = 1.55–4.77, p < 0.001) over time, compared to the patients in the KRAS G12/G13-wild type cfDNA group.
Furthermore, univariate analysis using KRAS G12/G13 mutational load in cfDNA as a continuous variable (log-transformed values of MAF and KRAS G12/G13 mutant copies/mL of plasma) corroborated the above-mentioned results. As shown in Table 4, the risk of relapse ((HR) = 1.97, (95% CI) = 1.25–3.11, p = 0.003) and death ((HR) = 2.04, (95% CI) = 1.28–3.24, p = 0.003) was significantly associated with KRAS G12/G13 MAF in cfDNA. Similarly, the risk of relapse ((HR) = 1.76, (95% CI) = 1.26–2.47, p = 0.001) and death ((HR) = 1.65, (95% CI) = 1.18–2.30, p = 0.003) was also significantly associated with KRAS G12/G13 mutant copies/mL of plasma.
Table 4.
Cox univariate regression analysis: prognostic performance of KRAS G12/G13 mutational load in ctDNA and clinicopathological variables of NSCLC patients.
| Univariate Analysis | ||
|---|---|---|
| Variable | PFS | OS |
| KRAS ctDNA status (MUT vs. WT) | ||
| HR a | 3.42 | 2.72 |
| 95% CI b | 1.92–6.07 | 1.55–4.77 |
| p-value | <0.001 | <0.001 |
| Log10 KRAS G12/G13 MAF | ||
| HR a | 1.97 | 2.04 |
| 95% CI b | 1.25–3.11 | 1.28–3.24 |
| p-value | 0.00 3 | 0.003 |
| Log10 KRAS G12/G13 copies/mL | ||
| HR a | 1.76 | 1.65 |
| 95% CI b | 1.26–2.47 | 1.18–2.30 |
| p-value | 0.00 1 | 0.003 |
| TNM stage (IV vs. III) | ||
| HR a | 2.13 | 2.44 |
| 95% CI b | 1.12–4.07 | 1.20–4.97 |
| p-value | 0.0 22 | 0.014 |
| Number of metastatic organs (>2 vs. ≤2) | ||
| HR a | 2.49 | 4.24 |
| 95% CI b | 1.17–5.28 | 2.00–8.99 |
| p-value | 0.018 | <0.001 |
| Performance status (1 or 2 vs. 0) | ||
| HR a | 1.89 | 1.78 |
| 95% CI b | 1.14–3.13 | 1.03–3.08 |
| p-value | 0.013 | 0.041 |
| Histologic subtype (ADC vs. Other) | ||
| HR a | 1.02 | 1.01 |
| 95% CI b | 0.57–1.82 | 0.54–1.90 |
| p-value | 0.950 | 0.964 |
| Smoking history (Smokers vs. No-smokers) | ||
| HR a | 0.67 | 0.919 |
| 95% CI b | 0.42–1.07 | 0.55–1.54 |
| p-value | 0.090 | 0.749 |
| PD-L1 status (Positive vs. Negative) | ||
| HR a | 0.79 | 0.787 |
| 95% CI b | 0.47–1.35 | 0.44–1.39 |
| p-value | 0.396 | 0.410 |
| Gender (Female vs. Male) | ||
| HR a | 0.885 | 1.11 |
| 95% CI b | 0.49–1.57 | 0.63–1.98 |
| p-value | 0.678 | 0.716 |
a Hazard ratio, estimated from Cox proportional hazard regression model. b Confidence interval of the estimated HR. The values in bold indicate statistical significance (p < 0.05).
Additional clinicopathological variables associated with poor patients’ prognosis included TNM stage, number of metastatic organs and performance status (All HR > 1.0, and p values < 0.05; Table 4).
3.5. Plasma KRAS G12/G13 MAF Independently Predicts for Unfavorable Prognosis in NSCLC
A multivariate Cox regression analysis was performed to evaluate whether KRAS G12/G13 mutations in plasma cfDNA bare independent prognostic implications in terms of both PFS and OS. We firstly developed a full multivariate model that included all statistically significant predictors detected in the univariate analysis (Table 5). In this multivariate model, the presence of plasma KRAS G12/G13 mutations, was found to be a strong independent predictor of adverse prognosis for both PFS ((HR) = 3.12, (95% CI) = 1.72–5.67, p < 0.001) and OS ((HR) = 2.53, (95% CI) = 1.40–4.56, p = 0.002) in NSCLC patients. We then repeated this analysis using KRAS G12/G13 MAF in cfDNA as a continuous variable. KRAS G12/G13 MAF in cfDNA retained its independent unfavorable prognostic nature with an HR of 1.99 for PFS ((95% CI) = 1.25–3.16, p = 0.004) and an HR of 1.85 ((95% CI) = 1.21–2.85, p = 0.005) for OS.
Table 5.
Cox multivariate analysis of KRAS G12/G13 MAF as continuous and as dichotomous variable, regarding PFS and OS.
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| Variable | HR a | 95% CI b | p-Value | HR a | 95% CI b | p-Value |
| Full multivariate Model | ||||||
| KRAS G12/G13 status | ||||||
| WT | 1.00 | |||||
| MUT | 3.12 | 1.72–5.67 | <0.001 | 2.53 | 1.40–4.56 | 0.002 |
| TNM stage | ||||||
| III | 1.00 | |||||
| IV | 1.60 | 0.82–3.13 | 0.170 | 1.97 | 0.94–4.15 | 0.073 |
| Metastatic organs | ||||||
| ≤2 | 1.00 | |||||
| >2 | 1.95 | 0.91–4.23 | 0.089 | 3.38 | 1.56–7.34 | 0.002 |
| Performance status | ||||||
| 0 | 1.00 | |||||
| 1 or 2 | 1.77 | 1.04–3.02 | 0.035 | 1.56 | 0.86–2.80 | 0.141 |
| KRAS G12/G13 MAF | ||||||
| Log10 (KRAS G12/G13 MAF) | 1.99 | 1.25–3.16 | 0.004 | 1.85 | 1.21–2.85 | 0.005 |
| TNM stage | ||||||
| III | 1.00 | |||||
| IV | 1.70 | 0.37–7.76 | 0.495 | 2.05 | 0.55–7.74 | 0.288 |
| Metastatic organs | ||||||
| ≤2 | 1.00 | |||||
| >2 | 1.32 | 0.48–3.63 | 0.590 | 2.88 | 1.07–7.80 | 0.037 |
| Performance status | ||||||
| 0 | 1.00 | |||||
| 1 or 2 | 2.29 | 0.99–5.30 | 0.030 | 1.88 | 0.80–4.43 | 0.148 |
| Reduced Multivariate Models | ||||||
| KRAS G12/G13 status c | ||||||
| MUT | 3.05 | 1.69–5.48 | <0.001 | 2.70 | 1.51–4.82 | 0.001 |
| KRAS G12/G13 MAF c | ||||||
| Log10 (KRAS G12/G13 MAF) | 1.82 | 1.16–2.86 | 0.009 | 1.93 | 1.23–3.03 | 0.004 |
| KRAS G12/G13 status d | ||||||
| MUT | 3.20 | 1.79–5.73 | <0.001 | 2.48 | 1.40–4.39 | 0.002 |
| KRAS G12/G13 MAF d | ||||||
| Log10 (KRAS G12/G13 MAF) | 1.92 | 1.22–3.04 | 0.005 | 1.95 | 1.23–3.08 | 0.004 |
| KRAS G12/G13 status e | ||||||
| MUT | 3.55 | 1.98–6.36 | <0.001 | 2.73 | 1.55–4.80 | 0.001 |
| KRAS G12/G13 MAF e | ||||||
| Log10 (KRAS G12/G13 MAF) | 2.08 | 1.32–3.28 | 0.002 | 1.89 | 1.21–2.95 | 0.005 |
a Hazard ratio, estimated from Cox proportional hazard regression model. b Confidence interval of the estimated HR. c Reduced multivariate model adjusted for TNM stage. d Reduced multivariate model adjusted for number of metastatic organs. e Reduced multivariate adjusted for patients’ performance status. The values in bold indicate statistical significance (p < 0.05).
In the second step, separate reduced multivariate models were developed for plasma KRAS G12/G13 mutational load and each of the variables used in the full model. Interestingly, this analysis showed that the impact of KRAS G12/G13 mutations in ctDNA, both as a binary and as a continuous variable, on patients’ outcome is independent from TNM stage (PFS: (HR) = 3.05, (95% CI) = 1.69–5.48, p < 0.001; OS: (HR) = 2.70, (95% CI) = 1.51–4.82, p = 0.001 as a dichotomous variable; PFS: (HR) = 1.82, (95% CI) = 1.16–2.86, p = 0.009; OS: (HR) = 1.93, (95% CI) = 1.23–3.03, p = 0.004 as a continuous variable), number of metastatic organs (PFS: (HR) = 3.20, (95% CI) = 1.79–5.73, p < 0.001; OS: (HR) = 2.48, (95% CI) = 1.40–4.39, p = 0.002 as a dichotomous variable; PFS: (HR) = 1.92, (95% CI) = 1.22–3.04, p = 0.005; OS: (HR) = 1.95, (95% CI) = 1.23–3.08, p = 0.004 as a continuous variable) and performance status (PFS: (HR) = 3.55, (95% CI) = 1.98–6.36, p < 0.001; OS: (HR) = 2.73, (95% CI) = 1.55–4.80, p =0.001 as a dichotomous variable; PFS: (HR) = 2.08, (95% CI) = 1.32–3.28, p = 0.002; OS: (HR) = 1.89, (95% CI) = 1.21–2.95, p = 0.005 as a continuous variable); Table 5.
3.6. Concordance between the Detection of KRAS G12/G13 Mutations in Plasma and Tissue Samples
In the next step of our study, we examined the concordance in KRAS G12/G13 mutation status as determined using ddPCR in plasma cfDNA and via Sanger sequencing in tissue samples. Among 35 patients with KRAS G12/G13 mutant tumor samples, 18 patients (51.43%) carried KRAS mutations in plasma and 17 (48.57%) were cfDNA KRAS wild-type. When different types of KRAS mutations in tissue were considered, the frequency of mutation detection in plasma was as follows: 37.50% (N = 6/16) for G12C, 40.0% (N = 4/10) for G12D, 80% (N = 4/5) for G12V, 100% (N = 2/2) for G13D and 100% (N = 1/1) for G12S and G12A.
Of the 96 NSCLC sample pairs analyzed, we found that 18 patients (18.75%) were positively concordant; that is, KRAS G12/G13 mutations were detected in both cfDNA by ddPCR and in tumor tissues via Sanger sequencing. Additionally, 54 patients (56.25%) were negatively concordant; that is, both plasma cfDNA samples and tumor DNA were KRAS G12/G13 wild-type. These results yield an overall inter-assay concordance in KRAS mutation detection of 75.0% (concordance in 72/96 paired samples). The Cohen’s kappa coefficient was 0.395 (SE, 0.098; 95% CI, 0.20–0.59; p < 0.001), suggesting a moderate agreement between the two methods. For the remaining 24 patients, results between ddPCR in plasma and Sanger sequencing in tissue were discordant. In more detail0, KRAS mutations were not identified in the plasma of 17 (17.71%) patients with KRAS mutant tumor tissue, whereas no mutations were detected in the tumor tissue of 7 (7.29%) patients with KRAS mutant ctDNA ( Table S1).
3.7. Serial Monitoring of KRAS G12/G13 Mutations in Plasma cfDNA
Τhe impact of variations of KRAS G12/G13 mutation load in cfDNA in the monitoring of tumor burden was investigated in a subset of 14 NSCLC patients with detectable plasma KRAS G12/G13 mutations at baseline (T0).
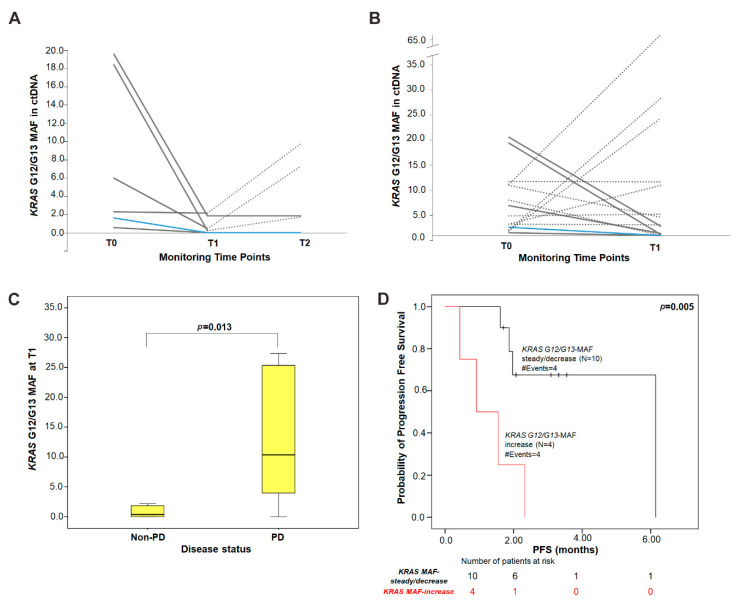
Using an arbitrary cut-off of 1.5-fold change from T0, KRAS G12/G13 MAF decreased in 7/14 patients, remained stable in 3/14 and increased in 4/14 at the time of the first response evaluation (T1). Among 10 patients with either decreased or stable KRAS G12/G13 MAF, 6 experienced disease control (5/10, SD; 1/10, PR) and 4 had progressive disease (4/10, PD), whereas all 4 patients presenting an increase in KRAS G12/G13 MAF experienced PD at T1. Of the six patients assessable at T2, one patient presented an increase and five had steady or decreased KRAS G12/G13 MAF as compared to T0. The patient with increased KRAS G12/G13 MAF had PD, whereas two out of five patients with steady or decreased MAF had disease control (1, SD; 1, PR) and three progressed (Figure 2).
Figure 2.
Serial monitoring of KRAS G12/G13 MAF in plasma cfDNA during treatment. Changes in KRAS G12/G13 MAF in (A) a subset of patients (N = 6) with available three time-points during treatment and (B) in all patients (N = 14) from baseline to T1. Each line represents one patient; dashed lines depict progressive disease (PD); solid grey lines indicate stable disease (SD) and the blue line indicates a patient with partial response (PR); (C) distribution of the absolute levels of KRAS G12/G13 MAF at T1 in patients experiencing PD (median = 10.38% MAF) compared to patients with non-PD (PR/SD; median = 0.35% MAF). p-value was calculated by the Mann–Whitney U-test; (D) Kaplan–Meier PFS survival curve according to KRAS G12/G13 MAF variation from baseline to T1 (p = 0.005). Patients were categorized into KRAS G12/G13 MAF-increase group and KRAS G12/G13 MAF-steady/decrease group according to the cut-off of ≥1.5-fold change in MAF from baseline. p values were calculated via the log-rank algorithm. T0: baseline; T1: time of first response evaluation; T2: disease progression or end of first-line treatment; PD: progressive disease; PR: partial response; SD: stable disease.
Furthermore, the absolute levels of KRAS G12/G13 MAF at T1 tended to be higher (p = 0.013; Mann–Whitney U test) in patients experiencing PD (median = 10.38%) compared to patients with disease control (non-PD; median = 0.35%) (Figure 2). Although a similar trend was observed for KRAS G12/G13 MAF at T2, this was not statistically significant (p = 0.533; median: 4.51% (PD) versus 0.93% (non-PD).
Kaplan–Meier analysis showed that patients classified in the KRAS G12/G13 MAF-increase group at T1 had shorter PFS intervals (median: 0.92 months; p = 0.005) when compared with patients in the KRAS G12/G13 MAF-steady/decrease group (median: 6.15 months) (Figure 2). In contrast, this did not reach statistical significance (p = 0.774) at T2. The same conclusions were drawn when KRAS G12/G13 copies/mL of plasma were used as a dichotomous variable (T1, p = 0.084; T2, p = 0.774).
3.8. Association of Tissue KRAS G12/G13 Mutation Status with Patient Prognosis
In our patients’ cohort, no significant relationships were observed between KRAS tumor mutation status and smoking history (p = 0.508), number of metastatic organs (p = 0.226), histologic subtype (p = 0.765), performance status (p = 0.340), gender (p = 0.791) and PD-L1 status (p = 0.214). The only positive association was found between KRAS tissue mutation status and TNM stage (p = 0.026), since KRAS mutations were more frequently found in stage IV (66.70%) as compared to stage III (33.30%) tumors.
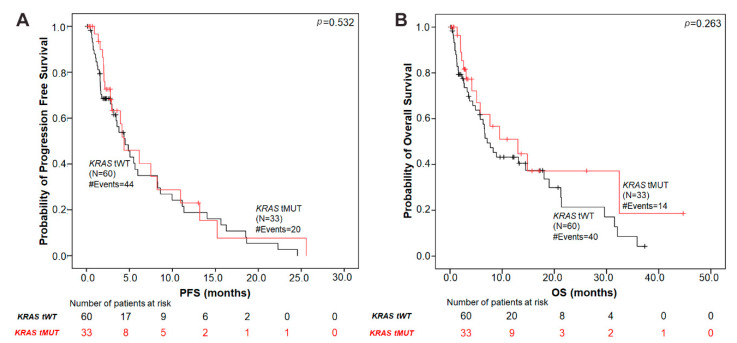
Using Kaplan–Meier survival analyses, no significant association was found between the presence of KRAS mutations in tumor tissue and patient’s prognosis in terms of both PFS (p = 0.532) and OS (p = 0.263) (Figure 3).
Figure 3.
Kaplan–Meier analysis in NSCLC patients according to tumor tissue KRAS-mutation status. (A) PFS by KRAS G12/G13 mutation status in tumor tissue. The median PFS in patients with tissue KRAS mutations was 4.37 months versus 4.50 months in the KRAS tissue wild-type group (p = 0.532). (B) OS according to tissue KRAS mutation status. The median OS in tissue KRAS mutant patients was 13.02 months versus 7.13 months in the KRAS tissue wild-type group (p = 0.263). p values were calculated via the log-rank algorithm. tMUT, tissue KRAS mutant; tWT, tissue KRAS wild-type.
Since the most frequently observed KRAS mutation was G12C, we evaluated patient outcomes according to the detection of this mutation in tumor tissue. No significant difference in terms of PFS (p = 0.406) or OS (p = 0.117) was observed between patients with KRAS G12C mutation subtype as compared to those with KRAS non-G12C mutation subtypes or with KRAS wild-type NSCLC (Figure S1). Similarly, among patients with KRAS mutant tumor tissue (N = 35), no difference in PFS (p = 0.297) was observed between those harboring KRAS G12C and KRAS non-G12C tumor mutations (p = 0.297). However, patients with the KRAS G12C mutation had a trend for longer OS (p = 0.049) but not for PFS (p = 0.217), compared to those with KRAS non-G12C mutation subtypes.
3.9. Survival Analysis According to KRAS G12/G13 Mutational Status in cfDNA and Paired tisSue Samples
Patients with available data (N = 96) were divided into four subgroups according to KRAS G12/G13 mutational status in cfDNA and paired tissue samples: (1) pMUT-tMUT patients carrying mutations in both plasma and tissue, (2) pMUT-tWT patients carrying mutations only in plasma, (3) pWT-tMUT patients harboring mutations only in tissue and (4) pWT-tWT patients that were KRAS G12/G13 wild type in both samples. The median PFS was 1.64 months (95% CI = 0.46–4.37 months) for patients with plasma KRAS mutations only (pMUT-tWT) and 10.95 months (95% CI = 2.79–25.58 months) in patients with tumor tissue mutations only (pWT-tMUT). Accordingly, the median OS was 1.64 months (95% CI = 0.46–8.91 months) for pMUT-tWT and 32.45 months (95% CI = 7.66–32.45 months) for patients with KRAS mutations in tumor tissue only. Kaplan–Meier curves for PFS and OS according to plasma and tissue mutation status are shown in Supplementary Figure S2.
The subgroup of patients with KRAS mutations detected in both plasma and matched tumor tissue (pMUT-tMUT) showed shorter PFS (p = 0.042) and a trend for shorter OS (p = 0.062) compared to patients with single specimen mutations (pMUT-tWT and pWT-tMUT) and patients that were KRAS G12/G13 wild type in both samples (pWT-tWT) (Figure S2).
4. Discussion
Overall, data concerning the prognostic and predictive role of KRAS mutations in cfDNA isolated from patients with NSCLC remain controversial, due to the relatively small number of studies performed to date [6]. In the present study, we applied a multiplex ddPCR-based assay to detect and quantify the seven most common hotspot mutations in codons 12 and 13 (G12A, G12C, G12D, G12V, G12R, G12S, and G13D) of KRAS in cfDNA from a large cohort of NSCLC patients, to explore the prognostic impact of KRAS G12/G13 MAF prior to first-line treatment for recurrent or metastatic disease. We used ddPCR, because it allows absolute quantification of tumor-derived mutations in cfDNA with superior sensitivity compared to conventional standard methods. The limit of detection (LoD) of the multiplex ddPCR assay, used in this study, was determined to be equal to 0.2%, which is in agreement with the reported assay sensitivity, demonstrating its applicability for rapid, robust and highly sensitive quantification of KRAS G12/G13 mutations in NSCLC patients’ plasma samples.
Our data demonstrate that plasma KRAS G12/G13 MAF is associated with unfavorable PFS ((HR) = 3.42, p < 0.001), and inferior OS ((HR) = 2.72, p < 0.001) in NSCLC patients. The same results were observed when KRAS G12/G13 MAF in cfDNA was expressed as a continuous variable ((HR) = 1.97, p = 0.003 for PFS; (HR) = 2.04; p = 0.003 for OS). Most importantly, we show here that KRAS G12/G13 MAF in plasma is an independent prognosticator of unfavorable outcome with respect to PFS ((HR) = 3.12, p < 0.001, as a binary variable; (HR) = 1.99, p = 0.004 as a continuous variable) and OS ((HR) = 2.53, p = 0.002, as a dichotomous variable; (HR) = 1.85, p = 0.005 as a continuous variable), irrespective of TNM stage, number of organs affected and patients’ performance status.
A number of previously published studies showed that the detection of KRAS G12/G13 mutations in cfDNA are negatively associated with survival in NSCLC patients, whereas other studies did not report such a correlation. Thus, the clinical significance of KRAS mutations detected in cfDNA has not been fully elucidated as of yet [6]. In particular, Nygaard et al. [16], using amplification refractory mutation system-quantitative PCR (ARMS-qPCR) to detect seven KRAS mutations in plasma samples from advanced NSCLC, showed the independent negative prognostic value of plasma KRAS mutations for OS, but not for PFS. Similarly, Gautschi et al. [17], using RFLP–PCR in combination with Sanger sequencing provided evidence that the detection of plasma KRAS mutations is associated with poor OS. In the same study, KRAS mutations were detected in only 9% of plasma samples (N = 16/175 patients), whereas the study cohort was heterogenous, including patients with both early and advanced disease stages [17]. On the contrary, the study by Camps et al., performed in patients’ serum [18] and plasma [19], did not show significant differences between KRAS wild-type and KRAS mutant patients, in terms of PFS and OS. This could be attributed to the fact that only two KRAS mutations at codon 12 (G12C and G12V) were examined [19].
In our study, we have used the state of the art ultrasensitive ddPCR-multiplex assay, which has been repeatedly reported to have superior sensitivity compared to standard quantitative PCR assays such as ARMS-qPCR and RFLP–PCR [13,14,20], and at the same time, enabled us to perform statistical analyses using KRAS G12/G13 MAF in ctDNA both as a categorical and as a continuous measure, thus permitting a more reliable measurement of the mutational load and its correlations with patient outcome.
We further evaluated the clinical significance of the variation in KRAS G12/G13 MAF in cfDNA in a subset of 14 NSCLC patients receiving systemic first-line therapy at two subsequent time points, at the time of treatment evaluation (T1) and on disease progression (T2). Notably, the absolute levels of KRAS G12/G13 MAF at T1 tended to be higher in patients experiencing disease progression compared to patients with objective response or stable disease. Survival analysis at T1 showed that patients classified in the KRAS G12/G13 MAF-increase group had significantly shorter PFS (p = 0.005) when compared with patients in the KRAS G12/G13 MAF-steady/decrease group. Although the number of patients used for longitudinal monitoring was rather small, our data are in agreement with the results reported by Guibert et al. [21]. In their study, by using ddPCR, they showed that in 16 patients treated with chemotherapy or targeted therapy, the variation of KRAS mutated ctDNA correlated with the response to treatment. Finally, a recent study by Zulato et al. [22] demonstrated in a larger cohort of 58 patients with advanced disease carrying KRAS mutations in tumor tissue that increasing KRAS MAF in plasma during treatment was associated with increased probability of PD and conferred a negative impact on both PFS and OS.
We next examined the concordance in KRAS G12/G13 mutation status as determined using ddPCR in plasma and via Sanger sequencing in FFPE samples. The overall inter-assay concordance was found to be 75.0% (N = 72/96 paired samples; k= 0.395; p < 0.001), which is similar to previous studies [23,24]. In 48.6% (N = 17/35 cases) of patients with KRAS mutations in tumor tissues, no mutations were detected in cfDNA. Possible causes that limit the detection sensitivity of ctDNA in liquid biopsy samples include the low contribution of ctDNA originating from tumor cells into plasma, as well as the short half-life time of ctDNA in the blood circulation [9,25]. Moreover, we also found seven cases (N = 7/61, 11.5%) that were KRAS wild-type in tumor tissue and KRAS mutant in plasma. Possible causes for this discordance might include [24,26] the intra-tumoral genetic heterogeneity [25,27] and/or the presence of mutations with low fractional abundance in tissue samples falling below the detection limit of Sanger sequencing [28,29]. Unfortunately, we were not able to provide matched tumor tissue KRAS analysis using ddPCR in these samples, due to inadequate FFPE tissue specimens. Additionally, a false-positive plasma genotyping result due to the presence of clonal hemopoiesis [30] cannot be excluded in these patients. Overall, our study indicates the feasibility of utilizing a cfDNA assay as an alternative source to determine KRAS status in cases where tissue samples are unavailable or not easily accessible.
Controversial results exist regarding the prognostic value of KRAS mutations in lung cancer tumor tissues [31]. In contrast to the prognostic associations obtained with plasma analysis, we showed that the presence of tumor KRAS G12/G13 mutations in tissue was not associated with patients’ PFS or OS. Interestingly, when KRAS tissue status was coupled with plasma results, pMUT-tMUT patients had shorter PFS (p = 0.042) and a trend for shorter OS (p = 0.062) compared to those with single specimen mutations and the wild-type subgroups. Different KRAS mutation subtypes in tumor tissue have been shown to exhibit distinct associations with patient prognosis and/or therapeutic outcomes [32,33]. In the same line, we found that patients harboring the KRAS G12C mutation in tumor tissue had a trend for longer OS, compared to those with KRAS non-G12C mutation subtypes (p = 0.049). Analysis of a larger patient cohort using the multiplex ddPCR-based assay is required to delineate the prognostic effect of different KRAS mutations in NSCLC tumor tissues.
In order to better apply these study results in clinical practice, there are several potential limitations that should be cautiously considered. Firstly, although our results are in agreement with previous reports, further validation in larger patient cohorts is needed. Secondly, we did not assess the impact of co-mutations, such as STK11/LKB1 [34], on clinical outcomes. Third, in the case of longitudinal samples, we evaluated only three time-points; additional early- or later- time points [22] in a larger patient cohort, could strengthen the significance of our findings. Other interesting perspectives would be the assessment of KRAS MAF changes in cfDNA of NSCLC patients treated with the novel targeted therapies such as the KRAS G12C specific inhibitor and the application of next-generation sequencing to track other tumor-specific co-mutations with potential prognostic significance in plasma samples.
5. Conclusions
Our study shows that KRAS G12/G13 mutations in cfDNA assessed using ddPCR are significantly associated with poor clinical outcomes, in terms of disease control, PFS and OS among patients with NSCLC treated with first-line systemic treatment, delineating the prognostic value of plasma KRAS mutations in NSCLC. Most importantly, KRAS G12/G13 mutational load in plasma represents an independent predictor of poor prognosis in these patients. We demonstrated that the dynamic changes in plasma KRAS MAF during treatment are correlated with disease progression thus underscoring the monitoring potential of this circulating biomarker. Therefore, exploration of KRAS G12/G13 mutational load in cfDNA has great potential in assisting clinicians not only in predicting patients’ outcomes but also for treatment monitoring, towards the optimal management of NSCLC patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/11/2514/s1, Figure S1: Kaplan-Meier survival curves according to tissue KRAS mutation subtypes, Figure S2: Kaplan-Meier survival curves according to KRAS G12/G13 mutational status in cfDNA and paired tissue samples, Table S1: Concordance in KRAS G12/G13 mutation status as determined using Sanger sequencing in tumor-tissues and via ddPCR in plasma ctDNA in 96 paired samples.
Author Contributions
Conceptualization, S.A.; Methodology, K.M. (Kleita Michaelidou); K.M. (Konstantinos Mavridis); C.K.; Clinical data acquisition, C.K., E.V., M.A.P.; Tumor samples characterization, A.V.K.; Data analysis, K.M. (Kleita Michaelidou); Writing—original draft preparation, K.M. (Kleita Michaelidou), K.M. (Konstantinos Mavridis), S.A.; Writing—review and editing, K.M. (Kleita Michaelidou), S.A., C.K., K.M. (Konstantinos Mavridis), D.M., A.V.K., E.V., M.A.P.; Supervision, S.A.; Funding acquisition, S.A., D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study has received funding from the European Commission Horizon 2020-Research and Innovation Program H2020-FETOPEN-1-2016-2017 (GA NUMBER-737212-CATCH-U-DNA). This work was also partly supported by the Hellenic Society of Medical Oncology (HESMO).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yuan M., Huang L.-L., Chen J.-H., Wu J., Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019;4:1–14. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Planchard D., Popat S., Kerr K., Novello S., Smit E., Faivre-Finn C., Mok T., Reck M., Van Schil P., Hellmann M., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018;29(Suppl. 4):iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 3.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood K., Hensing T., Malik R., Salgia R. Prognostic and Predictive Value inKRASin Non–Small-Cell Lung Cancer. JAMA Oncol. 2016;2:805–812. doi: 10.1001/jamaoncol.2016.0405. [DOI] [PubMed] [Google Scholar]
- 5.Yang H., Liang S.-Q., Schmid R.A., Peng R.-W. New Horizons in KRAS-Mutant Lung Cancer: Dawn After Darkness. Front. Oncol. 2019;9:953. doi: 10.3389/fonc.2019.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garzón M., Villatoro S., Teixidó C., Mayo C., Martínez A., de Gil M.L.L., Viteri S., Morales-Espinosa D., Rosell R. KRAS mutations in the circulating free DNA (cfDNA) of non-small cell lung cancer (NSCLC) patients. Transl. Lung Cancer Res. 2016;5:511–516. doi: 10.21037/tlcr.2016.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Román M., Baraibar I., López I., Nadal E., Rolfo C., Vicent S., Gil-Bazo I. KRAS oncogene in non-small cell lung cancer: Clinical perspectives on the treatment of an old target. Mol. Cancer. 2018;17:1–14. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janku F., Huang H.J., Fujii T., Shelton D.N., Madwani K., Fu S., Tsimberidou A.M., Piha-Paul S.A., Wheler J.J., Zinner R.G., et al. MultiplexKRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction. Ann. Oncol. 2017;28:642–650. doi: 10.1093/annonc/mdw670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liam C.-K., Mallawathantri S., Fong K.M. Is tissue still the issue in detecting molecular alterations in lung cancer? Respirology. 2020;25:933–943. doi: 10.1111/resp.13823. [DOI] [PubMed] [Google Scholar]
- 10.Rolfo C., Mack P.C., Scagliotti G.V., Baas P., Barlesi F., Bivona T.G., Herbst R.S., Mok T.S., Peled N., Pirker R., et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018;13:1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Guibert N., Pradines A., Favre G., Mazieres J. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur. Respir. Rev. 2020;29:190052. doi: 10.1183/16000617.0052-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 13.Rowlands V., Rutkowski A.J., Meuser E., Carr T.H., Harrington E.A., Barrett J.C. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci. Rep. 2019;9:12620. doi: 10.1038/s41598-019-49043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polivka J., Pesta M., Janku F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: Are we there yet? Expert Rev. Mol. Diagn. 2015;15:1631–1644. doi: 10.1586/14737159.2015.1110021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhauer E., Therasse P., Bogaerts J., Schwartz L., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Nygaard A.D., Spindler K.-L.G., Pallisgaard N., Andersen R.F., Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79:312–317. doi: 10.1016/j.lungcan.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Gautschi O., Huegli B., Ziegler A., Gugger M., Heighway J., Ratschiller D., Mack P., Gumerlock P., Kung H., Stahel R., et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett. 2007;254:265–273. doi: 10.1016/j.canlet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Camps C., Sirera R., Bremnes R., Blasco A., Sancho E., Bayo P., Safont M.J., Sanchez J.J., Tarón M., Rosell R. Is there a prognostic role of K-ras point mutations in the serum of patients with advanced non-small cell lung cancer? Lung Cancer. 2005;50:339–346. doi: 10.1016/j.lungcan.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Camps C., Jantus-Lewintre E., Cabrera A., Blasco A., Sanmartín E., Gallach S., Caballero C., Del Pozo N., Rosell R., Guijarro R., et al. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer. 2011;72:365–369. doi: 10.1016/j.lungcan.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Li C., He Q., Liang H., Cheng B., Li J., Xiong S., Zhao Y., Guo M., Liu Z., He J., et al. Diagnostic Accuracy of Droplet Digital PCR and Amplification Refractory Mutation System PCR for Detecting EGFR Mutation in Cell-Free DNA of Lung Cancer: A Meta-Analysis. Front. Oncol. 2020;10:290. doi: 10.3389/fonc.2020.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guibert N., Pradines A., Farella M., Casanova A., Gouin S., Keller L., Favre G., Mazieres J. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS -mutated lung adenocarcinoma. Lung Cancer. 2016;100:1–4. doi: 10.1016/j.lungcan.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 22.Zulato E., Attili I., Pavan A., Nardo G., Del Bianco P., Bragadin A.B., Verza M., Pasqualini L., Pasello G., Fassan M., et al. Early assessment of KRAS mutation in cfDNA correlates with risk of progression and death in advanced non-small-cell lung cancer. Br. J. Cancer. 2020;123:81–91. doi: 10.1038/s41416-020-0833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahangiri L., Hurst T. Assessing the Concordance of Genomic Alterations between Circulating-Free DNA and Tumour Tissue in Cancer Patients. Cancers. 2019;11:1938. doi: 10.3390/cancers11121938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S., An T., Wang J., Zhao J., Wang Z., Zhuo M., Bai H., Yang L., Zhang Y., Wang X., et al. Potential Clinical Significance of a Plasma-Based KRAS Mutation Analysis in Patients with Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2010;16:1324–1330. doi: 10.1158/1078-0432.CCR-09-2672. [DOI] [PubMed] [Google Scholar]
- 25.Keller L., Belloum Y., Wikman H., Pantel K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer. 2020:1–14. doi: 10.1038/s41416-020-01047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J.-M., Liu X.-J., Angelo P., Lin L., Wang Y., Sharma M.R., Liu Z.-Y., Tommasi S., Paradiso A. KRAS mutations in tumor tissue and plasma by different assays predict survival of patients with metastatic colorectal cancer. J. Exp. Clin. Cancer Res. 2014;33:1–8. doi: 10.1186/s13046-014-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marino F.Z., Bianco R., Accardo M., Ronchi A., Cozzolino I., Morgillo F., Rossi G., Franco R. Molecular heterogeneity in lung cancer: From mechanisms of origin to clinical implications. Int. J. Med. Sci. 2019;16:981–989. doi: 10.7150/ijms.34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Sun K., Jing C., Cao H., Ma R., Wu J. Comparison of droplet digital PCR and direct Sanger sequencing for the detection of the BRAFV600E mutation in papillary thyroid carcinoma. J. Clin. Lab. Anal. 2019;33:e22902. doi: 10.1002/jcla.22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McEvoy A.C., Wood B.A., Ardakani N.M., Pereira M.R., Pearce R., Cowell L., Robinson C., Grieu-Iacopetta F., Spicer A.J., Amanuel B., et al. Droplet Digital PCR for Mutation Detection in Formalin-Fixed, Paraffin-Embedded Melanoma Tissues. J. Mol. Diagn. 2018;20:240–252. doi: 10.1016/j.jmoldx.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y., Ulrich B.C., Supplee J., Kuang Y., Lizotte P.H., Feeney N.B., Guibert N.M., Awad M.M., Wong K.-K., Jänne P.A., et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin. Cancer Res. 2018;24:4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 31.Goulding R.E., Chenoweth M., Carter G.C., Boye M.E., Sheffield K.M., John W.J., Leusch M.S., Muehlenbein C.E., Li L., Jen M.-H., et al. KRAS mutation as a prognostic factor and predictive factor in advanced/metastatic non-small cell lung cancer: A systematic literature review and meta-analysis. Cancer Treat. Res. Commun. 2020;24:100200. doi: 10.1016/j.ctarc.2020.100200. [DOI] [PubMed] [Google Scholar]
- 32.Nadal E., Beer D.G., Ramnath N. KRAS-G12C Mutation is Associated with Poor Outcome in Surgically Resected Lung Adenocarcinoma. J. Thorac. Oncol. 2015;10:e9–e10. doi: 10.1097/JTO.0000000000000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei L., Wang W.-X., Yu Z.-Y., Liang X.-B., Pan W.-W., Chen H.-F., Wang L.-P., Fang Y., Wang M., Xu C., et al. A Real-World Study in Advanced Non–Small Cell Lung Cancer with KRAS Mutations. Transl. Oncol. 2020;13:329–335. doi: 10.1016/j.tranon.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skoulidis F., Goldberg M.E., Greenawalt D.M., Hellmann M.D., Awad M.M., Gainor J.F., Schrock A.B., Hartmaier R.J., Trabucco S.E., Gay L., et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.