Abstract
Wastewater-based epidemiology (WBE) is a useful tool that has the potential to act as a complementary approach to monitor the presence of SARS-CoV-2 in the community and as an early alarm system for COVID-19 outbreak. Many studies reported low concentrations of SARS-CoV-2 in sewage and also revealed the need for methodological validation for enveloped viruses concentration in wastewater. The aim of this study was to evaluate different methodologies for the concentration of viruses in wastewaters and to select and improve an option that maximizes the recovery of SARS-CoV-2. A total of 11 concentration techniques based on different principles were evaluated: adsorption-elution protocols with negatively charged membranes followed by polyethylene glycol (PEG) precipitation (Methods 1–2), PEG precipitation (Methods 3–7), aluminum polychloride (PAC) flocculation (Method 8), ultrafiltration (Method 9), skim milk flocculation (Method 10) and adsorption-elution with negatively charged membrane followed by ultrafiltration (Method 11). To evaluate the performance of these concentration techniques, feline calicivirus (FCV) was used as a process control in order to avoid the risk associated with handling SARS-CoV-2. Two protocols, one based on PEG precipitation and the other on PAC flocculation, showed high efficiency for FCV recovery from wastewater (62.2% and 45.0%, respectively). These two methods were then tested for the specific recovery of SARS-CoV-2. Both techniques could recover SARS-CoV-2 from wastewater, PAC flocculation showed a lower limit of detection (4.3 × 102 GC/mL) than PEG precipitation (4.3 × 103 GC/mL). This work provides a critical overview of current methods used for virus concentration in wastewaters and the analysis of sensitivity for the specific recovery of SARS-CoV-2 in sewage. The data obtained here highlights the viability of WBE for the surveillance of COVID-19 infections in the community.
Keywords: COVID-19 surveillance, Sewage, Wastewater-based epidemiology, Viral concentration methods
Graphical abstract
Highlights
-
•
Critical overview of current methods used for virus concentration in wastewaters
-
•
PEG precipitation and PAC flocculation recovered SARS-CoV-2 from sewage.
-
•
PAC flocculation showed a LOD of 4.3 × 102 GC/mL of SARS-CoV-2 in sewage.
-
•
PEG precipitation showed a LOD of 4.3 × 103 GC/mL of SARS-CoV-2 in sewage.
-
•
WBE is viable for the surveillance of COVID-19 infections in the community.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of coronavirus disease 2019 (COVID-19), is primarily transmitted through respiratory droplets, aerosols and fomites (WHO, 2020). Diarrhea is also reported in a significant proportion of the COVID-19 patients and recent reports show that infectious virions or viral RNA of SARS-CoV-2 have been detected in stool samples of COVID-19 cases and asymptomatic individuals (Chen et al., 2020; Ling et al., 2020; Wang et al., 2020). Indeed, individuals infected with SARS-CoV-2 have been reported to shed the virus in the stool for prolonged periods and with viral loads that can reach up to 1 × 106 genomic copies (GC) per gram of fecal material (Amirian, 2020; Yeo et al., 2020). Moreover, SARS-CoV-2 RNA has been found in sewage (Ahmed et al., 2020a; Medema et al., 2020), opening a new perspective in the survey of SARS-CoV-2 carriers.
Although in the COVID-19 pandemic several efforts are made to have a careful monitoring of the infected population, the precise number of infected people is difficult to assess, especially because of the high proportion of infected people that exhibit only few or no symptoms but could excrete and silently transmit the virus (Li et al., 2020; Rothe et al., 2020). Therefore, sewage surveillance could serve to monitor the circulation of the virus in the community, encompassing symptomatic and asymptomatic secretors, complementing thus current clinical surveillance. Moreover, wastewater-based epidemiology (WBE) could be useful as an early sign system of (re-) emergence of SARS-CoV-2, identification of outbreaks in the community and typing the strains circulating in the community (Polo et al., 2020).
Methods to recover SARS-CoV-2 from wastewater have varied widely in the recent reports with implications for their cost, access to reagents and scalability. In addition, prior investigations demonstrated that some standard virus concentration methods are inefficient to recover enveloped viruses from environmental water samples (Kitajima et al., 2020).
Some researchers concentrate and purify viral particles using ultrafiltration devices (Medema et al., 2020). Other groups had some success by filtrating the viral particles through electronegative membranes (Ahmed et al., 2020a). Different protocol variants of polyethylene glycol (PEG) precipitation are also used for viral concentration (Ahmed et al., 2020a; Wu et al., 2020). Another simple procedure reported is the ultracentrifugation of the wastewater, but it requires an equipment that is not often available in many laboratories (Wurtzer et al., 2020; Prado et al., 2020). Finally, aluminum-driven flocculation has consistently detected SARS-CoV-2 RNA in sewage samples when communicated cases in those regions were only incipient or not declared at all (Randazzo et al., 2020a; Randazzo et al., 2020b).
The goal of our investigation was to evaluate different methods for the concentration of viruses in wastewater in order to select and improve a concentrating option that maximizes the recovery of SARS-CoV-2 in this complex matrix.
2. Materials and methods
2.1. Sewage samples
Five raw sewage samples were collected between the 27th of March and the 20th of October 2020, from a wastewater treatment plant (WWTP) located at Neuquen city, in the province of Neuquen, Argentina. Two and a half liters of grab samples were collected from the WWTP influent, immediately stored at 4 °C, and dispatched to the Center of Research and Technological Assistance to the Industry (CIATI) for analysis.
2.2. Sewage seeding with feline calicivirus
Feline calicivirus (FCV), a non-enveloped, positive sense RNA genome virus (Etherington et al., 2006) member of the Caliciviridae family, genus Vesivirus, was used as a process control as it causes acute, self-lining oral and upper respiratory tract disease in cats and is not expected to be associated with human illnesses (Radford et al., 2007; Mattison et al., 2009). Briefly, subsamples of 200 mL of two sewage samples were seeded with 1.2 × 105 PCR units of FCV and subjected to viral concentration. One PCR Unit was defined as the last dilution of a sample from which the FCV genome could be amplified. Thus, the titer of viral RNA in a sample was the reciprocal of that dilution. One sewage sample was used for the first round of analysis, and the other sample for the duplicate.
2.3. Viral concentration
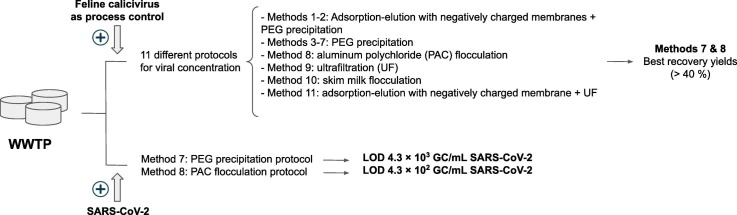
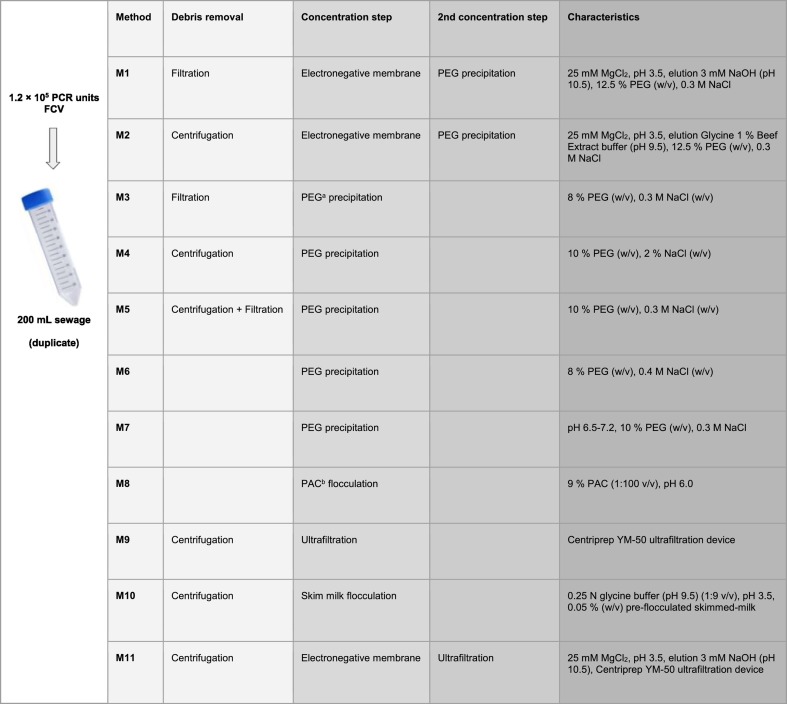
FCV was concentrated by duplicate from the seeded sewage samples using eleven previously published methods, with modifications as noted. These methods are referred as Methods 1–2 (adsorption-elution based protocols with negatively charged membranes followed by PEG precipitation), Methods 3–7 (PEG precipitation protocols), Method 8 (aluminum polychloride flocculation, PAC), Method 9 (ultrafiltration), Method 10 (skim milk flocculation) and Method 11 (adsorption-elution with negatively charged membrane followed by ultrafiltration) (Table 2).
Table 2.
Summary of the viral concentration methods used in this study.
aPEG: polyethylene glycol; bPAC: aluminum polychloride.
Method 1 (Katayama et al., 2002) began with the filtration of the sewage sample through a 0.2 μm pore-size membrane (Merck Millipore Ltd) to remove bacterial cells and debris. Then MgCl2 was added to a final concentration of 2.5 mM and pH was adjusted to 3.5. The conditioned sample was then passed through a 0.45 μm negatively charged membrane (Merck Millipore Ltd). Subsequently, 0.5 mM H2SO4 (pH 3.0) was passed through the membrane to remove cations prior to viral elution with 3 mM NaOH (pH 10.5). For neutralization, 50 μL of 100 mM H2SO4 (pH 1.0) and 100 μL 100 X Tris-EDTA buffer (pH 8.0) were added. The eluate was further concentrated by PEG precipitation by adding PEG 6000 to a final concentration of 12.5% (w/v) and NaCl to 0.3 M. The resulting suspension was stirred for 1 h at 4 °C and centrifuged at 10,000 ×g for 30 min. The PEG-containing supernatant was discarded and the pellet was suspended in 500 μL PBS and stored at −70 °C until further processing.
Method 2 is a modified version of Method 1. Briefly, the sewage sample was centrifuged at 5000 ×g for 5 min before membrane filtration. Then, as in Method 1, supernatants were passed through 2 μm membranes, conditioned with MgCl2 and passed through a 0.45 μm negatively charged membrane. Subsequently, 0.5 mM H2SO4 (pH 3.0) was passed through the membrane to remove cations prior to viral elution with Tris Glycine 1% Beef Extract buffer (pH 9.5). The eluate was stirred for 20 min at room temperature to release the membrane-adsorbed viruses and then pH was adjusted to 7.0. Viruses were further concentrated by PEG precipitation as in Method 1.
Method 3 (Wu et al., 2020) began with the filtration of sewage through a 0.2 μm membrane. Then, the eluate was concentrated by PEG precipitation by adding PEG 6000 to a final concentration of 8% (w/v) and NaCl to 0.3 M. Immediately after that, the sample was centrifuged at 12,000 ×g for 2 h. The PEG-containing supernatant was discarded and the pellet was suspended in 300 μL lysis buffer from the Direct-zol RNA MiniPrep Kit (Zymo Research) for further RNA extraction.
In Method 4 (Lewis and Metcalf, 1988) sewage was centrifuged at 4750 ×g for 20 min at 4 °C. Supernatant (S1) was maintained at 4 °C to be used later and the sediment was mixed with 3% Beef extract/2 M NaNO3 eluant (pH 5.5) and stirred for 1 h at 4 °C. Solids were then removed by centrifugation at 10,000 ×g for 20 min and the eluate was mixed with the first supernatant obtained (S1) and adjusted to pH 7.2. PEG 6000 was added to a final concentration of 10% (w/v) and NaCl to 2% (w/v). The resulting suspension was stirred for 2 h at 4 °C and centrifuged at 10,000 ×g for 25 min. The PEG-containing supernatant was discarded and the pellet was suspended in 2 mL PBS (pH 7.2), adjusted to pH 8.0, incubated for 1 h with occasional vortex, and centrifuged at 10,000 ×g for 20 min. The supernatant was stored at −70 °C.
Prior to viral concentration steps of Method 5 (Kocamemi et al., 2020), sewage was shaken at 4 °C at 100 rpm for 30 min to transfer viruses to the aqueous phase. Then, bacterial debris and large particles were removed from the samples by centrifugation at 7471 ×g for 30 min at 4 °C. Supernatant was filtered through a 0.45 μm membrane to remove remaining particles and the filtrate was mixed thoroughly with PEG 6000 (10% w/v) and NaCl (0.3 M) by shaking for 1 min. The mixture was incubated at 4 °C at 100 rpm for at least 2 h. Following incubation, viruses were precipitated by centrifugation at 7471 ×g for 2 h at 4 °C. Supernatant was removed carefully and pellets were suspended with 200 μL water. The supernatant was stored at −70 °C.
Method 6 (Iwai et al., 2009; Thongprachum et al., 2018) began with the addition of 16 g PEG 6000 (8% w/v) and 4.6 g NaCl (0.4 M) to the sewage. The suspension was stirred at 4 °C for 2 h and then centrifuged at 10,000 ×g for 30 min at 4 °C. The pellet was suspended in 2 mL of RNase-free distilled water and stored at −70 °C.
Method 7 (Lewis and Metcalf, 1988; Greening et al., 2002) began with the adjustment of pH to 6.5–7.2 and the addition of PEG 6000 (10% w/v) and NaCl (0.3 M). The solution was stirred for 2 h at 4 °C and then centrifuge at 10,000 ×g for 25 min at 4 °C. The PEG-containing suspension was discarded and the pellet was suspended in 1 mL PBS (pH 7.2). Then, pH was adjusted to 8.0 and the solution was incubated at room temperature for 1 h with occasional agitation. After incubation, the suspension was centrifuged at 10,000 ×g for 20 min and the supernatant was stored at −70 °C.
In Method 8 (Randazzo et al., 2020a) an Al(OH)3 precipitate was formed by adding 1:100 v/v of 9% aluminum polychloride solution, pH was adjusted to 6.0 and the solution was gently agitated for 30 min at room temperature. Then, the sample was centrifuged at 1700 ×g for 20 min. Pellets were resuspended into 10 mL of 3% beef extract (pH 7.4), shacked for 20 min at 80 rpm and centrifuged at 1900 ×g for 30 min. The pellet was resuspended in 1 mL of PBS and stored at −70 °C.
Method 9 was based on ultrafiltration. Sewage was centrifuged during 1 h at 10,000 ×g at 4 °C and the pellet was resuspended in 10 mL Tris Glycine 1% Beef Extract (pH 9.5). After 1 h slow agitation at 4 °C, the sample was centrifuged at 10,000 ×g for 30 min at 4 °C. Then the pH of the supernatant was adjusted at 7.0 and was subsequently concentrated using a Centriprep YM-50 ultrafiltration device (Merck Millipore Ltd) by centrifuging at 1500 ×g for 10 min. 1.5 mL of concentrated sample was collected from the filter device sample reservoir using a pipette and was stored at −70 °C.
Method 10 (Assis et al., 2017) was based on skim milk flocculation. The viruses in the sewage were precipitated by high speed centrifugation at 10,000 ×g for 1 h at 4 °C. The pellet was eluted by adding 0.25 N glycine buffer (pH 9.5) to achieve a relation 1:9 (v/v). The solution was stirred for 30 min and centrifuged at 8000 ×g for 30 min at 4 °C. The supernatant was transferred to a new 50 mL falcon tube and the pH was adjusted to 3.5. The pre-flocculated skimmed-milk solution (1% w/v) was prepared as previously described (Calgua et al., 2013) and was added to the supernatant in order to obtain a final concentration of 0.05% (w/v) of skimmed-milk. Samples were stirred slowly for 3 h at room temperature to allow floc formation and were precipitated by centrifugation at 8000 ×g for 30 min at 4 °C. The supernatants were carefully removed and the pellet was dissolved in 500 μL of PBS (pH 7.2). The final viral concentrates were stored at −70 °C.
Method 11 consisted of the combination of Methods 1, 4 and 9. Briefly, the sewage was first centrifuge at 4750 ×g for 20 min at 4 °C. Supernatant (S1) was maintained at 4 °C to be used later and the sediment was mixed with 3% Beef extract/2 M NaNO3 eluant (pH 5.5) and stirred for 1 h at 4 °C. Solids were then removed by centrifugation at 10,000 ×g for 20 min and the eluate was mixed with the first supernatant obtained (S1). 2 M MgCl2 was added to obtain a final concentration of 25 mM, pH was adjusted to 3.5 and was subsequently passed through a 0.45 μm negatively charged membrane (Merck Millipore Ltd). 0.5 mM H2SO4 (pH 3.0) was passed through the membrane prior to viral elution. Then, the membrane was placed in a 50 mL falcon tube with 3 mM NaOH (pH 10.5) and was stirred for 5 min. For neutralization, 50 μL of 100 mM H2SO4 (pH 1.0) and 100 μL 100 X Tris-EDTA buffer (pH 8.0) were added. The eluate was further concentrated by using a Centriprep YM-50 ultrafiltration device (Merck Millipore Ltd) by centrifuging at 1500 ×g for 10 min. 1 mL of concentrated sample was collected from the filter device sample reservoir using a pipette and was stored at −70 °C.
2.4. RNA extraction and molecular detection of FCV
200 μL of the viral concentrates were subjected to RNA extraction using the commercial kit Direct-zol RNA Miniprep (Zymo Research) to obtain 25 μL of RNA extract, according to the manufacturer's instructions. Five microliters of the viral RNA solution were subjected to one step real time RT-PCR assay for quantitative detection of the seeded FCV, using the kit WHATfinder Recovery Efficiency (Generon S.p.A.) and the primers and probe described by Di Pasquale et al. (2010). A standard curve (106 to 101 PCR units per reaction) was generated using tenfold serial dilutions of a FCV standard of known concentration.
2.5. Sewage seeding with SARS-CoV-2
Previous to viral seeding, subsamples of 200 mL of three sewage samples were concentrated by PEG precipitation (Method 7) and PAC flocculation (Method 8) and further analyzed for SARS-CoV-2 detection, as described below, to ensure that they did not have natural contamination with the virus. Then, 200 mL of the sewage samples were seeded with a clinical sample positive for SARS-CoV-2 to a final concentration of 4.3 × 104 genomic copies/mL (GC/mL). Serial dilutions (10−1, 10−2 and 10−3) of the SARS-CoV-2 sample were tested in triplicate in order to determine the limit of detection of the methodology. Samples were then concentrated following the methodology described above as Methods 7 (based on PEG precipitation) and 8 (based on PAC flocculation). As negative control sewage not seeded with the clinical SARS-CoV-2 sample were tested in parallel. To ensure occupational safety a certified type 2 biological safety cabinet and standard personal protective equipment (PPE), such as water-repellent romper, disposable coif, boots, shoe covers, disposable gowns, double pair of gloves, safety glasses and N95 respirator, were used.
2.6. RNA extraction and molecular detection of SARS-CoV-2
200 μL of the viral concentrates were subjected to proteinase K treatment and RNA extraction using the Maxwell® RSC 48 Extraction System (Promega) to obtain 50 μL of RNA extract, according to the manufacturer's instructions. Five microliters of the viral RNA solution were subjected to one-step real time RT-PCR assay with primers and TaqMan probe sequences that target regions of the SARS-CoV-2 nucleocapsid gene (N1 and N2), which are described by the US Centers for Disease Control and Prevention design (CDC, 2020). The internal control for the test is RNase P, which is a conserved nucleic acid sequence present in human samples. A standard curve (106 to 101 GC per reaction) was generated using tenfold serial dilutions of a SARS-CoV-2 plasmid control (2019-nCoV_N_Positive Control) provided by IDT (Integrated DNA Technologies) containing the complete nucleocapsid gene. The RT-PCR assays were performed using a Bio-Rad CFX96 thermal cycler (Bio-Rad Laboratories).
2.7. RT-qPCR controls and interpretation
All the samples were tested undiluted and diluted 10-fold to evaluate the effect of RT-PCR inhibitors. Positive and negative controls were included in each RT-qPCR run. A sample that had a cycle threshold (Ct) value below 43, with no evidence of amplification in the negative control (threshold not reached after 45 cycles) was considered positive. All the amplification reactions were run in duplicate in two independent assays.
2.8. Recovery efficiency
FCV and SARS-CoV-2 recovery efficiency for each concentration method was calculated based on the copies quantified by RT-qPCR as follows: Recovery Efficiency (%) = (Virus recovered / Virus seeded) × 100.
The mean and standard deviation for each concentration method was calculated.
2.9. Statistical analysis
The one-way analysis of variance (ANOVA) was used to determine whether there was a difference in FCV recovery among the concentration methods analyzed.
3. Results
3.1. Viral concentration methods efficiency
As preliminary screening of methods, eleven different protocols were initially evaluated. The mean efficiencies of the viral concentration methods tested for FCV recovery varied from 0% to 62.2% (Table 1 ), with an average of 10.7%. Methods 7 (a PEG precipitation approach) and 8 (based on PAC flocculation) revealed media recovery rates above 40%, Method 4 achieved a recovery rate of 9.9% and the other methods had media efficiency yields ≤1%.
Table 1.
Recoveries of feline calicivirus (FCV) from seeded sewage samples. 200 mL sewage was seeded by duplicate with 1.2 × 105 PCR units of FCV. Each concentrated sample was analyzed by RT-qPCR by duplicate.
| Concentration method | FCV recovered (PCR units) ± SDa | Mean recovery (%) (range) |
|---|---|---|
| 1 | NDb | 0 |
| 2 | 5.5 ± 9.3 | <1 |
| 3 | ND | 0 |
| 4 | 1.2 × 104 ± 1.6 × 104 | 9.9 (0.7–30.5) |
| 5 | ND | 0 |
| 6 | 2.7 ± 3.0 | <1 |
| 7 | 7.3 × 104 ± 3.7 × 104 | 62.2 (30.5–104.2) |
| 8 | 5.3 × 104 ± 2.3 × 104 | 45.0 (28.2–70.8) |
| 9 | ND | 0 |
| 10 | 1.1 × 103 ± 7.2 × 102 | 1.0 (0.2–1.6) |
| 11 | 6.5 × 101 ± 6.6 × 101 | <1 |
SD: standard deviation.
ND: Not detected.
3.2. Recovery of SARS-CoV-2 RNA from seeded wastewater samples
As Method 7 and 8 performed statistically better than the other evaluated methods (P < 0.05), SARS-CoV-2 was recovered from the seeded sewage samples after viral concentration with both methods. In order to determine the limit of detection of SARS-CoV-2 by these two methods, three serial dilutions of the viral stock were analyzed in triplicate. The internal control Rp was detected in all the samples analyzed. The minimum amount of the viral RNA detected was equivalent to 4.3 × 103 GC/mL by Method 7 and 4.3 × 102 GC/mL by Method 8 (Table 3). None of the methods could recover SARS-CoV-2 gene N1 or N2 at viral concentrations lower than 102 GC/mL in any sample. When wastewaters were seeded with SARS-CoV-2 concentration higher than 4.3 × 102 GC/mL PEG precipitation revealed a mean recovery of 8.4% and PAC flocculation of 24.0%.
Table 3.
SARS-CoV-2 recovery from sewage samples concentrated by PEG precipitation and PAC flocculation.
| Concentration method | SARS-CoV-2 seeded (GC/mL) | SARS-CoV-2 recovered |
Mean recovery (%) (range) | ||
|---|---|---|---|---|---|
| Detection (n detected/n tested) |
Mean concentration (GC/mL) ± SDa | ||||
| N1 gene | N2 gene | ||||
| PEG precipitation (Method 7) | 4.3 × 103 | 3/3 | 3/3 | 3.2 × 102 ± 3.4 × 102 | 7.4 (2.7–16.7) |
| 4.3 × 102 | 2/3 | 0/3 | 4.0 × 101 ± 6.3 × 101 | 9.4 (0–26.4) | |
| 4.3 × 101 | 0/3 | 0/3 | 0 | 0 | |
| PAC flocculation (Method 8) | 4.3 × 103 | 3/3 | 3/3 | 3.9 × 102 ± 4.5 × 102 | 9.2 (0.7–21.0) |
| 4.3 × 102 | 3/3 | 3/3 | 1.7 × 102 ± 2.0 × 102 | 38.8 (7.8–93.6) | |
| 4.3 × 101 | 0/3 | 0/3 | 0 | 0 | |
SD: standard deviation.
4. Discussion
WBE is a useful tool to monitor infectious diseases spread in the community. Lately, there has been growing evidence of the presence of SARS-CoV-2 genome in wastewater. However due to its high chemical and biological complexity, wastewater may result in low viral recovery yields, poorly reproducible yields or both (Shi et al., 2017). In addition, both particulate and dissolved constituents inherently present in wastewater get concentrated along with the target virus and can hinder the detection of viruses and influence the viral recovery yield of the concentration method (Michael-Kordatou et al., 2020). Therefore, a major challenge in SARS-CoV-2 detection in wastewater is to have an effective, standardized and optimized protocol for viral concentration (Bivins et al., 2020; Haramoto et al., 2020).
According to the available literature, there are numerous methodological approaches for concentrating viruses in sewage, such as filtration with charged membranes, precipitation, flocculation, ultrafiltration and ultracentrifugation (Stals et al., 2012). Some of these methodologies have already been used to recover SARS-CoV-2 from sewage (Ahmed et al., 2020b; Medema et al., 2020; Randazzo et al., 2020a; Wu et al., 2020), but limited information is available on the efficiency yields of the existing virus concentration methods for SARS-CoV-2 recovery (Ahmed et al., 2020b).
To evaluate the performance of the concentration techniques, virus substitutes are commonly used due to the risk associated with handling SARS-CoV-2. Since FCV is a virus with a single-stranded positive-sense RNA genome, similar to SARS-CoV-2, and testing FCV is simple and safe, it was chosen as a model for SARS-CoV-2 to evaluate the concentration efficiency. The use of a non-enveloped virus such as FCV, provides an estimation of the recovery efficiency of the methods and allows to evaluate whether the protocols worked correctly, but further evaluation with SARS-CoV-2 should be carried out. Based on the equipment available in our laboratory, eleven different methods were evaluated to concentrate viruses in wastewater.
Three of the tested methods were efficient to recover FCV from sewage, two based on PEG precipitation (Methods 4 and 7) and the other on PAC flocculation (Method 8). Of these three methodologies, Methods 7 and 8 showed mean recovery efficiencies higher than 40%.
Concentration methods based on viral filtration through charged membranes were not efficient for FCV recovery. It must be pointed out that high particulate and dissolved constituents present in the samples plugged the filters and more than 2 charged membranes were necessary for concentrating a 200 mL untreated sewage sample, even when pre-filtration or centrifugation steps were added in order to decrease the organic matter present in the samples. Our results disagree with Ahmed et al. (2020a) who have successfully used charged membrane filtration to recover murine hepatitis virus, a human coronavirus surrogate in sewage. The difference in the recovery yields may be related to the volume of sample processed. Larger volumes of wastewater were analyzed in this study, which resulted difficult to filter due to membrane clogging. Also, although no total inhibition of the RT-qPCR tests were noted in our study, the processing of larger volumes of samples could lead to the co-concentration of greater quantities of inhibitory substances and ultimately impact in the recovery yields.
Moreover, Ahmed et al. (2020a) analyzed the concentration of enveloped viruses while in our study electronegative membranes were tested for the recovery of a non-enveloped virus. Some authors reported that enveloped viruses have a greater adsorption efficiency to electronegative membranes than non-enveloped viruses, like FCV (Haramoto et al., 2009; Ye et al., 2016). However, Randazzo et al. (2020a) reported similar recovery yields for mengovirus (a non-enveloped virus) and porcine epidemic diarrhea virus (an enveloped virus member of the Coronaviridae family) when processing the samples by aluminum hydroxide adsorption-precipitation, and also their results are in line with the mengovirus recoveries reported for other concentration methodologies.
In the present study, we could not recover FCV by ultrafiltration using a centrifugal concentration device. It is probable that the high concentration of particulate matter could interfere with the viral concentration, although a centrifugation and viral elution steps were added before ultrafilter centrifugation. Moreover, it was also reported by others that viral recovery efficiency varied greatly based upon the centrifugal concentration device utilized and that not all centrifugal devices can effectively concentrate SARS-CoV-2 from wastewater (Ahmed et al., 2020a).
Methods 7 (a PEG precipitation approach) and 8 (based on PAC flocculation), which showed the best yields for FCV recovery from wastewater, were tested for the specific concentration of SARS-CoV-2. PAC flocculation showed a limit of detection of 4 × 102 GC/mL of SARS-CoV-2 in sewage, and the PEG precipitation approach revealed a limit of detection of 4 × 103 GC/mL. However, recoveries obtained by both concentration methods were variable, suggesting that the quantitative analysis is difficult and somewhat random. Although PAC flocculation showed a better efficiency for SARS-CoV-2 recovery, it turned out more tedious when sewage samples had high loads of particulate and dissolved constituents.
Different RNA extraction methods were used for FCV and SARS-CoV-2 in wastewaters because the analysis of FCV was a preliminary assay and then, for the specific recovery of SARS-CoV-2 different RNA extraction kits were assessed and evaluated to obtain the best recovery efficiencies. The nucleic acid extraction step is also a critical point in virus recovery from wastewaters which should be evaluated in depth.
Previous studies have suggested that coronaviruses may be bound to particulate matter in sewage (Gundy et al., 2009; Ye et al., 2016). It is likely that the two methodologies tested in this study were efficient in SARS-CoV-2 recovery as they incorporate the concentration of viral particles suspended in the liquid phase but also those bound to the solid fractions of the sewage.
Recently, Ahmed et al. (2020a) evaluated and compared different virus concentration techniques for the recovery of a surrogate of SARS-CoV-2 from untreated wastewater. Also, Rusiñol et al. (2020) published a review on viral concentration methods from wastewater using different surrogate viruses, some enveloped and other non-enveloped viruses. Although the use of surrogates provides an estimation of the recovery efficiency of the methods, this could be different for the virus of interest because every single virus will have a different behaviour during viral concentration. In this sense, to the best of our knowledge this is the first study that evaluates the efficiency of concentration methods for the specific recovery of SARS-CoV-2 and also the first in determining the limit of detection of the methodologies. The methods implemented in this study are being routinely used in our laboratory for the analysis of sewage from different cities of Argentina, revealing good performance for the early detection of SARS-CoV-2 in places with low viral circulation in the community (data not shown). The data obtained here highlights the viability of WBE for the surveillance of COVID-19 infections in the community.
5. Conclusions
A critical overview of current methods used for virus concentration in wastewaters was carried out in this study.
PEG precipitation and PAC flocculation protocols were efficient for SARS-CoV-2 concentration and recovery from wastewaters.
PAC flocculation showed a lower limit of detection (4.3 × 102 GC/mL) than PEG precipitation (4.3 × 103 GC/mL).
CRediT authorship contribution statement
Patricia Angélica Barril: Supervision, Investigation, Conceptualization, Methodology, Validation, Formal analysis, Writing - original draft. Luis Alfredo Pianciola: Supervision, Conceptualization, Resources, Writing - review & editing. Melina Mazzeo: Methodology, Validation. María Julia Ousset: Methodology, Writing - review & editing. María Virginia Jaureguiberry: Methodology. Mauricio Alessandrello: Methodology. Gloria Sánchez: Conceptualization, Writing - review & editing. Juan Martín Oteiza: Supervision, Conceptualization, Resources, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Centro de Investigación y Asistencia Técnica a la Industria (CIATI), Neuquén, Argentina.
Editor: Damia Barcelo
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirian S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis ASF, Otenio MH, Drumond BP, Fumian TM, Miagostovich MP, da Rosa E Silva ML. (2017). Optimization of the skimmed-milk flocculation method for recovery of adenovirus from sludge. Sci. Total Environ. 583:163–168. [DOI] [PubMed]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de Los Reyes F.L., 3rd, Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ Sci Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Calgua B., Rodriguez-Manzano J., Hundesa A., Sunen E., Calvo M., Bofill-Mas S., Girones R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J. Virol. Methods. 2013;187:215–221. doi: 10.1016/j.jviromet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Coronavirus disease 2019 (COVID-19) real-time RT-PCR panel primers and probes. 2020. www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223):507- 513. [DOI] [PMC free article] [PubMed]
- Di Pasquale S., Paniconi M., De Medici D., Suffredini E., Croci L. Duplex Real Time PCR for the detection of hepatitis A virus in shellfish using feline calicivirus as a process control. J. Virol. Methods. 2010;163(1):96–100. doi: 10.1016/j.jviromet.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Etherington GJ, Ring SM, Charleston MA, Dicks J, Rayward-Smith VJ, Roberts IN. (2006). Tracing the origin and co-phylogeny of the caliciviruses. J Gen Virol 87(Pt5):1229–1235. [DOI] [PubMed]
- Greening G.E., Hewitt J., Lewis G.D. Evaluation of integrated cell culture-PCR (C-PCR) for virological analysis of environmental samples. J. Appl. Microbiol. 2002;93:745–750. doi: 10.1046/j.1365-2672.2002.01741.x. [DOI] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ Virol. 2009;1(1):10. [Google Scholar]
- Haramoto E, Kitajima M, Katayama H, Ito T, Ohgaki S. (2009). Development of virus concentration method for detection of koi herpesvirus in water. J Fish Disease 32(3):297–300. [DOI] [PubMed]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M., Hasegawa S., Obara M., Nakamura K., Horimoto E., Takizawa T., Kurata T., Sogen S., Shiraki K. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyama, Japan (2006 to 2008) Appl. Environ. Microbiol. 2009;75(5):1264–1270. doi: 10.1128/AEM.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Shimasaki A., Ohgaki S. Development of a virus concentration methods and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002;68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M, Ahmed W, Bibby K, Carducci A, Gerba C, Hamilton K, Haramoto E, Rose J. (2020). SARS-CoV-2 in wastewater: state of the knowledge and research needs, Sci. Total Environ. 739(15):139076. [DOI] [PMC free article] [PubMed]
- Kocamemi BA, Kurt H, Sait A, Sarac F, Saatci AM, Pakdemirli B. (2020). SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. MedRxiv. doi: 10.1101/2020.05.12.20099358. [DOI]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54(8):1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. (2020). Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 6490(368):489–493. [DOI] [PMC free article] [PubMed]
- Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song Z.G., Huang W., Chen J., Hu B.J., Wang S., Mao E.Q., Zhu L., Zhang W.H., Lu H.Z. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K., Brassard J., Gagné M.J., Ward P., Houde A., Lessard L., Simard C., Shukla A., Pagotto F., Jones T.H., Trottier Y.L. The feline calicivirus as a sample process control for the detection of food and waterborne RNA viruses. Int. J. Food Microbiol. 2009;132(1):73–77. doi: 10.1016/j.ijfoodmicro.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I, Karaolia P, Fatta-Kassinos D. (2020). Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J Environ Chem Eng 8(5):104306. [DOI] [PMC free article] [PubMed]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Maranhão A.G., Siqueira M.M., Miagostovich M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A.D., Coyne K.P., Dawson S., Porter C.J., Gaskell R.M. Feline calicivirus. Vet. Res. 2007;38:319–335. doi: 10.1051/vetres:2006056. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230:113621. doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Pasco E.V., Tarabara V.V. Membrane-based methods of virus concentration from water: a review of process parameters and their effects on virus recovery. Environ Sci: Water Res Technol. 2017;3(5):778–792. [Google Scholar]
- Stals A., Baert L., Van Coillie E., Uyttendaele M. Extraction of food-borne viruses from food samples: a review. Int. J. Food Microbiol. 2012;153(1–2):1–9. doi: 10.1016/j.ijfoodmicro.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Thongprachum A., Fujimoto T., Takanashi S., Saito H., Okitsu S., Shimizu H., Khamrin P., Maneekarn N., Hayakawa S., Ushijima H. Detection of nineteen enteric viruses in raw sewage in Japan. Infect. Genet. Evol. 2018;63:17–23. doi: 10.1016/j.meegid.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. (2020). Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323(18):1843–1844. [DOI] [PMC free article] [PubMed]
- World Health Organization (WHO) Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. 2020. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations (Accessed date: 10 August 2020)
- Wu F, Zhang J, Xiao A, Gu X, Lee WL, Armas F, Kauffman K, Hanage W, Matus M, Ghaeli N, Endo N, Duvallet C, Poyet M, Moniz K, Washburne AD, Erickson TB, Chai PR, Thompson J, Alm EJ. (2020). SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 5(4):e00614–20. [DOI] [PMC free article] [PubMed]
- Wurtzer S, Marechal V, Mouchel JM, Maday Y, Teyssou R, Richard E, Almayrac JL, Moulin L. (2020). Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome 2 quantification in Paris wastewaters. medRxiv: doi: 10.1101/2020.04.12.2006267. [DOI] [PMC free article] [PubMed]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ Sci Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]