Abstract
The field of synthetic biology has used the engineered assembly of synthetic gene networks to create a wide range of function in biological systems. As part of this work, gene circuit-based sensors have primarily used optical proteins (e.g. fluorescent, colorimetric) as reporter outputs, which has limited the potential to measure multiple distinct signals. Here we present an electrochemical interface that permits expanded multiplexed reporting for cell-free gene circuit-based sensors. We have engineered a scalable system of reporter enzymes that cleave specific DNA sequences in solution, which results in an electrochemical signal when these newly liberated strands are captured at the surface of a nanostructured microelectrode. We describe the development of this interface and show its utility using a ligand-inducible gene circuit and toehold switch-based sensors, including the detection of multiple antibiotic resistance genes in parallel. This technology has the potential to expand the field of synthetic biology by providing an interface with materials, hardware and software.
Introduction
The field of synthetic biology uses genetically-encoded tools to create biological systems with new functions1,2. Work to date has generated organisms with engineered metabolic pathways for bioproduction3,4, embedded synthetic logic and memory5–7 and the capacity to sense and respond8,9. Despite being poised to revolutionize many aspects of modern life, this cell-based approach requires that all processes be laboriously encoded within a living organism10, and introduces significant complexity into the application of synthetic biology including limits to the distribution of these tools over concerns of biosafety. Recent efforts have aimed to tackle this long-standing challenge by creating cell-free synthetic biology applications that use the enzymes of transcription and translation11–13 to provide a biosafe format for applications ranging from point-of-care diagnostics to biomanufacturing to classroom education14–20. Cell-free systems are particularly advantageous as they can be freeze-dried for distribution without refrigeration and so the central motivation for many of these projects has been to provide portable diagnostics/sensors for global health, agriculture, national security and other applications that would benefit from sensing outside of laboratory settings. Sensors used in these and conventional synthetic biology studies have relied on the expression of optical reporter proteins (e.g. colorimetric, fluorescence), which, while successful, generally provide the capacity for one, or at most two or three, reporter signals from a single reaction.
Here we describe a direct gene circuit-to-electrode interface that allows for the output from engineered, cell-free gene circuits to be transformed into a signal that can be detected electrochemically. Electrochemical methods have previously been developed to detect nucleic acids21–25, small molecules26–29, and proteins30,31 with high sensitivity and specificity. However, by amalgamating programmable gene circuit-based sensors with electrochemical detection, we have created a biohybrid system that is adaptive, broadly capable and has the potential to allow 5–10 multiplexed sensors to operate with parallel but distinct signals. Importantly, this approach can be adapted to retrofit other gene circuit-based sensors by simply swapping the respective reporter proteins. We envision that electrochemical interfaces will enable multiplexed gene circuit-based portable diagnostics and, more broadly, will foster greater interaction between synthetic biology and electronic device development.
Using DNA-functionalized nanostructured microelectrodes as electrochemical detectors26,32, the activation of gene circuits is linked to specifically paired electrodes through the expression of orthogonal reporters (Fig. 1). Sequence-specific and scalable, this approach uses the production of restriction enzyme-based reporters to catalyze the release of methylene blue-labelled ssDNA (reporterDNA), which in turn interacts with complementary ssDNA (captureDNA) conjugated to the electrode surface. Upon hybridization of reporterDNA with captureDNA, methylene blue, a redox reporter molecule, is brought in close proximity to the electrode surface, allowing for a large increase in the measured current at that electrode22,27. It is this conversion of gene circuit-based sensor activation into sequence-specific DNA interactions that enables distinct and multiplexed signals to operate without crosstalk. Here, we demonstrate the power of this new electrochemical interface by detecting the activation of rationally designed toehold-switch-based RNA sensors, a small-molecule actuated synthetic gene network and demonstrate the multiplexed detection of colistin antibiotic resistance genes.
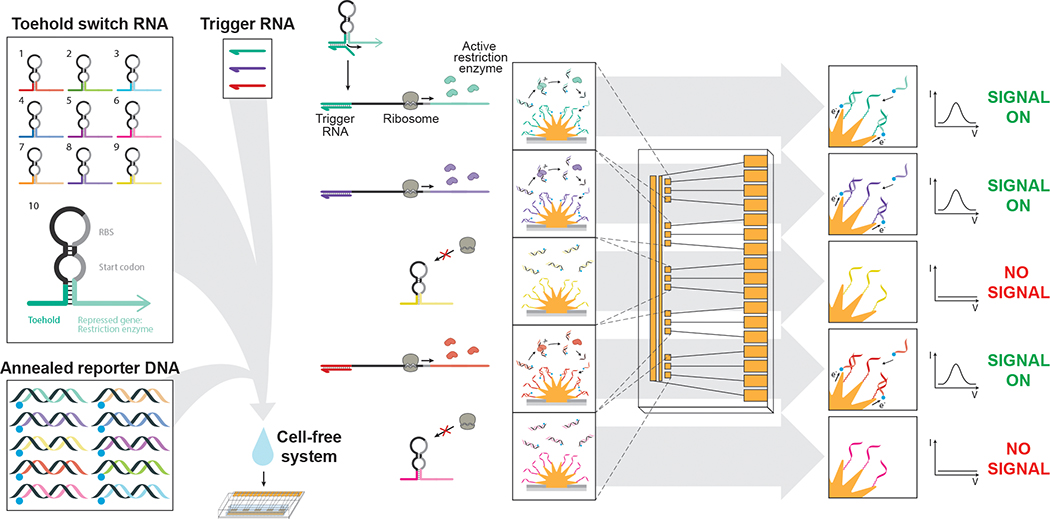
Figure 1. A gene circuit-electrode interface for cell-free synthetic gene networks.
By combining cell-free transcription and translation systems with engineered gene circuits on nanostructured microelectrodes, distinct and multiplexed output signals can be tracked in parallel. This approach uses toehold switch-based RNA sensors, which, in the presence of trigger RNA, express one of ten restriction enzyme-based reporters. Upon sensor activation, expressed restriction enzymes cleave annealed reporterDNA, which is free floating in cell-free reactions, releasing the redox reporter-labelled reporterDNA (blue circle). Nanostructured microelectrodes with conjugated captureDNA then recruit the redox-active reporterDNA to their surface, generating an electrochemical signal. Each toehold switch is engineered to produce a unique restriction enzyme-based reporter that is coupled to a distinct reporterDNA and captureDNA pair for multiplexed signaling
Results and Discussion
Screening for high-performing restriction enzyme-based reporters
The creation of this electrochemical approach to gene circuit-based sensor signaling required that we first identify a set of restriction enzymes capable of rapid and robust performance. We screened 66 commercially available restriction enzymes for cleavage activity under buffer conditions required for the cell-free transcription and translation system (Fig. 2a, b)11. The ability of restriction enzymes to cleave target DNA was evaluated using gel electrophoresis (data not shown). The commercially available PURExpress cell-free system (NEB) was selected for experiments because of its recombinant nature, meaning that it is free from background nuclease activity. Of the 37 restriction enzymes that were capable of cleavage, DNA sequences encoding for 26 restriction enzymes were designed and proteins were successfully expressed upon transcription by T7 RNA polymerase (RNAP) in the cell-free system (Supplementary Table 1, Supplementary Fig. 1). After a three-hour expression period, restriction enzyme activity was tested using gel-electrophoresis-based analysis (Supplementary Fig. 2, Supplementary Table 2).
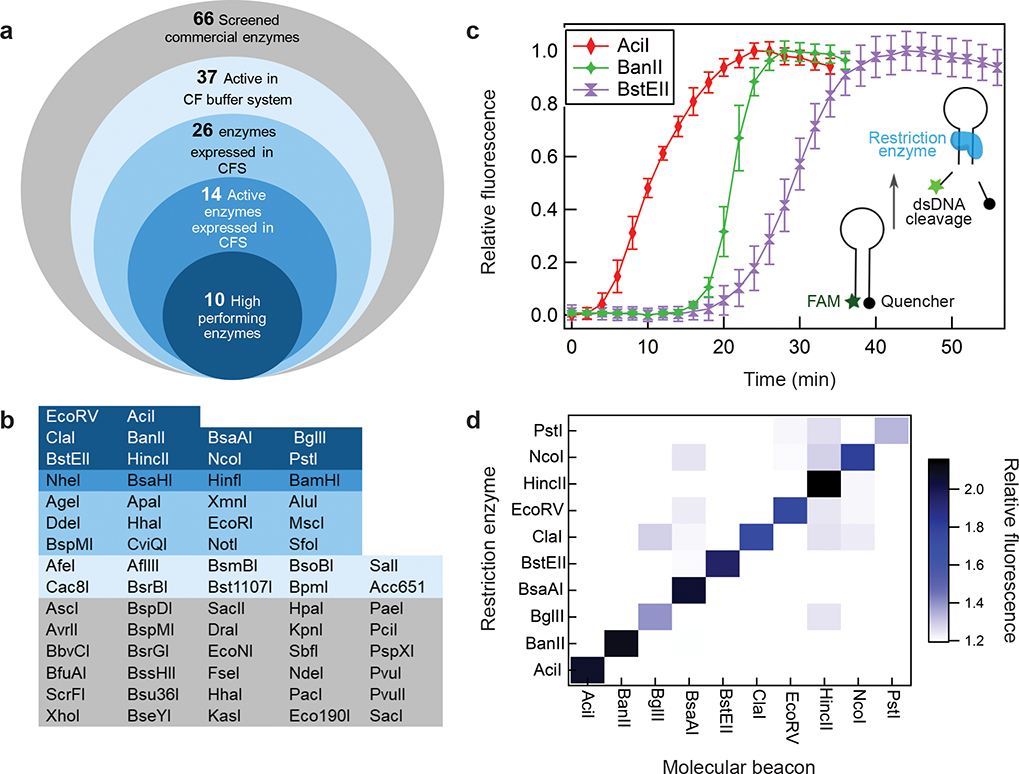
Figure 2. Development of orthogonal, restriction enzyme-based reporters.
a. Candidate restriction enzymes were evaluated through a four-step screening pipeline (i-iv) designed to find enzymes with high rates of expression and processivity in the cell-free expression system (CFS). i) 37 of 66 commercially available enzymes demonstrated activity in the cell-free (CF) buffer system that replicates the pH, buffer and salt composition found in the complete transcription and translation system, ii) 26 of the 37 above restriction enzymes were successfully expressed de novo in the cell-free system (Supplementary Fig. 1), iii) 14 of these 26 cell-free expressed enzymes showed high levels of cleavage activity (Supplementary Fig. 2), iv) 10 of the 14 enzymes demonstrated high rates of enzyme-mediated cleavage from de novo cell-free expression. b. A summary of the performance for screened restriction enzymes with colors matching the categories described in Figure. 2a. c. Representative data of three candidate restriction enzyme-based reporters in molecular beacon cleavage assays. Data presented as percent of maximum fluorescence for each molecular beacon, error bars represent SE (N=3, Supplementary Fig. 3). d. Heat map of specific enzyme activity. All combinations of restriction enzymes and molecular beacons were tested. Values are average of triplicates at 180 min.
Given that restriction enzyme expression and processivity are critical for reporter performance, we developed a fluorescence-based molecular beacon assay to monitor DNA cleavage in real-time as the restriction enzymes were expressed in vitro. The hairpin-based molecular beacons contain a DNA recognition site for each of the respective restriction enzymes and are designed with a 5’ FAM-6 fluorophore and a 3’ BHQ-1 quencher (Fig. 2c, Supplementary Table 3)33. Upon restriction-enzyme-mediated cleavage, the FAM-6 fluorophore is released, allowing for tracking of enzyme activity over time. De novo expression of candidate restriction enzymes resulted in significant cleavage activity by six restriction enzymes in as early as fifteen minutes (AciI, BanII, BsaAI, BstEII, ClaI, EcoRV) and by others at later time points (HincII, BglII, NcoI, PstI; Supplementary Fig. 3). The orthogonality of these ten restriction enzymes was then tested by expressing each enzyme in the presence of all ten respective molecular beacons individually. Results indicated good orthogonality, with little crosstalk between restriction enzymes (Fig. 2d).
Electrochemical detection of restriction enzyme-activated redox reporters
With a set of restriction enzymes established, we next developed the companion electrode and redox reporter systems. Each chip contains an array of fifteen micropatterned electrodes arranged in five sets of three, which were prepared using standard photolithography techniques (Fig. 3a, b). Briefly, gold electrodes were patterned on a glass wafer, followed by a layer of photoresist, to create six 400 μm x 20 μm openings over each electrode and to prevent nonspecific interactions (Supplementary Fig. 4). Electrodeposition in a gold chloride solution was then employed to create nanostructured microelectrode topologies, which were found to provide optimal speed of detection and sensitivity (Supplementary Fig. 5)34. Nanostructured electrodes were tested over time and found to perform stably, with little decrease in current, when stored under ambient atmosphere, humidity and temperature (Supplementary Fig. 6). After electrode preparation, captureDNA complementary to the reporterDNA for each restriction enzyme was conjugated to each of the five triplicate electrode sets via a terminal thiol group; 6-mercaptohexanol (MCH) was added as a co-adsorbent to minimize electrostatic repulsion on the electrode surface (Supplementary Table 4).
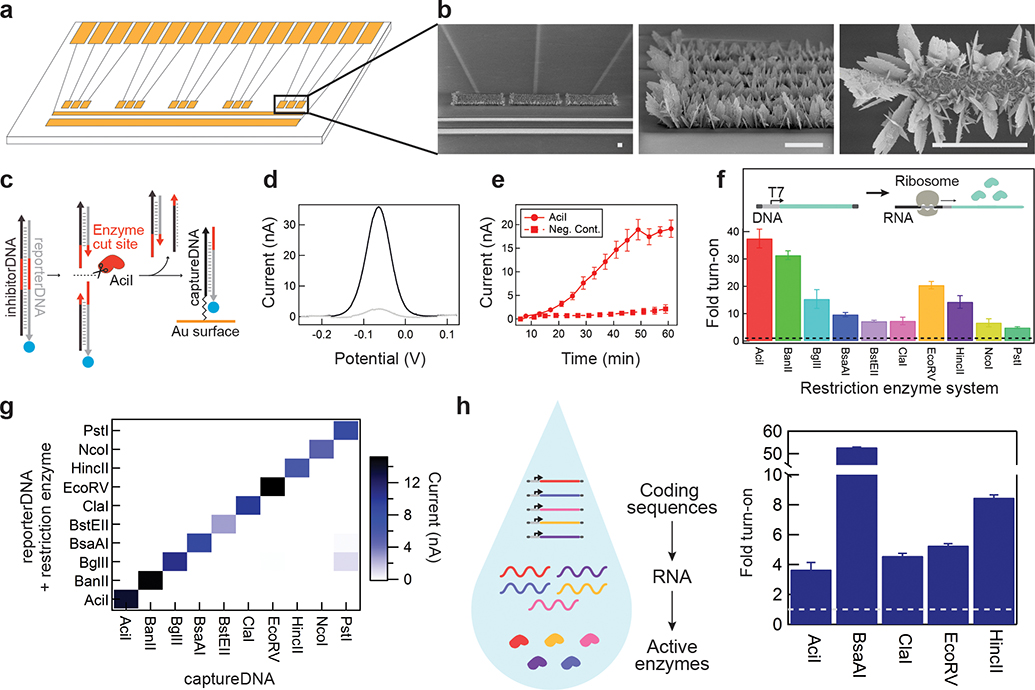
Figure 3. Electrochemical detection of restriction enzyme reporters.
a. A schematic of the electrochemical detection chip designed to detect (in triplicate) reporterDNA generated by five restriction enzyme-based reporters in parallel. b. Scanning electron microscopy images of nanostructured microelectrodes. Scale bar is 50 μm. c. In solution, restriction enzymes cleave a reporter/inhibitor DNA duplex. The reporterDNA strand carrying methylene blue (blue circle) is then recruited to the surface of the electrode through duplex formation with conjugated captureDNA, bringing the electrochemical reporter molecule to the electrode surface. d. Representative square wave voltammetry data showing the measured current with (black) and without (gray) restriction enzyme expression. e. On-chip square wave voltammetry measurements in real-time as the restriction enzyme AciI is expressed (Supplementary Fig. 8). f. Fold turn-on of measured peak current in the presence of restriction enzymes for each of the ten respective reporterDNA-captureDNA systems. Data was normalized to the measured current in the absence of DNA encoding each restriction enzyme, represented as dotted line (Supplementary Fig. 9). g. Electrochemical reporterDNA-captureDNA systems were tested to evaluate cross-reactivity between respective restriction enzyme reporters. Values on heat map represent the average current of triplicates at 30 min. h. Using methylated DNA, fold turn-on from the co-expression of five restriction enzyme reporters in a single solution and measured on a single chip. Data was normalized to the measured current in the absence of DNA encoding each restriction enzyme, represented as dotted line (Supplementary Fig. 10). Data represents the mean ±SE of three replicates. All electrochemical measurements were performed with square wave voltammetry and peak current is used for calculation of fold turn-on.
Release of the complementary reporterDNA is catalyzed by restriction enzyme-dependent cleavage of free-floating DNA duplexes in cell-free reactions (Fig. 3c). These stable DNA duplexes are comprised of a full-length reporterDNA strand and a complementary inhibitor DNA strand (iDNA), which prevents interaction with electrodes in the absence of cleavage (Supplementary Fig. 7). Conversely, upon duplex cleavage, the truncated reporterDNA is designed to hybridize with captureDNA and bring the terminal methylene blue label to the electrode surface, enabling electron transfer to create a restriction enzyme-dependent electrochemical signal (Fig. 3d). The preference of the truncated reporterDNA for the captureDNA is driven by mismatched base pairing with the iDNA, which leads to greater relative stability (higher melting temperature, ΔG) when hybridized to on-chip captureDNA (Supplementary Table 5). As in the molecular beacon assay, restriction enzyme specificity results from recognition of sequence-specific DNA cleavage sites, and electrochemical detection requires conjugation of the complementary captureDNA on the electrode surface.
Using T7-RNAP expression, we showed that the restriction enzyme-mediated electrochemical signal can be detected in as little as 20 minutes after transcription initiation using square wave voltammetry (Fig. 3e, Supplementary Fig. 8). We next determined that this model of restriction enzyme-mediated signaling is scalable with the demonstration of ten restriction enzyme-electrode pairs (Fig. 3f, Supplementary Fig. 9). Here expression constructs for each restriction enzyme were combined in cell-free reactions with their respective reporterDNA duplexes (reporter DNA-iDNA) and electrodes containing one of the ten captureDNA strands. After a 1-hour off-chip incubation reaction at 37 °C, all ten pairs led to significant electrochemical signal fold change at 30 minutes once applied to the electrodes. The orthogonality of all ten restriction enzymes was then tested by repeating the expression of each enzyme in the presence of the ten reporterDNA duplexes individually. The resulting electrochemical signaling demonstrated strong orthogonality and little crosstalk (Fig. 3g). A subset of these restriction enzymes could also be co-expressed in a single reaction mixture to generate clear electrochemical signals on chip. In this experimental set-up, all five restriction enzyme expression constructs and their corresponding reporterDNA duplexes were combined in a single cell-free reaction volume (Fig. 3h left). When expressing multiple restriction enzymes in a single pot, it is important to ensure that enzymes are not cross-reactive towards the DNA encoding the respective restriction enzymes. To address this, restriction enzyme expression was performed using methylated DNA to prevent restriction enzyme-mediated cleavage (Fig. 3h, Supplementary Figs. 10, 11) and, alternatively, by modifying their DNA sequences to remove restriction enzyme cleavage sites (Supplementary Fig. 11, Supplementary Table 6). These strategies effectively limited cross-reactivity and broadened the pool of restriction enzymes that could be used as reporters. With reporter validation complete, we now refer to these restriction enzymes simply as reporter enzymes.
Electrochemical detection of gene circuit driven expression: TetO and toehold switches
With the fundamental components for the electrochemical interface complete, we next developed applications to demonstrate electronic sensing of gene circuit activation. We began with TetO-regulated expression of a reporter enzyme (Fig. 4a, Supplementary Table 7). TetO is a 19-bp operator sequence that can be placed between a promoter and a gene of interest to provide tetracycline-responsive expression. In this system, transcription is regulated through Tet repressor (TetR) binding to the T7-TetO promoter region, which inhibits transcription, and the small molecule tetracycline analog anhydrotetrcycline (ATc), which relieves this inhibition (Fig. 4a, left). By placing the reporter enzyme AciI under TetO regulation, we were able to demonstrate the corresponding activation and inhibition of electrochemical signaling in the presence and absence of ATc, respectively (Fig. 4a, right). These signals corresponded closely with positive and negative controls.
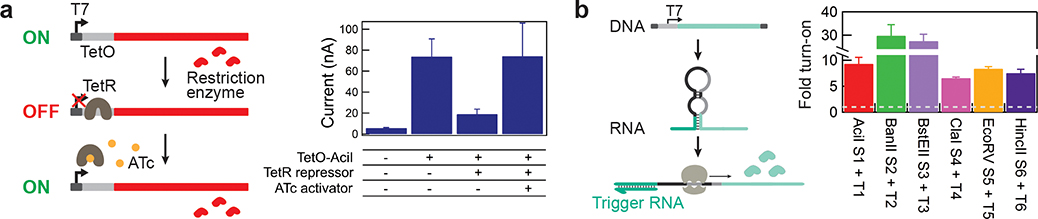
Figure 4. Application of the gene-circuit electrochemical interface for small molecule- and RNA-actuated electrochemical signaling.
a. Anhydrotetracycline (ATc)-mediated derepression of TetR-regulated TetO expression of the restriction enzyme-based reporter AciI (left). ATc-dependent induction (40 μM) of electrochemical signaling on-chip (right). Data represents the mean ±SE of three replicates. b. Toehold switches specific to synthetic RNA sequences were designed to control the expression of six different restriction enzyme-based reporters. RNA-dependent activation of toehold switches induces electrochemical signaling. Dotted line indicates switch alone negative controls. All electrochemical measurements were performed with square wave voltammetry and peak current is used for calculation of fold turn-on. Data represents the mean ±SE of three replicates.
The multiplexed linkage of gene circuits to electronics has the potential to enable automated and high-capacity biosensing. We have previously demonstrated that toehold switches can be designed to recognize specific RNA sequences and that these RNA sensors can be used to identify the presence of pathogens14,15. To explore the potential of multiplexing such sensing capacity, we used six toehold switches designed to recognize six synthetic model sequences (Supplementary Table 8)35. Cell-free reactions containing a toehold switch, with or without its respective RNA trigger sequence, were monitored electrochemically at 37 °C on microelectrode arrays containing the complementary captureDNA and free-floating reporterDNA duplex. This resulted in the sequence-specific RNA activation of toehold switches and electrochemical outputs that increased 7- to 30-fold (Fig. 4b) compared to background. The fold change values represent the maximum signal between 60 and 90 minutes, with significant signals observable in as little as 15–20 minutes (Supplementary Fig. 12). Importantly, as previously demonstrated14,35, the performance of riboregulators is switch-dependent (Supplementary Fig. 13), and here we show that this signal can also be modulated by the choice of reporter enzyme (Supplementary Fig. 14), providing a further layer of engineering control.
Application: Detection of colistin antibiotic resistance genes
The overuse of antibiotics has given rise to the growing threat of drug resistance. With the vast majority of antibiotic use related to agricultural settings, farms represent a significant risk for the emergence of resistance36,37. Here we developed toehold switch-based sensors specific to the coding regions of resistance genes for the key last-line antibiotic colistin (MCR-1, MCR-2, MCR-3 and MCR-4). These genes have recently been identified in livestock globally and represent a dangerous threat to the efficacy of an antibiotic of last resort.
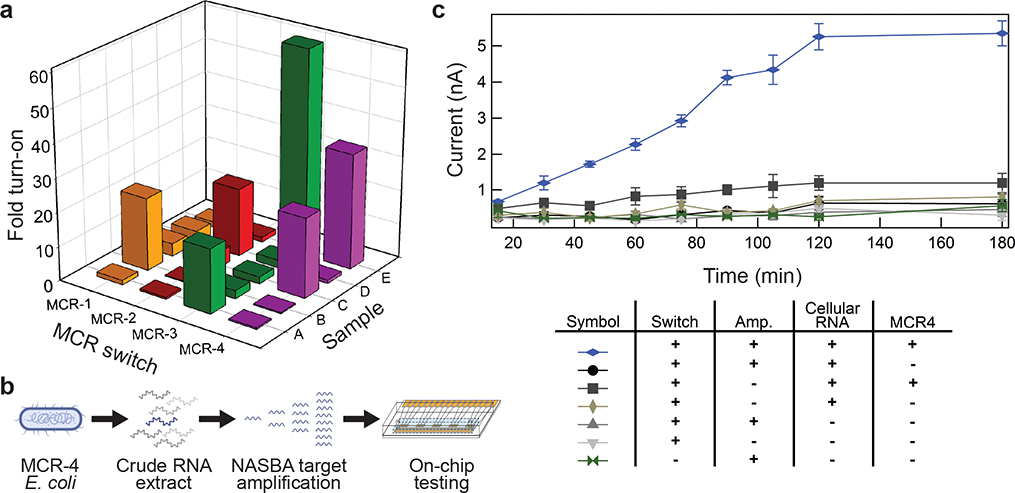
Using a purpose-built algorithm for toehold switch design15, each MCR gene was computationally screened for regions of low structural complexity that could be targeted by toehold switches. We then synthesized toehold switch designs complementary to the 24 top ranked binding sites within each MCR gene and ligated a unique reporter enzyme gene to each set of switches (MCR-1_EcoRV, MCR-2_AciI, MCR-3_BanII, MCR-4_ClaI). Reporter enzyme selection was based on the speed of DNA cleavage in time-course assays (Fig. 2c and Supplementary Fig. 3). The performance of the resulting 96 sensors was then screened against their respective RNA sequences for optimal ON/OFF ratios using molecular beacons (Supplementary Fig. 15, Supplementary Tables 9, 10). Top performing sensors were then validated on microelectrode arrays. With the activity of each MCR sensor linked to a unique reporter enzyme, we first demonstrated that the corresponding electrode array for all four MCR genes could detect each gene independently (Fig. 5a, samples A-D). Each of the MCR RNAs (1nM, samples A-D) were added to four cell-free reactions each containing one set of the toehold sensors (MCR-1_EcoRV, MCR-2_AciI, MCR-3_BanII, MCR-4_ClaI) and corresponding reporterDNAs, and incubated for 30 minutes off-chip at 37°C. After separate incubation, the four reactions were pooled together on-chip (e.g. samples A, B, C and D) for incubation at 37°C. The electrochemical response for each of the four MCR genes individually provided a clear signal enhancement of ~20-fold after 50 minutes. We then demonstrated that this capacity could be multiplexed, enabling the specific and simultaneous detection of RNA sequences from MCR-3 and MCR-4 (Fig. 5a, Supplementary Fig. 16). The full data sets with standard error can be found in Supplementary Fig. 16.
Figure 5. Detection of Mobilized Colistin Resistance (MCR) genes.
a. Toehold switch-based RNA sensors were designed and screened for the detection of four MCR genes. Five separate experiments were performed on-chip in the presence of all components except the corresponding MCR-specific trigger RNA(s). The first four experiments (samples A-D) test the detection of single MCR-related RNAs (1 nM) based on the electrochemical response (Sample A: MCR-3 RNA trigger, Sample B: MCR-1 RNA trigger, Sample C: MCR-4 trigger and Sample D: MCR-2 trigger). Sample E tests the co-detection of MCR-3 and MCR-4 RNA triggers (1 nM each) in parallel (for standard error data see Supplementary Fig. 16). Data was normalized to the measured current in the absence of trigger RNA (5 μM). Graphs represent peak current for methylene blue using square-wave voltammetry. Data represents the mean ± SE of three replicates. b. Work flow for detection of MCR-4 RNA from a complex sample. c. On-chip electrochemical signaling from activation of MCR-4_ClaI in the presence of MCR-4 RNA from complex whole cell RNA samples isolated from E. coli. Tested with a combination of inputs, the real-time signal is only detected in the presence of MCR-4 RNA and isothermal amplification. Cellular RNA was isolated from DH5α E. coli cells in the presence or absence of a plasmid expressing MCR-4. All electrochemical measurements were performed with square wave voltammetry and peak current is used for calculation of fold turn-on. Switch, MCR-4_ClaI; Amp, NASBA with primers (+) or without primers (−). Data represents the mean ± SE of three replicates.
As with other cell-free sensors15,20, we next used an isothermal amplification reaction (1 hour) to specifically amplify target sequences before adding the sample to cell-free reactions containing toehold sensors. The limit of detection (LOD) without an amplification step was determined through on-chip sensing of MCR-1 at RNA concentrations between 10 nM and 150 nM using 200 nM MCR-1_EcoRV switch following the experimental design described above. These measurements yielded a calculated LOD of 64.5 nM (Supplementary Fig. 17). With the addition of a Nucleic Acid Sequence Based Amplification (NASBA) step upstream of the electrochemical workflow, we were able to extend the detection of the antibiotic resistance gene MCR-4 to the low femtomolar range (1 fM), an improvement of over seven orders of magnitude (Supplementary Fig. 18). In these experiments, MCR-4 RNA at 1 fM was added to a NASBA reaction (1 hour) and this amplified RNA was then added to cell-free reactions at 14% of total cell-free reaction volume. Here the combined workflow led to a robust increase in electrochemical signal on-chip after 120 minutes of incubation at 37 °C. This level of performance compares well with the turnaround time and sensitivity of other recently published cell-free synthetic biology-based detection schemes (2–3 hours)15,38.
Finally, with a long-term goal of applying this approach to the interrogation of real-world samples, we wanted to determine the capacity of our on-chip detection for more complex samples (Fig 5b). Here the MCR-4 gene was expressed in E. coli and then total RNA was collected from the resulting culture. Total cellular RNA was then added to the NASBA reaction (1 hour) at a concentration of 30 ng/μl for isothermal amplification using MCR-4 specific primers. The amplified MCR-4 mix was added without purification to the cell-free reactions containing the MCR-4_ClaI switch, followed by 30 minutes of off-chip and 15 minutes of on-chip incubation at 37 °C prior to taking the first electrochemical measurement (Fig. 5c). Electrochemical activation of the system was specific to MCR-4 RNA in the presence of high background off-target RNA sequences and provided a strong, distinct signal against negative controls.
Conclusion
Taken together, we have presented a series of proof-of-concept experiments for a direct and scalable interface between engineered gene circuits and electronics. Interfacing cell-free synthetic biology with electronics will enable engineered gene networks to curate mixed molecular information and rapidly share data with computational tools, a capability that promises to drive more sophisticated and interactive applications. Potential uses include high-content multiplexing systems for decentralized sensing in health, agriculture, national security and industry, among others. Using low-cost electronics, ultimately, we envision this interface to enable dozens of diagnostics to operate for the cost of a single test in our current colorimetric format (<$1/test)15. Moreover, by simply modifying the upstream molecular sensor elements in the system, the same reporter enzyme-electrode pairs can be left unchanged, along with common microelectrode hardware, to serve any sensor application in principle. Toehold switches can be rationally designed35 and therefore the platform can be tailored to detect virtually any nucleic acid sequence. The stable and biosafe nature of the cell-free format also means that the technology can be used without the limitations of cellular systems, potentially enabling new applications and operating environments outside of the laboratory.
This approach also holds exciting technical implications for the field of synthetic biology. First, this work highlights the potential for chemistry to enable and mediate signaling for synthetic gene networks, creating a much-needed mechanism for increasing the bandwidth of sensor outputs. This contrasts with conventional reporters, which are optical and have a limited capacity for multiplexing9,15. While sensing arrays can be contemplated for optical detection (e.g. microarrays), the complexity of these devices and the optics needed for detection are major disadvantages. Second, tackling another key challenge in the field, here we demonstrate that rather than having to encode decision making and memory features genetically into gene circuits, we can off-load these features to attached electronics. The system continues to take advantage of biology’s incredible capacity to sense, but has the potential to dramatically reduce the time needed to develop synthetic biology applications. With this approach, the underlying connectivity of sensory outputs can be re-programmed at will, easily creating any number of logic calculations (e.g. AND gates, etc.) by simply modifying the code at the level of the software rather than at the level of the DNA or RNA. Looking forward, we see this bioelectrochemical approach as providing the field a new enabling venue and one that provides new opportunities for even greater interdisciplinarity and rational design of chemical and biological systems.
Methods:
Chip fabrication:
Microelectrode patterns including reference, counter, and working electrodes were generated using standard contact photolithography techniques from glass substrates layered with chrome, gold, and with or without positive photoresist (AZ1600) obtained from Telic Company or EMF. The working electrodes were nanostructured using electrodeposition in a solution of 50 mM AuCl3 in 0.5 M HCl. A standard three-electrode system with an Ag/AgCl reference electrode and a platinum counter electrode was run at a constant potential of 0 mV for 100 s using Bioanalytical Systems Epsilon potentiostat (West Lafayette, IN, USA). Finally, 100-μm high PDMS channels were fabricated and bonded to chips using standard soft lithography techniques. See supplementary methods for detailed protocols for each step.
CaptureDNA deposition:
CaptureDNA strands were obtained from Integrated DNA Technologies containing a 6-carbon linker with a terminal thiol. Final concentrations of 10.5 μM of captureDNA along with 5 μM of mercaptohexanol (MCH) were deposited on nanostructured working electrodes, and incubated in a humid environment at room temperature for approximately 14 hr. In order to deposit multiple DNA capture strands on a single chip, 3145 RTV silicone adhesive sealant (Dowsil, Midland, Michigan, USA) was used to create separate chambers for DNA deposition (Supplementary Fig. 4), and the glue was removed after overnight incubation with the DNA solutions. Chips were then washed 3x with 1x PBS. A solution of 1 mM MCH was added to cover the working electrodes of each chip to backfill any gold surface and prevent nonspecific interaction. After incubation for 3 hr at room temperature, chips were washed 3x with 1x Phosphate-Buffered Saline, pH 7.4 (PBS), then rinsed with ddH2O and dried under a stream of N2.
Reporter DNA preparation:
ReporterDNA was ordered from Integrated DNA Technologies with a terminal amine, which was used for labeling with a methylene blue NHS Ester (Glen Research, Sterling, VA, USA) according to manufacturer’s protocol. Labeled DNA was then purified using reverse phase HPLC, dried via lyophilization, and re-dissolved in 1x PBS. Then reporter and inhibitor DNA strands were annealed at ratio of 1:4 (for initial proof-of-concept experiments) or 1:10 (for multiplexed experiments) in 1x PBS incubated at 95 °C for 4 min. The solutions were then cooled slowly to room temperature.
Cell-Free expression of restriction enzymes:
All experiments were performed using the recombinant cell-free protein expression system (CFS), PURExpress (E6800S, NEB, Ipswich, MA, USA) following manufacturer’s procedures with an additional 0.5% by volume RNase inhibitor (M0314S, NEB). All DNA constructs and gene-circuits were designed to be compatible with this cell-free system. Restriction enzyme expression reactions were assembled using 10 nM linear DNA encoding for the restriction enzyme in CFS. To measure restriction enzyme expression and activity electrochemically, the reporterDNA-iDNA complex was added to a final concentration of 100–280 nM reporterDNA. If measurements were to be made in real-time, the solution was then added directly to the electrochemical chips for measurement during incubation at 37 °C. Unless otherwise stated, the reactions were incubated at 37 °C for 1 hr before addition to chips. The reactions were then incubated on the chips for 15–30 min at 37 °C before electrochemical measurements were obtained.
Electrochemical Measurements:
All measurements were performed using either a Bioanalytical Systems Epsilon potentiostat or a PalmSens PSTrace potentiostat, both with a three-electrode system. An on-board gold reference electrode was used in addition to a platinum wire auxiliary electrode. The multiplex chip was designed to house both an on-board gold reference and counter electrodes. Square wave voltammetry (SW) signals were obtained with a potential pulse step of 1 mV at a frequency of 60 Hz and an amplitude of 25 mV, with measurements taken from 100 to −450 mV.
Toehold switch-based multiplexed sensing of MCR antibiotic resistances:
For proof of concept experiments (Fig. 4b) cell-free reactions were set up as described above supplied with 10 nM DNA encoding the toehold switches. For multiplexed MCR detection, switches were supplied as RNA, with concentrations of 200 nM MCR-1_EcoRV, 25 nM MCR-2_AciI, 250 nM MCR-3_BanII, and 100 nM MCR-4_ClaI. Respective switches were triggered by 1 nM RNA trigger sequences in presence of 100 nM reporterDNA (Fig. 5a).
Data and materials availability
All raw data presented and used in the manuscript are available upon request from the corresponding authors.
Code availability
All custom computer code used in the manuscript are available upon request from the corresponding authors.
Supplementary Material
Acknowledgments:
Molecular components of ligand-inducible gene-circuit were kindly provided by the Doktycz lab. The plasmid pSB3C5-proD-B0032-E0051 was a gift from Joseph Davis & Robert Sauer (Addgene plasmid # 107241).
Funding: J.B.C. is funded by the Ontario Graduate Scholarship. This work was supported by the NSERC Discovery Grants Program (RGPIN-2016-06352), CIHR Foundation Grant Program (201610FDN-375469), The University of Toronto’s Connaught New Research Award and the CIHR Canada Research Chair Program (950-231075) to K.P.; the University of Toronto’s Medicine by Design initiative which receives funding from the Canada First Research Excellence Fund (C1TPA-2016-06) to K.P./S.O.K.; an NIH Director’s New Innovator Award (1DP2GM126892), an Arizona Biomedical Research Commission New Investigator Award (ADHS16-162400), an Alfred P. Sloan Research Fellowship (FG-2017-9108), Gates Foundation funds (OPP1160667), and Gordon and Betty Moore Foundation funds (#6984) to A.A.G. We would like to thank Seray Cicek for her help enhancing the high-throughput data analysis. We would like to thank Mahmoud Labib and Carine Nemr for their advice and support on the project.
Footnotes
Competing interests: Authors declare no competing interests.
References and Notes:
- 1.Cameron DE, Bashor CJ & Collins JJ A brief history of synthetic biology. Nat. Rev. Microbiol 12, 381–90 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Cheng AA & Lu TK Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng 14, 155–78 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Fossati E et al. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun 5, 3283 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Smanski MJ et al. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol 14, 135–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green AA et al. Complex cellular logic computation using ribocomputing devices. Nature 548, 117–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yehl K & Lu T Scaling Computation and Memory in Living Cells. Curr. Opin. Biomed. Eng 4, 143–151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitada T, DiAndreth B, Teague B & Weiss R Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao N, Cubillos-Ruiz A, Cameron DE & Collins JJ Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med 10, eaao2586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotula JW et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U. S. A 111, 4838–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keasling JD Synthetic biology and the development of tools for metabolic engineering. Metab. Eng 14, 189–95 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Shimizu Y et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol 19, 751–755 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ & Swartz JR An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol 4, 220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J, Jardine P & Noireaux V Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth. Biol 1, 408–413 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Pardee K et al. Paper-based synthetic gene networks. Cell 159, 940–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pardee K et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 165, 1255–1266 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Pardee K et al. Portable, On-Demand Biomolecular Manufacturing. Cell 167, 248–259.e12 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Huang A et al. BioBitsTM Explorer: A modular synthetic biology education kit. Sci. Adv 4, eaat5105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark JC et al. BioBitsTM Bright: A fluorescent synthetic biology education kit. Sci. Adv 4, eaat5107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen KY et al. A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa -Infected Respiratory Samples. ACS Synth. Biol 6, 2293–2301 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Takahashi MK et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun 9, 3347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alligrant TM, Nettleton EG & Crooks RM Lab on a Chip Electrochemical detection of individual DNA. Lab Chip 349–354 (2013). doi: 10.1039/c2lc40993c [DOI] [PubMed] [Google Scholar]
- 22.Fan C, Plaxco KW & Heeger AJ Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci 100, 9134–9137 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan HU et al. In situ, label-free DNA detection using organic transistor sensors. Adv. Mater. 22, 4452–4456 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Patolsky F, Lichtenstein A & Willner I Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol 19, 253–257 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Slinker JD, Muren NB, Gorodetsky AA & Barton JK Multiplexed DNA-modified electrodes. J. Am. Chem. Soc 132, 2769–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das J et al. An ultrasensitive universal detector based on neutralizer displacement. Nat. Chem 4, 642–8 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Zuo X, Xiao Y & Plaxco KW High Specificity, Electrochemical Sandwich Assays Based on Single Aptamer Sequences and Suitable for the Direct Detection of Small-Molecule Targets in Blood and Other Complex Matrices. J. Am. Chem. Soc 131, 6944–6945 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuang Z, Kim SN, Crookes-goodson WJ, Farmer BL & Naik RR Biomimetic Chemosensor : Designing Peptide Recognition Elements for Surface Functionalization of Carbon Nanotube Field Effect Transistors. ACS Nano 4, 452–458 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Xiang Y, Lu Y & Crooks RM Aptamer-Based Origami Paper Analytical Device for Electrochemical Detection of Adenosine. Angew. Chemie Int. Ed 51, 6925–6928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das J & Kelley SO Protein Detection Using Arrayed Microsensor Chips: Tuning Sensor Footprint to Achieve Ultrasensitive Readout of CA-125 in Serum and Whole Blood. Anal. Chem 83, 1167–1172 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Tang D, Yuan R & Chai Y Ultrasensitive Electrochemical Immunosensor for Clinical Immunoassay Using Thionine-Doped Magnetic Gold Nanospheres as Labels and Horseradish Peroxidase as Enhancer. Anal. Chem 80, 1582–1588 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Sage AT, Besant JD, Lam B, Sargent EH & Kelley SO Ultrasensitive electrochemical biomolecular detection using nanostructured microelectrodes. Acc. Chem. Res 47, 2417–25 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Li JJ, Geyer R & Tan W Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res. 28, E52 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soleymani L, Fang Z, Sargent EH & Kelley SO Programming the detection limits of biosensors through controlled nanostructuring. Nat. Nanotechnol 4, 844–848 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Green AA, Silver PA, Collins JJ & Yin P Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J et al. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep 8, 3705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García V et al. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 52, 104–108 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Gootenberg JS et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karig DK, Iyer S, Simpson ML, Doktycz MJ. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res 40, 3763–3774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM & Pierce NA, “NUPACK: Analysis and design of nucleic acid systems,” Journal of computational chemistry 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res 2011;39(3):1131–41. 10.1093/nar/gkq810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.