Abstract
Pinus massoniana Lamb, an economically important conifer tree, is widely distributed in China. WRKY transcription factors (TFs) play important roles in plant growth and development, biological and abiotic stress. Nevertheless, there is little information about the WRKY genes in P. massoniana. By searching for conserved WRKY motifs in transcriptomic RNA sequencing data for P. massoniana, 31 sequences were identified as WRKY TFs. Then, phylogenetic and conserved motif analyses of the WRKY family in P. massoniana, Pinus taeda and Arabidopsis thaliana were used to classify WRKY genes. The expression patterns of six PmWRKY genes from different groups were determined using real-time quantitative PCR for 2-year-old P. massoniana seedings grown in their natural environment and challenged by phytohormones (salicylic acid, methyl jasmonate, or ethephon), abiotic stress (H2O2) and mechanical damage stress. As a result, the 31 PmWRKY genes identified were divided into three major groups and several subgroups based on structural and phylogenetic features. PmWRKY genes are regulated in response to abiotic stress and phytohormone treatment and may participate in signaling to improve plant stress resistance. Some PmWRKY genes behaved as predicted based on their homology with A. thaliana WRKY genes, but others showed divergent behavior. This systematic analysis lays the foundation for further identification of WRKY gene functions to aid further exploration of the functions and regulatory mechanisms of PmWRKY genes in biological and abiotic stress in P. massoniana.
Keywords: P. massoniana, WRKY, biological and abiotic stress, expression pattern
1. Introduction
Plants are capable of managing various types of stress during their life cycle and have developed several mechanisms to adapt to biotic and abiotic stress. Families of transcription factors (TFs) play an important role in stress response by regulating the expression of target genes, interacting with specific cis-acting elements present in their promoter regions, and acting as activators or repressors in plant stress responses [1,2]. WRKY transcription factors (TFs) constitute one of the largest families of transcription regulators in plants. The most prominent feature of the WRKY protein is its highly conserved WRKY domain, which is composed of 60 amino acids and has a "WRKYGQK" amino acid motif at the N-terminus [3,4]. In a few WRKY proteins, the conserved “WRKYGQK” amino acid sequence can also be replaced by various other forms, such as “WRKYDHK”, “WRKYDQK” and “WRKYGKK” [5]. In addition, the WRKY protein contains a C2H2- or C2HC-type zinc finger structure [5]. Based on structural differences in the WRKY domain and zinc finger, WRKY TFs have been divided into three main groups [5]. Members of the first group contain two WRKY domains and one C2H2 zinc finger motif, those of the second group include a WRKY domain and a C2H2 zinc finger motif, and those of the third group have a C2HC zinc finger motif [6].
Since the initial report of the first WRKY protein in sweet potato, WRKYs have been widely identified and analysed throughout the plant lineage [7]. Previous studies have indicated that some WRKY TFs also play key roles in increasing or decreasing plant resistance to biological and abiotic stresses, and these TFs were found to differ from one another with respect to their inducibilities and functions [6]. For example, the StWRKY45 gene showed high expression under low-phosphorus stress in Stylosanthes guianensis, so it was speculated that the StWRKY45 gene might be involved in the response to low-phosphorus stress [8]. In transgenic Arabidopsis thaliana, AtWRKY15 and AtWRKY17 enhance tolerance to salt and osmotic stress [9]. AtWRKY6 was significantly induced by mechanical damage in A. thaliana [10]. Studies have shown that the WRKY protein affects the signal transduction process mediated by plant hormones, such as abscisic acid (ABA) and salicylic acid (SA) [10,11]. For instance, EgrWRKY70, EgrWRKY46 and EgrWRKY53 have synergistic effects with the hormones SA and methyl jasmonate (MeJA) and play a positive regulatory role in Eucalyptus grandis [12].
Pinus massoniana is a tree species of Pinaceae that is widely distributed and is an important raw material in industrial and agricultural production. P. massoniana is highly resistant to drought and barren land and exhibits high levels of secondary metabolic changes [13]. The biosynthetic regulators of secondary metabolism play a very important role in plant adaptation to environmental stress. P. massoniana is, therefore, a good specimen for exploring stress response mechanisms in woody plants. Hence, the identification of functional genes in P. massoniana is of great interest. In addition, as Pinus massoniana is widely distributed in a variety of environments, facing different stresses and lacking of management, the economic losses caused by biological and abiotic stresses are very serious. WRKY genes is one of the largest family of transcription factors in plants, and play a key role in the regulation of plant biotic and abiotic stress response. Therefore, functional identification of the WRKY family in P. massoniana is necessary. Some chromosomal WRKY homologues have evolved to perform different functions in different species, so it is difficult to draw conclusions by comparing WRKY genes and homologous chromosomes [6]. The increasing number of sequenced genomes has facilitated genome-wide identification and analysis of WRKY genes on a large scale in some species, aiding the understanding of their biological functions [14]. The genome of P. massoniana is more than 200 times larger than that of A. thaliana, so it is difficult to rapidly obtain the sequence; therefore, we used the transcriptome to identify the WRKY gene family here. The WRKY gene family may be crucial for the remarkable tolerance of P. massoniana, and we identified 31 PmWRKY genes based on RNA-seq, and the classification, domain structures, subcellular localization prediction, and expression patterns of PmWRKYs were analysed using bioinformatic methods. In this study, we analysed the transcript levels and expression patterns of six selected PmWRKY genes under the effects of stress and phytohormone treatments. This research will not only enrich the understanding of the molecular regulation mechanism of the WRKY gene family but also provide useful information for further exploring the function and regulatory mechanism of PmWRKY genes under biological and abiotic stress in P. massoniana.
2. Materials and Methods
2.1. Selection of PmWRKY Genes
Transcriptome data for Masson pine were derived from the previously determined CO2 stress transcriptome [15] and young shoots transcriptome (PRJNA655997). Based on the WRKY domain (PF03106), the hidden Markov model file was downloaded from the Pfam protein family database. HMMER 3.0 was selected to search for WRKY genes in the databases of Iso-Seq. The default parameters were used for screening, and the E value was set to E < 10−3 [14]. Pfam (http://pfam.xfam.org/) and CD-search (https://www.ncbi.nlm.nih.gov/cdd/) were used to screen out protein sequences of Masson pine with the WRKY domain. Finally, sequences with complete WRKY domains were selected and sequences more than 97% similarity between different databases were deleted.
2.2. Sequence Analysis
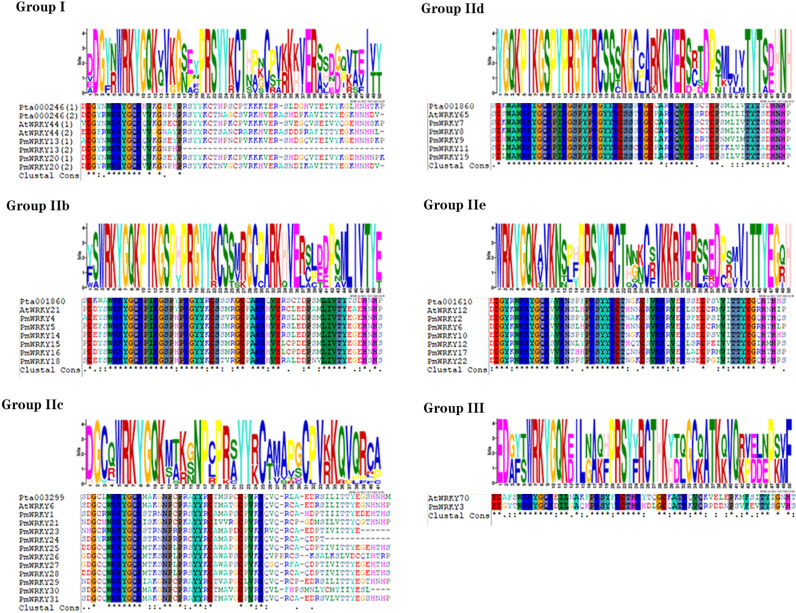
The molecular weights and isoelectric points of the identified PmWRKY proteins were obtained using tools from the ExPASy website. The subcellular localization of PmWRKY proteins was predicted and analyzed with CELLO (http://cello.life.nctu.edu.tw/) and PSORT (https://psort.hgc.jp/). In addition, we also located the NLS of sequences with cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/) and NLStradamus (http://www.moseslab.csb.utoronto.ca). Based on the AtWRKY classification in A. thaliana, 65 AtWRKY proteins from groups I, II, or III were selected and downloaded from the Arabidopsis Information Resource (TAIR) to analyze the phylogenetic relationships among the AtWRKY proteins, the 21 PtWRKY proteins and the 31 PmWRKY proteins in this study [6,16]. The deduced amino acid sequences of PmWRKY domains spanning ~60 amino acids were aligned with AtWRKYs and PtWRKYs using the Clustal program (version 2.1). A phylogenetic tree of the deduced amino acid sequences from WRKY domains in A. thaliana, P. taeda and P. massoniana was constructed using the maximum likelihood method, and a graphical representation was produced with the help of MEGA-X software using 1000 bootstrap replicates [4,17]. The multiple sequence alignment (MSA) of the 60-amino-acid conserved region of P. massoniana was visualized using DNAMAN. The MSA included conserved regions of WRKY members representing each group and subgroup from A. thaliana and P. taeda as references [18,19]. The conserved motifs of PmWRKY proteins were analyzed using Multiple Expectation Maximization for Motif Elicitation (MEME: http://meme-suite.org/tools/meme) with the following parameters: minimum and maximum motif widths, 6 and 50, respectively; and maximum number of motifs, 10 [4,20].
2.3. Subcellular Localization
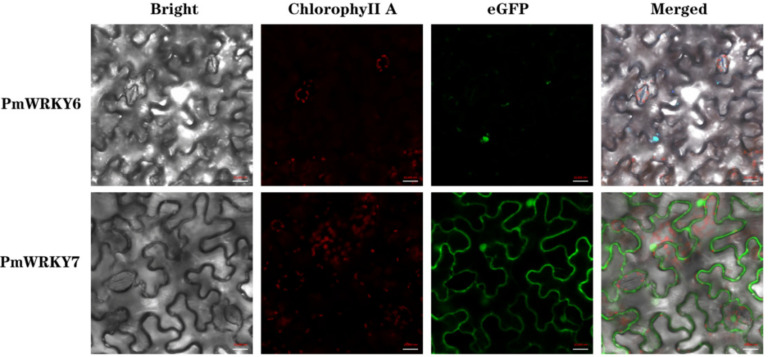
PmWRKY6 and PmWRKY7 were selected for a transient expression experiment. The coding DNA sequence (CDS) regions were inserted into a pJIT166-GFP expression vector. Transient expression vectors (35S::PmWRKY6-GFP and 35S::PmWRKY7-GFP) containing green fluorescent protein (GFP) were transferred into the leaves of Nicotiana benthamiana following the method of Li [21]. The fluorescence signals were observed with an LSM 710 confocal microscope (Zeiss, Jena, Germany).
2.4. Plant Material and Treatments
Two-year-old Masson pine seedlings obtained from the seed orchard of the Baisha state-owned forest farm, Shanghang, Fujian Province, China, were used in this study. To study the expression level of PmWRKY, we subjected the Masson pine seedlings to five treatments. The five treatments included abiotic stress and hormone treatments, namely, mechanical injury, 10 mM H2O2, 100 M methyl jasmonate (MeJA), 500 μM ethephon (ETH) and 1 mM salicylic acid (SA). The mechanical damage treatment involved cutting the upper half of the needle, and the rest of the treatments were spray treatments. Three uniformly growing seedlings were selected for each treatment as three biological replicates. The needles were taken as samples every 0 h, 3 h, 6 h, 12 h, 24 h and 48 h, immediately frozen in liquid nitrogen and stored at −80 ℃. Samples collected at 0 h without any treatment were used as controls.
2.5. RNA-Seq Data Analysis of PmWRKY Genes
Information about CO2 treatment of P. massoniana for Illumina RNA-seq is provided below. In brief, one-year-old P. massoniana seedlings were used in this study. Individuals of the same clones with similar heights, uniform growth and strong growth potential were selected as the test materials and subsequently moved into a growth chamber. The growth conditions were 10 h light/14 h dark cycles at 25 °C in the chamber. Air containing approximately twice the CO2 concentration in the chamber before the experiment was aerated into the growth chamber constantly for at least 24 h. The CO2 concentration in the chamber was monitored by an infrared CO2 analyzer (SenseAir, Delsbo, Sweden). The seedlings were sampled after 6 h, 12 h and 24 h of treatment with the high CO2 concentration [15]. For each treatment, leaves collected from three triangular bottles were sampled as three biological replicates for RNA-seq. The mixed RNA sample (mixture combining treated plant samples from all time points) was sent for Iso-Seq. Fragments per kilobase of exon model per million reads mapped (FPKM) values were calculated to estimate the abundance of P. massoniana WRKY gene transcripts. The relative expression of control check(CK) was set as "1". Online OmicShare tools were used to create heatmaps based on the values of log2 (FPKM +0.01).
2.6. RNA Extraction and Quantitative Real-Time Reverse Transcription PCR
Total RNA was extracted using the RNAprep Pure Kit (DP441, Tiangen Biotech, Beijing, China). RNA concentration and purity were measured with a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was estimated by 1.2% agarose gel electrophoresis [22]. First-strand cDNA was synthesized using the One-step gDNA Removal and cDNA Synthesis Kit (AT311, TransGen Biotech, Beijing, China). Primers for quantitative real-time reverse transcription PCR (qRT-PCR) were designed using Primer 5.0 (Data S1). SYBR Green reagents were used to detect the target sequence. Each PCR mixture (10 µL) contained 1 µL of diluted cDNA (20× dilution), 5 µL of SYBR Green Real-time PCR Master Mix, 0.4 µL of each primer (10 µM), and 3.2 µL of ddH2O. The PCR program had six stages: (1) 95 °C for 60 s (preincubation); (2) 95 °C for 15 s, (3) 60 °C for 15 s and (4) 72 °C for 10 s, repeated 40 times (amplification); (5) 95 °C for 0.5 s; and (6) 60 °C for 1 min (melt). The PCR quality was estimated based on melting curves. TUB (tubulin beta) was used as the internal control [22]. Three independent biological replicates and three technical replicates for each biological replicate were examined. Quantification was achieved using comparative cycle threshold (Ct) values, and gene expression levels were calculated using the 2-∆∆Ct method. The significance was determined by the t-test using SPSS statistical software (IBM, New York, NY, USA) (* p < 0.05, ** p < 0.01).
3. Results
3.1. Verification of WRKY Proteins in P. massoniana
Initially, 55 gene candidates corresponding to the WRKY family were obtained from the Pfam database. Further verification was performed using the CD-search program based on these gene models to ensure the presence of the WRKY domain. Following the removal of incorrectly predicted WRKY genes and redundant sequences, 31 genes were selected and annotated as P. massoniana WRKY genes. These WRKY genes were named PmWRKY1–PmWRKY31 (Table 1). The coding sequences of validated PmWRKY genes are available in Supplementary Data S2. The number of amino acids in the predicted protein products varied from 99 to 788. Among the 31 PmWRKY proteins, PmWRKY15, with 330 amino acids, was the smallest, while the largest protein was PmWRKY20 (788 amino acids). The range of protein molecular weights was 11.3–86.2 kDa, and the isoelectric point values ranged from 4.68 (PmWRKY8) to 9.99 (PmWRKY26). According to the results of CELLO and PSORT prediction of the subcellular localization of PmWRKY1-PmWRKY31 proteins, almost all the proteins have a predicted nuclear localization. The prediction results show that 31 sequences have NLS or similar sequences. The prediction results for these proteins are shown in Table 1 and Table S2. In order to verify the nuclear localization, we transiently expressed PmWRKY6 and PmWRKY7 fused to GFP in N. benthamiana leaves and we analyzed their subcellular localization. The fluorescence signals of PmWRKY6/7:GFP were observed on the nucleus (Figure 1).
Table 1.
Summary of PmWRKY Sequences.
| Gene ID | Gene | cDNA Length | Aa | MW(kDa) | pl | Group | WRKY Domain | Subcellular Localization | NLS |
|---|---|---|---|---|---|---|---|---|---|
| m.15387 | PmWRKY1 | 996 | 331 | 37.6 | 8.9 | IIb | WRKYGQK | Nucleus |

|
| m.7589 | PmWRKY2 | 1428 | 475 | 50.9 | 5.82 | IIc | WRKYGQK | Nucleus |

|
| m.7738 | PmWRKY3 | 1434 | 477 | 52.5 | 5.56 | III | WRKYGQK | Nucleus |

|
| m.6876 | PmWRKY4 | 1347 | 448 | 48.8 | 9.41 | IId | WRKYGQK | Nucleus |

|
| m.8763 | PmWRKY5 | 1092 | 364 | 40.4 | 9.68 | IId | WRKYGQK | Nucleus |

|
| m.256914 | PmWRKY6 | 783 | 261 | 29.8 | 6.49 | IIc | WRKYGQK | Nucleus |

|
| m.293152 | PmWRKY7 | 1134 | 377 | 41.0 | 4.75 | IIe | WRKYGQK | Nucleus |

|
| m.293159 | PmWRKY8 | 1269 | 422 | 45.7 | 4.68 | IIe | WRKYGQK | Nucleus |

|
| m.199902 | PmWRKY9 | 1377 | 458 | 50.1 | 6.22 | IIe | WRKYGQK | Nucleus |

|
| m.140205 | PmWRKY10 | 1359 | 452 | 50.5 | 6.43 | IIc | WRKYGQK | Nucleus |

|
| m.257305 | PmWRKY11 | 1332 | 443 | 48.0 | 6.47 | IIe | WRKYGQK | Nucleus |

|
| m.115513 | PmWRKY12 | 969 | 322 | 36.4 | 9.18 | IIc | WRKYGQK | Nucleus |

|
| m.62912 | PmWRKY13 | 1578 | 525 | 55.7 | 8.95 | I | WRKYGQK X2 | Nucleus |

|
| m.156607 | PmWRKY14 | 1095 | 364 | 40.4 | 9.68 | IId | WRKYGQK | Nucleus |

|
| m.423632 | PmWRKY15 | 330 | 233 | 26.2 | 9.89 | IId | WRKYGQK | Nucleus Cytoplasmic |

|
| m.423648 | PmWRKY16 | 546 | 181 | 20.1 | 9.55 | IId | WRKYGQK | Nucleus |

|
| m.59196 | PmWRKY17 | 843 | 280 | 32.3 | 6.83 | IIc | WRKYGQK | Nucleus |

|
| m.416602 | PmWRKY18 | 1041 | 346 | 37.4 | 9.42 | IId | WRKYGQK | Nucleus |

|
| m.318109 | PmWRKY19 | 1425 | 475 | 50.8 | 5.37 | IIe | WRKYGQK | Nucleus |

|
| m.198227 | PmWRKY20 | 2367 | 788 | 86.2 | 9.02 | I | WRKYGQK X2 | Nucleus |

|
| m.175245 | PmWRKY21 | 1914 | 637 | 70.1 | 6.77 | IIb | WRKYGQK | Nucleus |

|
| m.394091 | PmWRKY22 | 1380 | 459 | 50.2 | 6.28 | IIc | WRKYGQK | Nucleus |

|
| m.282372 | PmWRKY23 | 705 | 234 | 27.1 | 8.62 | IIb | WRKYGQK | Nucleus |

|
| m.282362 | PmWRKY24 | 651 | 216 | 24.9 | 9.66 | IIb | WRKYGQK | Nucleus |

|
| m.50488 | PmWRKY25 | 633 | 210 | 23.8 | 9.47 | IIb | WRKYGQK | Nucleus |

|
| m.50492 | PmWRKY26 | 417 | 138 | 15.7 | 9.99 | IIb | WRKYGQK | Nucleus |

|
| m.50499 | PmWRKY27 | 654 | 217 | 24.4 | 9.27 | IIb | WRKYGQK | Nucleus |

|
| m.50472 | PmWRKY28 | 756 | 251 | 28.6 | 9.43 | IIb | WRKYGQK | Nucleus |

|
| m.57136 | PmWRKY29 | 2202 | 733 | 79.5 | 6.39 | IIb | WRKYGQK | Nucleus |

|
| m.252813 | PmWRKY30 | 1293 | 430 | 47.5 | 5.74 | IIb | WRKYGQK | Nucleus |

|
| m.252861 | PmWRKY31 | 1929 | 642 | 69.4 | 6.06 | IIb | WRKYGQK | Nucleus |

|
Figure 1.
Subcellular localization of PmWRKY6 and PmWRKY7 in Nicotiana benthamiana.
3.2. Phylogenetic Analysis of PmWRKY Proteins
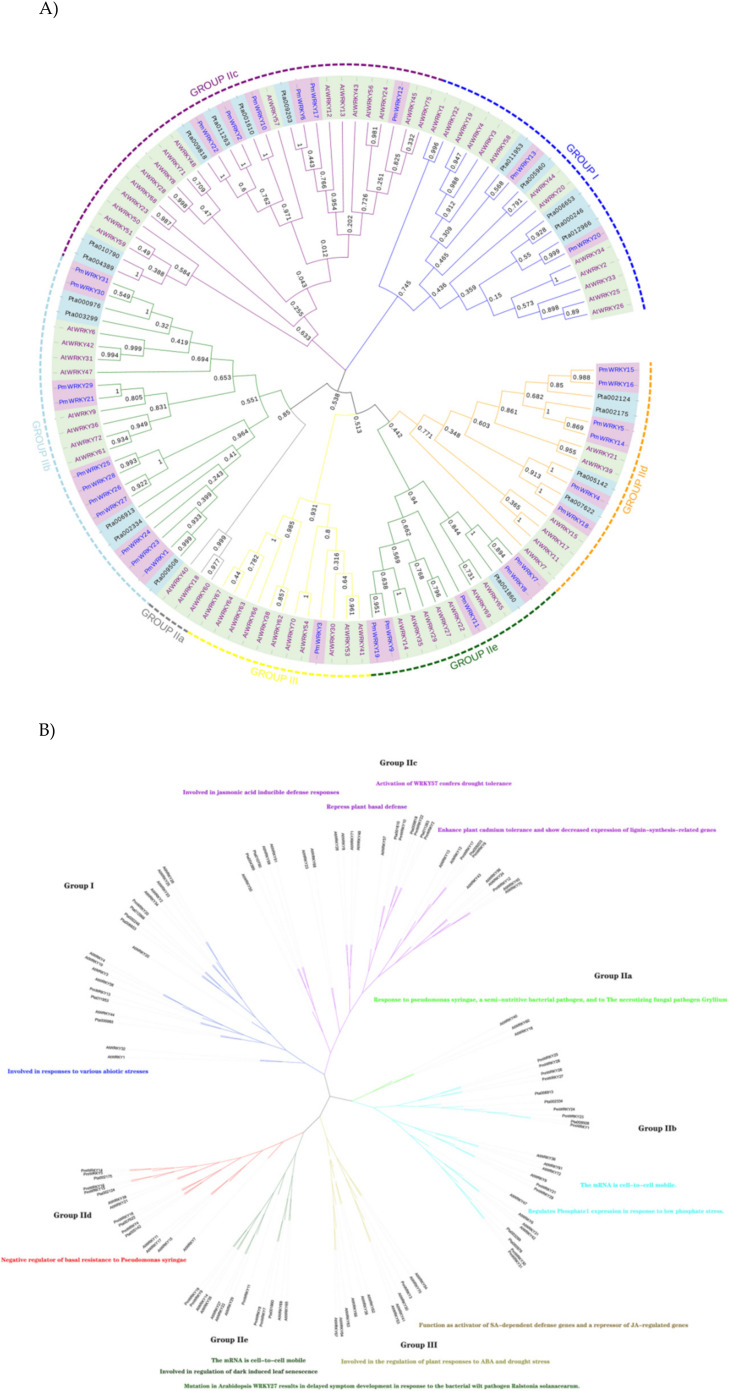
Based on the classification in Arabidopsis, 65 AtWRKY proteins from various groups or subgroups were randomly selected as representatives for comparison with P. massoniana and P. taeda based on previous studies [14,23]. The most prominent feature of WRKY proteins is the 60-amino-acid WRKY domain, which comprises the highly conserved signature "WRKYGQK" followed by a C2H2- or C2HC-type zinc finger motif [4]. As a result, the alignment of the PmWRKY proteins was examined based on their WRKY domains, which span ~60 amino acids. According to the combined alignment results, PmWRKY basically contains the conserved heptapeptide "WRKYGQK" in its domain sequence (Figure 2). The phylogenetic relationship among the WRKY domains of the PmWRKY, PtWRKY and AtWRKY proteins was studied as shown in Figure 3. The phylogenetic analysis indicated that PmWRKY proteins can be divided into three major groups, corresponding to groups I, II and III, when compared with the proteins from A. thaliana [3]. Of the 31 PmWRKY proteins, 2 belong to group I, 28 to group II, and 1 to group III. In group II, WRKY proteins can further be classified into five subgroups: 0 PmWRKY proteins belong to IIa, 11 to IIb, 6 to IIc, 6 to IId and 5 to IIe (Table 2).
Figure 2.
Multiple sequence alignment of PmWRKY domains.
Figure 3.
Classification of different groups of PmWRKYs, AtWRKYs and PtWRKYs. Different color branches and strips are used to distinguish different subgroups. In addition, the purple background is P. massoniana, the blue background is P. taede and the yellow background is A. thaliana. (A) The maximum likelihood method was used to analyze the evolutionary trees of 31 PmWRKYs, 65 AtWRKYs and 21 PtWRKYs. (B) The functions of each AtWRKY group marked in the evolutionary tree [24,25,26,27,28].
Table 2.
Comparison of groups between P. massoniana and P. taeda.
| Group | Subgroup | Gene Number | |
|---|---|---|---|
| PmWRKY | PtWRKY | ||
| I | 2 | 5 | |
| II | IIa | 0 | 0 |
| IIb | 11 | 5 | |
| IIc | 6 | 6 | |
| IId | 6 | 4 | |
| IIe | 5 | 1 | |
| III | 1 | 0 | |
| Total | 31 | 21 | |
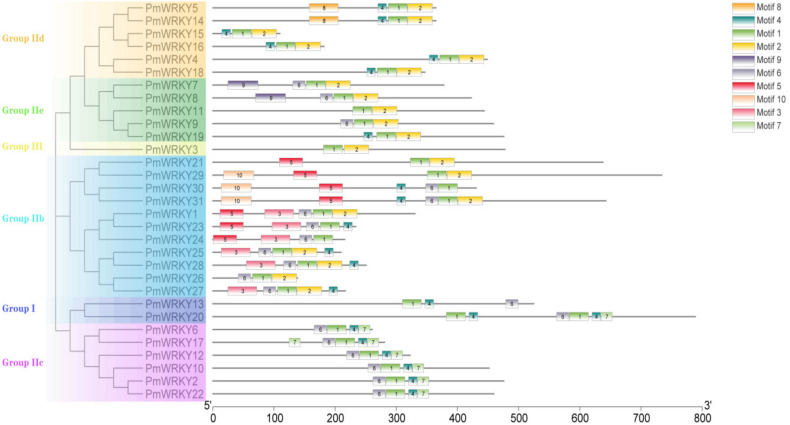
3.3. Compositions of PmWRKY Protein Motifs
The conserved shared motifs were determined using the full-length open reading frames (ORFs) of PmWRKY proteins by the MEME program. Ten motifs were identified in the 31 PmWRKY protein sequences. The amino acid length of the 10 motifs ranged from 15 to 50. The pattern of the conserved motifs is listed in Table 3 and illustrated in Figure 4. Motif 1 and Motif 2 formed the main structure of the WRKY domain ("WRKYGQK"); motif 1 was identified in all PmWRKY proteins, while motifs 3, 5 and 10 were observed only in group IIb. As shown in Figure 4, in addition to the widely distributed WRKY domain motifs 1, 2 and 4, the same group of PmWRKY members usually had similar motif compositions (Table 3). Motif 8 was exclusively found in group IId, whereas motif 9 was observed only in group IIe. Motif 6 was observed in groups I, IIb, IIc, and IIe, and motif 7 was observed in groups I and IIb. The functions of most of these motifs remain to be elucidated. A similar motif arrangement for PmWRKY proteins in a subpopulation indicated that particular subfamilies had conserved structures. Coupled with the results from the phylogenetic tree, the conserved composition of PmWRKY proteins in the same groups or subgroups strongly supports the reliability of these group classifications.
Table 3.
Details of conserved motifs from PmWRKY proteins.
| Motif | Width | Motif Sequence |
|---|---|---|
| 1 | 32 | SEADIPSDGYRWRKYGQKPVKGSPYPRSYYRC |
| 2 | 41 | SSARGCPARKQVERCATDPSILITTYEGEHNHSWPLSANAS |
| 3 | 48 | WDCLEQGWEKDNKNAKFMDDQQLPSSKRTLNYFQSAQIENRINSSTDD |
| 4 | 15 | RVKKRVERTIDDPAI |
| 5 | 39 | QVEINRMKEENQNLKSMLSRMINNYHNLQMHMMSVMQQQ |
| 6 | 21 | KKHKVKGRRTIRVPRFIVSTR |
| 7 | 19 | VITTYEGQHTHPSPALLRS |
| 8 | 48 | ADTNRHQQLHPQMHYPPLQLQHLSPQPEVMFRNGYMQLDNSMSCTATI |
| 9 | 50 | RCAATCLGGVAALYPEKQENSCNQRNEGEFMFGTSIVKQELEDQLDFVQP |
| 10 | 50 | VRELLDTELKQKCRRKGDFMADAPRVDRLGGIDLSVKLEETENEEKLMTD |
Figure 4.
Visualization of classification of 31 PmWRKY proteins. The motifs present in the PmWRKY proteins based on MEME analysis. Architecture of 10 conserved protein motifs in PmWRKYs. Each motif is represented in a different color (Motif 1–10).
3.4. Analysis of the Transcriptional Profiles of PmWRKY Genes
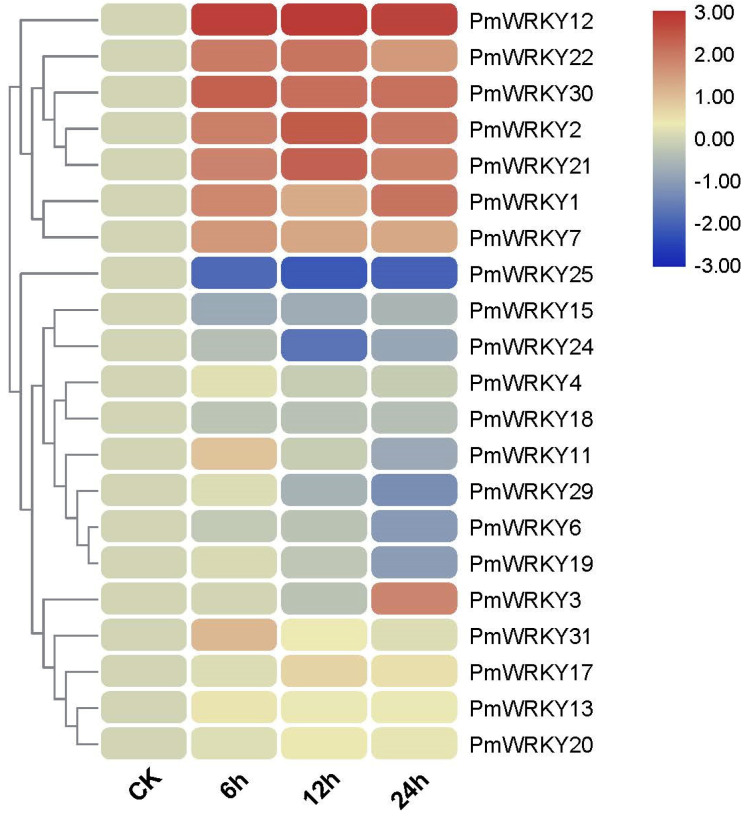
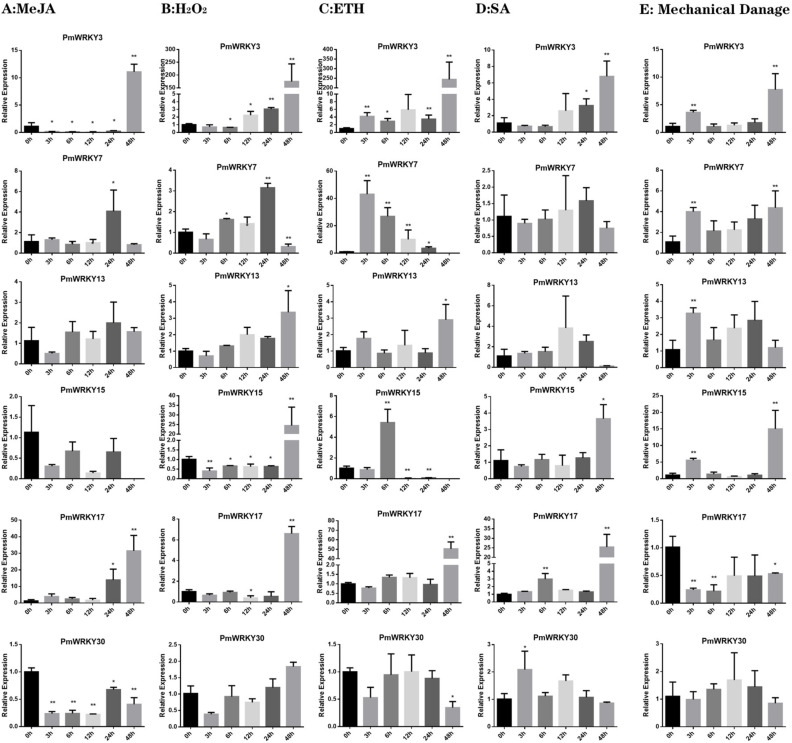
The sequence and heat map data of this experiment were based on CO2 stress transcriptome (Figure 5). The expression levels of some genes in the transcription group under CO2 stress were too low to be detected, so only 21 expressions of PmWRKY were given. The expression of some genes exhibited significant trends during treatment with CO2 stress. For instance, the expression of PmWRKY25 under CO2 stress was kept lower than control; whereas, the transcriptions of PmWRKY1, PmWRKY2, PmWRKY7, PmWRKY12, PmWRKY21, PmWRKY22 and PmWRKY30 were increased under CO2 stress treatment. In addition, the expression levels of some genes change significantly at some point in time. The expression level of PmWRKY24 was also significantly lower than that of the control at 12h. The expression level of PmWRKY3 was up-regulated at 24h. The results indicate that these genes may be regulated by CO2 stress. To obtain insights into the potential roles of PmWRKY genes, the expression levels of PmWRKYs were further determined by quantitative real-time PCR in leaves under treatments with three hormones (i.e., ETH, MeJA and SA), one abiotic stressor (i.e., H2O2) and mechanical damage stress. In order to analyze the transcription level of PmWRKY genes under different stresses, six PmWRKY genes with potential of resistance to stresses and high expression level in the transcriptome were selected for qRT-PCR through homologous clustering of AtWRKY genes related to stress resistance (Table S3).
Figure 5.
Transcriptional profiles of WRKY family members in P. massoniana under elevated CO2 stress, which includes four sets of data: 0 h(CK), 6 h, 12 h and 24 h after treatment. The relative expression of CK was set as "1". The color scale represents relative expression levels based on the values of log2 fold change scale.
Under MeJA treatment, the general trend of expression of PmWRKY3, PmWRKY7 and PmWRKY17 were increasing. It is worth noting that the expression of PmWRKY3 decreased at 3–24 h, and the expression level of PmWRKY7 peaked at 24 h and then decreased. The expressions of PmWRKY13 and PmWRKY15 were relatively stable, and that of PmWRKY30 was generally down-regulated. Among these genes, PmWRKY3, PmWRKY7, PmWRKY17 and PmWRKY30 were more sensitive to MeJA induction. In general, H2O2 treatment induced the expression of PmWRKYs(PmWRKY3, PmWRKY7, PmWRKY13, PmWRKY15 and PmWRKY17) to be up-regulated and peak at 48 h (Except that PmWRKY7 peaks at 24 h). In addition, the expression of PmWRKY30 was relatively stable and had no significant tendency. Under ETH treatment, the expression trends of PmWRKYs were diverse (Figure 6C). The expressions of PmWRKY3, PmWRKY13 and PmWRKY17 were up-regulated after treatment. However, the expressions of PmWRKY13 and PmWRKY17 were relatively stable during 3–24 h, while PmWRKY3 was up-regulated steadily from 3 h. The expression of PmWRKY7 and PmWRKY15 first increased to the highest level and then decreased. The expression of PmWRKY30 was stable in the early stage and decreased at 48 h. Under SA treatment, the expression levels of PmWRKY3, PmWRKY15 and PmWRKY17 showed an overall increasing trend, while the expression level of PmWRKY7 and PmWRKY13 showed a stable trend. However, that of PmWRKY30 increased to its maximum at 3 h and then restored stability. Under mechanical damage stress, the expression levels of the PmWRKY3, PmWRKY7, PmWRKY13 and PmWRKY15 increased at 3 h and then recovered to be stable, but the expression levels of PmWRKY3, PmWRKY7 and PmWRKY15 increased again and reached their maxima at 48 h. The expression of PmWRKY17 was always lower than that of control after treatment, while the expression of PmWRKY30 showed no obvious trend of change.
Figure 6.
Differential transcription of PmWRKY genes in P. massoniana leaves under abiotic treatments and exogenous hormone supplies. (A) MeJA, (B) H2O2, (C) ETH, (D) SA, (E) Mechanical damage stress. The color scale represents relative expression levels based on the values of log2 (2−∆∆CT). Asterisks represent significant differences between each time point and 0 h (* p < 0.05, ** p < 0.01). MeJA, methyl jasmonate; H2O2, hydrogen peroxide; ETH, ethephon; SA, salicylic acid. Color images are available online.
4. Discussion
Since the genome of P. massoniana is 200 times larger than that of A. thaliana, it is difficult to obtain the sequence in a short time, so we used the transcriptome to identify the WRKY gene family here. PmWRKY genes were searched for based on the Illumina RNA-seq and Iso-Seq databases in this study. A total of 31 genes, which were designated PmWRKY1 to PmWRKY31, were identified. With the aim of improving P. massoniana, this systematic analysis lays the foundation for further research into the functions of WRKY genes.
In this study, the same group of PmWRKY proteins in the phylogenetic tree shared common motifs based on the MEME analysis results with default parameters, indicating that the proteins were highly conserved and strongly supporting the reliability of the group classifications [29,30]. The number of WRKY gene family members in plants increased rapidly as the systems evolved from lower single-celled eukaryotic algae to higher multicellular angiosperm plants. Group I may be the ancestor, transitioning to group II by losing or splitting the structural domain. Most of the studied WRKY TFs belong to group II [3]. Group III may have been produced from group II through substitution of the H (histidine) of the zinc finger structure to a C residue. As a result, groups I and II are more highly conserved [31]. Among the 31 PmWRKY proteins identified, only one domain belonged to group III. The possible reasons are that the databases are not complete and that PmWRKY protein sequences belonging to group III were incomplete and manually discarded. Another reason for the lack of group III proteins may be that the screening criteria were too stringent and the E value cut-off (set to 10−3) was too strict. Previous studies have reported that the woody plants evolved a lower number of WRKY genes than herbaceous plants [32], and the evolutionary loss of the WRKY domain in dicotyledons was less than that in monocotyledons [33]. We can speculate further that P. massoniana, which is a gymnosperm, underwent a high degree of evolutionary loss of the WRKY domain, similar to P. taeda (Table 3). As shown in Figure 1, the PmWRKY proteins from groups IIa and IIb present a close relationship, while groups IId and IIe are closer together. Since the split of IIa and IIb and the split of IId and IIe occurred much later than those of other groups in the ancestor of land plants [3], IIa and IIb should likely be merged into a single subfamily that incorporates subgroups IId and IIe [34]. With evolution from lower plants to higher plants and from the aquatic environment to the terrestrial environment, plants have established signal transduction pathways related to growth, development, morphogenesis, metabolic regulation and stress resistance.
In order to verify the nuclear localization, we randomly selected two genes for subcellular localization, PmWRKY6 and PmWRKY7, which have localization on the nucleus, whereas PmWRKY7 also seems to be localized in other locations that are compatible with the cytoplasm (Figure 1). We speculated that N. benthamiana has a huge vacuole that sometimes displaces/pushes the cytoplasmic proteins against the membrane, which means that GFP-cytoplasmic proteins could be observed as "false membrane localized proteins." In addition, the possible reason is that the function or localization of transcription factors may be affected by other transcription factors. For example, GL1 is located in the nucleus. However, in the co-localization study, the interaction between AtMYC and GL1 leads to GL1 localization to the cytoplasm [35].
Since domains and motifs are related to transcriptional activity and protein interactions, generally, the function and characteristics of TFs can be determined by domain and motif analyses [36]. The results of conservative domain analysis showed that motifs 1 and 2 are related to a complete WRKY domain including a WRKY heptapeptide domain and a zinc finger structure and motifs 1 exist in all WRKY subfamily members. This sequence is essential for WRKY transcription factor recognition and binding to the W-box element at the target gene promoter [37]. Previous studies have reported variations in WRKYGQK sequences in different plants, which may lead to WRKY transcription factor recognition and binding to cis-regulatory elements other than W-Box [38]. There are three types of mutations in the WRKY heptapeptide domain. The first type is the second site variation of heptapeptide domain, usually from Rrg(R) to Lys(K). Namely, WRKYGQK mutates to WKKYGQK. The second type is the sixth site from Glu(Q) mutation to Glu(E) or Lys(K), namely WRKYGQK mutation to WRKYGEK or WRKYGKK. The third type shows the overall loss of C-terminal WRKY heptapeptide domain. According to the study, the second type of mutation is the common mutation type of WRKY gene family, which has been reported in many species, such as Prunus persica [39], Populus [40] and Cunninghamia lanceolata [41]. There is ample evidence that the WRKY gene family is important in the regulation of plant growth and development [42,43,44,45]. In addition, some WRKY proteins act as negative regulators of plant defense by binding to the DNA sequence TGAC [46]. Since gene expression patterns can provide important clues regarding gene function, we used qPCR to detect the expression of PmWRKY genes in the leaves of 2-year-old P. massoniana seedlings. The expression profiles showed variations in PmWRKY expression in different treatments. Our study showed that six PmWRKYs (PmWRKY3, PmWRKY7, PmWRKY13, PmWRKY15, PmWRKY17 and PmWRKY30) responded to at least two types of stress, among which PmWRKY3 was significantly elevated by induction with MeJA, SA, ETH, H2O2, and mechanical stress. Interestingly, the expression of PmWRKY3 peaked at 48 h under all the treatments. This suggests that the expression of these genes may be “time-specific”. In addition, some pairs of WRKY homologues have evolved different functionalities among different species [6], so little can be concluded by comparing WRKY genes with homologues. For example, the expression of WRKY42 was significantly reduced under 10 mM H2O2 and 500 μM SA treatments at 1, 3 and 12 h, whereas PmWRKY30 was not inhibited under MeJA and H2O2 treatment. Therefore, it is necessary to verify the function of genes through experiments in the later stage. In this study, we obtained four candidate genes (PmWRKY3, PmWRKY7, PmWRKY15, PmWRKY17) that are highly expressed in multiple stressors and deserve further exploration. The regulatory mechanism of WRKY genes under hormone, abiotic and mechanical stressors is complex, and the study of PmWRKY gene expression profiles will further promote functional research on the stress responses and signaling pathways of conifer trees in the future.
5. Conclusions
Comprehensive analysis of the PmWRKY genes was carried out in P. massoniana. Thirty-one PmWRKY genes were identified and divided into three major groups based on the presence of certain amino acid motifs. PmWRKY genes are important for the growth and development of P. massoniana, as shown by their expression levels under various treatments. PmWRKY gene expression is not completely consistent with the expression pattern of the corresponding genes in A. thaliana. This phylogenetic and gene expression analysis sheds light on the functions of PmWRKY genes in this specific conifer tree. This study will provide a theoretical basis for functional studies of the WRKY gene family, be very useful for understanding the biological role of individual WRKY gene in P. massoniana and provide a potential strategy for further breeding P. massoniana.
Acknowledgments
The authors appreciate the support from Ziyuan Hao, Peihuang Zhu, Yu Chen, The College of Forestry of Nanjing Forestry University. We would like to thank Major biotechnology corporation (Shanghai, China) for assistance with sequencing services.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/11/1386/s1, Table S1: Primer pairs for quantitative polymerase chain reaction, Table S2: Subcellular localization prediction of PmWRKY proteins, Table S3: Homologous genes in Arabidopsis thaliana.
Author Contributions
Conceptualization, S.Y.; software, S.Y. and Q.H.; investigation, S.Y. and F.W.; resources, F.W.; writing—original draft preparation, S.Y.; writing—review and editing, S.Y. and K.J.; visualization, S.Y. and Q.H. All authors have read and agreed to the published version of the manuscript.
Funding
The research was financially supported by the National Key R&D Program of China (2017YFD0600304) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reddy V.A., Purangaiah M., Sudhakar C., Lokesh U., Kirankumar T.V., Venkatesh B., Nareshkumar A., Kiranmai K., Gunupuru L.R. Expression Analysis of WRKY Transcription Factor Genes in Response to Abiotic Stresses in Horsegram (Macrotyloma uniflorum(lam.) Verde.) Am. J. Mol. Biol. 2016;6:125–137. [Google Scholar]
- 2.Zhang L.L., Cheng J., Sun X.M., Zhao T.T., Li M.J. Overexpression of VaWRKY14 increases osmotic tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep. 2018;37:1159–1172. doi: 10.1007/s00299-018-2302-9. [DOI] [PubMed] [Google Scholar]
- 3.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 4.Tao X., Chen C., Li C., Liu J., Liu C., He Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018;19:490. doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M.Y., Xu Z.S., Tian C., Huang Y., Wang F., Xiong A.S. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 2016;6:23101. doi: 10.1038/srep23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song A.P., Li P.L., Jiang J.F., Chen S.M., Li H.Y. Phylogenetic and Transcription Analysis of Chrysanthemum WRKY Transcription Factors. Int. J. Mol. Sci. 2014;8:15. doi: 10.3390/ijms150814442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ülker B., Somssich I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Devaiah B.N., Karthikeyan A.S., Raghothama K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amjad A.M., Farrukh A., Amjad N.M., Tuba A., Amjad A., Muhammad I.Q., Hussain S.K., Mamoon R.H., Gyuhwa C., Hwan Y.S., et al. Transcription WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018;226:12–21. doi: 10.1016/j.jplph.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Robatzek S., Somssich I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001;28:123–133. doi: 10.1046/j.1365-313X.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 11.Phukan U.J., Jeena G.S., Shukla R.K. WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants. Front. Plant Sci. 2016;7:760. doi: 10.3389/fpls.2016.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jing L., Günter B., Tapio P.E. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang L.Y., Chen Z.C., Wang Q.M., Sun J.J., Yao L.Q. Pinus Massoniana Morphological Architecture Analysis and 3D Visualization. J. Syst. Simul. 2006;S1:315–318. [Google Scholar]
- 14.Wang Z.Q., Ni L.J., Guo J.B., Liu L.Q., Li H.G., Yin Y.L., Gu C.S. Phylogenetic and Transcription Analysis of Hibiscus hamabo Sieb. et Zucc. WRKY Transcription Factors. DNA Cell Biol. 2020;39:1141–1154. doi: 10.1089/dna.2019.5254. [DOI] [PubMed] [Google Scholar]
- 15.Wu F., Sun X., Zou B., Zhu P., Ji K. Transcriptional Analysis of Masson Pine (Pinus massoniana) under High CO2 Stress. Genes. 2019;10:804. doi: 10.3390/genes10100804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He C.M., Teixeira D.S.J.A., Tan J.W., Zhang J.X., Pan X.P., Li M.Z., Luo J.P., Duan J.A. Genome-Wide Identification of the WRKY Family Genes and a Survey of Potential WRKY Target Genes in Dendrobium officinale. Sci. Rep. 2017;7:9200. doi: 10.1038/s41598-017-07872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 18.Kinuthia K.B., Fan L.X., Xu L., Wang Y., Zhu X.W., Tang M.J., Wang R.H., Zhang F., M’mbone M.E., Liu L.W. Genome-wide characterization of the WRKY gene family in radish (Raphanus sativus L.) reveals its critical functions under different abiotic stresses. Plant Cell Rep. 2017;36:1757–1773. doi: 10.1007/s00299-017-2190-4. [DOI] [PubMed] [Google Scholar]
- 19.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J.Y., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan C., Yao H., Qiu Z., Ma H., Zeng B. Genome-wide analysis of Eucalyptus grandis WRKY genes family and their expression profiling in response to hormone and abiotic stress treatment. Gene. 2018;678:38–48. doi: 10.1016/j.gene.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Li Z.Q., Wang C.J., Yang A.G., Ding A.M., Feng Q.F., Xu J., Jiao H.P., Shang K.Y. Cloning and Subcellular Localization of DXS Gene in Tobacco. J. Anhui Agric. Sci. 2013;41:11957–11960. [Google Scholar]
- 22.Zhu P.H., Ma Y.Y., Zhu L.Z., Chen Y., Ji K.S. Selection of Suitable Reference Genes in Pinus massoniana Lamb. Under Different Abiotic Stresses for qPCR Normalization. Forests. 2019;10:632. doi: 10.3390/f10080632. [DOI] [Google Scholar]
- 23.Goyal P., Manzoor M.M., Vishwakarma R.A., Sharma D., Dhar M.K., Gupta S. Comprehensive Transcriptome-Wide Identification and Screening of WRKY Gene Family Engaged in Abiotic Stress in Glycyrrhiza glabra. Sci. Rep. 2020;10:373. doi: 10.1038/s41598-019-57232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Q.T., Yu D.Q. Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Hereditas (Beijing) 2010;32:848–856. doi: 10.3724/SP.J.1005.2010.00848. [DOI] [PubMed] [Google Scholar]
- 25.Kloth K.J., Wiegers G.L., Busscher-Lange J. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 2016;67:3383–3396. doi: 10.1093/jxb/erw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L.Y., Zhou B., Li Y.H. Roles of WRKY Transcription Factors in Abscisic Acid Signal Transduction. Mol. Plant Breed. 2014;12:404–410. [Google Scholar]
- 27.Ulker B., Shahid M.M., Somssich I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta. 2007;226:125–137. doi: 10.1007/s00425-006-0474-y. [DOI] [PubMed] [Google Scholar]
- 28.Wenke K., Wanke D., Kilian J., Berendzen K., Harter K., Piechulla B. Volatiles of two growth-inhibiting rhizobacteria commonly engage AtWRKY18 function. Plant J. Cell Mol. Biol. 2012;70:445–459. doi: 10.1111/j.1365-313X.2011.04891.x. [DOI] [PubMed] [Google Scholar]
- 29.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee A., Roychoudhury A. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015;2015:807560. doi: 10.1155/2015/807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H.J., Watanabe K.A., Zhang L.Y., Shen Q.X.J. WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes. 2016;23:311–323. doi: 10.1093/dnares/dsw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z.J., Li X.H., Liu Z.W., Li H., Wang Y.X., Zhuang J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol. Genet. Genom. MGG. 2016;291:255–269. doi: 10.1007/s00438-015-1107-6. [DOI] [PubMed] [Google Scholar]
- 33.Wei K.F., Chen J., Chen Y.F., Wu L.J., Xie D.X. Molecular Phylogenetic and Expression Analysis of the Complete WRKY Transcription Factor Family in Maize. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei C., Yue H., Alessandro V., Wu K.C., Cai H.Y., Qin Y., Alison M., Lin Z.G., Zhang L.S. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2017;36:311–335. [Google Scholar]
- 35.Yu N., Cai W.J., Wang S., Shan C.M., Wang L.J., Chen X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010;22:2322–2335. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L., White M.J., MacRae T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999;262:247–257. doi: 10.1046/j.1432-1327.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y.J., Wang L.J. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X.Z., Li H., Yang Y.C. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus) PLoS ONE. 2018;13:146–149. doi: 10.1371/journal.pone.0191308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Y.B., Ji Z.R., Chi F.M. Genome-wide identification and expression analysis of the WRKY gene family in peach. Hereditas (Beijing) 2016;38:77–93. doi: 10.16288/j.yczz.15-235. [DOI] [PubMed] [Google Scholar]
- 40.Liu T.T., Fan D., Ran L.Y. Highly efficient CRISPR/Cas9-mediated targeted mutagenesis of multiple genes in Populus. Hereditas (Beijing) 2015;37:1044–1052. doi: 10.16288/j.yczz.15-303. [DOI] [PubMed] [Google Scholar]
- 41.Zeng M., Gao W.J., Shuai P. Identification of WRKY Gene Family Members in Chinese Fir and Its Expression Analysis under Low Phosphorus Stress. J. Northeast For. Univ. 2019;47:12–20. [Google Scholar]
- 42.Yao L., Wang J., Sun J., He J., Paek K.Y., Park S.Y., Huang L., Gao W. A WRKY transcription factor, PgWRKY4X, positively regulates ginsenoside biosynthesis by activating squalene epoxidase transcription in Panax ginseng. Ind. Crop. Prod. 2020;154:112671. doi: 10.1016/j.indcrop.2020.112671. [DOI] [Google Scholar]
- 43.Singh D., Debnath P., Roohi, Sane A.P., Sane V.A. Expression of the tomato WRKY gene, SlWRKY23, alters root sensitivity to ethylene, auxin and JA and affects aerial architecture in transgenic Arabidopsis. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2020;26:1187–1199. doi: 10.1007/s12298-020-00820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun W.J., Ma Z.T., Chen H., Liu M.Y. Genome-wide investigation of WRKY transcription factors in Tartary buckwheat (Fagopyrum tataricum) and their potential roles in regulating growth and development. PeerJ. 2020;8:e8727. doi: 10.7717/peerj.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui X., Zhao P.Y., Liang W.W., Cheng Q., Mu B.B., Niu F.F., Yan J.L., Liu C.L., Hua X., Kav N.N.V., et al. A rapeseed WRKY transcription factor phosphorylated by CPK modulates cell death and leaf senescence by regulating the expression of ROS and SA-synthesis-related genes. J. Agric. Food Chem. 2020;68:7348–7359. doi: 10.1021/acs.jafc.0c02500. [DOI] [PubMed] [Google Scholar]
- 46.Sun Y.D., Yu D.Q. Activated expression of AtWRKY53 negatively regulates osmotic tolerance by mediating stomatal movement. Plant Cell Rep. 2015;34:1295–1306. doi: 10.1007/s00299-015-1787-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.