Abstract
Simple Summary
In this study, we comprehensively and synthetically analyzed mutations in lung cancer based on the next generation sequencing data of lung tumors surgically removed from the patients, and identified the mutation-related factors that can affect clinical outcomes. Detailed understanding of the genomic landscape of lung cancers will establish the ideal model for best surgical outcomes in the era of “precision medicine”.
Abstract
Findings on mutations, associated with lung cancer, have led to advancements in mutation-based precision medicine. This study aimed to comprehensively and synthetically analyze mutations in lung cancer, based on the next generation sequencing data of surgically removed lung tumors, and identify the mutation-related factors that can affect clinical outcomes. Targeted sequencing was performed on formalin-fixed paraffin-embedded surgical specimens obtained from 172 patients with lung cancer who underwent surgery in our hospital. The clinical and genomic databases of the hospital were combined to determine correlations between clinical factors and mutation profiles in lung cancer. Multivariate analyses of mutation-related factors that may affect the prognosis were also performed. Based on histology, TP53 was the driver gene in 70.0% of the cases of squamous cell carcinoma. In adenocarcinoma cases, driver mutations were detected in TP53 (26.0%), KRAS (25.0%), and epidermal growth factor receptor (EGFR) (23.1%). According to multivariate analysis, the number of pathogenic mutations (≥3), presence of a TP53 mutation, and TP53 allele fraction >60 were poor prognostic mutational factors. The TP53 allele fraction tended to be high in caudally and dorsally located tumors. Moreover, TP53-mutated lung cancers located in segments 9 and 10 were associated with significantly poorer prognosis than those located in segments 1–8. This study has identified mutation-related factors that affect the postoperative prognosis of lung cancer. To our knowledge, this is the first study to demonstrate that the TP53 mutation profile varies with the site of lung tumor, and that postoperative prognosis varies accordingly.
Keywords: lung cancer, next generation sequencing, mutation, TP53, survival
1. Introduction
Along with the technological advancement in next generation sequencing (NGS), accumulated findings on mutations, associated with lung cancer, have led to the development of mutation-based precision medicine [1]. In fact, novel therapies, based on information regarding cancer antigens and cancer mutations, such as immune checkpoint blockade and molecular-targeted therapy, have recently been developed, and treatment outcomes for lung cancer, have improved dramatically [2,3]. Therefore, patient-based clinicogenomic datasets may significantly accelerate the advancement of clinical practice and the development of novel therapeutics.
Although, the postoperative prognosis of lung cancer has been conventionally and stochastically predicted, based on the histological classification and the tumor-node-metastasis (TNM) stage [4,5], a prognostic model specifically applicable for each case has not been established. The criteria for adjuvant chemotherapy are also not clear. Hence, accurate criteria for adjuvant chemotherapy based on appropriate prognostic models in the future should be urgently established [6,7].
Our study group has continuously analyzed the NGS data of patients with lung cancer since January 2014. Hence, this study aimed to synthetically analyze the correlation between mutation profiles of patients with lung cancer and clinical factors, using the integrated results of this NGS analysis, and identify mutational factors affecting clinical outcomes.
2. Results
2.1. Patient Characteristics
We studied surgical samples from 172 patients with lung cancer who underwent surgery at our hospital between June 2014 and June 2019. The characteristics of the enrolled patients are summarized in Table 1. Among the 172 patients, 116 were men and 56 were women, and 137 were smokers and 35 were non-smokers. The age of patients ranged between 44 and 90 (mean ± SD, 71.1 ± 10.8) years. Histologically, there were 103 cases of adenocarcinoma, 40 cases of squamous cell carcinoma, 4 cases of adenosquamous carcinoma, 10 cases of small cell carcinoma, and 15 cases with other histology. In terms of pathological stage, there were 79 stage I, 21 stage II, 33 stage III, and 39 stage IV cases. One patient received four cycles of preoperative cisplatin + vinorelbine chemotherapy. The median (range) follow-up period for all censored cases was 973 (21–2056) days.
Table 1.
Patient characteristics.
| Parameters | Variables | Total Number | Percentages |
|---|---|---|---|
| Total number | 172 | ||
| Age (years), median (range) | 71 (44–90) | ||
| Sex | |||
| Male | 116 | 67.4% | |
| Female | 56 | 32.6% | |
| Histology | |||
| Adeno | 103 | 59.9% | |
| Squamous | 40 | 23.3% | |
| Adenosquamous | 4 | 2.3% | |
| Small | 10 | 5.8% | |
| other | 15 | 8.7% | |
| Pathological stage | |||
| Ⅰ | 79 | 45.9% | |
| Ⅱ | 21 | 12.2% | |
| Ⅲ | 33 | 19.2% | |
| IV | 39 | 22.7% | |
| Smoking status | |||
| Smoker | 137 | 79.7% | |
| Non-smoker | 35 | 20.3% |
Adeno, adenocarcinoma; Squamous, squamous cell carcinoma; Adenosquamous, adenosquamous carcinoma; Small, small cell carcinoma.
2.2. Clinicogenomic Features and Associations
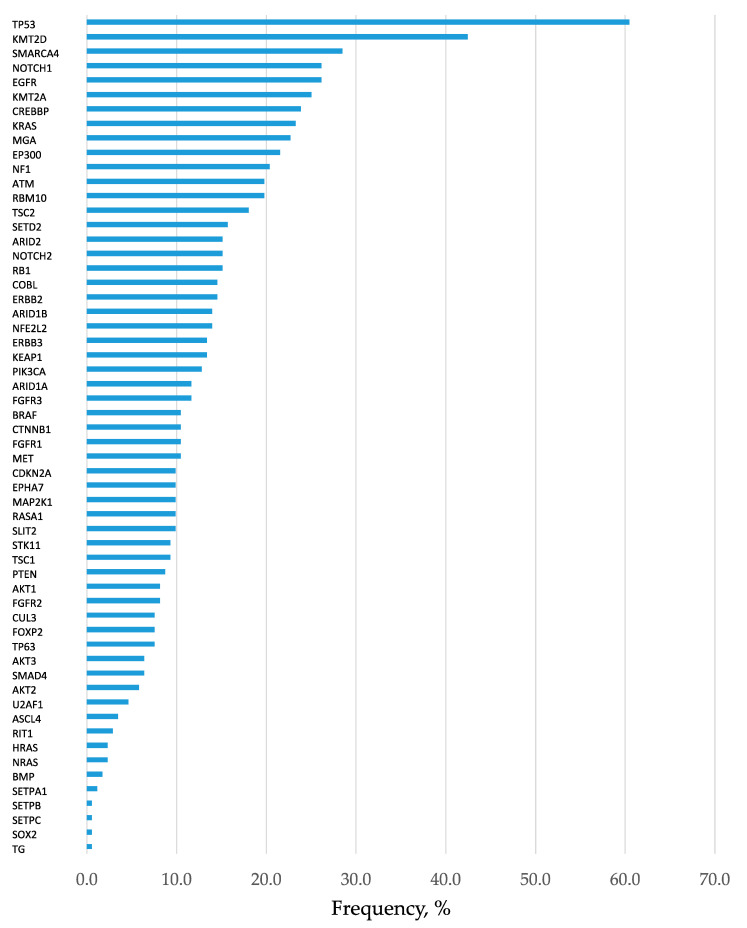
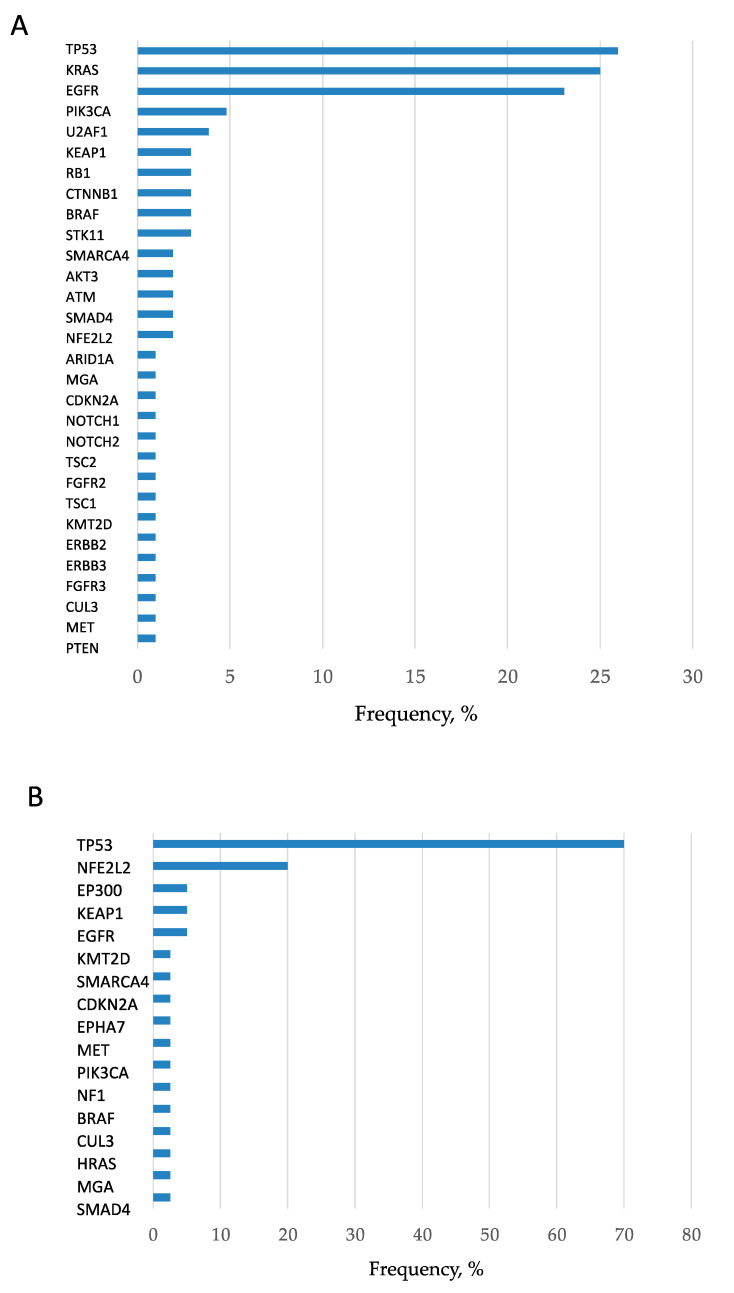
In total, 2372 mutations were detected at an allele fraction (AF) of >1%, and the mean number of mutations per cancer lesion was 13.8 ± 2.9. Of these, 447 mutations were annotated as pathogenic (or oncogenic) mutation (Table S1), and the mean number of pathogenic mutations per cancer lesion was 2.6 ± 0.4. The frequency of short variant mutations is shown in Figure 1. The most common mutation was in the gene encoding tumor protein p53 (TP53), which was detected in approximately 60% of patients with lung cancer. When only pathological mutations at an AF of >20% were examined, common mutations were detected in genes encoding TP53, KRAS, and EGFR in adenocarcinoma and in the TP53 gene in squamous cell carcinoma (Figure 2A,B). Pathogenic mutations with AF of >20 were regarded as driver mutations, and 89.5% of patients with lung cancer were found to harbor driver mutations in EGFR, KRAS, or TP53, which were found to be the three major mutations in lung cancer. Fusion gene screening through transcriptome sequencing revealed that ROS1, ALK, and RET fusion genes were not detected in any of the 172 patients who enrolled in this study.
Figure 1.
Frequency of detection of gene mutations. Analysis of all 172 patients. All mutations with allele fraction (AF) of >1% are presented in the order of frequency. Mutations in the tumor protein p53 (TP53) was the most common kind of mutation, present in approximately 60% of lung cancers.
Figure 2.
Frequency of detection of pathogenic mutations in adenocarcinomas and squamous cell carcinomas. (A) Pathogenic mutations with AF of >20 in adenocarcinomas are presented. The frequencies of TP53, KRAS, and epidermal growth factor receptor (EGFR) mutations were remarkably high, and hence they can be considered the three major mutations. (B) Pathogenic mutations with AF of >20 in squamous cell carcinomas are presented. The frequency of TP53 mutation was remarkably high.
2.3. Pathogenic vs. Nonpathogenic Mutations in EGFR, KRAS, and TP53
For EGFR and TP53, the number of pathogenic mutations was 1.24 and 1.38 times higher than the number of nonpathogenic mutations, respectively, whereas, for KRAS, the former was 7.40 times higher and significantly higher than the latter (Figure S1). In other words, most mutations detected in KRAS were pathogenic mutations.
2.4. Multivariate Analysis of Progression-Free Survival (PFS) after Surgery
When multivariate analysis was performed with age, gender, histology, stage, number of pathogenic mutations, presence of EGFR, KRAS, and TP53 mutations, and AF of EGFR, KRAS, and TP53 mutations to identify factors affecting PFS, the stage, number of pathogenic mutations, presence of TP53 mutations, and AF of TP53 mutations were found to be significant prognostic factors (p < 0.05).
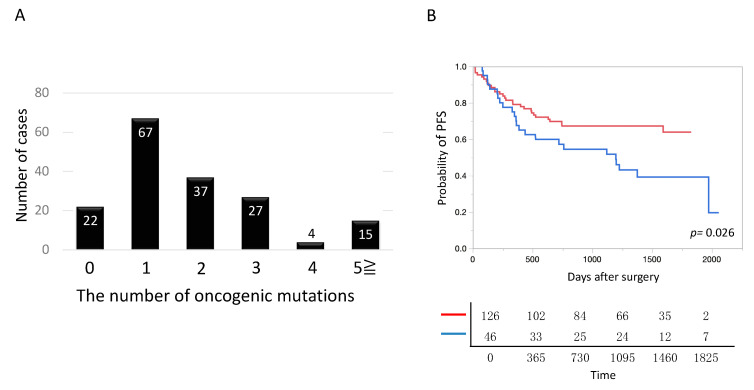
2.5. Number of Oncogenic Mutations in Lung Cancer and Patient Prognosis
The largest proportion of patients (67/172 patients) harbored one oncogenic mutation. The mean number of oncogenic mutations per tumor was 2.6 ± 0.4 (Figure 3A). The prognosis was significantly poorer in tumors with three or more oncogenic mutations than in tumors with two or less mutations (Figure 3B).
Figure 3.
Number of pathogenic mutations in lung cancer. (A) The number of mutations harbored by each tumor is shown. Tumors harboring one mutation were most common. On average, tumors harbored 2.6 mutations. (B) The postoperative prognosis was significantly poorer in lung cancer, harboring three or more oncogenic mutations, than harboring 2 or less oncogenic mutations. Blue and red lines denote lung cancer harboring three or more oncogenic mutations (n = 46) and that harboring 2 or less oncogenic mutations (n = 126), respectively.
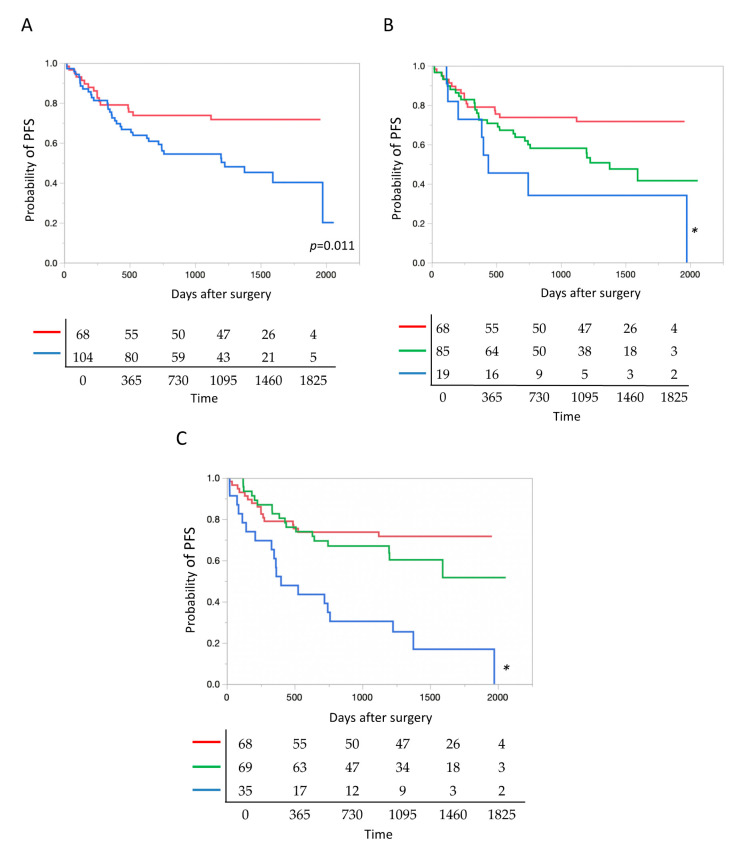
2.6. Presence of TP 53 Mutation and Patient Prognosis
Irrespective of oncogenic and non-oncogenic mutations, patients with TP53 mutations had significantly poorer prognosis than those without TP53 mutations (Figure 4A). Next, we compared the prognosis among patients with different numbers of TP53 mutations, including oncogenic and non-oncogenic mutations. Prognosis was significantly poorer in patients with tumors harboring 0, 1, and 2 or more mutations (in this order) (Figure 4B).
Figure 4.
TP53 mutation as a prognostic factor after surgery. (A) Postoperative prognosis was significantly poorer in lung cancer with tumor protein p53 (TP53) mutations than in lung cancer without TP53 mutations. Blue and red lines denote cancers with, and without TP53 mutations (n = 104 and 68), respectively. (B) The prognosis deteriorated significantly as the number of TP53 mutations increased, i.e., in the ascending order of tumors with 0, 1, and 2 or more mutations. Red, green and blue lines denote cancers with 0, 1, and 2 or more mutations (n = 68, 85 and 19), respectively. * p = 0.014. (C) According to allele fractions (AFs) of TP53 mutations, the prognosis of lung cancer was significantly poorer in the AF > 60 group than in the other two groups with AF = 0 and 0 < AF < 60. Blue, green and red lines denote cancers with the AF > 60, 0 < AF < 60, and AF = 0 (n = 35, 69 and 68), respectively. * p < 0.0001.
2.7. Allele Fraction of TP 53 Mutation and Patient Prognosis
According to AF, TP53 mutations, including oncogenic and non-oncogenic mutations, were classified into three groups: AF = 0, 0 < AF < 60, and AF > 60. The TP53 mutation with AF = 0 represented cancer without any TP53 mutations. In tumors with multiple TP53 mutations, the highest AF was used to classify each tumor. PFS was significantly shorter in the group of TP53 mutations with AF > 60 than in the two other groups (hazard ratio (HR) 2.84, 95% confidence interval (CI) 1.48–5.48, compared to AF = 0; HR 2.19, 95% CI 1.05–4.78, compared to 0 < AF < 60) (Figure 4C).
2.8. Mutation Profiles According to Cancer Location
We investigated whether the mutation profiles differed with tumor sites. Between tumors located at the cranial side (segments 1, 2, 3, and 1 + 2, n = 84) and the caudal side (segments 7, 8, 9, and 10, n = 56), no correlation was observed in the presence or absence of EGFR, KRAS, or TP53 mutations (Table S2). No correlation was also observed in the presence or absence of EGFR, KRAS, or TP53 mutations between tumors located at the ventral side (segments 3, 4, and 5, n = 29) and the dorsal side (segments 2, 6, 9, and 10, n = 59) (Table S2). These findings revealed that tumor sites were not associated with the presence or absence of specific gene mutations.
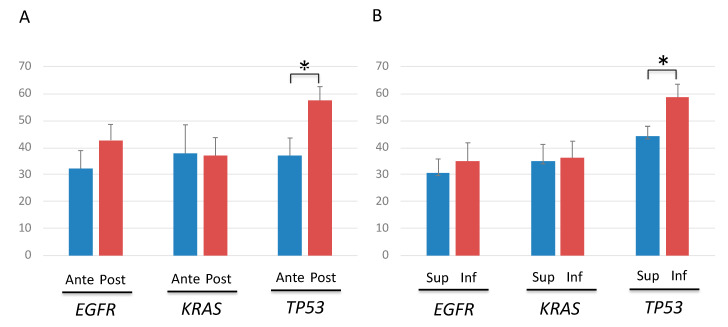
No significant differences were observed in the AFs of EGFR or KRAS mutations when the AFs of TP53, EGFR, and KRAS mutations were compared between tumor sites. However, the AFs of TP53 mutations were significantly higher in tumors located at the caudal and dorsal sides (Figure 5A,B).
Figure 5.
Allele fraction (AF) of TP53, EGFR and KRAS mutations according to the cancer location. (A) Comparison of AFs between tumors located at the ventral and dorsal sides showed significantly higher AFs only in tumor protein p53 (TP53)-mutated tumors located at the dorsal side. Blue and red lines denote anteriorly-, and posteriorly-located cancers, respectively. * p < 0.05. (B) Comparison of AFs between tumors located at the cranial and caudal sides showed significantly higher AFs only in TP53-mutated tumors located at the caudal side. Blue and red lines denote superiorly- and inferiorly-located cancers, respectively. * p < 0.05.
2.9. Characteristics of TP53-Mutated Lung Cancer in Segments 9 and 10
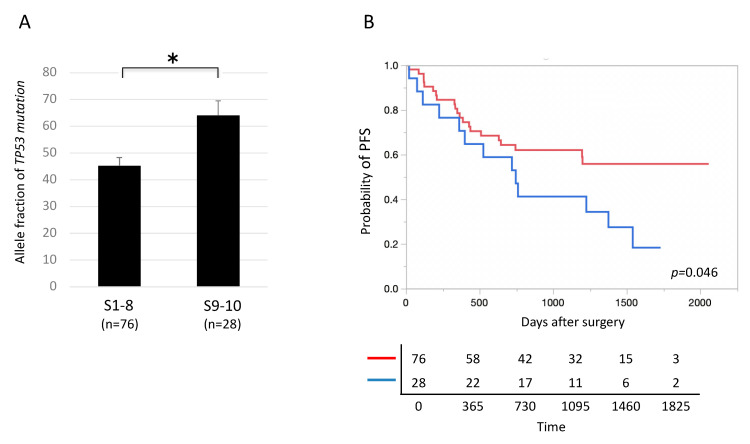
Among TP53-mutated lung cancers, AFs of TP53 mutations, including oncogenic and non-oncogenic mutations, were significantly higher in lung cancers located in segments 9 and 10, the dorsal inferior lobes, than in those located in the other segments (1 to 8) (Figure 6A, Figure S2). In addition, among TP53-mutated lung cancers, PFS was significantly shorter in patients with lung cancers located in segments 9 and 10 than in those where tumors were located in the other segments (1 to 8) (Figure 6B). In contrast, among lung cancers without TP53 mutations, no significant difference was observed in postoperative prognosis between lung cancers located in segments 9 and 10 and those located in segments 1 to 8 (Figure S3). Among patients with TP53-mutated lung cancer, no significant differences in either the proportion of smokers (segments 1–8: 86.8%, segments 9–10: 82.1%, p = 0.55) or the smoking index, defined as number of cigarettes smoked per day multiplied by years smoked (segments 1–8: 280 ± 54, segments 9–10: 234 ± 62, p = 0.83), were observed between those with lung cancers located in segments 9 and 10 and those with lung cancers located in segments 1 to 8.
Figure 6.
Locational variation in prognosis in lung cancers with TP53 mutations. (A) Allele fractions of tumor protein p53 (TP53) mutations were significantly higher in TP53-mutated lung cancers located in segments 9 and 10 than in those located in segments 1 to 8. * p < 0.05. (B) The prognosis was significantly poorer in TP53-mutated lung cancers located in segments 9 and 10 than in those located in segments 1 to 8. Blue and red lines denote tumors located in segments 9–10 and 1–8 (n = 28 and 76), respectively.
2.10. Correlation between Smoking and TP53 Mutations
To further investigate the correlation between smoking and TP53 mutations, we compared the prevalence of TP53 mutations between smokers (n = 137) and non-smokers (n = 35) with lung cancer. The results showed a significantly higher prevalence of TP53 mutations in smokers with lung cancer (smokers: 65.0% versus non-smokers: 42.9%, p = 0.018). Comparison of the AFs of TP53 mutations between smokers and non-smokers with lung cancer revealed that the AFs were significantly higher in smokers with lung cancer (smokers: 32.1 ± 3.1 versus non-smokers: 12.5 ± 5.6, p = 0.003).
3. Discussion
In lung cancer treatment, gene sequencing has been performed mainly as a companion diagnostic approach for identifying mutations in genes such as EGFR and anaplastic lymphoma kinase (ALK) and subsequent selection of molecular target drugs [8,9,10]. Currently, targeted sequencing using NGS is being increasingly performed at clinical facilities worldwide [1,11]. The advancement in NGS technology has realized comprehensive detection of many gene mutations in lung cancer. However, the correlation between the detected mutation profiles and the pathology of lung cancer has not been sufficiently analyzed. The availability of NGS is still limited, due to its cost, cumbersome device operations and its supposedly limited clinical benefits. Furthermore, even in clinical facilities performing NGS, the amount of data generated at each facility is not sufficient for scientific analysis. In this study, we aimed to analyze the effect of gene mutation profiles on the prognosis of lung cancer. Toward this objective, examination of many surgical samples collected from a large patient cohort with similar clinical characteristics was required for effective analysis of prognosis. As gene sequencing on surgical samples is being aggressively performed at our hospital since July 2014, we had access to sufficient number of sequenced samples [12,13,14,15,16,17,18,19]. In this context, association between clinical and genomic characteristics was analyzed in this study. Among the mutational factors, the number of pathogenic mutations, presence of TP53 mutation, and AF of TP53 mutation were found to be the prognostic factors.
Our study revealed that, on average, one cancer lesion harbors 2.6 pathogenic mutations, which can be explained by branched evolution model [20]. Furthermore, several studies have suggested that even if one mutation pathway is inhibited, the presence of other functional pathways of driver mutations can drive proliferation of tumor cells and render the cancer aggressive [9,21,22,23]. Therefore, when multiple driver mutations are incorporated into cancer during the course of tumor evolution, the proliferation drive is activated at multiple stages, which enhances the aggressiveness of cancer. Our study also showed that PFS was shorter in tumors with a large number of pathogenic mutations, validating the above-mentioned hypothesis from another perspective. For lung cancer harboring multiple pathogenic mutations, radical treatment may be recommended considering tumor aggressiveness. Meanwhile, cases of lung cancer harboring no oncogenic mutation include, (i) those harboring mutations that cannot be sequenced via NGS for reasons, such as presence of fusion genes, DNA amplification, DNA methylation, and histone modification; (ii) those harboring mutations located at gene sites other than the targeted regions, which can be sequenced on our cancer panel.
The correlation between the presence of TP53 mutations and poor postoperative prognosis has been suggested by previous studies involving immunostaining [24,25,26]. Our study validated this observation. However, owing to low specificity, immunostaining can only detect the presence or absence of abnormal TP53 proteins, but does not directly detect TP53 mutations [27]. Furthermore, it does not reflect the details of TP53 mutations (e.g., mutational pattern and AF) [28]. In relation to the prognostic effects of TP53 mutations on non-small cell lung cancer (NSCLC) detected via direct sequencing, inconsistent results have been reported previously. Aisner et al. [29] reported that TP53 mutations are associated with shorter survival. Whereas, Devarakonda et al. [30] and Ma et al. [31] reported that TP53 mutations exhibited no such prognostic effect. Meanwhile, some other studies have revealed that TP53 mutations frequently exhibit intra-tumor heterogeneity (ITH) in NSCLC [32,33]. Based on this finding, Lee et al. [34] reported that TP53 mutations with ITH exhibit worse survival when compared to wild-type TP53, whereas TP53 mutations without ITH did not exhibit such an extreme phenotype. Our study, in which TP53 mutations were directly detected by deep targeted sequencing, revealed that not only the number, but also the AF of TP53 mutations correlated with prognosis. Therefore, we clearly demonstrated that TP53 mutations are additively involved in tumor aggressive behavior. In our study, tumor cells were selected using laser capture microdissection (LCM) before DNA extraction. Although, measurement bias is inevitable for AF measurement because of the effect of contamination with normal cells, a single technician (Kenji Amemiya) has performed LCM as uniformly as possible at our institute. The result of this analysis, showing that AF of TP53 mutations affects prognosis, is an interesting and novel finding in oncology.
The hypothesis that aggressiveness of lung cancer may vary with tumor sites has not been verified scientifically till date. In our study, AFs of TP53 mutations were significantly high in tumors located at the dorsal and caudal sides, and the prognosis of TP53-mutated lung cancer located in segments 9 and 10 was poor. To date, there has been no reports on the effect of the sites of lung cancer on prognosis. This is possibly the first report providing scientific verification of this effect. Interestingly, the common sites of lung cancer with poor prognosis, the dorsal and caudal regions of the lung, corresponded to the common sites of idiopathic pulmonary fibrosis. The dorsal and caudal regions of the lung may be vulnerable to smoke. Owing to the lack of significant difference in the smoking status and the smoking index between patients with TP53-mutated lung cancer located in segments 9 and 10 and that located in segments 1 to 8, we concluded that even a comparable amount of smoking may exert a strong effect on cancer initiation and progression in segments 9 and 10 in the dorsal inferior lobe. Detailed investigation regarding the association between tumor sites and carcinogenesis associated with TP53 mutations is warranted. In the future, survival after surgery may be improved by changing treatment strategies and the extent of resection based on the mutations harbored and the location of the tumors.
A limitation of this study is that it is a single-institution study with a small sample size. Furthermore, unlike whole exome sequencing, panel sequencing does not provide information regarding all exons, and thus, it is possible that each tumor may actually possess much more pathogenic mutations. However, the lung cancer panel that we developed covers the main hot spots of gene mutations in lung cancer, and we assume that almost all significantly mutated genes associated with lung cancer can be detected. In addition, as the objective was to investigate the postoperative prognosis, we did not investigate the correlation between mutations and responses to medical treatment (drug therapy). Currently, the development of novel therapeutic strategies for lung cancer is rapidly progressing. In the future, various therapeutics, in addition to EGFR-tyrosine kinase inhibitors, ALK-inhibitors, programmed cell death 1 antibody, and cytotoxic T lymphocyte antigen-4 antibody, will be developed [2,35,36,37]. The correlation between these novel therapies and presence of mutations should be determined to develop precision medicine. Moreover, the treatment for postoperative recurrence has to be substantially changed to improve both PFS and overall survival. The establishment of a large-scale clinicogenome database at the international level, and the promotion of large-scale clinical studies using such a database, are warranted.
4. Methods
4.1. Patients and Sample Preparation
The study involved 172 patients who underwent surgery in our Department between June 2014 and June 2019. These patients provided written informed consent for the genetic research studies, which were performed in accordance with protocols approved by the Institutional Review Board of Yamanashi Central Hospital (approval date is 21 May 2014, although no number or any ethics code was allocated). Patient data were collected from our institutional cancer registry database and from patient follow-up visits to our outpatient office. The information collected from the records included preoperative patient characteristics, disease status, operative procedure, pathological diagnosis, and follow-up data. Histological typing was performed according to the World Health Organization classification [38], and cancer staging was based on the TNM classification of the International Union Against Cancer, 8th edition [39].
The serial section of formalin-fixed, paraffin-embedded (FFPE) tissue was stained with hematoxylin-eosin and then micro-dissected using an ArcturusXT laser capture microdissection system (Thermo Fisher Scientific, Tokyo, Japan). Tumor DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, Tokyo, Japan). DNA fragmentation in FFPE DNA was assessed using The TaqMan RNase P Detection Reagents Kit and the FFPE DNA QC Assay on ViiA7 Real-Time PCR instrument (Thermo Fisher Scientific). Human control genomic DNA (included with TaqMan RNase P Detection Reagents Kit) was serially diluted 4 times for a 5-point for a standard curve and the absolute DNA concentrations were determined. Assessment of DNA fragmentation was estimated with the ratio of DNA (relative quantification, RQ) obtained for the long amplicon (256 bp) to the short amplicon (87 bp). RQ value was an indicator of the degradation level of genomic DNA. A peripheral blood sample was drawn from each patient immediately prior to surgery and was collected into EDTA-2Na tubes. The buffy coats were isolated following centrifugation of these samples at 820× g at 25 °C for 10 min. DNA was extracted from the buffy coats using the QIAamp DNA blood mini kit (Qiagen, Tokyo, Japan).
4.2. Targeted Deep Sequencing and Data Analysis
A panel targeting the exons of 53 lung cancer-associated genes (see Table S3) was selected to perform targeted sequencing. We searched the literature and selected these genes based on the following criteria: (a) Genes often involved in lung cancer reported in The Cancer Genome Atlas [40] and other projects; or (b) genes frequently mutated in lung cancer in the Catalogue of Somatic Mutations in Cancer (COSMIC) database [41]. The primers for targeted sequencing were designed to cover the hot-spot mutations present in the lung cancer-associated genes, using the Ion AmpliSeq designer software (Thermo Fisher Scientific), as reported previously [42,43,44,45,46,47,48]. Sequencing libraries were prepared using Ion AmpliSeq Library Kit Plus (Thermo Fisher Scientific), according to the manufacturer’s instruction. Multiplex PCR was performed with two primer pools. After PCR reaction, primer sequences were partially digested with 2 μL of FuPa reagent (Thermo Fisher Scientific), and then barcoded using Ion Xpress Barcode Adapters (Thermo Fisher Scientific). The library samples were purified using Agencourt AMPure XP reagent (Beckman Coulter, Brea, CA, USA) and subsequently quantified using Ion Library Quantitation Kit (Thermo Fisher Scientific). Each library was diluted to 60 pM, and the same amount of libraries was pooled for one sequence reaction. Emulsion PCR and chip loading were performed on the Ion Chef with the Ion PI Hi-Q Chef kit. Sequencing was performed using the Ion PI Hi-Q Sequencing Kit on the Ion Proton Sequencer (Thermo Fisher Scientific).
The sequence data were processed using standard Ion Torrent suite software running on the Torrent server version 4.4 (Thermo Fisher Scientific). Raw signal data were analyzed using Torrent suite version 4.0. The pipeline included signal processing, base calling, quality score assignment, read alignment to the human genome 19 reference (hg19), and quality control of mapping and coverage analysis. Following data analysis, annotation of single nucleotide variants, insertions, and deletions were performed using the Ion Reporter server system (Thermo Fisher Scientific), and lymphocytes from peripheral blood DNA were used as controls for detecting any variants (tumor-normal pair analysis). We used the following filtering parameters for variant calling on Ion Reporter Local Server: (i) the minimum number of variant allele reads was ≥10, (ii) the coverage depth was ≥50, (iii) variant allele fraction (AF) ≥ 0.01, (iv) UCSC Common SNPs = Not In, (v) Confident Somatic Variants = In. Sequence data were visually confirmed using the Integrative Genomics viewer. The allele fractions represent the number of mutant reads divided by the total number of reads (coverage) at a specific genomic position. The Functional Analysis through Hidden Markov Models prediction in the COSMIC database was used to estimate oncogenic function of single nucleotide variations [49]. In order to estimate oncogenic function of indel mutations, OncoKB, a comprehensive and curated precision oncology knowledge base, was utilized [50]. Thus, all detected mutations were annotated by either COSMIC or OncoKB database [49].
4.3. Statistics
Continuous variables were presented as mean ± standard deviation (SD), and compared using unpaired Student’s t tests. One-way analysis of variance and the Tukey-Kramer multiple comparison test were used to detect significant differences between groups. Chi-square tests were used to compare the categorical data between groups. Progression-free survival was defined as the period from the day of operation to the day of recurrence or the day of final follow-up. Survival was assessed using the Kaplan-Meier method, and comparisons among the survival curves were made using the log-rank test. To determine the predictors of survival within the cohort, we constructed Cox proportional hazards model including each variable of interest. Multivariate analyses were performed using the JMP function of the SAS software (JMP 15.1.0, SAS Institute, Cary, NC, USA). p-values less than 0.05 in the two-tailed analyses were considered to denote statistical significance.
5. Conclusions
The number of pathogenic mutations, presence of TP53 mutations, and AF of TP53 mutations were identified as mutation factors affecting postoperative prognosis of lung cancer. Importantly, the TP53 mutation profiles varied with the site of lung cancer, and the postoperative prognosis changed accordingly. Developing a detailed understanding of the genomic landscape of lung cancers will establish the ideal model for best surgical outcomes.
Acknowledgments
The authors greatly appreciate Yumiko Kakizaki, Toshiharu Tsutsui, and Yoshihiro Miyashita for their helpful scientific discussion.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/11/3472/s1, Figure S1: Pathogenic versus nonpathogenic mutations. The distribution of pathogenic and nonpathogenic mutations in epidermal growth factor receptor (EGFR), KRAS, and tumor protein p53 (TP53) is shown. KRAS mutations were significantly more frequently pathogenic than EGFR and TP53, Figure S2: Explanatory schemes of the anatomical lung segments. Segments 9 and 10, the dorsal inferior lobes, were highlighted in yellow, Figure S3: Locational variation in prognosis of lung cancers without TP53 mutations. The prognosis was not significantly different between lung cancers without TP53 mutations located in segments 9 and 10 and those located in segments 1 to 8, Table S1: Pathogenic variants identified in our assay, Table S2: Occurrence of mutations according to the location of cancer, Table S3: The genes targeted in the cancer panel.
Author Contributions
T.G., K.K., Y.H. and R.H. wrote the manuscript. T.G., T.N., R.H., Y.Y. and S.O. performed the surgery. T.O., K.A. and R.H. carried out the pathological examination. K.K., Y.H., K.A., T.G., T.N., H.M., R.H., S.O. and M.O. participated in the genomic analyses. M.O. and T.G. edited the final manuscript. All authors have read and agreed to the final version of this manuscript.
Funding
This study was supported by a Grant-in-Aid for Genome Research Project from Yamanashi Prefecture (to Y.H. and M.O.).
Conflicts of Interest
The authors declare no conflict of interest. The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singal G., Miller P.G., Agarwala V., Li G., Kaushik G., Backenroth D., Gossai A., Frampton G.M., Torres A.Z., Lehnert E.M., et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA. 2019;321:1391–1399. doi: 10.1001/jama.2019.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunimasa K., Goto T. Immunosurveillance and Immunoediting of Lung Cancer: Current Perspectives and Challenges. Int. J. Mol. Sci. 2020;21:597. doi: 10.3390/ijms21020597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J., Li C., Seery S., Yu J., Meng X. Identifying optimal first-line interventions for advanced non-small cell lung carcinoma according to PD-L1 expression: A systematic review and network meta-analysis. Oncoimmunology. 2020;9:1746112. doi: 10.1080/2162402X.2020.1746112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asamura H., Aokage K., Yotsukura M. Wedge Resection Versus Anatomic Resection: Extent of Surgical Resection for Stage I and II Lung Cancer. Am. Soc. Clin. Oncol Educ. Book. 2017;37:426–433. doi: 10.14694/EDBK_179730. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita T., Ohtsuka T., Yotsukura M., Asakura K., Goto T., Kamiyama I., Otake S., Tajima A., Emoto K., Hayashi Y., et al. Prognostic impact of preoperative tumor marker levels and lymphovascular invasion in pathological stage I adenocarcinoma and squamous cell carcinoma of the lung. J. Thorac. Oncol. 2015;10:619–628. doi: 10.1097/JTO.0000000000000480. [DOI] [PubMed] [Google Scholar]
- 6.Broderick S.R. Adjuvant and Neoadjuvant Immunotherapy in Non-small Cell Lung Cancer. Thorac. Surg. Clin. 2020;30:215–220. doi: 10.1016/j.thorsurg.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Liang H., Deng H., Liang W., Guo K., Gao Z., Wiesel O., Flores R.M., Song K., Redwan B., Migliore M., et al. Perioperative chemoimmunotherapy in a patient with stage IIIB non-small cell lung cancer. Ann. Transl. Med. 2020;8:245. doi: 10.21037/atm.2020.01.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunimasa K., Hirotsu Y., Amemiya K., Nagakubo Y., Goto T., Miyashita Y., Kakizaki Y., Tsutsui T., Otake S., Kobayashi H., et al. Genome analysis of peeling archival cytology samples detects driver mutations in lung cancer. Cancer Med. 2020 doi: 10.1002/cam4.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin K., Hou H., Liang Y., Zhang X. Prognostic value of TP53 concurrent mutations for EGFR- TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: A meta-analysis. BMC Cancer. 2020;20:328. doi: 10.1186/s12885-020-06805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reckamp K.L. Molecular Targets Beyond the Big 3. Thorac. Surg. Clin. 2020;30:157–164. doi: 10.1016/j.thorsurg.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Reis D., Marques C., Dias M., Campainha S., Cirnes L., Barroso A. Mutational profile of non-small cell lung cancer patients: Use of next-generation sequencing. Pulmonology. 2020;26:50–53. doi: 10.1016/j.pulmoe.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Goto T., Hirotsu Y., Nakagomi T., Shikata D., Yokoyama Y., Amemiya K., Tsutsui T., Kakizaki Y., Oyama T., Mochizuki H., et al. Detection of tumor-derived DNA dispersed in the airway improves the diagnostic accuracy of bronchoscopy for lung cancer. Oncotarget. 2017;8:79404–79413. doi: 10.18632/oncotarget.18159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirotsu Y., Otake S., Ohyama H., Amemiya K., Higuchi R., Oyama T., Mochizuki H., Goto T., Omata M. Dual-molecular barcode sequencing detects rare variants in tumor and cell free DNA in plasma. Sci. Rep. 2020;10:3391. doi: 10.1038/s41598-020-60361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iijima Y., Hirotsu Y., Amemiya K., Higuchi R., Nakagomi T., Goto T., Uchida Y., Kobayashi Y., Tsutsui T., Kakizaki Y., et al. Endotracheal or Endobronchial Metastasis of Lung Squamous Cell Carcinoma. J. Bronchol. Interv. Pulmonol. 2019;26:e46–e50. doi: 10.1097/LBR.0000000000000595. [DOI] [PubMed] [Google Scholar]
- 15.Iijima Y., Hirotsu Y., Amemiya K., Ooka Y., Mochizuki H., Oyama T., Nakagomi T., Uchida Y., Kobayashi Y., Tsutsui T., et al. Very early response of circulating tumour-derived DNA in plasma predicts efficacy of nivolumab treatment in patients with non-small cell lung cancer. Eur. J. Cancer. 2017;86:349–357. doi: 10.1016/j.ejca.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Kunimasa K., Hirotsu Y., Nakamura H., Tamiya M., Iijima Y., Ishida H., Hamamoto Y., Maniwa T., Kimura T., Nishino K., et al. Rapid progressive lung cancers harbouring multiple clonal driver mutations with big bang evolution model. Cancer Genet. 2020;241:51–56. doi: 10.1016/j.cancergen.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Miyashita Y., Hirotsu Y., Tsutsui T., Higashi S., Sogami Y., Kakizaki Y., Goto T., Amemiya K., Oyama T., Omata M. Analysis of significantly mutated genes as a clinical tool for the diagnosis in a case of lung cancer. Respir. Med. Case Rep. 2017;20:171–175. doi: 10.1016/j.rmcr.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagomi T., Goto T., Hirotsu Y., Shikata D., Amemiya K., Oyama T., Mochizuki H., Omata M. Elucidation of radiation-resistant clones by a serial study of intratumor heterogeneity before and after stereotactic radiotherapy in lung cancer. J. Thorac. Dis. 2017;9:E598–E604. doi: 10.21037/jtd.2017.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagomi T., Goto T., Hirotsu Y., Shikata D., Yokoyama Y., Higuchi R., Amemiya K., Okimoto K., Oyama T., Mochizuki H., et al. New therapeutic targets for pulmonary sarcomatoid carcinomas based on their genomic and phylogenetic profiles. Oncotarget. 2018;9:10635–10649. doi: 10.18632/oncotarget.24365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto T., Hirotsu Y., Amemiya K., Mochizuki H., Omata M. Understanding Intratumor Heterogeneity and Evolution in NSCLC and Potential New Therapeutic Approach. Cancers. 2018;10:212. doi: 10.3390/cancers10070212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M., Xu Y., Zhao J., Zhong W., Zhang L., Bi Y., Wang M. Concurrent Driver Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive Non-Small Cell Lung Cancer. EBioMedicine. 2019;42:304–310. doi: 10.1016/j.ebiom.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y., Che N., Yu Y., Gao Y., Shi H., Feng Q., Wei B., Ma L., Gao M., Ma J., et al. Co-occurring genetic alterations and primary EGFR T790M mutations detected by NGS in pre-TKI-treated NSCLCs. J. Cancer Res. Clin. Oncol. 2020;146:407–416. doi: 10.1007/s00432-019-03065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono A., Isaka M., Serizawa M., Omae K., Kojima H., Nakashima K., Omori S., Wakuda K., Kenmotsu H., Naito T., et al. Genetic alterations of driver genes as independent prognostic factors for disease-free survival in patients with resected non-small cell lung cancer. Lung Cancer. 2019;128:152–157. doi: 10.1016/j.lungcan.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Fukuyama Y., Mitsudomi T., Sugio K., Ishida T., Akazawa K., Sugimachi K. K-ras and p53 mutations are an independent unfavourable prognostic indicator in patients with non-small-cell lung cancer. Br. J. Cancer. 1997;75:1125–1130. doi: 10.1038/bjc.1997.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uramoto H., Sugio K., Oyama T., Nakata S., Ono K., Nozoe T., Yasumoto K. Expression of the p53 family in lung cancer. Anticancer Res. 2006;26:1785–1790. [PubMed] [Google Scholar]
- 26.Carbone D.P., Mitsudomi T., Chiba I., Piantadosi S., Rusch V., Nowak J.A., McIntire D., Slamon D., Gazdar A., Minna J. p53 immunostaining positivity is associated with reduced survival and is imperfectly correlated with gene mutations in resected non-small cell lung cancer. A preliminary report of LCSG 871. Chest. 1994;106:377s–381s. doi: 10.1378/chest.106.6.377S. [DOI] [PubMed] [Google Scholar]
- 27.Bodner S.M., Minna J.D., Jensen S.M., D’Amico D., Carbone D., Mitsudomi T., Fedorko J., Buchhagen D.L., Nau M.M., Gazdar A.F., et al. Expression of mutant p53 proteins in lung cancer correlates with the class of p53 gene mutation. Oncogene. 1992;7:743–749. [PubMed] [Google Scholar]
- 28.Mitsudomi T., Oyama T., Nishida K., Ogami A., Osaki T., Nakanishi R., Sugio K., Yasumoto K., Sugimachi K. p53 nuclear immunostaining and gene mutations in non-small-cell lung cancer and their effects on patient survival. Ann. Oncol. 1995;6(Suppl. 3):S9–S13. doi: 10.1093/annonc/6.suppl_3.S9. [DOI] [PubMed] [Google Scholar]
- 29.Aisner D.L., Sholl L.M., Berry L.D., Rossi M.R., Chen H., Fujimoto J., Moreira A.L., Ramalingam S.S., Villaruz L.C., Otterson G.A., et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2) Clin. Cancer Res. 2018;24:1038–1047. doi: 10.1158/1078-0432.CCR-17-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devarakonda S., Rotolo F., Tsao M.S., Lanc I., Brambilla E., Masood A., Olaussen K.A., Fulton R., Sakashita S., McLeer-Florin A., et al. Tumor Mutation Burden as a Biomarker in Resected Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018;36:2995–3006. doi: 10.1200/JCO.2018.78.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X., Le Teuff G., Lacas B., Tsao M.S., Graziano S., Pignon J.P., Douillard J.Y., Le Chevalier T., Seymour L., Filipits M., et al. Prognostic and Predictive Effect of TP53 Mutations in Patients with Non-Small Cell Lung Cancer from Adjuvant Cisplatin-Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J. Thorac. Oncol. 2016;11:850–861. doi: 10.1016/j.jtho.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L.L., Kan M., Zhang M.M., Yu S.S., Xie H.J., Gu Z.H., Wang H.N., Zhao S.X., Zhou G.B., Song H.D., et al. Multiregion sequencing reveals the intratumor heterogeneity of driver mutations in TP53-driven non-small cell lung cancer. Int. J. Cancer. 2017;140:103–108. doi: 10.1002/ijc.30437. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.Y., Jeon H.S., Hwangbo Y., Jeong J.Y., Park J.Y., Lee E.J., Jin G., Shin K.M., Yoo S.S., Lee J., et al. The influence of TP53 mutations on the prognosis of patients with early stage non-small cell lung cancer may depend on the intratumor heterogeneity of the mutations. Mol. Carcinog. 2015;54:93–101. doi: 10.1002/mc.22077. [DOI] [PubMed] [Google Scholar]
- 35.Dankner M., Rose A.A.N., Rajkumar S., Siegel P.M., Watson I.R. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene. 2018;37:3183–3199. doi: 10.1038/s41388-018-0171-x. [DOI] [PubMed] [Google Scholar]
- 36.Schulze A.B., Evers G., Kerkhoff A., Mohr M., Schliemann C., Berdel W.E., Schmidt L.H. Future Options of Molecular-Targeted Therapy in Small Cell Lung Cancer. Cancers. 2019;11:690. doi: 10.3390/cancers11050690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan M., Huang L.L., Chen J.H., Wu J., Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019;4:61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbs A.R., Thunnissen F.B. Histological typing of lung and pleural tumours: Third edition. J. Clin. Pathol. 2001;54:498–499. doi: 10.1136/jcp.54.7.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chansky K., Detterbeck F.C., Nicholson A.G., Rusch V.W., Vallieres E., Groome P., Kennedy C., Krasnik M., Peake M., Shemanski L., et al. The IASLC Lung Cancer Staging Project: External Validation of the Revision of the TNM Stage Groupings in the Eighth Edition of the TNM Classification of Lung Cancer. J. Thorac. Oncol. 2017;12:1109–1121. doi: 10.1016/j.jtho.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 40.The Cancer Genome Atlas Program. [(accessed on 22 May 2014)]; Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
- 41.Catalogue of Somatic Mutations In Cancer. [(accessed on 24 May 2014)]; Available online: http://cancer.sanger.ac.uk/cancergenome/projects/cosmic.
- 42.Goto T., Hirotsu Y., Mochizuki H., Nakagomi T., Oyama T., Amemiya K., Omata M. Stepwise addition of genetic changes correlated with histological change from “well-differentiated” to “sarcomatoid” phenotypes: A case report. BMC Cancer. 2017;17:65. doi: 10.1186/s12885-017-3059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goto T., Hirotsu Y., Oyama T., Amemiya K., Omata M. Analysis of tumor-derived DNA in plasma and bone marrow fluid in lung cancer patients. Med. Oncol. 2016;33:29. doi: 10.1007/s12032-016-0744-x. [DOI] [PubMed] [Google Scholar]
- 44.Amemiya K., Hirotsu Y., Goto T., Nakagomi H., Mochizuki H., Oyama T., Omata M. Touch imprint cytology with massively parallel sequencing (TIC-seq): A simple and rapid method to snapshot genetic alterations in tumors. Cancer Med. 2016;5:3426–3436. doi: 10.1002/cam4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higuchi R., Nakagomi T., Goto T., Hirotsu Y., Shikata D., Yokoyama Y., Otake S., Amemiya K., Oyama T., Mochizuki H., et al. Identification of Clonality through Genomic Profile Analysis in Multiple Lung Cancers. J. Clin. Med. 2020;9:573. doi: 10.3390/jcm9020573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagomi T., Goto T., Hirotsu Y., Shikata D., Yokoyama Y., Higuchi R., Otake S., Amemiya K., Oyama T., Mochizuki H., et al. Genomic Characteristics of Invasive Mucinous Adenocarcinomas of the Lung and Potential Therapeutic Targets of B7-H3. Cancers. 2018;10:478. doi: 10.3390/cancers10120478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagomi T., Hirotsu Y., Goto T., Shikata D., Yokoyama Y., Higuchi R., Otake S., Amemiya K., Oyama T., Mochizuki H., et al. Clinical Implications of Noncoding Indels in the Surfactant-Encoding Genes in Lung Cancer. Cancers. 2019;11:552. doi: 10.3390/cancers11040552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goto T., Hirotsu Y., Mochizuki H., Nakagomi T., Shikata D., Yokoyama Y., Oyama T., Amemiya K., Okimoto K., Omata M. Mutational analysis of multiple lung cancers: Discrimination between primary and metastatic lung cancers by genomic profile. Oncotarget. 2017;8:31133–31143. doi: 10.18632/oncotarget.16096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34:511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H., et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.