Abstract
The prospective multicenter COMET trial followed a cohort of 306 consecutive metastatic breast cancer patients receiving bevacizumab and paclitaxel as first-line chemotherapy. This study was intended to identify and validate reliable biomarkers to better predict bevacizumab treatment outcomes and allow for a more personalized use of this antiangiogenic agent. To that end, we aimed to establish risk scores for survival prognosis dichotomization based on classic clinico-pathological criteria combined or not with single nucleotide polymorphisms (SNPs). The genomic DNA of 306 patients was extracted and a panel of 13 SNPs, covering seven genes previously documented to be potentially involved in drug response, were analyzed by means of high-throughput genotyping. In receiver operating characteristic (ROC) analyses, the hazard model based on a triple-negative cancer phenotype variable, combined with specific SNPs in VEGFA (rs833061), VEGFR1 (rs9582036) and VEGFR2 (rs1870377), had the highest predictive value. The overall survival hazard ratio of patients assigned to the poor prognosis group based on this model was 3.21 (95% CI (2.33–4.42); p < 0.001). We propose that combining this pharmacogenetic approach with classical clinico-pathological characteristics could markedly improve clinical decision-making for breast cancer patients receiving bevacizumab-based therapy.
Keywords: breast cancer, precision medicine, pharmacogenetics, bevacizumab, SNP, VEGF, overall survival
1. Introduction
The vascular endothelial-derived growth factor A (VEGFA) is a positive regulator of angiogenesis and a coordinator of vascular homeostasis. In addition, the capacity of VEGFA to act as an immunosuppressor is well-established [1]. Therefore, targeting VEGFA and/or its receptors (VEGFR1, 2 and 3) constitutes a direct means to influence the tumoral environment. Bevacizumab, a VEGFA targeting monoclonal antibody, entered clinical practice more than 15 years ago [2] and is currently the most prescribed antiangiogenic treatment. Indeed, bevacizumab is indicated for colorectal cancer, breast cancer, non-small cell lung carcinoma (NSCLC), glioblastoma, renal cell carcinoma, ovarian cancer and cervical cancer [3]. The immunosuppressive properties of VEGFA have recently opened new perspectives regarding bevacizumab’s immunomodulatory potential. Indeed, bevacizumab has recently demonstrated clinical benefits for NSCLC and hepatocellular carcinoma in combined cancer immunotherapy [2,4]. However, despite intense investigations, reliable biomarker signatures that would allow us to pinpoint the individuals most susceptible to benefit from bevacizumab-based treatments remain to be identified and validated. Germline genetic polymorphisms may influence the outcome of cancer treatments in several aspects, such as individual variability in drug metabolism, immune response [5] and target availability [6,7].
The French multicentric cohort study COMET aimed to identify biological factors that could predict the clinical benefit of a bevacizumab–paclitaxel combination therapy as first-line treatment in metastatic breast cancer. The COMET study was specifically designed to survey key candidate genes potentially linked to the pharmacogenetics of both bevacizumab and paclitaxel. Notably, the elimination of paclitaxel is known to be influenced by CYP2C8 and ABCB1 polymorphisms [8,9]. In addition, by following a small cohort of metastatic breast cancer patients receiving another bevacizumab-based combination (bevacizumab–taxane), we previously established the prognostic value of the single nucleotide polymorphism (SNP) 936 C > T (rs3025039) in VEGFA [10]. The ECOG 2100 trial demonstrated the importance of polymorphisms in the VEGFA and VEGFR2 genes for predicting the outcome of patients treated with the bevacizumab–paclitaxel combination compared with paclitaxel alone [11].
In the present study, 13 different SNPs across six genes were analyzed in an effort to identify a specific SNP signature significantly associated with bevacizumab-paclitaxel treatment outcome in terms of survival among metastatic breast cancer patients. The risk score analyses evaluating their predictive value as stand-alone variables versus when combined with common pathophysiologic criteria used in clinical practice are presented.
2. Results
2.1. Study Population and Clinical Characteristics of Tumors
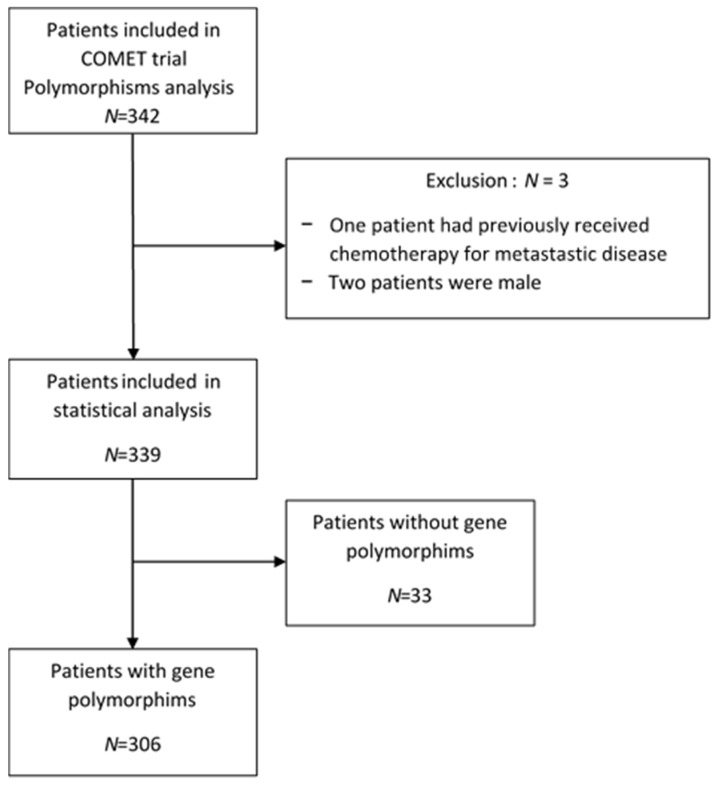
Out of the 342 patients included, 306 genomic DNA samples were available for gene polymorphism analysis. The patients’ disposition is presented in Figure 1. The patients’ characteristics, the disease and the treatment baselines are summarized in Table 1. The mean age at inclusion was 55 years (range: 28–80). At initial diagnosis, the main histological type was invasive ductal carcinoma (79.5%). Most patients had received prior neoadjuvant/adjuvant chemotherapy (n = 210, 68%). Patients presented a small number of metastatic sites (90% had less than three metastatic sites); 62.5% of patients had a metastasis-free interval greater than 24 months. One hundred and thirty-five patients (48.5%) had histological Grade II tumors and 43.5% had Grade III tumors. The vast majority of patients had a positive hormone receptor status (n = 206, 76.5%) and 23.5% had triple-negative subtype tumors. Twenty-eight patients (9.35%) showed a complete clinical response (CR), 139 (46.5%) a partial response (PR), 101 (33.8%) a stable disease (SD) and 131 (10.35%) disease progression (PD).
Figure 1.
Flow diagram of the study participants.
Table 1.
Patients and tumor characteristics at baseline.
| Characteristics | Number of Patients (N = 306) | % |
|---|---|---|
| Mean age (min–max) at inclusion | 55.528–80 | |
| Histology | ||
| Invasive ductal carcinoma | 237 | 79.5 |
| Invasive lobular carcinoma | 38 | 12.5 |
| Mixed (ductal and lobular) | 10 | 3.5 |
| Other | 13 | 4.5 |
| Menopausal status at inclusion | ||
| Premenopausal | 88 | 29 |
| Postmenopausal | 213 | 71 |
| Unknown | 5 | |
| Performance status at inclusion | ||
| 0 | 172 | 56 |
| 1/2 | 134 | 43 |
| Histological grade | ||
| I | 23 | |
| II | 135 | 8 |
| III | 121 | 48.5 |
| Unknown | 27 | 43.5 |
| Subtype | ||
| Triple-negative | 64 | |
| Hormonal receptor-positive | 206 | 23.5 |
| Unknown | 36 | 76.5 |
| Tumor stage at initial diagnosis | ||
| pT0/pT1 | 109 | |
| pT2 | 84 | 46.5 |
| pT3/pT4 | 41 | 36 |
| Unknown | 72 | 17.5 |
| Axillary lymph node status at initial diagnosis | ||
| pN0 | 83 | |
| pN+ | 153 | 35 |
| Unknown | 68 | 65 |
| Prior (neo)adjuvant chemotherapy | ||
| Yes | 210 | 68 |
| No | 96 | 32 |
| Number of metastatic sites | ||
| <3 | 211 | |
| ≥3 | 24 | 90 |
| Unknown | 71 | 10 |
| Metastasis-free interval | ||
| 0 months | 17 | 7 |
| 0–24 months | 71 | 30.5 |
| 24 months | 147 | 62.5 |
| Unknown | 71 | |
| Metastatic sites | ||
| Liver | 97 | 41 |
| Distant node | 60 | 25.5 |
| Bone | 37 | 15.5 |
| Skin | 13 | 5.5 |
| Lung | 19 | 8.0 |
| Other 1 | 9 | 4.5 |
| Unknown | 71 |
1: Soft tissue, central nervous system, breast, peritoneum, glands.
2.2. Association between Progression-Free Survival and Clinico-Pathological Features and SNPs
The median follow-up was 50 months (95% CI (47–54)). The 5-year overall survival (OS) was 21% (16–28) and the 5-year progression-free survival (PFS) was 4% (2–7). The median OS and PFS were equal to 32 months (95% CI (28.5–36)) and 11 months (95% CI (9.5–11.5)), respectively.
The relationship between PFS and patient characteristics and 13 SNPs was assessed by univariate analyses (Tables S1 and S2). PFS was significantly associated (Table 2 and Figure S1) with histological Grade III (HR = 1.6, 95% CI (1.2–2.0); p < 0.001), triple-negative subtype (HR = 1.9, 95% CI (1.4–2.5); p < 0.001), VEGFA rs699947 A/A (HR = 1.3, 95% CI (1–1.8); p = 0.042), VEGFA rs833061 C/C (HR = 1.40, 95% CI (1.0–1.8); p = 0.028), VEGFA rs2010963 G/C or C/C (HR = 0.77, 95% CI (0.61–0.97); p = 0.026) and VEGFR1 rs9582036 C/A or C/C (HR = 1.4, 95% CI (1.10–1.70); p = 0.01). As shown in Figure S2, patients with the triple-negative subtype and histological Grade III tumors had a median PFS of 5.9 months (95% CI (4.50–7.85)), while patients with hormonal receptor-positive and histological Grade I or II tumors had a median PFS of 12.94 months (95% CI (11.83–14.82); p < 0.001).
Table 2.
Univariate analysis for the polymorphisms and clinical characteristics according to progression-free survival (PFS) and overall survival (OS).
| Progression-Free Survival | Overall Survival | ||||
| Significant Gene Polymorphisms | HR 95% CI | p-Value | HR 95% CI | p-Value | |
| VEGFA | rs699947 | ||||
| C/C or A/C | 1 | Referent | 1 | - | |
| A/A | 1.3 (1–1.8) | 0.042 | 1.5 (1.1–2.1) | 0.007 | |
| rs833061 | |||||
| T/T or T/C | 1 | Referent | 1 | - | |
| C/C | 1.4 (1–1.8) | 0.028 | 1.6 (1.1–2.1) | 0.005 | |
| rs2010963 | |||||
| G/G | 1 | Referent | - | - | |
| G/C or C/C | 0.77 (0.6–0.97) | 0.026 | - | - | |
| VEGFR1 | rs9582036 | ||||
| A/A | 1 | Referent | 1 | - | |
| C/A or C/C | 1.4 (1.1–1.7) | 0.010 | 1.4 (1.1–1.9) | 0.010 | |
| VEGFR2 | rs1870377 | ||||
| T/A or A/A | - | - | 1 | - | |
| T/T | - | - | 1.4 (1–1.8) | 0.021 | |
| Significant Clinical Characteristics | HR 95% CI | p-Value | HR 95% CI | p-Value | |
| Histological grade | |||||
| Grade I or II | 1 | Referent | 1 | - | |
| Grade III | 1.6 (1.2–2) | <0.001 | 1.4 (1.1–1.9) | 0.011 | |
| Subtype | |||||
| Hormonal receptor-positive | 1 | Referent | 1 | - | |
| Triple-negative | 1.9 (1.4–2.5) | <0.001 | 2.4 (1.7–3.3) | <0.001 | |
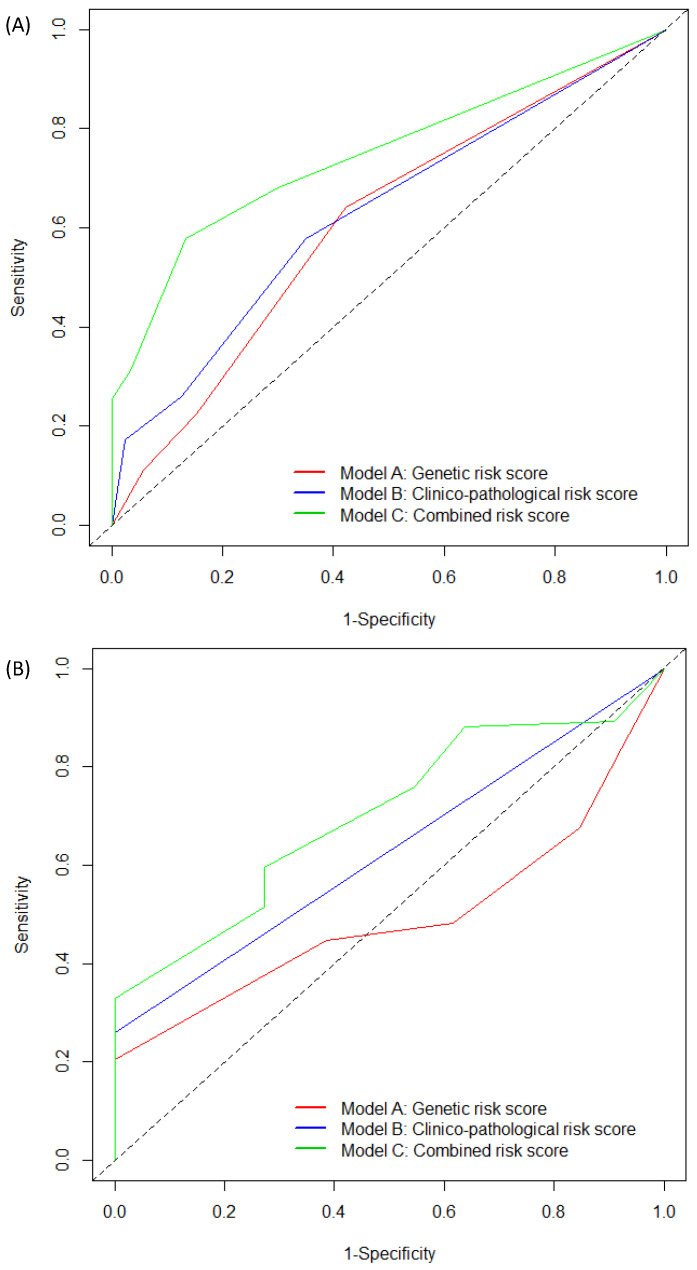
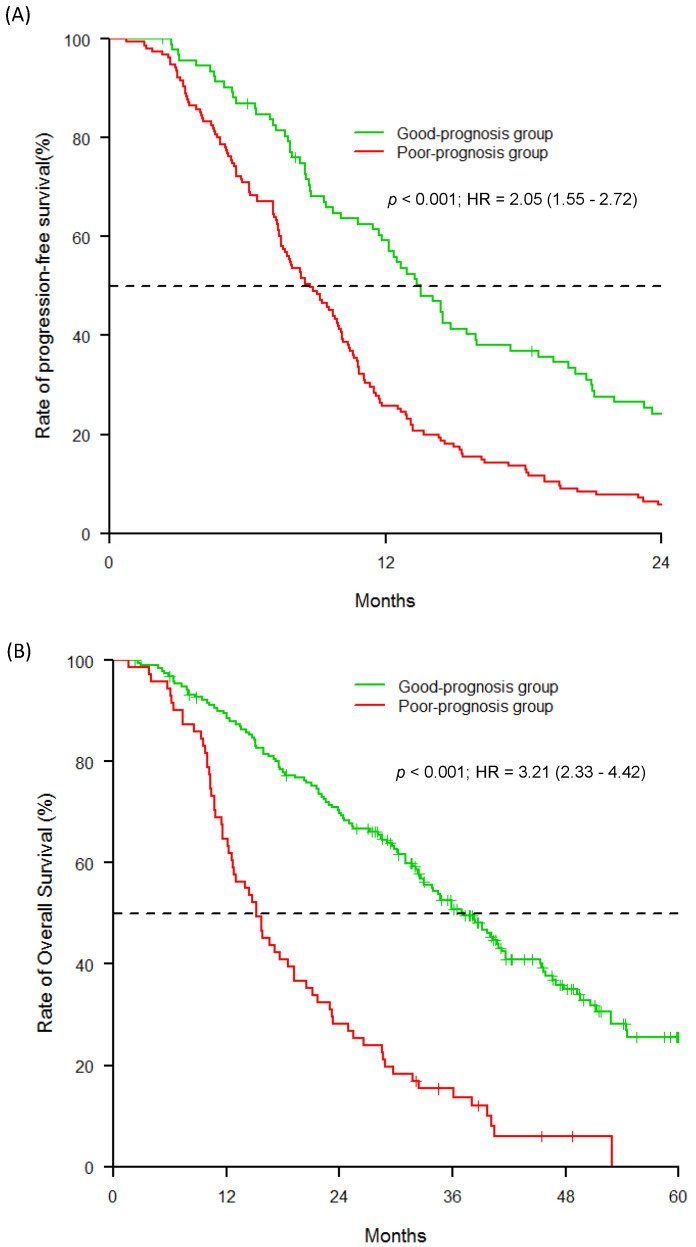
Through multivariable analyses, three distinct predictive models (Model A: genetic; Model B: clinico-pathological; Model C: combined) were evaluated for PFS (Table 3). Two significant variables remained predictive of a poor prognosis: VEGFA rs833061 C/C (HR = 1.35, 95% CI (1.02–1.80); p = 0.032) and VEGFR1 rs9582036 C/A or C/C (HR = 1.34, 95% CI (1.06–1.71); p = 0.011) for Model A, and histological Grade III (HR = 1.5, 95% CI (1.15–1.95); p < 0.001) and triple-negative subtype (HR = 1.75, 95% CI (1.29–2.37); p < 0.001) for Model B. For Model C, histological Grade III (HR = 2.0, 95% CI (1.50–2.65); p < 0.001), triple-negative subtype (HR = 2.26, 95% CI (1.64–3.12); p < 0.001) and VEGFA rs833061 C/C (HR = 1.39, 95% CI (1.01–1.91); p = 0.037) were associated with a shorter PFS. As shown in Figure 2A, the AUC for the risk score of Model C at 24 months was higher compared with Models A and B (AUCmodel C: 74.75 (95% CI (67.98–81.51)); AUCmodel B: 63.12 (95% CI (55.22–71.03)); AUCmodel A: 60.97 (95% CI (53.08–68.87)). Dichotomization risk score was then carried out using the receiver operating characteristic (ROC) curve at 24 months (optimal cut-off value: −0.22) separating the poor from good prognosis groups of patients with a sensitivity of 58% and a specificity of 87%. As shown in Figure 3A, patients assigned to the poor prognosis group (N = 155) presented a median PFS of 8.71 months (95% CI (7.46–9.92)) and patients in the good prognosis group (N = 93) presented a median PFS of 13.53 months (95% CI (11.70–15.90); p < 0.001). The Cox regression analysis indicated that patients assigned to the poor prognosis group were significantly more prone to disease progression (HR = 2.05, CI 95% (1.55–2.72); p < 0.001).
Table 3.
Multivariate analysis according to PFS.
| Significant Gene Polymorphisms | Progression-Free Survival | ||||||
|---|---|---|---|---|---|---|---|
| Model A: Genetic | Model B: Clinico-Pathological | Model C: Combined | |||||
| HR [95% CI] | p-Value | HR [95% CI] | p-Value | HR [95% CI] | p-Value | ||
| VEGFA | rs833061 | ||||||
| T/T or T/C | 1 | - | - | - | 1 | - | |
| C/C | 1.35 (1.02–1.80) | 0.032 | - | - | 1.39 (1.01–1.91) | 0.037 | |
| VEGFR1 | rs9582036 | ||||||
| A/A | 1 | - | - | - | - | - | |
| C/A or C/C | 1.34 (1.06–1.71) | 0.011 | - | - | - | - | |
| Significant clinical characteristics | |||||||
| Histological grade | Grade I or II | - | - | 1 | - | 1 | - |
| Grade III | - | - | 1.50 (1.15–1.95) | <0.001 | 2.0 (1.50–2.65) | <0.001 | |
| Subtype | Hormonal receptor-positive | - | - | 1 | - | 1 | - |
| Triple-negative | - | - | 1.75 (1.29–2.37) | <0.001 | 2.26 (1.64–3.12) | <0.001 | |
Figure 2.
Receiver operating characteristic (ROC) curves and AUC for risk scores for three models. (A) PFS at 24 months; (B) OS at 60 months.
Figure 3.
PFS and OS of the patients within different risk groups based on the predictive model. (A) PFS; (B) OS.
2.3. Associations among Overall Survival, SNPs and Patient Characteristics
Univariate analyses of OS were performed (Tables S3 and S4). Among all tested variables, OS was significantly correlated (Table 3 and Figure S3) with histological grade III (HR = 1.4, 95% CI (1.1–1.9); p = 0.011), triple-negative subtype (HR = 2.4, 95% CI (1.7–3.3); p < 0.001), VEGFA rs699947 A/A (HR = 1.5, 95% CI (1.1–2.1); p = 0.007), VEGFA rs833061 C/C (HR = 1.6, 95% CI (1.1–2.1); p = 0.005), VEGFA rs1870377 T/T (HR = 1.4, 95% CI (1–1.8); p = 0.021) and VEGFR1 rs9582036 C/A or C/C (HR = 1.4, 95% CI (1.1–1.9); p = 0.01). As shown in Figure S4, triple-negative subtype and hormonal receptor-positive status were predictive of the worst and best prognosis, respectively. More precisely, triple-negative subtype patients had a median OS of 14.4 months (95% CI (12–21.7)) when having a histological Grade III tumor and a median OS of 15.7 months (95% CI (11.6–NA)) with histological Grade I or II tumors. In contrast, hormonal receptor-positive patients had a median OS of 38 months (95% CI (33.9–45.6)) when having a histological Grade I or II tumor and a median OS of 31.8 months (95% CI (25.3–41.7)) for histological Grade III patients.
As for PFS, three distinct predictive models (i.e., Model A: genetic; Model B: clinico-pathological; Model C: combined) were evaluated (Table 4). Three significant variables remained predictive of poor prognosis for Model A: VEGFA rs833061 C/C (HR = 1.59, 95% CI (1.15–2.20); p = 0.003), VEGFR1 rs9582036 C/A or C/C (HR = 1.34, 95% CI (1.01–1.78); p = 0.034) and VEGFR2 rs1870377 T/A or A/A (HR = 1.39, 95% CI (1.04–1.86); p = 0.02). Only the histological Grade III variable remained predictive for Model B (HR = 3.42, 95% CI (2.44–4.81); p < 0.001). With regard to Model C, the triple-negative subtype (HR = 4.22, 95% CI (2.96–6.01); p < 0.001), VEGFA rs833061 C/C (HR = 1.40, 95% CI (1.00–1.98); p = 0.049), VEGFR1 rs9582036 C/A or C/C (HR = 1.56, 95% CI (1.15–2.10); p = 0.003) and VEGFR2 rs1870377 T/A or A/A (HR = 1.69, 95% CI (1.24–2.3); p < 0.001) were associated with shorter OS. As shown in Figure 2B, at 60 months, the AUC for the risk score of Model C was better than the ones from Models A or B (AUCmodel C: 70.23 (95% CI (57.84–82.62)); AUCmodel B: 62.99 (95% CI (59.94–66.05)); AUCmodel A: 49.53 (95% CI (38.43–60.63))). At 60 months and based on Model C, the optimal cut-off value was 0.06 and risk groups were associated with a sensitivity of 33% and a specificity of 100%, respectively.
Table 4.
Multivariate analysis according to OS.
| Significant Gene Polymorphisms | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|
| Model A: Genetic | Model B: Clinico-Pathological | Model C: Combined | |||||
| HR [95% CI] | p-Value | HR [95% CI] | p-Value | HR [95% CI] | p-Value | ||
| VEGFA | rs833061 | ||||||
| T/T or T/C | 1 | - | - | - | 1 | - | |
| C/C | 1.59 (1.15–2.20) | 0.003 | - | - | 1.40 (1.00–1.98) | 0.049 | |
| VEGFR1 | rs9582036 | ||||||
| A/A | 1 | - | - | - | 1 | - | |
| C/A or C/C | 1.34 (1.01–1.78) | 0.034 | - | - | 1.56 (1.15–2.10) | 0.003 | |
| VEGFR2 | rs1870377 | ||||||
| T/T | 1 | - | - | - | 1 | - | |
| T/A or A/A | 1.39 (1.04–1.86) | 0.020 | - | - | 1.69 (1.24–2.30) | <0.001 | |
| Significant clinical characteristics | |||||||
| Histological grade | Grade I or II | - | - | - | - | - | - |
| Grade III | - | - | - | - | - | - | |
| Subtype | Hormonal receptor-positive | - | - | 1 | - | 1 | - |
| Triple-negative | - | - | 3.42 (2.44–4.81) | <0.001 | 4.22 (2.96–6.01) | <0.001 | |
As shown in Figure 3B and as defined by Model C, patients falling into the good prognosis group had a longer median OS (15.2 months; 95% CI (12.3–19.2)) compared with patients assigned to the poor prognosis group (37.1 months; 95% CI (32.6–41.2); p <0.001) (Figure 3B). Notably, patients assigned to the poor prognosis group had a death hazard ratio of 3.21 (95% CI (2.33–4.42); p < 0.001).
3. Discussion
The present pharmacogenetic study was based on a homogeneous group of 306 patients with respect to cancer type and advanced disease status with an identical treatment protocol. An optimal predictive model for OS was obtained with the tumor subtype (triple-negative) and polymorphisms within the gene encoding the target of bevacizumab itself: VEGFA (rs833061 C/C), as well as within the genes encoding its receptors (VEGFR1; rs9582036 C/A or C/C and VEGFR2; rs1870377 T/T). The germline genome is sometimes considered to have limited applications regarding cancer therapy optimization [7]. However, the present study supports the idea that gene polymorphisms may help predict bevacizumab-based treatment outcomes. Bevacizumab is increasingly used outside its classical clinical area of prescription [12]. Moreover, the immunomodulatory properties of bevacizumab have recently led to very promising associations in combined immunotherapies for NSCLC and hepatocellular carcinomas [2]. Thus, the present data may further encourage the identification and validation of reliable and much needed bevacizumab biomarkers. The clinical impact of VEGFA gene polymorphisms was recently examined in patients with advanced colorectal cancer treated with bevacizumab in association with chemotherapy. So far, the prognostic value of VEGFA rs699947 is unclear [13,14]. In contrast, VEGFA rs833061 demonstrated strong prognostic value in advanced colorectal cancer patients and, to a lesser extent, in breast cancer patients [13]. Notably, our data further support the prognostic value of VEGFA rs833061 for breast cancer patients.
Considering that rs833061 is located in the 5’-UTR of VEGFA, we then sought to investigate whether this specific polymorphism might have an impact on VEGFA expression, especially considering that this SNP is associated with nine SNPs (r2 > 0.8) all located in the promoter region or in the first introns of VEGFA. Together, these SNPs were associated with the high expression of VEGFA in thyroid tissue or different alternative splicing events in the left heart ventricle vs. muscle (according to GTEX portal query results). These observations warrant further investigation to establish whether this SNP is functionally linked to the phenotype (poor outcomes) observed in the context of the present study.
rs9582036 is located on the FLT1 gene (also known as VEGFR1) and is part of a cluster of 15 SNPs with close linkage disequilibrium (0.6 < r2 < 0.8). Fourteen of these SNPs are intronic, including the synonymous variant rs9582036 associated with an alternative splicing event in adipose tissue and the thyroid (according to the GTEX portal). All SNPs are located between the 25th and the 28th intron (30 exons in total). An in silico analysis did not show evidence of reliable variations. Nevertheless, the presence of a shorter isoform (ENST0000061 7835.4) that starts at Exon 28 and codes for the last 85 amino acids of the protein could be regulated differently due to the surrounding SNPs. Further phenotypic analyses would be required to understand the relationship between the SNP and the isoform (ENST00000617835.4). The rs1870377 T/A variant located on the KDR gene (also known as VEGFR2) introduces a missense glutamine (Gln) to histidine (His) substitution at the amino acid position 472, potentially impacting VEGFR2 degradation.
The present study contains a certain number of limitations, as we aimed to focus on a predefined set of SNPs. GWAS would be required in order to gain a broader insight into the SNPs involved in bevacizumab-based treatment outcomes. However, GWAS necessitates a larger number of patients. The impact in terms of OS conferred by the specific SNPs touching the targets of bevacizumab and its cellular receptors merits further investigation in the case where bevacizumab is associated with immunotherapy. In this respect, recent experimental data [15] highlighted a direct effect of VEGFA on endothelial receptors that modifies T cell diapedesis and allows TRegs rather than T cytotoxic cells to migrate to the tumor bed, thus decreasing the tumor’s immune capacity. The SNPs described herein that have an impact on the outcome of bevacizumab-based treatment may thus be of potential prognostic value in the context of bevacizumab–checkpoint inhibitor combinations, a particularly promising domain of application for this widely used antiangiogenic and immunomodulatory agent.
4. Materials and Methods
4.1. Study Design and Patients
This prospective, multicenter COMET trial covered a cohort of 342 consecutive patients included between 2012 and 2015. The inclusion criteria were: age > 18 years, histologically confirmed metastatic HER2- breast cancer, ECOG ≤ 2, life expectancy ≥ 12 weeks, patients receiving first-line chemotherapy with bevacizumab (10 mg/kg every 2 weeks) and paclitaxel (90 mg/m2 at Day 1, Day 8 and Day 15). Treatment was repeated every 4 weeks, according to routine practice, until disease progression or unacceptable toxicity. The exclusion criteria were prior chemotherapy for metastatic breast cancer, concomitant endocrine therapy or radiation therapy with curative intent for oligometastatic disease. The tumor response was defined according to Response Evaluation Criteria in Solid Tumours (RECIST 1.1) criteria as complete response (CR), partial response (PR), stable disease (SD) or progressive disease (PD). The objective response rate (ORR) was defined as the proportion of patients whose best overall response over the entire treatment period was CR or PR. Informed written consent was obtained from each patient. The French Institutional Ethics Committee approved the study in June 2012 and it was registered (NCT01745757).
4.2. SNP Selection and Genotyping
Genomic DNA was extracted from blood samples using the commercially available Maxwell® 16 LEV Blood DNA Kit (#AS1290, Promega©, Madison, WI, USA). High-throughput genotyping of germline DNA was performed by MassARRAY [16] (AGENA Bioscience®, San Diego, CA, USA) using the custom panel of 18 SNPs covering 7 objectively preselected genes corresponding to the drug response: VEGFA (rs833061, rs2010963, rs3025039, rs1570360, rs699947), VEGFR1 (rs9582036), VEGFR2 (rs2071559, rs2305948, rs1870377), IL8 (rs4073), CYP2C8 (rs11572080, rs10509681), CYP450 (rs2740574, rs776746) and ABCB1 (rs2229109, rs2032582, rs1045642, rs2032582). Independent controls were performed by conducting SNPs’ analyses either at another center (rs1045642 and 1128503) or by a different technique (RFLP: rs833061, rs2010963, rs1570360). For 18 SNPs, the minor allele frequency was ≥5% in Caucasians, according to SNPpedia (http://www.snppedia.com) and the Ensemble database (http://www.Ensembl.org).
Five SNPs were excluded from analyses, 4 SNPs (VEGFA rs1570360; ABCB1 rs2032582; CYP450 rs2740574, rs776746) for not respecting Hardy–Weinberg equilibrium and 1 SNP (CYP2C8 rs10509681) for pairwise linkage disequilibrium (LD) higher than 0.8 (CYP2C8 rs11572080) [17]. Overall, the analyses were performed on 13 SNPs in 6 genes (Table S5).
4.3. In Silico Analysis
The potential functional impact of SNPs was investigated using several in silico applications. HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) was used to collect all linkage disequilibrium and to examine the haplotype blocks within the genes. Regulome DB (http://www.regulomedb.org/) was accessed to explore the chromatin status, conservation and regulatory motif alterations within sets of genetically-linked variants. The GTEX Portal (https://gtexportal.org/home/) served to identify all cis-eQTL SNPs affecting the expression of genes of interest and as a microRNA binding-site prediction tool. SNPMIR (https://www.genomique.info/joomla/) was used to predict whether a SNP within the 3’-UTR of the genes of interest would disrupt/eliminate or enhance/create a microRNA binding site.
4.4. Statistical Analysis
An evaluation of the missing data rate was performed on 13 SNPs. All SNPs included in the analysis had a missing data rate of less than 10%. Missing genotypes were assigned using multiple imputations by chained equations (MICE) [18]. Dominant and recessive models were investigated to test possible associations between SNPs and progression-free survival (PFS) and overall survival (OS). For each SNP, risk alleles were coded as 1 and non-risk alleles as 0. For each SNP, the hazard ratio (HR) and 95% confidence intervals were calculated by Cox regression for associations between the genotype and PFS or OS. PFS was defined as the time between inclusion and the date of disease progression or death due to any cause. OS was defined as the time between inclusion and death due to any cause. Patients showing no event (death or progression) or lost to follow-up were censored at the date of last contact. PFS and OS were performed using the Kaplan–Meier method. Median follow-up with a 95% confidence interval was calculated with the reverse Kaplan–Meier method. For PFS and OS, we constructed a genetic and a clinico-pathological predictive model using a multivariable Cox regression method with backward elimination (p-value ≤ 0.05). The variables selected in each of the previous two predictive models were included in a third multivariable Cox regression with backward elimination to create a combined predictive model. For each model, the proportional hazard assumption was checked using statistical tests and graphical diagnostics based on the scaled Schoenfeld residuals [19]. For each multivariable model, a classifier predicting the risk of progression or death was based on the linear predictor given by the model. The predictive ability of each model was compared using the area under the ROC curve (AUC). For PFS and OS, the clinical endpoint was fixed at t = 24 months and t = 60 months, respectively. Risk groups for predictive models were obtained using the best threshold value to obtain the highest sensitivity with a specificity of at least 80%. The sensitivity was defined as the proportion of patients experiencing progression or death before the time t attributed to the poor prognosis group. The specificity was defined as the proportion of event-free patients beyond the time t attributed to the good prognosis group. A p-value of <0.05 was considered statistically significant and all tests were two-sided. All statistical analyses were performed with R.3.5.2 software on Windows® and the survMisc [20], timeROC [21], Survminer [22] and LDcorSV packages [23].
5. Conclusions
This study identified genetic polymorphisms related to a bevacizumab–paclitaxel treatment combination and established a genetic risk score in association with the clinico-pathological characteristics of patients. The highlighted risk groups identified patients with a poor or good prognosis and could be used to improve clinical decision-making.
Acknowledgments
We are grateful to the patients who participated in the study. This research was funded by Roche. UNICANCER is the sponsor of the study. The authors acknowledge support from the University Côte d’Azur, Centre Antoine Lacassagne, Oncopharmacology Unit, France.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-8247/13/11/414/s1: Table S1: Univariate analysis for the 13 SNPs according to PFS; Table S2: Univariate analysis for clinico-pathological features according to PFS; Table S3: Univariate analysis for the 13 SNPs according to OS; Table S4: Univariate analysis for clinico-pathological features according to OS; Table S5: Summary of genotyping results by MassARRAY (AGENA) of 306 patients; Figure S1: Effect of significant clinical characteristics and polymorphisms according to PFS; Figure S2: Effect of subtype and histological grade combination according to PFS; Figure S3: Effect of significant clinical characteristics and polymorphisms according to OS; Figure S4: Effect of subtype and histological grade combination according to OS.
Author Contributions
G.M., J.G. (Jocelyn Gal), P.B., J.G. (Julia Gilhodes), M.-C.E.-G. and J.-Y.P. participated in the writing and were involved in critical revision of this manuscript for important intellectual content. All authors participated in the writing and were involved in critical revision of this manuscript for important intellectual content. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the French Government (National Research Agency, ANR) through the "Investments for the Future" programs LABEX SIGNALIFE ANR-11-LABX-0028-01 and IDEX UCA Jedi ANR-15-IDEX-01, Cancéropôle PACA; INCA Plan Cancer.
Conflicts of Interest
Gerard Milano is a member of an advisory board at Bristol-Myers Squibb (BMS), Merck Sharp and Dohme (MSD) and Roche.
Ethical Conduct or Research
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (French National Commission for Informatics and Liberties N°17010).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Apte R.S., Chen D.S., Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia J., Hurwitz H.I., Sandler A.B., Miles D., Coleman R.L., Deurloo R., Chinot O.L. Bevacizumab (Avastin®) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat. Rev. 2020;86:102017. doi: 10.1016/j.ctrv.2020.102017. [DOI] [PubMed] [Google Scholar]
- 3.Gerriets V., Kasi A. Bevacizumab. StatPearls Publishing; Treasure Island, FL, USA: 2020. [PubMed] [Google Scholar]
- 4.Pinter M., Scheiner B., Peck-Radosavljevic M. Immunotherapy for advanced hepatocellular carcinoma: A focus on special subgroups. Gut. 2020 doi: 10.1136/gutjnl-2020-321702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim Y.W., Chen-Harris H., Mayba O., Lianoglou S., Wuster A., Bhangale T., Khan Z., Mariathasan S., Daemen A., Reeder J., et al. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc. Natl. Acad. Sci. USA. 2018;115:E11701–E11710. doi: 10.1073/pnas.1804506115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna D.L., Lenz H.-J. Novel therapeutics in metastatic colorectal cancer: Molecular insights and pharmacogenomic implications. Expert Rev. Clin. Pharmacol. 2016;9:1091–1108. doi: 10.1586/17512433.2016.1172961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertz D.L., Rae J. Pharmacogenetics of Cancer Drugs. Annu. Rev. Med. 2015;66:65–81. doi: 10.1146/annurev-med-053013-053944. [DOI] [PubMed] [Google Scholar]
- 8.Daily E.B., Aquilante C.L. Cytochrome P450 2C8 pharmacogenetics: A review of clinical studies. Pharmacogenomics. 2009;10:1489–1510. doi: 10.2217/pgs.09.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamidovic A., Hahn K., Kolesar J. Clinical significance of ABCB1 genotyping in oncology. J. Oncol. Pharm. Pr. 2010;16:39–44. doi: 10.1177/1078155209104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etienne-Grimaldi M., Formento P., Degeorges A., Pierga J.-Y., Delva R., Pivot X., Dalenc F., Espié M., Veyret C., Formento J.-L., et al. Prospective analysis of the impact of VEGF-A gene polymorphisms on the pharmacodynamics of bevacizumab-based therapy in metastatic breast cancer patients. Br. J. Clin. Pharmacol. 2011;71:921–928. doi: 10.1111/j.1365-2125.2010.03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider B.P., Wang M., Radovich M., Sledge G.W., Badve S., Thor A., Flockhart D.A., Hancock B., Davidson N., Gralow J., et al. Association of Vascular Endothelial Growth Factor and Vascular Endothelial Growth Factor Receptor-2 Genetic Polymorphisms With Outcome in a Trial of Paclitaxel Compared With Paclitaxel Plus Bevacizumab in Advanced Breast Cancer: ECOG 2100. J. Clin. Oncol. 2008;26:4672–4678. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haunschild C., Tewari K.S. Bevacizumab use in the frontline, maintenance and recurrent settings for ovarian cancer. Futur. Oncol. 2020;16:225–246. doi: 10.2217/fon-2019-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui W., Li F., Yuan Q., Chen G., Chen C., Yu B. Role of VEGFA gene polymorphisms in colorectal cancer patients who treated with bevacizumab. Oncotarget. 2017;8:105472–105478. doi: 10.18632/oncotarget.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papachristos A., Kemos P., Katsila T., Panoilia E., Patrinos G.P., Kalofonos H.P., Sivolapenko G.B. VEGF-A and ICAM-1 Gene Polymorphisms as Predictors of Clinical Outcome to First-Line Bevacizumab-Based Treatment in Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2019;20:5791. doi: 10.3390/ijms20225791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motz G.T., Coukos G. The parallel lives of angiogenesis and immunosuppression: Cancer and other tales. Nat. Rev. Immunol. 2011;11:702–711. doi: 10.1038/nri3064. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel S., Ziaugra L., Tabbaa D. SNP Genotyping Using the Sequenom MassARRAY iPLEX Platform. Curr. Protoc. Hum. Genet. 2009;60:2.12.1–2.12.18. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 17.Mangin B., Siberchicot A., Nicolas S., Doligez A., This P., Cierco-Ayrolles C. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity. 2012;108:285–291. doi: 10.1038/hdy.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White I.R., Royston P., Wood A.M. Multiple imputation using chained equations: Issues and guidance for practice. Stat. Med. 2011;30:377–399. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- 19.Grambsch P.M., Therneau T.M. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- 20.Dardis C., Dardis M.C. Package ‘survMisc’. [(accessed on 1 October 2020)]; Available online: https://CRAN.R-project.org/package=survMisc.
- 21.Blanche P. TimeROC: Time-Dependent ROC Curve Estimation. [(accessed on 1 October 2020)]; Available online: https://CRAN.R-project.org/package=timeROC.
- 22.Kassambara A., Kosinski M., Biecek P., Fabian S. Package ‘Survminer’. Drawing Survival Curves Using ‘ggplot2’. [(accessed on 1 October 2020)]; R Package Version 0.3.1. Available online: https://CRAN.R-project.org/package=survminer.
- 23.Desrousseaux D., Sandron F., Siberchicot A., Cierco-Ayrolles C., Mangin B. LDcorSV: Linkage Disequilibrium Corrected by the Structure and the Relatedness. [(accessed on 1 October 2020)]; R Package Version 1.3.1. Available online: https://CRAN.R-project.org/package=LDcorSV.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.