Abstract
Earlier we have shown important roles of MYB in pancreatic tumor pathobiology. To better understand the role of MYB in the tumor microenvironment and identify MYB-associated secreted biomarker proteins, we conducted mass spectrometry analysis of the secretome from MYB-modulated and control pancreatic cancer cell lines. We also performed in silico analyses to determine MYB-associated biofunctions, gene networks, and altered biological pathways. Our data demonstrated significant modulation (p < 0.05) of 337 secreted proteins in MYB-silenced MiaPaCa cells whereas 282 proteins were differentially present in MYB-overexpressing BxPC3 cells, compared to their respective control cells. Alteration of several phenotypes such as cellular movement, cell death, and survival, inflammatory response, protein synthesis, etc. was associated with MYB-induced differentially expressed proteins (DEPs) in secretomes. DEPs from MYB-silenced MiaPaCa PC cells were suggestive of the downregulation of genes primarily associated with glucose metabolism, PI3K/AKT signaling and oxidative stress response, among others. DEPs from MYB-overexpressing BxPC3 cells suggested enhanced release of proteins associated with glucose metabolism and cellular motility. We also observed that MYB positively regulated the expression of four proteins with potential biomarker properties, i.e. FLNB, ENO1, ITGB1, and INHBA. Mining of publicly available databases using Oncomine and UALCAN demonstrated that these genes are overexpressed in pancreatic tumors and associated with reduced patient’s survival. Altogether, these data provide novel avenues for future investigations on diverse biological functions of MYB, specifically in the tumor microenvironment, and could also be exploited for biomarker development.
INTRODUCTION
Pancreatic cancer (PC) incidence and mortality have been on the rise for past several years. Recently, it became the third leading cause of cancer-related death in the United States and is expected to become the second by 2030 or earlier. About 56,770 new cases of PC are expected to be diagnosed and nearly 45,750 deaths will occur this year as compared to 55,440 new cases and 44,330 estimated deaths in 2018 (1, 2). Clearly, there is an urgent need to develop novel and effective strategies to manage this lethal malignancy. Identification of novel biomarkers and a better understating of mechanisms involved in aggressive and therapy-resistant progression of PC could be of great help to develop effective approaches for early diagnosis and treatment.
The development of tumors is a multi-stage process that requires intricate cross-talk between tumor and stromal cells. These interactions could occur either through direct cell-cell contacts or mediated through the tumor-derived secretomes that include proteins actively released by the cells through the classical secretory pathway or vesicular secretion (3–7). The proteins secreted by the tumor cells play important roles in the remodeling of stroma within the tumor microenvironment (8, 9). Moreover, they are also shown to help build supportive environment for disseminated tumor cells at distant metastatic sites (10–12). Their noticeable presence in blood serum and other biofluids has also been exploited clinically as potential biomarkers of disease existence, progression, and subtype (13–15). We recently demonstrated that MYB, an oncogenic transcription factor, is overexpressed in PC and plays important roles in tumor growth, metastasis and desmoplasia (16–18). Deep-sequencing of transcriptome of MYB-overexpressing and -silenced PC cells suggested that MYB potentially regulates growth and genomic stability of pancreatic cancer cells via targeting of complex gene networks and signaling pathways (16).
In the present study, we analyzed the secretomes of two sets of paired PC cell lines where MYB was either exogenously overexpressed (in BxPC3) or silenced (in MiaPaCa) through RNA interference. Differentially expressed proteins (DEPs) were identified by mass spectrometry in MYB-modulated and control cells and subjected to in silico analyses to predict their impact on biological pathways and functions. Analyses also suggested the clinical utility of some of the DEPs in secretomes as potential biomarkers. Collectively, these observations provide novel avenues for future research to understand broader pathobiological implications of MYB in PC and/or improve clinical management through biomarker development.
MATERIALS AND METHODS
Reagents
Roswell Park Memorial Institute Medium-1660 (RPMI-1660), phosphate buffer saline (PBS), penicillin (10,000 U/ml), streptomycin (10,000 μg/ml), fetal bovine serum (FBS) and trypsin-EDTA were purchased from Hyclone Laboratory (Logan, UT, USA); phenol-free RPMI-1660 was from Life Technologies (Carlsbad, CA); MycoAlert mycoplasma detection kit from Lonza (Rockland, ME); HPLC purified acetonitrile (ACN), and water were from Fisher Scientific (Waltham, MA, USA), and urea, ammonium bicarbonate, trifluoroacetic acid (TFA), EDTA, tris(2-carboxy-ethyl)phosphine hydrochloride (TCEP) were from Sigma-Aldrich (St. Louis, MO, USA). Sequencing grade modified porcine trypsin was procured from Promega (Madison, WI). LDHA and ENO1 ELISA kit were procured from BioVision Inc. (Milpitas, CA). Anti-HSP90 (rabbit monoclonal) and anti-SRSF1 (mouse monoclonal, clone 103) were from Cell Signaling Technologies (Danvers, MA) and Zymed laboratories, Invitrogen (Carlsbad, CA), respectively. Species specific secondary antibodies were from Santa Cruz Technologies (Dallas, TX).
Cell lines and collection of conditioned media
Stable MYB knockdown (MiaPaCa-shMYB) and MYB overexpressing (BxPC3-MYB) and their respective control cell lines; non-targeting scrambled (MiaPaCa-Scr) and empty vector control (BxPC3-Neo), respectively; were generated previously in our lab and maintained as described (18). All cells were routinely tested for mycoplasma. MYB expression in the paired cell lines (silencing and forced-overexpression in MiaPaCa and BxPC3, respectively) was also verified intermittently and at the beginning of the experiments. For the collection of conditioned media (CM), three different populations of each cell type varying in passage number were grown under similar culture conditions in 100 mm cell culture dishes (Corning, NY, USA) until they reached 70% confluence. Thereafter, cells were washed with PBS (x3, 5 min each) and FBS- phenol- and antibiotics (SPA) -free RPMI-1660 medium after 15 min incubation at 37 °C with intermittent gentle shaking followed by another rinse. Fresh SPA-free RPMI-1660 medium (3.5 ml) was added in each culture dish and cells incubated for another 16 h. Subsequently, conditioned media (CM) was collected and pooled for each cell type to account for biological variations. Cell number and viability were also determined at the time of CM collection using trypan blue exclusion assay.
Processing of the conditioned media
After collection, 10 ml of conditioned media (CM) was acidified with 200 μl of acetic acid and centrifuged at 800 × g for 10 min under cold (4 °C) temperature. The secreted proteins were then isolated using a C2 solid-phase extraction column (Sep-Pak, 0.3 × 5 mm (Waters, Milford, MA)). The column was first washed with 3-column volumes (3ml) of 100% acetonitrile (ACN) and equilibrated with 3ml of 0.1% trifluoroacetic acid (TFA) in water with a flow rate of 1 ml/min. After equilibration, CM was loaded onto the column and washed with 0.1% TFA in water to remove salts. The protein fraction was then eluted with 50% acetonitrile in 0.1% TFA and dried in a speed vac concentrator (Thermo Fisher Scientific, Waltham, MA, USA). Dried protein samples were further dissolved in 50μl of 50mM ammonium bicarbonate (Sigma-Aldrich, St Louis, MO) and 10mM tris(2-carboxyethyl)phosphine (Alfa-Aesar,Ward Hill, MA, USA) and digested overnight at 37°C with using 1μl of 10 μM sequencing grade trypsin (Promega, Madison,WI, USA).
Mass-spectrometry analysis
The processed samples were subjected to three technical replicates LC-MS/MS analyses with an Agilent 1200 series nano-liquid HPLC coupled to a linear ion trap/Orbitrap hybrid MS (LTQ-Orbitrap XL, Thermo Fisher Scientific, Waltham, MA, USA). The HPLC mobile phases consisted of 3% ACN and 0.2% formic acid in water (solvent A), 3% water and 0.2% formic acid in acetonitrile (solvent B). The sample was loaded onto a C18 pre-column (5μM; 5 by 0.3mm Zorbax, Agilent Technologies, Santa Clara, CA) with a 4μl/minute flow rate of 5% solvent B. A flow rate of 1μl/minute was used to elute the sample from the pre-column onto a Hypersil Gold C18 column (30mm by 0.18mm; ThermoFisher Scientific, Waltham, MA). A linear solvent gradient was used to slowly ramp to 40% B over 70 minutes for peptide elution from the column, maximizing separation and coverage. The gradient was then ramped to 90% B for 20 minutes to wash the column at the end of each run. The total run time for each sample injection was two hours. Blanks of 0.1% TFA were run in-between each set of triplicate injections to minimize and monitor for carryover between samples. Peptide ionization was achieved using electrospray ionization in positive mode. Samples were analyzed in a data-dependent manner, with full MS1 scans from 400–2000 m/z at a resolution of 60,000 in the Orbitrap mass analyzer, collecting at least 10 points across each peak for accurate integration. The top five multiply charged parent ions per MS1 scan were selected for MS2 scans in the LTQ mass analyzer with CID fragmentation. Selected ions were then added to a dynamic exclusion list to prevent repeated sampling of abundant peptides for 120 seconds to allow more in-depth coverage of the sample. Thermo’s Xcalibur software was used to generate RAW files of each MS experiment.
Processing of LC-MS/MS-raw data
The raw files were loaded into Proteome Discoverer (PD) (version 2.1.081; Thermo Fisher Scientific) to provide protein identification across all samples. Within the processing workflow, the nodes for both the Mascot and Sequest search engines were used to search a custom database (consisting of 71485 sequences and 45902068 residues) created from the non-redundant NCBI RefNCBInr database (version 12/08/14), which was combined the human immunoglobulin A, G, and M, lambda, and kappa constant region sequences from the SwissProt dataset (version 11/28/12). Identification was limited to unmodified semi-tryptic peptides only (cleaved at arginine and lysine residues) with a maximum of two missed cleavage sites, excluding those outside of a fragment mass tolerance of 0.6Da and a precursor mass tolerance of 10 ppm. Within the percolator node, the target FDR (strict) was 0.01, and target FDR (relaxed) was 0.05, with validation based on q-value. Within the consensus workflow, the peptide and protein quantifier node used both unique and razor peptides for quantification purposes, using the top 3 peptides of each protein for area calculation. Peptides were further filtered using the peptide and protein filter node to only include those of high confidence with a minimum peptide length of 6. Proteins were grouped by master protein using the strict parsimony principle parameter, to decrease redundancy based on sequence homology. The protein areas from three technical LC-MS/MS replicates of each sample were averaged to provide one area per protein per sample. The protein areas from each identified protein were then used for comparison between conditions. The areas of individual proteins present in triplicate injections of all four samples of the MiaPaCa and BxPC3 variants (BxPC3-Neo and BxPC3-MYB; MiaPaCa-Scr and MiaPaCa-shMYB) were compared statistically using the t-test function in Microsoft Excel 2010. Relative fold changes were calculated by dividing the average peak area of the identified proteins of MiaPaCa-Scr by the MiaPaCa-shMYB or BxPC3-MYB by BxPC3-Neo, respectively, while the proteins present only in one sample was divided by one. Proteins identified with at least 3 peptides per protein in two of the three technical replicates with differential expression of |2| fold change and p-value ≤ 0.05 were used for further data analysis.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was conducted to determine the level of lactate dehydrogenase A and alpha-enolase in the conditioned media of MYB-modualted and control pancreatic cancer cell lines. Conditioned media (CM) was collected from cells cultured in SPA-free RPMI-1660 media for 16 h and cleared for cells and cell debris.. Subsequently, 10 μL and 100 μL of CM was used to determine ENO1 and LDHA levels, respectively, and the assay performed as per the manufacturer’s instructions.
Immunoblotting
Processed conditioned media (900 μL) was concentrated (≈10X) by passing through an Amicon filter (3 kDa cutoff; Millipore Sigma, Burlington, MA) at 13000 rpm for 30 minutes at 4°C. Concentrated CM (30 μL) was then mixed with 6X gel loading dye and subjected to immunoblotting as previously described (18). Band intensitites were quantified using ImageJ software and data presented as relative fold-differences.
In silico analyses
Cellular localization, networks and canonical pathways of the identified proteins from BxPC3 and MiaPaCa groups were generated through the Ingenuity pathway analysis software (IPA; Qiagen, http://www.ingenuity.com/products/ipa) with the cutoff p-value ≤ 0.05 and ≥|2| fold change. Protein GI accessions were used as the identifier, and the IPA based Ingenuity Knowledge Base was used as a reference to perform core analysis. Network algorithm was used to generate the network of genes based on their connectivity and were assigned a score based on the number of focus genes. Canonical pathway analyses identified pathways from the IPA library that were of significance to the datasets. Proteins with potential ‘biomarker properties’ were identified using the IPA-biomarker filter tool. IPA-biomarker identifies the most promising and relevant biomarker candidates by prioritizing proteins according to characteristics that can qualify it as a biologically plausible candidate biomarker. Biomarker properties take into account whether a protein is detectable in sentinel tissues, bodily fluids or urine and if it has a strong association with disease processes. Oncomine microarray datasets were accessed to evaluate the expression profile of identified proteins in human pancreatic cancer (www.oncomine.org) (19). Patient survival correlation was evaluated using UALCAN online portal (20), and immunohistochemical data for protein expression in pancreatic cancer tissues was scrutinized from the human protein atlas (HPA; https://www.proteinatlas.org/) (21).
RESULTS
Mass spectrometry analysis identifies MYB-dependent differential protein levels in the secretome of pancreatic cancer cells
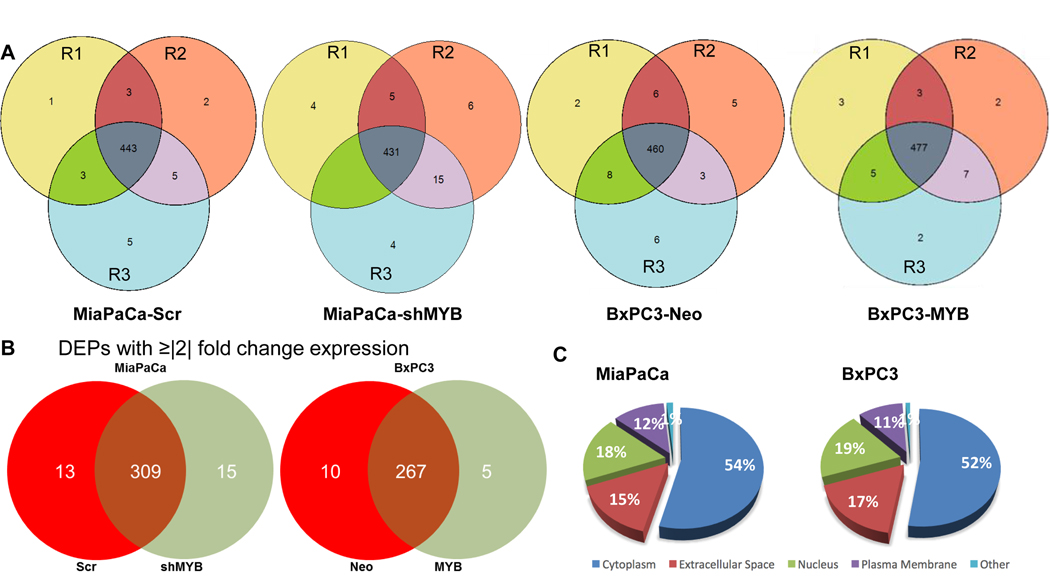
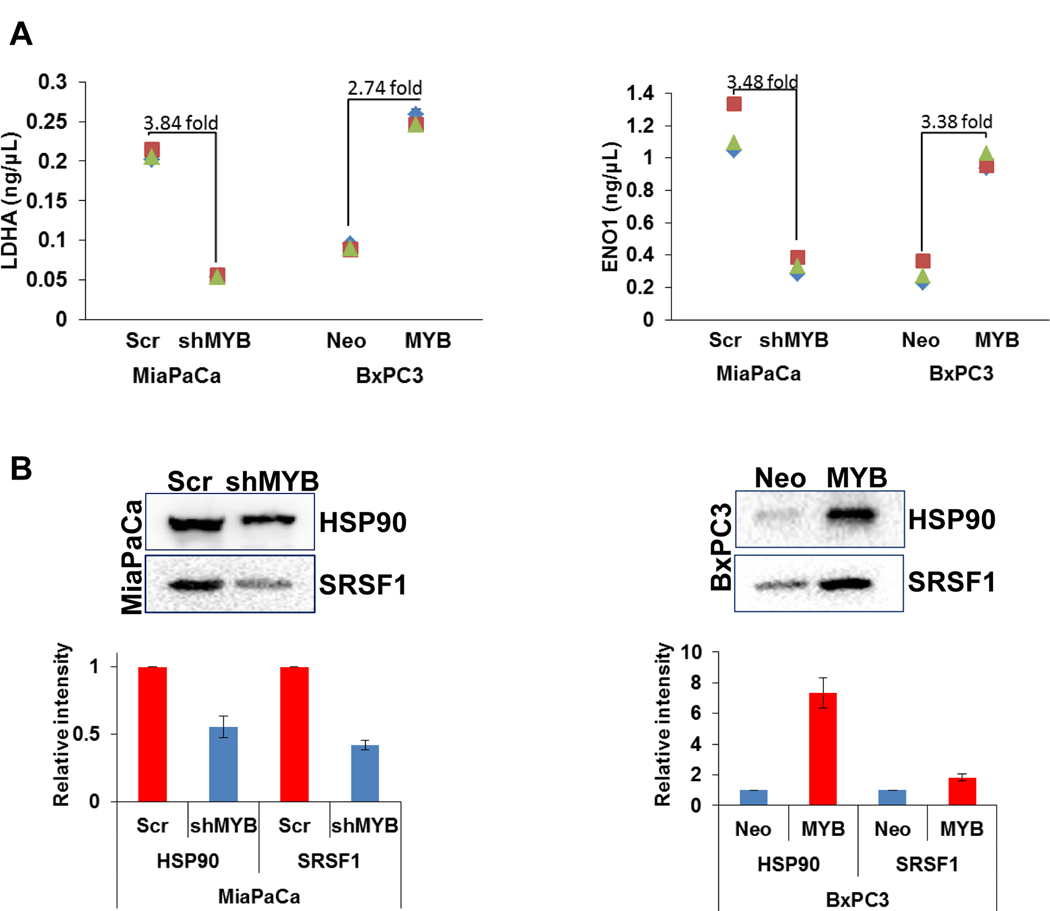
All the cells at the time of collection of conditioned media (CM) appeared healthy, and their viability was determined to be in the range of 95–97%. Proteomic analyses of CM collected from paired genetically-engineered pancreatic cancer cell lines (BxPC3-Neo, low MYB; BxPC3-MYB, high MYB, and MiaPaCa-Scr, high MYB; MiaPaCa-shMYB, low MYB) led to the identification of several proteins (Supplementary Table 1). A list of differentially-expressed proteins (DEPs) was generated by comparing the isotype cells with expression change of ≥|2| fold and p-value of ≤0.05 (Supplementary Table 2). In MiaPaCa-Scr and MiaPaCa-shMYB cells, 454 and 451 proteins were identified respectively, whereas 471 and 492 proteins were identified in BxPC3-Neo and BxPC3-MYB cells, respectively (Figure 1A). Among the total number of identified proteins, 337 were differentially expressed in MiaPaCa-Scr vs. –shMYB, of which 13 and 15 were exclusively detected in MiaPaCa-Scr and MiaPaCa-shMYB secretomes, respectively. Similarly, 10 and 5 proteins were exclusively identified in secretomes of BxPC3-Neo and BxPC3-MYB cells, respectively, among a total of 282 that were differentially present in their secretomes (Figure 1B). Next, we subjected our list of DEPs to Ingenuity Pathway Analysis (IPA) to determine their predominant subcellular localization. Of the 337 DEPs in MYB-overexpressing and –silenced MiaPaCa cells, analysis suggested that 181 proteins were predominantly cytoplasmic, 59 nuclear, 42 plasma-membrane associated, and 50 reported to be in the extracellular space. 5 DEPs remained unspecified in terms of predominant subcellular localization. Likewise, from the secretomes of control and high exogenous MYB-expressing BxPC3 cells, 148 DEPs were assigned to the cytoplasm, 53 to the nucleus, 48 to the extracellular space, and 30 to the plasma membrane, while 3 remained unspecified (Figure 1C). To test the valididaty of our pooling strategy and confirm the reproduicibility MS data, we further evaluated the levels of of four proteins that were identified to be differentially present in the secretomes of MYB-modulated cell lines. Levels of LDHA and ENO1 were determined by ELISA, whereas immunoblotting assays were performed for HSP90 and SRSF1 in three separate biological replicates. We found that in corcodance with MS data, all the four proteins examined were differentially present in the secretomes of paired (control and MYB-modulated) MiaPaCa and BxPC3 cells and this data was consistent in three biological replicates (Figure 2).
Figure 1: Identification of differentially expressed proteins in the secretome of MiaPaCa (-Scr and -shMYB) and BxPC3 (-Neo and -MYB).
(A) Conditioned media from three populations of MiaPaCa (-Scr and –shMYB), BxPC3 (-Neo and –MYB) differing in passage number, but strictly cultured under similar growth conditions were collected, pooled and processed for LC-MS/MS analysis as mentioned in the “Materials and methods” section. R1, R2, and R3 represent the three technical replicates of three pooled biological replicates. (B) Proteins identified with at least 3 peptide hits with p-value≤ 0.05 and present in at least 2 two out of three technical replicates of MiaPaCa (-Scr and –shMYB) and BxPC3 (-Neo and –MYB) cell lines were selected for the analysis. Data represent the differentially expressed proteins (DEPs) with ≥|2| fold change between paired MYB-modulated pancreatic cancer cell lines. (C) The pie chart represents the classically associated cellular localization of the total DEPs identified in MiaPaCa (-Scr and –shMYB) and BxPC3 (-Neo and –MYB).
Figure 2: Measurement of differential levels of proteins by ELISA and immunoblooting in the conditioned media of MYB-modulated cell lines.
(A) Conditined media from cells was cleared for cells and debris and subjected to ELISA for LDHA and ENO1 meaurement as per manufacturer’s instructions. Experment was performed in triplicates in three biological replicates. Data (mean ± SD, n=3) is presented in a graph. (B) Conditioned media was concentrated (≈10X) from three biological replicates using size-based (3K cutoff) centrifugal flitration and subjected to western blot analyses for HSP90 and SRSF1. Representative image from one replicate is shown and the data from the densitometric analysis of three replicates (mean ± SD, n=3) has been plotted in the graph. * reflects statistically significant differences (p-value < 0.05).
Prediction of functional properties associated with MYB-dependent differentially-expressed secreted proteins
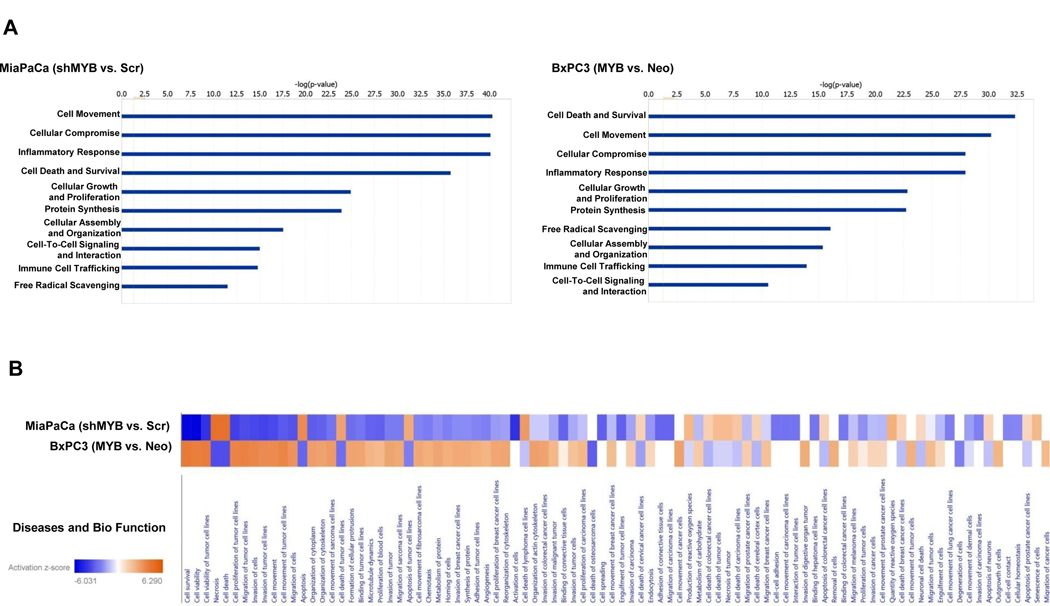
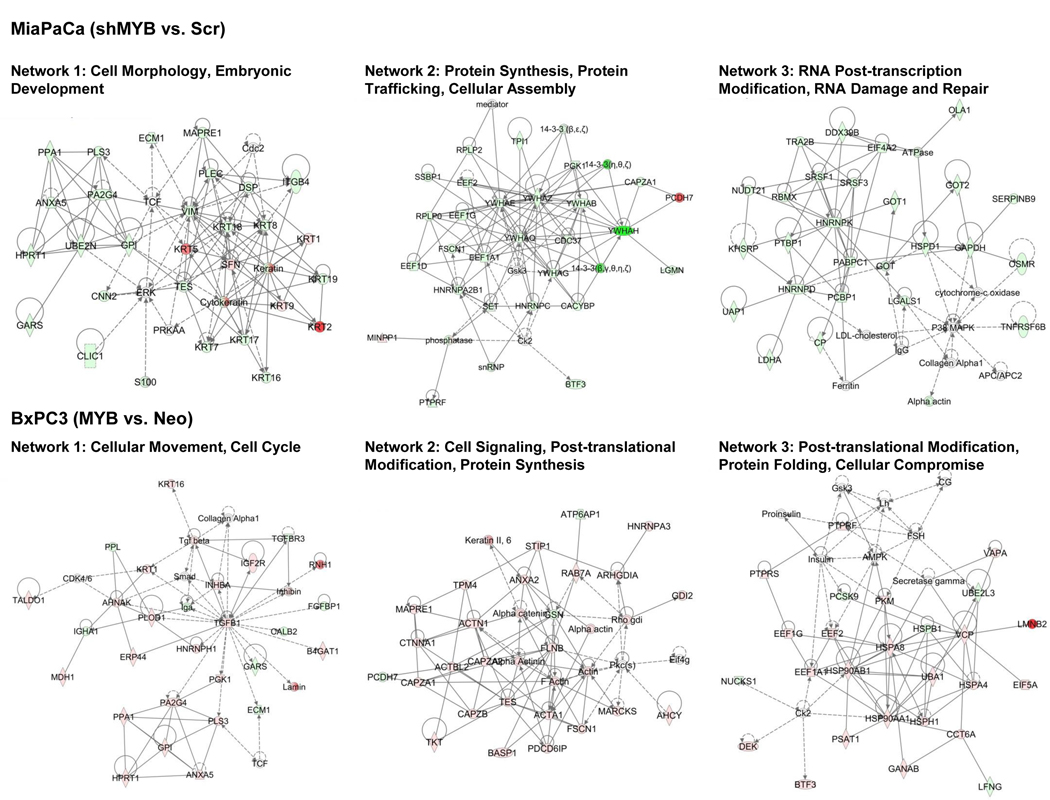
Since proteins in the secretion can impart the properties of the tumor cells as well as also serve as a surrogate of the status of the cells from which they are derived (4, 8), we subjected our dataset of DEPs to IPA analysis. Several key biological processes were identified. Among these, the top ten are presented, which include those associated with cellular movement, cellular compromise, cell death and survival, and inflammatory response in both MiaPaCa and BxPC3 cells resulting from MYB alterations (Figure 3A). A side by side comparison of the altered bio-functions upon MYB-silencing in endogenously overexpressing cells (MiaPaCa-shMYB vs. MiaPaCa–Scr) or with exogenous MYB overexpression in low expressing cells (BxPC3-MYB vs. BxPC3–Neo) shows opposite changes as expected for most bio-functions (Figure 3B). MYB-dependent profile of DEPs in MiaPaCa cells suggested compromised cellular invasion and migration of cancer cells as well as endothelial cells and decreased chemotaxis upon MYB-silencing. In BxPC3 cells, DEPs obtained by overexpression of MYB were suggestive of a decrease in apoptosis. These DEPs were also predicted to potentially favor cellular movement through induction of migration and invasion, and inflammatory response. All the bio-functions with predicted change in activity are listed in Supplementary Table 3. We next took advantage of IPA’s network analysis to determine MYB-regulated networks using our secretome datasets. This analysis places DEPs in various networks utilizing published records in the literature. MYB-silencing and overexpression led to a change in 19 networks in both MiaPaCa and BxPC3 respectively of which top 3 networks, their functionality, and directionality of connections between nodes for each condition, are presented (Figure 4). These networks correspond to (i) cell morphology, (ii) protein synthesis/trafficking and cellular assembly and (iii) RNA post-transcription modification/damage and repair in MYB-silenced MiaPaCa cells. Networks altered in BxPC3 cells upon MYB-overexpression are also similar wherein one Network (cellular movement/cell cycle), TGFβ1 occupies the central node. Other networks correspond to cell signaling/post-translational modification/protein synthesis and protein folding. A detailed list of proteins, score and focus molecules in each network is presented in Supplementary Table 4.
Figure 3: Determination of function of MYB-associated secreted protein in pancreatic cancer.
(A) Top-10 cancer-relevant functions altered upon MYB-modulation have been represented. (B) Bio-functions altered due to the DEPs upon MYB-silencing (MiaPaCa shMYB vs. Scr) and MYB-overexpression (BxPC3 MYB vs Neo) using Ingenuity Pathway Analysis (IPA) indicate a reciprocal effect of silencing and overexpression.
Figure 4: Top 3 networks modulated upon MYB-alteration based on DEPs in MiaPaCa and BxPC3 cell secretome.
Nodes upregulated are in red, while the ones downregulated are in green. Solid lines represent direct regulatory relationships, and dashed lines indicate indirect regulatory networks.
MYB-modulated secretome is indicative of major alterations in glucose metabolic pathways
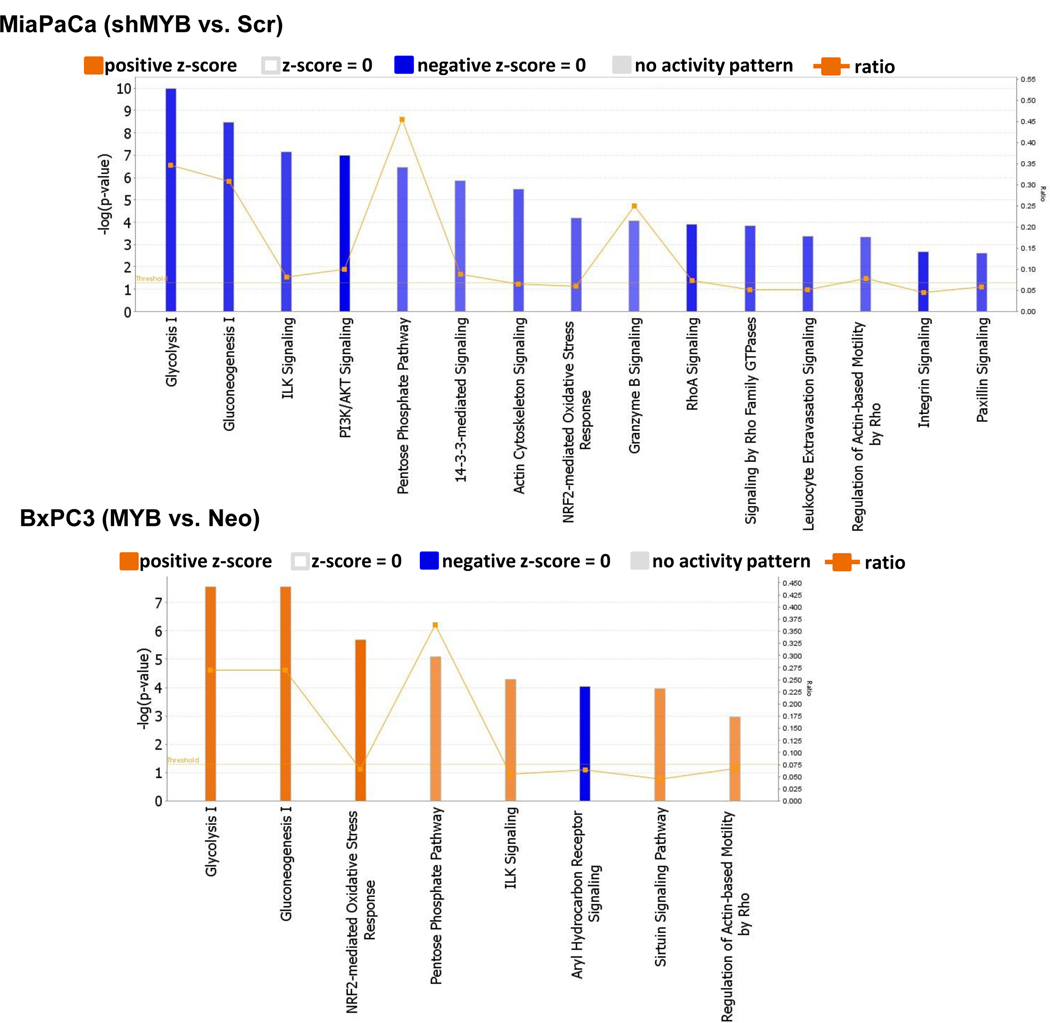
We utilized the datasets of MYB-modulated DEPs in secretomes of MiaPaCa and BxPC3 as an indirect measure to predict canonical pathways that could be altered in these cells. Pathways with a –log(p-value) of > 2 and activation z-score of absolute value ≥ 2 were considered (Figure 5). Glucose metabolism was observed to be the most commonly altered pathway between the both sets of (MiaPaCa vs. BxPC3) cell lines. Silencing of MYB in MiaPaCa predicted a decrease in glycolysis, gluconeogenesis and the pentose phosphate pathway, while its overexpression in BxPC3 led to a predicted increase in the activity of these pathways. The other canonical pathways altered upon MYB-silencing were related to cell cycle, cell survival and growth such as the ILK, PI3K/AKT, 14–3-3-mediated, NRF-2-mediated oxidative stress response, actin cytoskeleton, integrin signaling, and paxillin signaling. MYB-silencing also affected cellular assembly and organization and cell motility as Rho-based signaling pathways were also predicted to be down-regulated. Leukocyte extravasation signaling indicative of the potential migration of leukocytes from blood to tissue during inflammation was also downregulated upon MYB-silencing. On the other hand, along with the predicted activation of glucose metabolism pathway due to MYB-overexpression in BxPC3, pathways associated with cell survival, such as the ILK signaling, Sirtuin signaling, and the NRF-2-mediated oxidative stress response, were also activated. The list of altered proteins in these canonical pathways, activation z-score, and - log(p-value) is presented in Supplementary Table 5.
Figure 5: Identification of MYB-regulated canonical pathways based on the Ingenuity Knowledge Base.
Altered pathways predicted by IPA after MYB-silencing and MYB over-expression in PC cells MiaPaCa and BxPC3, respectively.
Identification of potential MYB-associated diagnostic and/or prognostic biomarkers
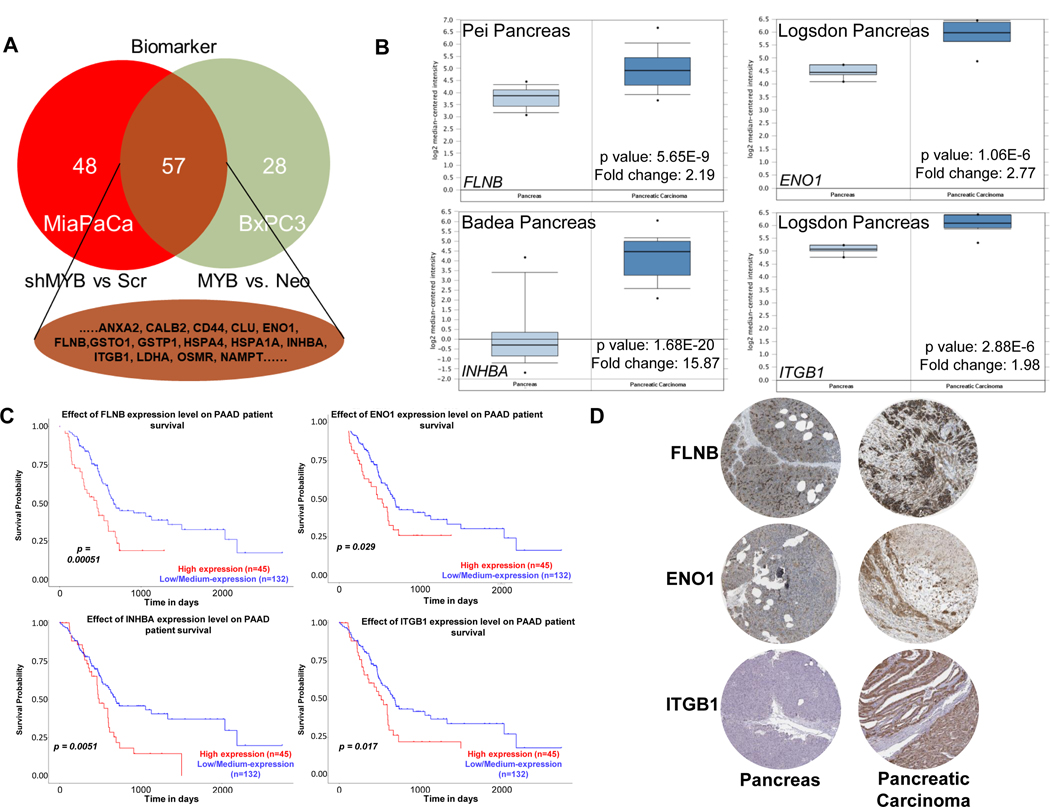
Next, to identify potential biomarkers reflective of MYB levels in pancreatic cancer cells, we conducted a comparative analysis of DEPs from MYB-silenced and –overexpressing cells using the biomarker filter tool of IPA. Potential candidates were shortlisted by filtering DEPs for ‘Human’ biomarker proteins potentially present in the serum/plasma. This led to the identification of a total of 57 proteins between the two DEP groups (Figure 6A). These proteins were then interrogated for their expression status and prognostic value between normal vs. control. Gene expression profile was obtained using the ONCOMINE database and expression changes observed were highly significant for four DEPs, INBHA (15.87 fold with p-value 1.68E-20), ENO1 (2.77 with p-value 1.06E-6), FLNB (2.19 fold with p-value 5.65E-9), ITGB1 (1.98 1-fold with p-value 2.88E-6) (Figure 6B). Next, The Cancer Genome Atlas was interrogated using the UALCAN portal for the association of genes encoding these proteins with patients’ survival. The analyses showed that the expression of these genes correlated significantly with poor survival suggesting their prognostic value in pancreatic cancer (Figure 6C). As our data comprises of protein changes and the functional unit of a gene is a protein, we also looked at the immunohistochemistry data of these proteins in normal pancreas and pancreatic cancer available at the Human Protein Atlas (HPA) website. ENO1, FLNB, and ITGB1 showed higher expressing in pancreatic cancer tissue than normal; data for INHBA was being curated at this time (Figure 6D).
Figure 6: Prediction of the potential prognostic marker and their association with patient survival.
The differentially expressed proteins (DEPs) identified from MiaPaCa (shMYB vs. Scr) and BxPC3 (MYB vs. Neo) were subjected to biomarker comparison using IPA. (A) 57 proteins with potential biomarker properties were identified common to both the subsets. (B) Gene expression of 4 DEPs between pancreas and pancreatic carcinoma was determined using the Oncomine database. (C) Association of expression with survival of pancreatic cancer patients was identified from the TCGA database using UALCAN. (D) Immunohistochemistry data of the selected protein in pancreas and pancreatic cancer was obtained from Human Protein Atlas. Data for INHBA was being curated at this time.
DISCUSSION
The secreted cellular proteins play important roles in the modeling of the extracellular environment and have great impact on cellular physiology and behavior (3, 4, 9, 22). Here we characterized MYB-associated changes by mass spectrometry in pancreatic cancer secretome by using two established cell lines and two modes (exogenous overexpression and silencing) of MYB modulation. The analysis showed the presence of several proteins in the secretomes that were not the classically secreted proteins (those having signal peptide). Using in silico analyses, we also identified associated biofunctions, gene networks, and canonical pathways to assess the pathobiological significance of these proteins. In addition, we utilized available gene expression databases to predict their potential as biomarkers for diagnosis and prognosis. Further, since we used a sample pooling strategy, we confirmed the resultant MS data on four randomly selected differentially-expressed proteins using ELISA and immunoblotting in three separate biological replicates.
Mass spectrometry has been extensively used for the analyses of differentially-secreted proteins (23, 24). Further, sample pooling aqpproach has also been employed to save time and resources with out adding a systematic bias (25, 26). In sample pooling approach, it is assumed that the measurements taken on the pool are equal to the average of measurments on individual samples. Concerns; however, have been raised against the pooling strategy particularly when dealing with samples obtained from the human subjects (27, 28). Samples from human subjects exhibit large individual-to-individual variations that may lead to skewing of data towards the outliers. Furthermore, considerable biological differences may also be observed in samples from the same individuals as well. However, pooling of samples (biological replicates) from cultured cancer cell lines appears acceptable as also demonstrated by our data where we observed only minor differences between replicates in confirmatory ELISA and immunoblot analyses. This is likely due to the fact that the exprements are well controlled under laboratory settings that minimize biological variations. Thus, sample type, experimental design and benefits/limitation assessment should be taken into consideration in exploratory proteomic analyses.
Among the MYB-associated classical secreted proteins were inflammatory response proteins, growth factors, cytokines, proteases, and inhibitors. For example, MYB positively influenced the release of Long Pentraxin family protein, Pentraxin-3 (PTX3), which is also known as TNF-inducible gene 14 (TSG14). PTX3 is a humoral pattern recognition molecule that participates in inflammation and mediates differentiation of fibroblasts. Interestingly, while the precise role of PTX3 has not been elucidated, it’s cancer-site specific (depending upon the cellular source and the microenvironmental tumor milieu) tumor-promoting or suppressing activities have been suggested (29–32). In pancreatic cancer, its increased expression is associated with increased migratory activity of cells and poor prognosis (33). Similarly, release of glucose phosphate isomerase (GPI), a member of the glycolytic pathway, correlated with the expression of MYB in our findings. GPI, also known as autocrine motility factor, moonlights as tumor-secreted cytokine and angiogenic factors promoting the growth of pancreatic tumors (34). The preproprotein INHBA was also found to be positively regulated by MYB. Proteolytic processing of INHBA generates a subunit of the dimeric activin and inhibin protein complexes and is suggested to be associated with cancer cachexia (35).
Non-classical secreted proteins identified in secretomes included those that are generally localized in the plasma membrane, cytoplasm, and/or nucleus. The plasma membrane proteins accounted for 12% and 11% of the observed proteins in the conditioned media of MiaPaCa and BxPC3 cells modulated for MYB, respectively. Whereas cytoplasmic and nuclear proteins constituted 54% and 52%, and 18% and 19% in MYB modulated MiaPaCa and BxPC3 cell secretomes, respectively. The presence of these proteins in the secretomes could have resulted from extravesicular secretion (36). Indeed, when we analyzed the ExoCarta and Vesiclepedia, we observed a major portion of these proteins to have been reported in these databases. In addition, the contribution of these proteins to the secretome could be from dying cells or the fragmented apoptotic bodies (37). The presence of membrane proteins in the conditioned media could also occur because of proteolytic cleavage, as has been reported previously (38, 39). Once released, these proteins could have functional impact by acting as ligands to surface receptors or upon their uptake by the recipient cells (40, 41). Indeed, extravesicular secretion of proteins and other bioactive molecules has been extensively studied lately and shown to play important physiological and pathological functions.
Released DEPs can have autocrine and/or paracrine functions and can impact the cells in the micro- or macro-environment. Our data suggested cell movement as the most altered “biofunction” associated with the secreted DEPs from MYB-silenced MiaPaCa cells while an increase in survival or growth imparting properties were associated with DEPs from MYB-overexpressing BxPC3 cells. This was in line with our previous studies (16, 18) and is suggestive of cell-specific differences in preferential functional influences. However, it would be interesting to examine how these DEPs would affect these phenotypes in non-cancerous cells, e.g. stromal cells. Our network analysis also suggested that the MYB-associated DEPs could modulate different aspects of cell development and morphology. Furthermore, the data suggested that MYB-associated proteins could positively promote protein integrity by regulating synthesis, folding, post-translational modifications, and trafficking. Of particular interest were MYB-associated DEPs that could potentially modulate RNA post-transcriptional modifications, RNA damage, and repair, a network that we also found to be altered upon at the transcriptomic level upon MYB modulation (16). Among the canonical pathways, glucose metabolism was the primary pathway that was significantly altered upon MYB-modulation through the regulation of key metabolic enzymes involved in glycolysis and gluconeogenesis. Some of these enzymes, GPI and enolase, have been suggested to have other functions as well in tumor progression depending upon their cellular localization (34, 42–44). Additionally, the positive regulation of structural proteins by MYB suggests novel ways through which it could impact cellular signaling.
The proteins identified in the secretomes could also serve as biomarkers that could serve as surrogates of disease status to help in disease diagnosis, subtyping, treatment planning and as targets of therapy as well (13, 37). In the IPA’s biomarker analysis, we identified 57 proteins to be common between the two MYB-modulated datasets. Of these DEPs, four proteins namely FLNB, ENO1, INHBA, and ITGB1 were suggested to be the candidate biomarkers based Ingenuity’s knowledge base and because of their association with patient survival and tumor expression (19, 20). These proteins have also been demonstrated to be involved in cancer pathogenesis. Alpha-enolase, a key glycolytic enzyme, is upregulated in a number of cancers. It’s moonshining properties include its activity as a plasminogen receptor regulating pancreatic cancer cell migration and invasion through the activation of plasminogen to plasmin (45). Enolase is also shown to promote growth and aggressiveness of pancreatic tumor cells in cooperation with integrins (46). The proteolytically cleaved inhibin subunit beta A (INHBA) acts as a subunit of the dimeric activin and inhibin protein complexes. Upregulated INHBA expression is associated with poor survival in lung adenocarcinoma and is an independent predictor of outcomes in patients with stage II/III gastric cancer that undergoes adjuvant chemotherapy (47, 48). ITGB1 has been associated with poor prognosis in pancreatic cancer and mediate migratory and invasive characteristics to the cancer cells (49, 50). FLNB along with other mechano-responsive proteins, myosin IIA, myosin IIC, α-actinin 4, mediates the structural rearrangement of the actin cytoskeleton to impact pancreatic cancer cell mechanics and facilitate metastasis (51). Of note, FLNB, ENO, and ITGB1 are not the classically secreted proteins, but are known to be constituents of extracellular vesicles (52–55). Thus, our findings provide further support to the currently prevailing notion that extracellular vesicles could be a good resource for the development of cancer biomarkers.
In summary, we have characterized MYB-regulated secretome from pancreatic cancer cells that could potentially be implicated in broader pathobiological functions of MYB and serve as surrogate biomarkers. The relevance of these observations needs to be evaluated in future functional and mechanistic studies. Further, the profile of these proteins needs to be examined in patient’s biofluids to explore their practical utility in pancreatic cancer diagnosis and prognosis.
Supplementary Material
Acknowledgments
We thank Ms. Lindsay Schambeau and Dr. Lewis Pannell, University of South Alabama Mitchell Cancer Institute Mass Spectrometry Core Facility for their assistance with the generation of Mass Spectrometry data. This work was supported, in part, by funding from NIH/NCI [R01CA224306, R01CA175772, and U01CA185490 (to APS)] and USA MCI. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- CM
Conditioned media
- PC
Pancreatic cancer
- SPA
Fetal bovine serum- phenol- and antibiotics- free
- TFA
Trifluoroacetic acid
Footnotes
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (56) partner repository with the dataset identifier PXD015126. Data on patient survival, gene expression, and immunohistochemistry can be accessed through publicly available databases of UALCAN, Oncomine and the Human Protein Atlas. All other data can be made available upon reasonable request.
References
- 1.Siegel RL, Miller KD, and Jemal A (2019) Cancer statistics, 2019. CA: a cancer journal for clinicians 69, 7–34 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, and Jemal A (2018) Cancer statistics, 2018. CA: a cancer journal for clinicians 68, 7–30 [DOI] [PubMed] [Google Scholar]
- 3.Hartwig T, Montinaro A, von Karstedt S, Sevko A, Surinova S, Chakravarthy A, Taraborrelli L, Draber P, Lafont E, Arce Vargas F, El-Bahrawy MA, Quezada SA, and Walczak H (2017) The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2. Molecular cell 65, 730–742 e735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Unver N (2019) Macrophage chemoattractants secreted by cancer cells: Sculptors of the tumor microenvironment and another crucial piece of the cancer secretome as a therapeutic target. Cytokine & growth factor reviews [DOI] [PubMed] [Google Scholar]
- 5.Mazzeo E, Hehlgans S, Valentini V, Baumann M, and Cordes N (2012) The impact of cell-cell contact, E-cadherin and EGF receptor on the cellular radiosensitivity of A431 cancer cells. Radiation research 178, 224–233 [DOI] [PubMed] [Google Scholar]
- 6.Patel GK, Khan MA, Bhardwaj A, Srivastava SK, Zubair H, Patton MC, Singh S, Khushman M, and Singh AP (2017) Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. British journal of cancer 116, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee P, and Mani S (2013) Methodologies to decipher the cell secretome. Biochimica et biophysica acta 1834, 2226–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez EO, Chirino YI, Delgado-Buenrostro NL, Lopez-Saavedra A, Meraz-Cruz N, and Lopez-Marure R (2016) Secretome derived from breast tumor cell lines alters the morphology of human umbilical vein endothelial cells. Molecular membrane biology 33, 29–37 [DOI] [PubMed] [Google Scholar]
- 9.Jin K, Pandey NB, and Popel AS (2017) Crosstalk between stromal components and tumor cells of TNBC via secreted factors enhances tumor growth and metastasis. Oncotarget 8, 60210–60222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, Hoffmann AD, Liu H, and Liu X (2018) Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ precision oncology 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y, Bado I, Wang H, Zhang W, Rosen JM, and Zhang XH (2019) Metastasis Organotropism: Redefining the Congenial Soil. Developmental cell 49, 375–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung T, Castellana D, Klingbeil P, Cuesta Hernandez I, Vitacolonna M, Orlicky DJ, Roffler SR, Brodt P, and Zoller M (2009) CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 11, 1093–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Q, Lim HS, Lin HL, Tan HT, Lim TK, Cheong WK, Cheah PY, Tang CL, Chow PK, and Chung MC (2015) Analysis of colorectal cancer glycosecretome identifies laminin beta-1 (LAMB1) as a potential serological biomarker for colorectal cancer. Proteomics 15, 3905–3920 [DOI] [PubMed] [Google Scholar]
- 14.Marimuthu A, Chavan S, Sathe G, Sahasrabuddhe NA, Srikanth SM, Renuse S, Ahmad S, Radhakrishnan A, Barbhuiya MA, Kumar RV, Harsha HC, Sidransky D, Califano J, Pandey A, and Chatterjee A (2013) Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochimica et biophysica acta 1834, 2308–2316 [DOI] [PubMed] [Google Scholar]
- 15.Piersma SR, Fiedler U, Span S, Lingnau A, Pham TV, Hoffmann S, Kubbutat MH, and Jimenez CR (2010) Workflow comparison for label-free, quantitative secretome proteomics for cancer biomarker discovery: method evaluation, differential analysis, and verification in serum. Journal of proteome research 9, 1913–1922 [DOI] [PubMed] [Google Scholar]
- 16.Azim S, Zubair H, Srivastava SK, Bhardwaj A, Zubair A, Ahmad A, Singh S, Khushman M, and Singh AP (2016) Deep sequencing and in silico analyses identify MYB-regulated gene networks and signaling pathways in pancreatic cancer. Scientific reports 6, 28446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhardwaj A, Srivastava SK, Singh S, Tyagi N, Arora S, Carter JE, Khushman M, and Singh AP (2016) MYB Promotes Desmoplasia in Pancreatic Cancer through Direct Transcriptional Up-regulation and Cooperative Action of Sonic Hedgehog and Adrenomedullin. The Journal of biological chemistry 291, 16263–16270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava SK, Bhardwaj A, Arora S, Singh S, Azim S, Tyagi N, Carter JE, Wang B, and Singh AP (2015) MYB is a novel regulator of pancreatic tumour growth and metastasis. Br J Cancer 113, 1694–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, and Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, and Varambally S (2017) UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindskog C (2016) The Human Protein Atlas - an important resource for basic and clinical research. Expert review of proteomics 13, 627–629 [DOI] [PubMed] [Google Scholar]
- 22.Lee YC, Gajdosik MS, Josic D, Clifton JG, Logothetis C, Yu-Lee LY, Gallick GE, Maity SN, and Lin SH (2015) Secretome analysis of an osteogenic prostate tumor identifies complex signaling networks mediating cross-talk of cancer and stromal cells within the tumor microenvironment. Molecular & cellular proteomics : MCP 14, 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S, Jeong HM, Chung SE, Kim TH, Thapa SK, Lee DY, Song CH, Lim JY, Cho SM, Nam KY, Kang WH, Choi YW, and Shin BS (2019) Simultaneous analysis of acetylcarnitine, proline, hydroxyproline, citrulline, and arginine as potential plasma biomarkers to evaluate NSAIDs-induced gastric injury by liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal 165, 101–111 [DOI] [PubMed] [Google Scholar]
- 24.Bosse K, Haneder S, Arlt C, Ihling CH, Seufferlein T, and Sinz A (2016) Mass spectrometry-based secretome analysis of non-small cell lung cancer cell lines. Proteomics 16, 2801–2814 [DOI] [PubMed] [Google Scholar]
- 25.Karp NA, and Lilley KS (2009) Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics 9, 388–397 [DOI] [PubMed] [Google Scholar]
- 26.Diz AP, Truebano M, and Skibinski DO (2009) The consequences of sample pooling in proteomics: an empirical study. Electrophoresis 30, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 27.Sadiq ST, and Agranoff D (2008) Pooling serum samples may lead to loss of potential biomarkers in SELDI-ToF MS proteomic profiling. Proteome Sci 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenthal L, and Schisterman E (2008) Pooling data when analyzing biomarkers subject to a limit of detection. Methods Mol Biol 477, 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang WC, Wu SL, Huang WC, Hsu JY, Chan SH, Wang JM, Tsai JP, and Chen BK (2015) PTX3 gene activation in EGF-induced head and neck cancer cell metastasis. Oncotarget 6, 7741–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Locatelli M, Ferrero S, Martinelli Boneschi F, Boiocchi L, Zavanone M, Maria Gaini S, Bello L, Valentino S, Barbati E, Nebuloni M, Mantovani A, and Garlanda C (2013) The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. Journal of neuroimmunology 260, 99–106 [DOI] [PubMed] [Google Scholar]
- 31.Rathore M, Girard C, Ohanna M, Tichet M, Ben Jouira R, Garcia E, Larbret F, Gesson M, Audebert S, Lacour JP, Montaudie H, Prod’Homme V, Tartare-Deckert S, and Deckert M (2019) Cancer cell-derived long pentraxin 3 (PTX3) promotes melanoma migration through a toll-like receptor 4 (TLR4)/NF-kappaB signaling pathway. Oncogene [DOI] [PubMed] [Google Scholar]
- 32.Rubino M, Kunderfranco P, Basso G, Greco CM, Pasqualini F, Serio S, Roncalli M, Laghi L, Mantovani A, Papait R, and Garlanda C (2017) Epigenetic regulation of the extrinsic oncosuppressor PTX3 gene in inflammation and cancer. Oncoimmunology 6, e1333215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo S, Ueno H, Hosoi H, Hashimoto J, Morizane C, Koizumi F, Tamura K, and Okusaka T (2013) Clinical impact of pentraxin family expression on prognosis of pancreatic carcinoma. British journal of cancer 109, 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kathagen-Buhmann A, Maire CL, Weller J, Schulte A, Matschke J, Holz M, Ligon KL, Glatzel M, Westphal M, and Lamszus K (2018) The secreted glycolytic enzyme GPI/AMF stimulates glioblastoma cell migration and invasion in an autocrine fashion but can have anti-proliferative effects. Neuro-oncology 20, 1594–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Kumar R, Underwood K, O’Connor AE, Loveland KL, Seehra JS, and Matzuk MM (2007) Prevention of cachexia-like syndrome development and reduction of tumor progression in inhibin-deficient mice following administration of a chimeric activin receptor type II-murine Fc protein. Molecular human reproduction 13, 675–683 [DOI] [PubMed] [Google Scholar]
- 36.Jabalee J, Towle R, and Garnis C (2018) The Role of Extracellular Vesicles in Cancer: Cargo, Function, and Therapeutic Implications. Cells 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown KJ, Formolo CA, Seol H, Marathi RL, Duguez S, An E, Pillai D, Nazarian J, Rood BR, and Hathout Y (2012) Advances in the proteomic investigation of the cell secretome. Expert review of proteomics 9, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy MP, Hickman LJ, Eckman CB, Uljon SN, Wang R, and Golde TE (1999) gamma-Secretase, evidence for multiple proteolytic activities and influence of membrane positioning of substrate on generation of amyloid beta peptides of varying length. The Journal of biological chemistry 274, 11914–11923 [DOI] [PubMed] [Google Scholar]
- 39.Lichtenthaler SF, Lemberg MK, and Fluhrer R (2018) Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam A, Adamik B, Hawari FI, Ma G, Rouhani FN, Zhang J, and Levine SJ (2006) Extracellular TNFR1 release requires the calcium-dependent formation of a nucleobindin 2-ARTS-1 complex. The Journal of biological chemistry 281, 6860–6873 [DOI] [PubMed] [Google Scholar]
- 41.Merilahti JAM, and Elenius K (2019) Gamma-secretase-dependent signaling of receptor tyrosine kinases. Oncogene 38, 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X, and Li S (2017) Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene 36, 2629–2636 [DOI] [PubMed] [Google Scholar]
- 43.Seki SM, and Gaultier A (2017) Exploring Non-Metabolic Functions of Glycolytic Enzymes in Immunity. Frontiers in immunology 8, 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsutsumi S, Yanagawa T, Shimura T, Kuwano H, and Raz A (2004) Autocrine motility factor signaling enhances pancreatic cancer metastasis. Clin Cancer Res 10, 7775–7784 [DOI] [PubMed] [Google Scholar]
- 45.Principe M, Ceruti P, Shih NY, Chattaragada MS, Rolla S, Conti L, Bestagno M, Zentilin L, Yang SH, Migliorini P, Cappello P, Burrone O, and Novelli F (2015) Targeting of surface alpha-enolase inhibits the invasiveness of pancreatic cancer cells. Oncotarget 6, 11098–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Principe M, Borgoni S, Cascione M, Chattaragada MS, Ferri-Borgogno S, Capello M, Bulfamante S, Chapelle J, Di Modugno F, Defilippi P, Nistico P, Cappello P, Riganti C, Leporatti S, and Novelli F (2017) Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J Hematol Oncol 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB, and Beer DG (2009) Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia 11, 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katayama Y, Oshima T, Sakamaki K, Aoyama T, Sato T, Masudo K, Shiozawa M, Yoshikawa T, Rino Y, Imada T, and Masuda M (2017) Clinical Significance of INHBA Gene Expression in Patients with Gastric Cancer who Receive Curative Resection Followed by Adjuvant S-1 Chemotherapy. In Vivo 31, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, Yamada Y, Arai T, Kita Y, Kijima Y, Mataki Y, Maemura K, and Natsugoe S (2018) Involvement of anti-tumor miR-124–3p and its targets in the pathogenesis of pancreatic ductal adenocarcinoma: direct regulation of ITGA3 and ITGB1 by miR-124–3p. Oncotarget 9, 28849–28865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y, Tang L, Zhang Z, Li S, Liang S, Ji L, Yang B, Liu Y, and Wei W (2018) Long Noncoding RNA TUG1/miR-29c Axis Affects Cell Proliferation, Invasion, and Migration in Human Pancreatic Cancer. Disease markers 2018, 6857042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surcel A, Schiffhauer ES, Thomas DG, Zhu Q, DiNapoli KT, Herbig M, Otto O, West-Foyle H, Jacobi A, Krater M, Plak K, Guck J, Jaffee EM, Iglesias PA, Anders RA, and Robinson DN (2019) Targeting Mechanoresponsive Proteins in Pancreatic Cancer: 4-Hydroxyacetophenone Blocks Dissemination and Invasion by Activating MYH14. Cancer research 79, 4665–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, and Clayton A (2010) Proteomics analysis of bladder cancer exosomes. Molecular & cellular proteomics : MCP 9, 1324–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, and Gho YS (2007) Proteomic analysis of microvesicles derived from human colorectal cancer cells. Journal of proteome research 6, 4646–4655 [DOI] [PubMed] [Google Scholar]
- 54.Chettimada S, Lorenz DR, Misra V, Dillon ST, Reeves RK, Manickam C, Morgello S, Kirk GD, Mehta SH, and Gabuzda D (2018) Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Scientific reports 8, 7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Pasa-Tolic L, and Jenster G (2013) Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PloS one 8, e82589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, and Vizcaino JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.