Graphical abstract
Keywords: ACE2, COVID-19, SARS-CoV-2, Host range, Host tropism
Highlights
-
•
Association of ACE2 with S is the molecular determinant of SARS-CoV-2 infection.
-
•
Diversify in ACE2 leads to differences in SARS-CoV-2 infection outcomes.
-
•
The host range SARS-CoV-2 is dependent on the ACE2 expression profile.
-
•
The cellular tropism of SARS-CoV-2 is dependent on the ACE2 expression profile.
Abstract
COVID-19, which is caused by SARS-CoV-2, has been declared a global pandemic. Although effective strategies have been applied to treat the disease, much is still unknown about this novel virus. SARS-CoV-2 enters host cells through ACE2, which is a component of the angiotensin-regulating system. Binding of the SARS-CoV-2 S protein to ACE2 is a prerequisite for SARS-CoV-2 infection. Many studies have indicated a close relationship between ACE2 expression and SARS-CoV-2 infection. The structural basis of receptor recognition by SARS-CoV-2 has been analyzed in detail. The diversification of the ACE2 sequence due to ACE2 polymorphisms and alternative splicing has to a large extent affected the susceptibility of different species. Differential ACE2 expression makes specific populations more prone to be infected, and ACE2 also plays a role in the broad tropism of SARS-CoV-2 in human organs and tissues. In this review, we comprehensively summarize how the ACE2 expression profile affects the host range and tropism of SARS-CoV-2, which will provide mechanistic insights into the susceptibilities and outcomes of SARS-CoV-2 infection.
1. Introduction
COVID-19, which emerged in the winter of 2019, has been declared a pandemic by the WHO and spread to more than 200 countries around the world. Over 57 million people have been affected as of Nov 19, 2020. Although the overall mortality rate of COVID-19 is lower than that of SARS, which broke out in 2002–2003, COVID-19 has aroused global concern as a huge threat to public health and the international economy.
Not long after the outbreak of COVID-19, researchers identified the causal pathogen to be 2019-nCoV [1], [2], which was later named severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) by ICTV (International Committee on Taxonomy of Viruses). Phylogenetic analysis revealed that SARS-CoV-2 belongs to the subgenus Sarbecovirus of Coronaviridae and is closely related to SARS-CoV, with 77.2% amino acid similarity [2]. After SARS-CoV and MERS-CoV (Middle East Respiratory Syndrome Coronavirus), SARS-CoV-2 is the third coronavirus that has threatened human life in the new century [3]. The high similarity of the sequences of SARS-CoV-2 and SARS-CoV has allowed researchers and physicians to study the mechanisms of pathogenesis and identify treatment strategies based on the knowledge of SARS-CoV. Patients infected by SARS-CoV-2 exhibit fever and cough, which are also observed in SARS and are typical symptoms of respiratory infectious disease [4]. However, there are obvious differences in their epidemiological and clinical features. SARS-CoV-2 shows stronger transmission ability and targets more organs, indicating the urgent need for comprehensive study of SARS-CoV-2.
The genome of SARS-CoV-2 is a single-strand positive RNA that produces ORF1a, ORF1b, and at least six accessory proteins and structural proteins, including spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) [5]. S protein causes the entry of SARS-CoV-2 into the target cell. Upon entry, S needs to be primed for further fusion with the membrane by cellular proteases, which cleave S at the linkages of S1/S2 and within S2. Similar to SARS-CoV, SARS-CoV-2 mainly utilizes angiotensin converting enzyme II (ACE2) as the receptor for entry and TMPRSS2 for S protein priming [6], [7]. Several other proteins are proposed to mediate SARS-CoV-2 entry, while the evidence is relatively lack and need to be strengthened. CD147, GRP78, and heparan sulfate (HS) interact with SARS-CoV-2, making them candidate routes for SARS-CoV-2 entry. ACE2, which was originally identified as a component of the angiotensin (Ang) regulating system, played an important role in those two coronavirus-caused epidemic diseases. It is speculated that the distribution and expression level of ACE2 largely affects the susceptible species, targeted organs, and severity of disease. In this review, we aim to explain the SARS-CoV-2 infection from the view of ACE2 expression profile. We will first describe the function of the ACE2 receptor in SARS-CoV-2 entry. Then, we will summarize the differential expression of ACE2 among species, within the human body and in specific populations with complications. We will focus on not only the mechanisms of infection but also the implications for treating this disease.
2. ACE2: from vascular regulator to viral receptor
ACE2 was first discovered in 2000 and shown to share 40% amino acid similarity with ACE1 [8]. Unlike ACE1, which converts Ang I into Ang II, the primary function of ACE2 is to cleave Leu or Phe in Ang II to produce Ang (1–7). Ang (1–7) counteracts the vaso/broncho-constriction effect of Ang II, which may alleviate acute respiratory distress syndrome (ARDS) and inflammation caused by SARS-CoV and SARS-CoV-2 [9]. ACE2 was experimentally determined to be a functional receptor for SARS-CoV [7]. ACE2 consists of a peptidase domain in the N-terminus and a collectrin-like domain in the C-terminus, with a single transmembrane helix and a short intracellular segment [10]. The structure of SARS-CoV S in complex with ACE2 has been solved, and the important amino acids (AAs) residing within the interface between two proteins were identified [11].
Receptor-mediated viral entry is a prerequisite for infection. Considering the close relationship between SARS-CoV-2 and SARS-CoV, ACE2 was immediately predicted as a potential receptor, which was later verified by in vitro experiments. Using HeLa cells that did not express ACE2 per se, Zhou et al. showed that the introduction of the human ACE2 gene facilitated SARS-CoV-2 infection [12]. Based on ACE2, other species supporting SARS-CoV-2 infection included Chinese horseshoe bats, civets, and pigs, with mice being the exception. In addition, they found that SARS-CoV-2 did not use aminopeptidase N (APN) and dipeptidyl peptidase 4 (DPP4) for entry, which are the receptors used by HCoV-229E and MERS, respectively [13], [14]. This conclusion was validated by another work [6]. Using S protein expressed by vesicular stomatitis virus, they confirmed that cell lines lacking ACE2 can be infected when human/bat ACE2 but not DPP4 or APN is expressed. They also confirmed the dependence on proteases for infection and found that camostat mesylate, a clinically approved inhibitor of TMPRSS2, effectively inhibited viral entry. There are alternative proteins to be predicted as potential receptors for SARS-CoV-2. The glycoprotein CD147 and ER chaperone GRP78 attracted attention, as both of the two have higher mRNA and protein levels than ACE2 in lung [15]. CD147 showed direct association with SARS-CoV-2 S in vitro. For GRP78, it was proposed to bind S through cyclic structure constituted by disulfide bonds. However, direct evidence supporting their receptor identity need to be strengthened.
Importantly, the AAs within the receptor binding domain (RBD) critical for ACE2 binding are conserved in SARS-CoV and SARS-CoV-2 [16]. A series of studies provided structural information on receptor binding. Since ACE2 also functions as a chaperone, Yan et al. obtained a high-resolution (2.9 Å) structure of the full-length ACE2 dimer in the presence of its trafficking cargo B0AT1 [10]. Furthermore, they resolved the complex of the RBD of SARS-CoV-2 and ACE2-B0AT1. Structural analysis indicated that an ACE2 dimer simultaneously binds to two S trimers, with one trimer binding with one ACE2 monomer. Specifically, they showed the detailed structure of the interface between the SARS-CoV-2 RBD and ACE2. The α1 helix is the main component of the interaction, with the α2 helix and linker between the β3 and β4 sheets also contributing to binding. Wrapp et al. provided the atomic-level structure of the SARS-CoV-2 S protein in the prefusion conformation and compared it with that of SARS-CoV [17]. The S trimer primarily adopted a state in which one RBD was exposed for receptor recognition. When measuring the kinetics of the interaction, the SARS-CoV-2 RBD was found to bind ACE2 with a 10- to 20-fold higher affinity (15 nM) than SARS-CoV. This result provided a clue to explain the rapid transmission of SARS-CoV-2. The structure of the SARS-CoV-2-CTD in complex with ACE2 at a resolution of 2.5 Å was resolved. Further investigation of the interface demonstrated that SARS-CoV-2-CTD has an increase in the number of residues that directly bind with ACE2 [18]. The dynamics of the binding of the ACE2 extracellular domain to the SARS-CoV/SARS-CoV-2 S proteins were studied by biolayer interferometry [19]. It is worth noting that neutralizing antibodies against SARS-CoV S were shown to potently inhibit SARS-CoV-2 pseudovirus entry, which was contrary to the results of other studies [17], [18]. Two independent studies elucidated the interaction of the SARS-CoV-2 RBD and ACE2 by X-ray crystallography [1], [20]. Structural analysis and protein pull-down assays illustrated that the residue changes in SARS-CoV-2 increase the stability of the binding of the virus with ACE2 [1]. This information on the SARS-CoV-2 sequence and structure provided insights helpful in identifying potential host and high-risk species and developing antiviral strategies targeting the S-RBD/ACE2 interaction.
3. Diversified ACE2 leads to differences in SARS-CoV-2 infection outcomes
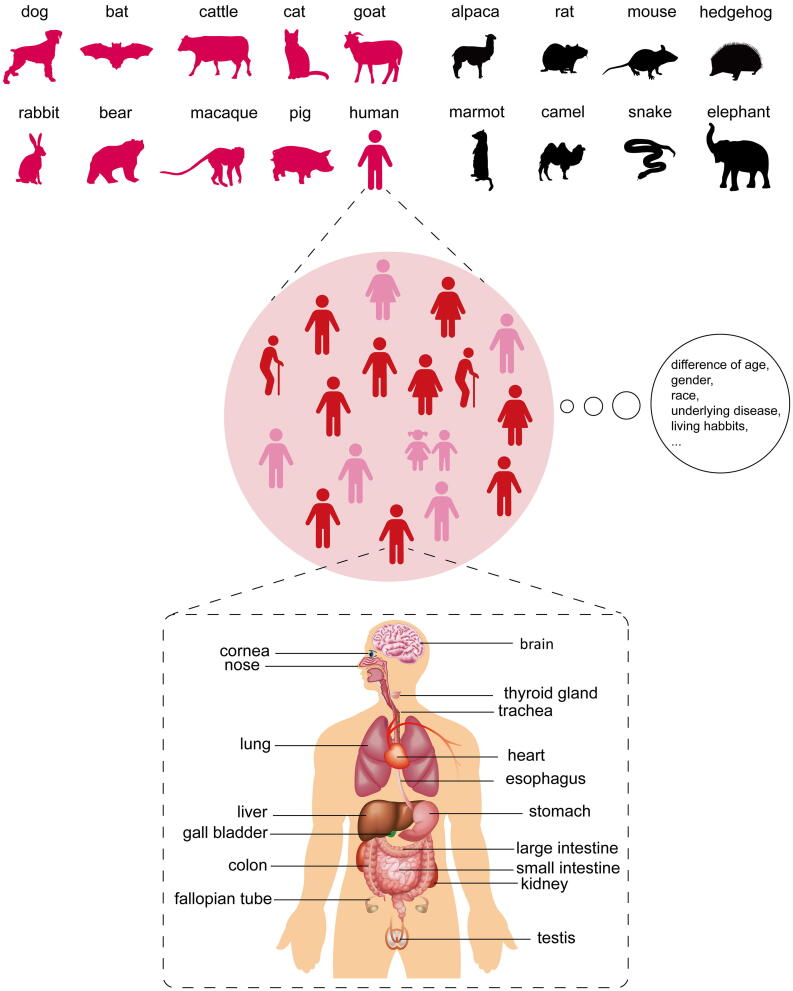
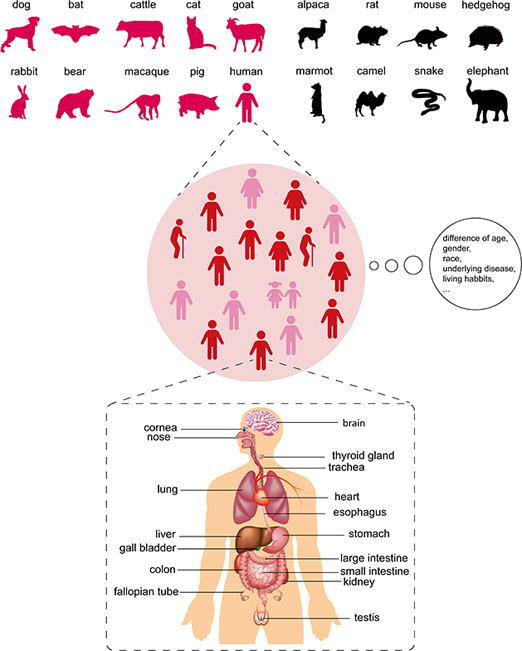
Recognition and binding of ACE2 is a critical step for SARS-CoV-2 entry into cells. In this regard, any difference in ACE2, including sequence alterations and expressional regulation, will affect the susceptibility of the host. To a large extent, ACE2 determines the potential host range of SARS-CoV-2. For individuals, many factors associated with ACE2 affect susceptibility to the virus. Furthermore, the organs targeted by SARS-CoV-2 are determined by the differential expression of ACE2. Next, we will discuss the relationship of ACE2 with SARS-CoV-2 infection at levels of species, population and organ/tissue (Fig. 1).
Fig. 1.
Susceptibility and outcoming of SARS-CoV-2 infection is dependent on ACE2 expression profile. Upper: SARS-CoV-2 host range could be predicted based on ACE2 orthologs. Animal species which possess ACE2 that has binding capacity to SARS-CoV-2 S (in wine red) are potential hosts of SARS-CoV-2. Animals of which the ACE2 is not suitable for SARS-CoV-2 S binding (in black) are resistant to this virus. Middle: Although humans are generally susceptible to this novel virus, the outcoming of infection is partially affected by different race, gender, age, underlying disease, and even living habits. Bottom: Clinical data showed that multiple organs other than lung could be injured by SARS-CoV-2. Researches about organ and cellular tropism of SARS-CoV-2 (marked in the Figure) are accumulating especially with the application of single-cell sequencing. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Sequence changes in ACE2 and host range
Given that close and stable contact is essential for RBD binding of ACE2, residue changes in the binding interface will have an effect on receptor recognition. Based on the resolved structure of the RBD-ACE2 complex, ACE2 residues that are critical for binding have been identified. These residues are located at the surface of the claw-like structure of ACE2. Lan et al. used a 4 Å distance cutoff and determined that 20 AAs of ACE2 were in contact with the SARS-CoV-2 RBD [1]. Wang et al. analyzed the interface using a distance cutoff of 4.5 Å and identified additional involved residues (24 AAs) compared to Lan et al. [18]. Critical AAs are clustered into two hot spots: one extends from AA 20 to 40 and the other from AA 330 to 360. It is believed that these conserved AAs determine cross-species and human-to-human transmission. ACE2 in potential hosts of SARS-CoV-2 must possess most of the key residues necessary for ligand binding. Therefore, many groups set out to screen ACE2 sequences from multiple species and compare them with that of human ACE2 to identify potential hosts of SARS-CoV-2. The palm civet is thought to be the intermediate host of SARS-CoV, and is a potential susceptible species to SARS-CoV-2. Wan et al. analyzed critical residue changes in ACE2 from civets [21]. While the hot spot residue K31 was changed to T31 and no longer able to form a salt bridge with E35 in the SARS-CoV-2 RBD, the overall surface was still capable of ligand binding. Hence, the civet can also be infected by the novel coronavirus. This group also analyzed critical changes in hotspots in ACE2 in other animals and speculated based on these changes that pigs, ferrets, cats, orangutans and monkeys were potential hosts. Another hotspot residue, K353, was changed to H in mice and rats, leading to unfavorable contact with the SARS-CoV-2 RBD. According to the relative synonymous codon usage bias, snakes had most similarity with SARS-CoV-2 [22]. Another paper indicated that turtles can function as a natural viral host because the interaction of residues 41 and 353 with the RBD showed more resemblance to that of human ACE2 than that of bat ACE2 [23]. However, these results were in contrast to the consensus that reptiles are resistant to SARS-like coronavirus and were disproved by other studies. Luan et al. performed two successive studies on ACE2. They collected the sequences of 40 mammalian ACE2 and constructed a phylogenetic tree. Potential hosts were predicted by matching the five most important AAs, including K31, E35, D38, M82 and K353. Through homogeneous modeling, they found that the change of M82 to N82 in the Chinese hamster shortened the distance between it and F486 in the SARS-CoV-2 RBD [16]. In another work, they expanded the analysis to all 20 key AAs involved in RBD-ACE2 binding [24]. Sequence alignment showed that nearly half of the critical AAs in turtles, snakes and birds were different from those in humans, indicating that they were hardly permissive for SARS-CoV-2 entry. By combining bioinformatic analysis and functional assays of ACE2 from various species, Liu et al. characterized 44 ACE2 orthologs from 49 species that could mediate SARS-CoV-2 entry, suggesting a broad host range of this novel coronavirus [25]. One of the results of this work showed that although they shared more than 90% similarity with human ACE2, ACE2 orthologs from three New World monkeys, including Callithrix jacchus, exhibited little, if any, permission for SARS-CoV-2 infection. This is consistent with another study that aimed to establish a nonhuman primate model for COVID-19 [26]. In this research, two Old World monkeys (Macaca mulatta and Macaca fascicularis) and Callithrix jacchus were inoculated with SARS-CoV-2. While the viral genome was successfully detected in swabs and blood from Callithrix jacchus, none of the monkeys had an increased body temperature, and there was no detection of the viral genome in any organs by necropsy [26]. The reason why Callithrix jacchus is resistant to SARS-CoV-2 and whether it is due to the deficiency of marmoset ACE2 remains to be clarified.
A detailed analysis of the susceptibility of multiple domesticated animals was performed [27]. SARS-CoV-2 replicated effectively in ferrets and cats. Dogs exhibited low susceptibility, and other livestock animals are insensitive to SARS-CoV-2. Since the animals used in this research are in close contact with humans, their susceptibility to viruses would have profound effects on human health. It is noticeable that dog and cat varied in response to SARS-CoV-2, notwithstanding both of them have canonical ACE2 that is capable of binding S. Alternative splicing is a way to increase protein complexity, which produces protein isoforms of different sizes. Different from human and ferret which have the only ACE2 isoform, both cat and dog have more than one isoform of ACE2. However, all three isoforms of cat are of the same length and have functional domains. As for dogs, one of the five isoforms lack the predicted transmembrane domain and so exists as a soluble ACE2. We think that this soluble dog ACE2 could play a competitive inhibitory role for interaction between SARS-CoV-2 S and canonical ACE2. We also made prediction of host susceptibility based on ACE2 isoform diversity [28]. Natural soluble ACE2 has been developed as an antiviral strategy. A human recombinant soluble ACE2 (hrsACE2) protein, which contains the catalytic ectodomain of human ACE2, has undergone phase 1 and phase 2 clinical testing for treating patients with acute respiratory distress syndrome (ARDS) [29]. Recently, this clinical-grade soluble ACE2 protein was found to significantly reduce viral growth in Vero cells. In human blood vessel organoids and kidney organoids, hrsACE2 markedly inhibited SARS-CoV-2 infection at the early stage [30]. Other studies studied the neutralization effect by fusing the extracellular domain of human ACE2 to the Fc region of human immunoglobulin with or without mutation. Li et al. developed ACE2-Fc variants of different lengths and found that ACE2-Fc containing the 18–740 residue fragment performed better than that containing the 18–615 residue fragment in blocking SARS-CoV-2 entry [31]. Another study also confirmed that both the SARS-CoV RBD and the SARS-CoV-2 RBD bound ACE2-Fc with high affinity and were neutralized by this fusion protein [32]. By combining structural analysis and docking experiments, Renzi et al. found a minimal ACE2 fragment consisting of two α-helices [33]. This soluble ACE2 retained its binding capacity with SARS-CoV-2 as well as the physiological ligand angiotensin II. The cross reactivity and specific inhibition of coronavirus indicated that soluble ACE2 is a potent therapeutic strategy.
3.2. Expression of ACE2 in specific populations and their susceptibility
Susceptibility to SARS-CoV-2 among different species is dependent on ACE2 orthologs. Although human beings are generally permissive to this novel virus at present, factors related to race, age, gender, underlying disease, or even living habits still lead to different susceptibilities and severities of infection. Obviously, it cannot be excluded that these factors also affect the immune response. However, an increasing number of studies have demonstrated that the ACE2 variation may be a critical factor.
ACE2 polymorphisms also affect individual susceptibility. Based on structural information about the SARS-CoV-2 S RBD/ACE2 complex, Calcagnile et al. searched the dbSNP and UNIPROT databases and identified 301 ACE2 SNPs leading to missense mutations [34]. Through in silico analysis, two SNPs were found to alter RBD binding. K26R may increase the affinity for the RBD, while S19P does the opposite, indicating that they may promote or inhibit infection, respectively. Interestingly, K26R is most common in European people, and S19P is common in Africa. Researchers speculated that the geographical difference in susceptibility to SARS-CoV-2 may be partially explained by this genetic factor. A similar study was performed based on 290,000 samples from more than 400 population groups [35]. It found that ACE2 variants were relatively rare, which was perhaps due to a lack of selection pressure. However, some SNPs may affect protein interactions, according to structural analysis. In summary, the SNPs that were predicted to strengthen binding to the SARS-CoV-2 RBD included S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P and H378R. In contrast, K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L and D509Y resulted in weak binding and hence may have a protective function.
Patients with cancer, diabetes, and hypertension are prone to infection and progress to more severe stages of COVID-19 than other patients. A tertiary care hospital in Wuhan city investigated 1524 cancer patients in the Department of Radiation and Medical Oncology and found that 12 of them had been diagnosed with COVID-19. This infection rate (0.79%) was higher than the cumulative incidence in this city (0.37%) during the same period [36]. In another larger survey, researchers reported a nationwide analysis of SARS-CoV-2 infection in cancer patients. Among the 1590 COVID-19 cases collected from 575 hospitals in 31 Chinese provincial administrative regions, 18 people (1%) had a history of cancer, which was higher than that in Chinese people overall (0.29%) [37]. In addition, a higher proportion of COVID-19 patients with cancer were admitted to the intensive care unit (39% of patients with cancer vs 8% of patients without cancer). The deterioration process was also shorter, with a median time to severe events of 13 days and 43 days in patients with or without cancer, respectively. The close relationship between cancer and SARS-CoV-2 can be attributed to many factors. Abnormal immune systems due to cancer either fail to protect patients against viruses or induce cytokine storms, which can cause lethal damage to many organs. Repeated visits to the hospital for cancer treatment increase the chance of infection within the hospital environment. Another important issue is the changes in the ACE2 expression level. Recently, a systematic analysis was performed with the help of bioinformatical tools such as cBioPortal, UALCAN, and gepia2. More than 10,000 samples of 30 kinds of cancers in TCGA database were used for an analysis focusing on the genetic variation, expression level and epigenetic changes of ACE2 in cancers [38]. Compared to its expression in normal tissue, ACE2 was upregulated in six tumors, including colon adenocarcinoma (COAD), kidney renal papillary cell carcinoma (KIRP), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), and lung adenocarcinoma (LUAD). Mutation and DNA copy variation was observed but was not relevant to ACE2 expression. Five of the tumors with upregulated ACE2 expression exhibited decreased methylation of ACE2. Considering that LUAD is the most frequent cancer in COVID-19 patients with cancer, this analysis provided valuable evidence that LUAD patients have high risk and should be given more personal protection.
It is not difficult to understand the regulation of cancer by ACE2 because previous studies have determined that ACE2 is differentially expressed in tumors [39]. In Zhang’s study on breast cancer, ACE2 was found to be downregulated in patients. An in vitro study verified that ACE2 downregulated VEGFA expression in breast cancer cells and inhibited cell migration. The causal relation between ACE2 and VEGFA reminds us of its original function in the cardiovascular system. Hypertension was listed as a top comorbidity in patients with confirmed COVID-19. In a study on ICU patients, cerebrovascular diseases were the most distinctive comorbidity [40]. In another study involving 1099 COVID-19 patients, 23.7% had hypertension [4]. It seems that people with hypertension are at higher risk for SARS-CoV-2 infection. However, Fang et al. pointed out that this may result from the treatment of hypertension rather than high blood pressure per se [41]. Their reasoning stemmed from a statement in a review that hypertension is often treated by ACE inhibitors (ACEis) and angiotensin receptor blockers (ARBs), while ACE2 was found to be upregulated [42]. They suggested that these ACE2-stimulating drugs result in a high risk of developing severe COVID-19, and alternative treatment for hypertension could be applied. Their assertion was questioned by other researchers. Rossi et al. disagreed with them for two reasons [9]. First, the causal link between the use of drugs and increased ACE2 was not very solid. Second, ACEis and ARBs as well as Ang (1-7) counteract vaso-/bronchoconstriction, inflammation, and the development of acute respiratory distress syndrome (ARDS). Thus, ACE is or ARBs administration and the ACE-2/Ang (1-7) axis play protective roles against ARDS caused by COVID-19.
Another distinctive comorbidity is diabetes. In studies by Yang and Guan, diabetes was reported in 22% and 16.2% of patients, respectively [4], [40]. Chronic hyperglycemia can lead to immune deficiency in patients. Meanwhile, ACEis and ARBs were also used for treating diabetes. Therefore, the effect of diabetes on COVID-19 remains to be further studied.
SARS-CoV-2 infection is also related to obesity. Heialy et al. investigated the underlying molecular mechanisms [43]. By analyzing recently uploaded transcriptomics data, they found that lipid metabolism was changed by SARS-CoV-2 infection, leading to an increase in sterol-response element binding protein (SREBP). To uncover the effect of dysregulated lipogenesis on SARS-CoV-2 infection, they further searched the available transcriptomics data for the effect of ACE2 expression after inducing obesity. One dataset from high-fat diet-induced obese mice showed that Ace2 was significantly upregulated in the lung. The results of this study coincide with the clinical characteristics that obesity confers patients with a high risk of SARS-CoV-2 infection.
Clinical data also correlated living habits with outcomes in COVID-19 patients. The clinical characteristics collected by Guan et al. showed that a higher proportion of patients with severe disease were former or current smokers than those with nonsevere disease [4]. On the other hand, smokers seem to have a higher probability of developing severe disease. This clinical characteristic may be partially explained by a previous study [44]. Researchers analyzed data from six independent studies performed in healthy people, patients with lung disease and smokers to determine ACE2 expression in multiple organs. Surprisingly, the ACE2 level was significantly higher in smokers than in healthy people, while there was no obvious difference between healthy people and those with chronic respiratory disease. This analysis indicated that smoking may pose a risk for people in terms of infection with SARS-CoV and SARS-CoV-2. The underlying mechanism may be attributed to epigenetic regulation. Two studies mentioned hypomethylation of ACE2 in malignancies and lupus [38], [45]. Long-term smoking may affect ACE2 by changing the epigenetic characteristics. Clinical data show a gender imbalance in the incidence of the disease. More patients are men, and the disease seems to be more serious in men, as men are more likely to be admitted to the ICU and have longer disease duration [4], [46]. This effect can be partially attributed to higher rates of hypertension and smoking, which are risk factors for SARS-CoV-2. However, the differential expression of ACE2 in men and women should not be ignored, which will be described in detail below.
3.3. Differential expression of ACE2 among human organs
The predominant clinical symptom of COVID-19 is in the respiratory system. However, clinical features affecting other tissues, such as inappetence, conjunctivitis, thrombus, and even mental abnormalities, were observed in a subset of patients. Dysfunction of multiple systems implies that COVID-19 is not merely pneumonia. Moreover, SARS-CoV-2 RNA was detected in samples collected from the pharynx, rectum, and even semen [47], indicating that many organs other than the lung facilitate SARS-CoV-2 infection. The delayed viral clearance in men compared to that in women was thought to be associated with high ACE2 protein expression in the testes [48]. Although treatment-induced secondary symptoms partially accounted for dysfunction, direct infection by SARS-CoV-2 of multiple organs should be considered.
For the potential targeted organ, expression of ACE2 receptor is a prerequisite for virus entry. Two cutting-edge technologies aid in the investigation of ACE2 distribution. One is single-cell RNA sequencing (scRNA-seq), and the other is human organoids. Although ACE2 has previously been identified in the respiratory tract as a gateway for SARS-CoV entry, the exact cell type expressing ACE2 that mediates viral infection remains largely unknown. By using single nuclei and single cell RNA sequencing, Lukassen et al. analyzed ACE2 as well as the priming protease TMPRSS2 in lung tissue and cells of subsegmental bronchial branches [49]. AT2 cells showed the highest ACE2 expression in the lung. In the subsegmental bronchial branches, a subtype of transient secretory cells was found to have the strongest ACE2 expression. In addition, the ACE2-positive cell population was also enriched for TMPRSS2. When the expression of FURIN was included in the analysis, an obvious enrichment of double- or triple-positive cells for ACE2, TMPRSS2, and FURIN was observed, indicating a coexpressional tendency of SARS-CoV-2 entry factors.
Several studies examined the host tropism of SARS-CoV-2 in other organs. Sungnak et al. surveyed scRNA-seq datasets from multiple organs and tissues, including the respiratory tree, cornea, retina, esophagus, ileum, colon, heart, skeletal muscle, spleen, liver, placenta/decidua, kidney, testis, pancreas, prostate gland, brain, skin and fetal tissues [50]. ACE2 and TMPRSS2 exhibited different general expression profiles. TMPRSS2 had a broader distribution with an overall high expression level, while ACE2 expression was generally low in the analyzed datasets. Cells with ACE2 expression were found in the airways, cornea, esophagus, ileum, colon, liver, gallbladder, heart, kidney and testis. Qi et al. conducted similar research by using scRNA-seq data from 31 organs from nine major human systems [51]. The ratio of TMPRSS2/ACE2 expression in cells and the TMPRSS2/ACE2 expression level in cell clusters were analyzed in each organ. This analysis indicated that the brain, gallbladder, and fallopian tubes were permissive for SARS-CoV-2 infection. All susceptible organs were ranked into three risk levels based on TMPRSS2 expression. Zhou et al. also reported a systematic analysis of 36 tissues and ranked the cell types that were vulnerable to SARS-CoV-2 [52]. They pointed out the discrepancy between the scRNA-seq data and the clinical manifestations and attributed it to a lack of protein expression information. Therefore, they combined the protein expression data for ACE2, TMPRSS2, and FURIN with the scRNA-seq data to develop so-called protein-proven single-cell RNA (pscRNA) profiling. Systematic pscRNA profiling revealed that lung AT2 cells and macrophages were at top of the high-risk list, followed by cardiomyocytes, adrenal gland stromal cells, stromal cells in testis, ovary and thyroid cells. It is worth noting that all these scRNA-seq data analyses incorporated TMPRSS2 and/or FURIN expression information when predicting organ susceptibility. Considering that TMPRSS2 and FURIN are two important cofactors assisting SARS-CoV-2 entry, it is necessary to evaluate the coexpression of ACE2 with TMPRSS2 and FURIN.
The predicted target organs based on scRNA-seq data can be partially verified by experiments in organoids. Lamers et al. established human small intestinal organoids (hSIOs) under different culture conditions and exposed them to SARS-CoV-2 [53]. By quantifying the viral sequence and titering live virus, SARS-CoV-2 was confirmed to productively infect hSIOs. Infection of intestinal enterocytes was further demonstrated by confocal and electron microscopy imaging, which was consistent with a previous study showing that the brush border of intestinal enterocytes had relatively high ACE2 expression in the human body [53], [54]. Organoids were also used in research of Monteil et al. [30], in which they generated human blood vessels and kidney organoids and demonstrated that both can be infected by SARS-CoV-2.
When analyzing the correlation coefficients, ACE2 was found to have a close association with genes involved in immune functions, including the antiviral innate response [50]. Through RNA-seq analysis, Ziegler et al. made a striking discovery that ACE2 is a human interferon-stimulated gene (ISG) in primary human nasal epithelial cells, which means that this cellular target could be maintained or strengthened by the host IFN response against SARS-CoV-2 [55]. Recent study provided specific evidence that it was a truncated isoform of ACE2 (dACE2) not the full-length ACE2 that was induced by IFN [56]. Since the dACE2 lacks the N-terminal 356 AAs, it fails to bind SARS-CoV-2 S RBD and affects receptor binding of full-length ACE2, which has been demonstrated by in vitro experiments [56]. It also raises a possibility that immune response rather than ACE2 per se determines the clinical outcome of SARS-CoV-2 infection. Zhang et al. performed a large genetic study about 659 patients with life-threatening COVID-19 and 534 subjects with asymptomatic or weak infection to discover the intrinsic factor of susceptibility of COVID-19. They focused on Toll-like receptor 3 (TLR3) and interferon regulatory factor 7 (IRF7) dependent type I interferon (IFN) pathway, which plays an important role in response to SARS-CoV-2 and other viruses. By analyzing 13 loci of type I IFN pathway, whose mutants have been described to confer other viruses’ susceptibility, 3.5% (23/659) of patients were found possessing deleterious variants, indicating a causal role of intrinsic type I IFN pathway in severe COVID-19 [57].
4. Conclusion
Currently, SARS-CoV-2 is spreading around the world at a rapid speed. Clinical and basic researchers are in a race with SARS-CoV-2 to save patients and uncover the pathogenic mechanisms. This novel virus has many features in common with the closely related virus SARS-CoV, but there are clear differences between them, such as the tighter binding of SARS-CoV-2 to the common receptor ACE2. Although other proteins, such as CD147, were inferred to mediate SARS-CoV-2 entry [58], ACE2 is well accepted as the functional receptor for both SARS-CoV-2 and SARS-CoV. Host susceptibility and the severity of COVID-19 are largely dependent on ACE2.
In this review, we summarized current studies on ACE2 from two dimensions: ACE2 itself and the susceptibility of its targets (Fig. 1). For ACE2, its association with the RBD of the spike protein from SARS-CoV-2 is the molecular determinant of infection, and the different AAs in binding hot spots determines its closer association with SARS-CoV-2. There are ACE2 orthologs with variations in AAs among multiple species and even in the same individual. In addition to sequence variations, the expression level is also variable. The difference in host susceptibility occurred at three levels, species, population and tissues, and was shown to have a close relationship with ACE2 variance.
However, when we evaluate the importance of ACE2 in SARS-CoV-2 infection, there is something that needs to be paid special attention to. Although SARS-CoV-2 entry is dependent on ACE2, there is no evidence showing a linear correlation between the severity of the disease and the expression level of ACE2. One consensus is that the existence of functional ACE2 is a prerequisite for SARS-CoV-2 infection. However, the negative results for ACE2 expression in some organs should be interpreted carefully. As Sungnak et al. mentioned in their paper, the expression level may be below the detection threshold. Moreover, the lack of ACE2 may be due to the sample isolation and the analysis methods [50]. Regardless, the rapidly developed scRNA-seq technology provides exquisitely detailed information about ACE2 expression, which greatly benefits antiviral studies.
As a regulating protein of angiotensin, ACE2 distributed broadly in human body. Apart from mRNA and scRNA-seq information introduced above, several groups detected ACE2 protein expression especially in respiratory system by immunohistochemistry (IHC) and western blot. Right after SARS-CoV outbreak, ACE2 expression in human organs was examined and showed positive signal in alveolar epithelial cells of lung and enterocytes of the small intestine, supporting infectious route for SARS-CoV [59]. Jennifer et al. analyzed in situ localization of ACE2 from publicly available dataset and performed immunoblot and IHC as well. Weak ACE2 signal was observed in epithelium of airway and lung alveoli [15]. The work of Hikmet et al. came to the same results. In their large-scale analysis comprising more than 150 types of cells covering major human organs, ACE2 was found none or low in respiratory system, limiting in a subset of ciliated cells in a few individuals [60]. Although ACE2 showed robust expression in airway epithelial cells and within their motile cilia, those results were from only one of six antibodies utilized in the study of Lee et al., leaving other antibodies with negligible or less staining [61]. The discrepancy of protein analysis could result from antibodies recognizing distinct epitopes and antigen retrieval of samples. However, it can be concluded that the expression level of ACE2 in airway especially in lung is relatively low, considering the consistent and high expression in other organs, such as intestine and kidney [15], [60], [61]. The low basal expression of ACE2 may be upregulated after viral infection or in specific condition of comorbidities. It also raised question about the existence of other receptors or co-receptors which mediate SARS-CoV-2 entry. The negatively-charged polysaccharide HS, which has been proven to be involved in entry of human coronavirus NL63 and eastern equine encephalitis virus [62], [63], was speculated as co-factor of SARS-CoV-2. Binding of S protein to HS stabilized it in the open conformation, which facilitated interaction with ACE2 [64].
In fact, ACE2 alone was not sufficient for SARS-CoV-2 infection. ACE2-mediated virus entry is assisted by TMPRSS2 and is affected by FURIN. Many studies have demonstrated the preference for the coexpression of ACE2 and TMPRSS2 [49], [50], [55]. Combining analysis of the expression profiles of ACE2 and TMPRSS2 will give us a more comprehensive and objective conclusion. ACE2 function may be affected by other factors. As several groups have reported, soluble human ACE2 could function as a decoy for SARS-CoV-2 S protein [30], [32], [33]. This natural competitor can effectively inhibit virus entry mediated by membrane-targeted ACE2. Moreover, we found that both SARS-CoV-2 S and ACE2 have RGD/KGD motifs, which can be recognized by integrin for binding. Thus, we speculated that integrin could hinder the receptor targeting of S by shielding both S and ACE2 [65].
In summary, the host range and cellular tropism of SARS-CoV-2 are dependent on the ACE2 expression profile. As the SARS-CoV-2 receptor, ACE2 is not only a gateway for SARS-CoV-2 entry into target host cells, but also a key to the understanding of SARS-CoV-2 pathogenesis.
5. Ethics statement
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. No ethical approval was required as this is a review article.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Plan for Research and Development of China [2016YFD0500300], National Natural Science Foundation of China [81871663 and 81672035], and Academic promotion programme of Shandong First Medical University [2019LJ001].
References
- 1.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet Microbiol. 2020;244:108693. doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W.-J., Ni Z.-Y., Hu Y.U., Liang W.-H., Ou C.-Q., He J.-X. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275(43):33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 9.Rossi G.P., Sanga V., Barton M. Potential harmful effects of discontinuing ACE-inhibitors and ARBs in COVID-19 patients. Elife. 2020;9:e57278. doi: 10.7554/eLife.57278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 12.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano T., Murakami M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity. 2020;52(5):731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguiar J.A., Tremblay B.J., Mansfield M.J., Woody O., Lobb B., Banerjee A. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J. 2020;56(3):2001123. doi: 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun. 2020;526(1):165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science:abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(281–292) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94(7):e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji W., Wang W., Zhao X., Zai J., Li X. Cross‐species transmission of the newly identified coronavirus 2019‐nCoV. J Med Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. J Med Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan J., Jin X., Lu Y., Zhang L. SARS‐CoV‐2 spike protein favors ACE2 from Bovidae and Cricetidae. J Med Virol. 2020;92(9):1649–1656. doi: 10.1002/jmv.25817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Hu G., Wang Y., Zhao X., Ji F., Ren W. Functional and Genetic Analysis of Viral Receptor ACE2 Orthologs Reveals Broad Potential Host Range of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.22.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. 2020 doi: 10.1101/2020.04.08.031807. [DOI] [Google Scholar]
- 27.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science:abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S., Luan J., Cui H., Zhang L. ACE2 isoform diversity predicts the host susceptibility of SARS-CoV-2. Transbound Emerg Dis. 2020 doi: 10.1111/tbed.13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan A., Benthin C., Zeno B., Albertson T.E., Boyd J., Christie J.D. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21(1):234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181(4):905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Wang H., Tang X., Ma D., Du C., Wang Y. Potential host range of multiple SARS-like coronaviruses and an improved ACE2-Fc variant that is potent against both SARS-CoV-2 and SARS-CoV-1. bioRxiv. 2020 doi: 10.1101/2020.04.10.032342. [DOI] [Google Scholar]
- 32.Lei C., Qian K., Li T., Zhang S., Fu W., Ding M. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun. 2020;11(1):2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renzi F., Ghersi D. ACE2 fragment as a decoy for novel SARS-Cov-2 virus. bioRxiv. 2020 doi: 10.1101/2020.04.06.028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. ACE2 polymorphisms and individual susceptibility to SARS-CoV-2 infection: insights from an in silico study. bioRxiv 2020.doi:10.1101/2020.04.23.057042.
- 35.Stawiski E., Diwanji D., Suryamohan K., Gupta R., Fellouse F., Sathirapongsasuti F. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. bioRxiv. 2020 doi: 10.1101/2020.04.07.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J., Ouyang W., Chua M.L.K., Xie C. SARS-CoV-2 Transmission in Patients With Cancer at a Tertiary Care Hospital in Wuhan, China. JAMA Oncol. 2020;6(7):1108. doi: 10.1001/jamaoncol.2020.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang W., Guan W., Chen R., Wang W., Li J., Xu K. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21(3):335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J Hematol Oncol. 2020;13:43. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q., Lu S., Li T., Yu L., Zhang Y., Zeng H. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J Exp Clin Cancer Res. 2019;38(1):173. doi: 10.1186/s13046-019-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L.u., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? The Lancet Respiratory Medicine. 2020;8(4):e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X.C., Zhang J., Zhuo J.L. The vasoprotective axes of the renin-angiotensin system: Physiological relevance and therapeutic implications in cardiovascular, hypertensive and kidney diseases. Pharmacol Res. 2017;125:21–38. doi: 10.1016/j.phrs.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heialy S.A., Hachim M., Senok A., Tayoun A.A., Hamoudi R., Alsheikh-Ali A. Regulation of angiotensin converting enzyme 2 (ACE2) in obesity: implications for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.04.17.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G., He X., Zhang L., Ran Q., Wang J., Xiong A. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J Autoimmun. 2020;112:102463. doi: 10.1016/j.jaut.2020.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clinical Immunology. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D., Jin M., Bao P., Zhao W., Zhang S. Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019. JAMA Netw Open. 2020;3(5):e208292. doi: 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shastri A., Wheat J., Agrawal S., Chaterjee N., Pradhan K., Goldfinger M. Delayed clearance of SARS-CoV2 in male compared to female patients: High ACE2 expression in testes suggests possible existence of gender-specific viral reservoirs. medRxiv. 2020 doi: 10.1101/2020.04.16.20060566. [DOI] [Google Scholar]
- 49.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10):e105114. doi: 10.15252/embj.20105114:e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sungnak W., Huang N.i., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi J., Zhou Y., Hua J., Zhang L., Bian J., Liu B. The scRNA-seq expression profiling of the receptor ACE2 and the cellular protease TMPRSS2 reveals human organs susceptible to COVID-19 infection. bioRxiv. 2020 doi: 10.1101/2020.04.16.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L., Niu Z., Jiang X., Zhang Z., Zheng Y., Wang Z. Systemic analysis of tissue cells potentially vulnerable to SARS-CoV-2 infection by the protein-proofed single-cell RNA profiling of ACE2, TMPRSS2 and Furin proteases. bioRxiv. 2020 doi: 10.1101/2020.04.06.028522. [DOI] [Google Scholar]
- 53.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science:abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526(1):135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziegler C.K., Allon S.J., Nyquist S.K. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Onabajo O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52(12):1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4570. doi: 10.1126/science:abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16(7):e9610. doi: 10.15252/msb:20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee I.T., Nakayama T., Wu C.-T., Goltsev Y., Jiang S., Gall P.A. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun. 2020;11(1):5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang A. Entry of Human Coronavirus NL63 into the Cell. J Virol. 2018;92(3):e01933–17. doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C.-L., Hasan S.S., Klose T., Sun Y., Buda G., Sun C., Klimstra W.B., Rossmann M.G. Cryo-EM structure of eastern equine encephalitis virus in complex with heparan sulfate analogues. Proc Natl Acad Sci USA. 2020;117(16):8890–8899. doi: 10.1073/pnas.1910670117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020;183:1043-1057 e1015. [DOI] [PMC free article] [PubMed]
- 65.Luan J., Lu Y., Gao S., Zhang L. A potential inhibitory role for integrin in the receptor targeting of SARS-CoV-2. J Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]