Abstract
Background: Well-differentiated gastroenteropancreatic neuroendocrine neoplasms are rare tumors with a slow proliferation. They are virtually resistant to many DNA-damaging therapeutic approaches, such as chemo- and external beam therapy, which might be overcome by DNA damage inhibition induced by proteasome inhibitors such as bortezomib. Methods and results: In this study, we assessed several combined treatment modalities in vitro and in vivo. By cell-based functional analyses, in a 3D in ovo and an orthotopic mouse model, we demonstrated sensitizing effects of bortezomib combined with cisplatin, radiation and peptide receptor radionuclide therapy (PRRT). By gene expression profiling and western blot, we explored the underlying mechanisms, which resulted in an impaired DNA damage repair. Therapy-induced DNA damage triggered extrinsic proapoptotic signaling as well as the induction of cell cycle arrest, leading to a decreased vital tumor volume and altered tissue composition shown by magnetic resonance imaging and F-18-FDG-PET in vivo, however with no significant additional benefit related to PRRT alone. Conclusions: We demonstrated that bortezomib has short-term sensitizing effects when combined with DNA damaging therapy by interfering with DNA repair in vitro and in ovo. Nevertheless, due to high tumor heterogeneity after PRRT in long-term observations, we were not able to prove a therapeutic advantage of bortezomib-combined PRRT in an in vivo mouse model.
Keywords: Bortezomib, Neuroendocrine tumors, Peptide receptor radionuclide therapy (PRRT), DNA repair, Combination therapy, Proteasome inhibition
Abbreviations: AP-sides, apurinic or apyrimidinic sites; BER, base excision repair; BRCA1/2, breast cancer protein 1/2; BRIP1, BRCA1 Interacting Protein C-Terminal Helicase 1; CAM, chorioallantoic membrane; CK2, casein kinase 2; d, day(s); DDR, DNA damage repair; DSB, double-strand breaks; DSMZ, Deutsche Sammlung für Mikroorganismen und Zellen; ER, endoplasmic reticulum; ERK, extracellular-signal-regulated kinase; F-18-FDG-PET, 2-deoxy-2-(F-18)fluoro-D-glucose positron emission tomography; FOXM1, forkheadbox protein M1; FOXO3a, forkheadbox protein O3a; g, gram; G0/1/2 phase, gap phase 0/1/2; G1/2/3, grade 1/2/3; Ga-68, Gallium-68; GEP-NEN, gastroenteropancreatic neuroendocrine neoplasms; GO, Gene Ontology; h, hour(s); H2AX, histone H2AX; HR, homologous recombination; HTCA, human tumor colony assay; i.p., intraperitoneal; i.v., intravenous; IC50, dose of half-maximum inhibitory effect; JC-1, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide; Lu-177, Lutetium-177; M, molar; M phase, mitosis phase; (M)Bq, (Mega-)Becquerel; MMR, mismatch repair; MRI, magnetic resonance imaging; n, number; NEN, neuroendocrine neoplasms; NER, nucleotide excision repair; NHEJ, non-homologous end joining; p70S6K, ribosomal protein S6 kinase; PARP1, poly(ADP-ribose) polymerase 1; PI3K, phosphoinositide 3-kinase; PRRT, peptide receptor radionuclide therapy; PTEN, phosphatase and tensin homolog; Rb, retinoblastoma protein; ROS, reactive oxygen species; RPA, replication protein A; (RT)-PCR, (real-time) polymerase chain reaction; S phase, synthesis phase; SCLC, small cell lung cancer; SKP2, S-Phase Kinase Associated Protein 2; SSB, single-strand breaks; SSTR2, somatostatin receptor 2; TGF, transforming growth factor; TLS, translesion synthesis
Introduction
Well-differentiated neuroendocrine neoplasms (NEN) of the pancreas and the intestine are rare tumors, characterized by slow proliferation and the absence of common driver mutations. Although mutations within the phosphoinositide 3-kinase (PI3K) pathway are only present in up to 14% of pancreatic NENs [1], primarily epigenetic dysregulation results in the loss of mTOR-associated tumor suppressors and renders the tumors accessible for mTOR inhibitors, such as everolimus [2,3]. Moreover, ATRX, DAXX or MEN1 mutations trigger aberrant chromatin structure and genetic instability, which impairs the function of other tumor suppressors, such as TP53, ATM or CDKN2A [4], [5], [6], [7].
Based on the NETTER-1 randomized clinical trial, peptide receptor radiotherapy (PRRT) using Lu-177-DOTATATE has been approved for somatostatin receptor 2 (SSTR2)-positive, nonresectable or metastatic, progressive, well-differentiated G1 and G2 NENs due to significantly increased progression-free survival (median time-to-progression was 36 months) [8], [9], [10]. The suggested mechanism is that after binding of the Lu-177 labeled ligand to SSTR, the emitted beta particles induce DNA single- and double-strand breaks (SSB and DSB) in SSTR2-positive tumor cells [11]. Accumulated SSBs and DSBs block transcription and/or DNA replication until repaired or finally induce cell death in the case of repair failure [12]. Although disease control rates are comparably satisfying and can improve patients’ quality of life, relatively low OR rates indicate mechanisms of baseline or acquired resistance to PRRT from a molecular-biological point of view.
Highly proliferative cancers frequently develop therapy-resistant tumor areas by selecting subclones, which are able to clear therapy-induced damage at an increased pace. In contrast, the slow proliferation of well-differentiated neoplasms per se provides extended time for DNA damage repair (DDR). The maintenance of DNA damage by inhibition of damage repair might therefore be a novel treatment approach in both, fast progressing cancers and slowly proliferating tumors such as well-differentiated pancreatic and intestinal NENs.
Accordingly, chemotherapeutic agents have limited application potential in well-differentiated NENs, where combined regimens containing streptozotocin (STZ) and 5-fluorouracil (5-FU), or capecitabine (CAP) and temozolomide (TEM) are deployed for the therapy of progressive or metastatic pancreatic NENs. Platinum-based regimens are only recommended for NEC and highly proliferative, progressive G3 tumors according to the current guidelines [13,14], yet with very limited effects in well-differentiated tumors [15]. Although there are canonical guidelines for the treatment of well-differentiated G1 and G2 NENs as well as poorly differentiated G3 NECs, there is no established therapy for G3 NETs so far. This produces the need to adapt regimens that are used for G2 NENs or G3 NECs (such as PRRT or Chemotherapy) to fit to the specific features of high proliferative and well-differentiated G3 NETs.
For in vitro experiments, the use of radiopharmaceuticals is limited due to radiation safety issues during long experimental hands-on times. To mimic PRRT in these experiments, we chose cisplatin and radiation, since their expected DNA damage response profiles were most likely to be comparable to those after Lu-177-DOTATATE. Whereas antimetabolites (such as 5-FU and its prodrug CAP) inhibit DNA and RNA metabolism by inhibition of thymidylate synthetase, or through incorporation into RNA and DNA, monofunctional alkylating agents (such as STZ and TEM) generate DNA adducts, and bifunctional platinum-based regimens (such as cisplatin) induce inter- and intrastrand DNA crosslinks. Thereby, alkylating and crosslinking drugs interfere with DNA replication by steric interference, which leads to inhibition of replication and DNA strand breaks, eventually requiring DNA repair mechanisms similar to those of PRRT repair.
In this study, we discuss the targeting of proteins that are involved in mechanisms, which repair cisplatin- and PRRT-induced DNA damage. This includes accumulation of intermediates, such as single-strand breaks (SSB). Accumulated double-strand breaks (DSB) and SSB finally induce apoptosis directly or by blocking transcription and/or DNA replication [12]. If cell death is not induced and cell cycle progresses, DNA repair defects lead to severe replication failure and chromosomal instability. Besides a major role in degradation of disposable proteins, the ubiquitin-proteasome-system is highly involved in the regulation of DNA damage response. On the one hand, the stabilization of wild type p53 toward its negative regulator MDM2 (which is frequently upregulated in pancreatic NENs [6,16]) might lead to a reactivation of “sensing” mechanisms and induction of a proapoptotic damage response in p53 wildtype neoplasms [7]. On the other hand, (poly-) ubiquitination strongly regulates the activity and specificity of a very high number of DNA-damage repair proteins and postreplicative bypass mechanisms [17], [18], [19]. Furthermore, ubiquitination modulates histone conditions and thereby controls the accessibility of DNA damage loci to DNA repair protein complexes [19].
The proteasome inhibitor bortezomib has been shown to inhibit advanced stages of DNA-damage repair while maintaining early stage damage response mechanisms, such as phospho-H2AX and replication protein A (RPA) foci accumulation. The preservation of early steps in the context of damage repair inhibition might be essential for the induction of controlled cell death rather than uncontrolled events [20]. Bortezomib has further demonstrated chemotherapy-sensitizing effects in several cancers in preclinical settings, where we were able to show an increased antitumoral effect in cisplatin plus bortezomib-treated small cell lung cancer in vivo [21,22]. Nevertheless, bortezomib has been withdrawn from clinical assessment as monotherapy after a very small clinical phase II study including 14 patients with mixed neuroendocrine tumor entities due to failure of objective therapy response [23] in 2004. Thus, its potential in combined drug therapy of GEP-NENs has not been analyzed so far. In the study presented herein, we analyzed the effect of the proteasome inhibitor bortezomib in preventing the repair of cisplatin-, radiation- and Lutetium-177-induced DNA-damage in pancreatic and intestinal neuroendocrine tumors in different preclinical models. Thereby, we explored clinically relevant and potential agents with comparable damage signatures for combined therapy.
Material and Methods
Cell lines
The following GEP-NEN cell lines were used for in vitro experiments: pancreatic: BON [24] and QGP-1 [25] (obtained from Japanese Collection of Research Bioresources), ileal: KRJ-I [26] and colonic: LCC-18 [27]. BON cells were a generous gift from CM Townsend (University of Texas, Galveston). KRJ-I midgut NET cells were generated by R. Pfragner (Medical University of Graz) and kindly provided by I. Modlin (Yale University, New Haven). The neuroendocrine origin and phenotype of KRJ-I cells has recently been questioned [28]. However, these cells have been shown to share a number of relevant properties with other GEP-NEN cell lines [29,30].
Cells were cultured and handled as previously published [31]. All cell lines were authenticated (if indicated as unique) by genetic STR typing at the DSMZ, Braunschweig, Germany in 2012, 2013 and 2015 (in vitro experiments were finished in 2017). Cells were not passaged longer than 20 passages after receipt. Cells were further tested periodically for maintained cell line specific expression of neuroendocrine and differentiation markers (chromogranin A, synaptophysin, cytokeratin, vimentin, syntaxin) by immunofluorescence microscopy.
Immunocytochemistry and establishment of SSTR2-positive BON cells
Native BON cells showed a low SSTR2 expression, which was verified by immunofluorescence microscopy using a SSTR2-antibody (Supplementary material and methods) and a standard immunocytochemistry protocol as previously published [16].
BON cells were transfected with human SSTR2 to generate BON-SSTR2+, a stably expressing SSTR2-positive BON cell line [32]. Briefly, BON-SSTR2+ cells were produced by transfecting wild type BON cells using the plasmid pcDNA3.1-huSSTR2 (#SSTR200000, cDNA Resource Center, Bloomsberg, PA, USA; www.cdna.org) and jetPEI transfection reagent (Polyplus-transfection, Illkirch, France). Positive clones were selected by addition of 600 µg/mL G-418 to the media and were tested for their SSTR2 expression by qPCR [32], immunofluorescence and radioligand binding assay.
Lu-177-DOTATOC radioligand binding assay
Cellular uptake of Lu-177-DOTATOC by native BON and BON-SSTR2+ cells was measured as described previously [32]. Briefly, cells were seeded in 24-well plates (2.5 × 105 cells/well). Radioligand binding assay including the washing steps, were performed in Hanks’ balanced salt solution (HBSS, Invitrogen), containing 10 mM HEPES, 0.5% BSA at pH 7.3. The HBSS incubation buffer further contained approx. 0.0074 MBq/mL Lu-177-DOTATOC. After one hour of incubation with HBSS/Lu-177-DOTATOC at 37 °C, wells were rapidly washed with ice-cold washing buffer (50 mM Tris-HCl pH 7.4, 125 mM NaCl, 0.05% BSA), before lysis with 1 N NaOH. Cell lysate measurements (quintuplicates) were performed in γ-counter (Wallac 1470 Wizard, Perkin Elmer, USA)
Western blot
SDS page and western blot of NP-40 lysed primary tumor or cell line material (cells were synchronized in 0.01% FBS for 24 h before treatment) was performed using a standard protocol and documented by ponceau S staining. Primary antibodies are listed in Supplementary material and methods. Secondary antibodies were obtained from Dako (swine anti rabbit IgG-HRP and goat anti-mouse IgG-HRP; Agilent, Santa Clara, USA). Antibody binding was documented by Fujifilm LAS-4000 luminescent image analyzer using ECL prime Western Blotting detection reagent (Amersham GE healthcare). For reprobing, membranes were treated with acidic glycine buffer as published [31]. Chemiluminescence signals were densitometrically analyzed with Multi Gauge V3.1 or Image J 1.53. Values of ≥3 independent experiments were normalized to internal controls and mean values were statistically assessed by using IBM SPSS Statistics v22.0.0.0 (based on Python 2.7) or Graphpad Prism 8.4.1.
Substances and radiopharmaceuticals
Bortezomib (velcade) and cisplatin were obtained in physiological saline from the Charité Universitätsmedizin Berlin dispensary. DOTA-D-Phe1-Tyr3-octreotide (Lu-177-DOTATOC) was obtained from ABX advanced biochemical compounds GmbH, Germany. Lutetium-177 was purchased from ITG Isotope Technologies (Garching, Germany) and synthesized to Lu-177-DOTATOC in our Laboratories at the Berlin Experimental Radionuclide Imaging Center (BERIC).
Viability assay
WST-1 (Roche; Basel, Switzerland) or AlamarBlue (Thermo Fisher, Waltham, USA) assays were performed according the manufacturer's instructions in quintuplets after treatment with decreasing concentrations of the substances or their combination. Signal density was colorimetrically quantified with a multi-well spectrophotometer (TECAN sunrise) and analyzed with MS Excel 2013 and GraphPad Prism 7.
Combined concentration-response curves were analyzed as recommended by the Chou-Talalay method [33]. Data was analyzed by use of Prism 7 and CompuSyn 1.0 [34].
Human tumor clonogenic assay
Sterile methylcellulose solution (MC) was prepared as follows: 4% MC was dissolved in 50:50 sterile water/Iscove´s MDM. 0.06% 2-Mercaptoethanol (BME) solution in sterile PBS and 3% (w/v) agar solution in sterile water were prepared. Prewarmed agar-solution was diluted 1:3680 in cell culture medium. Cells were harvested, counted and diluted to 30,000 cells/ mL. MC solution was prewarmed and working solution was prepared including 57% MC (4%) solution, 43% FBS, 0.001% BME (0.06%) solution, 0.01% cell culture medium, 0.005% cell suspension. Working solution was mixed with diluted agar solution and 1 mL/disc was transferred in Human tumor clonogenic assay (HTCA) disks and incubated in the cell incubator. Colonies were counted and data was analyzed by use of GraphPad Prism 7 software.
Radiation
KRJ-I and BON cells were irradiated with 0, 5, or 10 Gy and treated with increasing concentrations of bortezomib (Bon: 1–50 nM, KRJ-I 0.1–20 nM) for 48 h. Viable KRJ-I (floating) cells were quantitated by alamarBlue Cell Viability Assay by Thermo Fisher Scientific (Waltham, USA) according to the manufacturer's instructions. BON (adherent) cells were counted after DAPI staining. Data was analyzed using GraphPad Prism 7 software.
Combined concentration-response curves were studied as recommended by the Chou-Talalay method [33]. Data was analyzed by use of CompuSyn 1.0 [34].
In vivo models
Chicken chorioallantoic membrane xenografts [35]
Fertilized pathogen free chicken eggs (VALO Biomedia, Cuxhafen, Germany) were maintained as described earlier [36].
On day 7 of embryonic development, BON-SSTR2+ cells (1 × 107 cells in an equal volume of RPMI1640 [Invitrogen, Germany] and matrigel [basement membrane matrix high concentration from Corning, USA]) were grafted onto the chorioallantoic membrane (CAM) and maintained for 5 d to allow angiogenic connection of the tumors to the CAM and degradation of matrigel [37]. For treatment, CAMs were either injected intravenously with approximately 20 MBq Lu-177-DOTATOC or tumors were treated by dropping 80 µL bortezomib (25 nM) or NaCl (0.9%, negative control) onto microtumors. A second dose of 25 nM bortezomib was applied to the bortezomib only and combined treatment tumors on day 14. On day 18, tumor plaques were explanted and analyzed by Western blot. For details on multimodal imaging of the in ovo model see Supplemental materials & methods.
Final concentrations of bortezomib were calculated by a dose escalation study (data not shown) with respect to the toxicity of bortezomib to the chicken fetus assuming a blood volume of approximately 1.2 mL on the day of treatment [38].
Orthotopic pancreatic NET mouse model and multimodal imaging
For the development of an orthotopic, pancreatic NET xenograft mouse model, BON-SSTR2+ cells were implanted into the pancreas of 12-wk-old SCID mice as previously described by Aristizabal Prada et al. [39].
Tumor growth was monitored once a week by a preclinical 1T PET/MRI (nanoScan PET/MRI, Mediso, Hungary) using a mouse whole body coil. For tumor delineation and volumetry, a high-resolution T2w 2D fast spin echo (FSE) sequence was established.
When the tumor size reached ∼1000 mm3, the metabolic activity and tumor heterogeneity of the engrafted tumors was determined by simultaneous F-18-FDG PET/MRI (approximately 15 MBq injected i.v.). Animals were scanned after 30 min to ensure tracer accumulation in the tumor tissue.
When tumors reached a size of ∼1400 mm3, the experiment was terminated. Animal experiments were performed in accordance with the national and local guidelines for animal welfare and approved by the ethics committee of the state Berlin (G0011/16).
For detailed imaging parameters, please refer to Supplementary material and methods.
Data analysis assessing tumor heterogeneity
The viable tumor area was determined as previously described [40], using PMOD software version 3.505 (PMOD Technologies, Zürich, Switzerland). Briefly, a whole tumor region of interest (ROI) was placed onto the T2w images and copied to fused PET/MRI images. Using a threshold of 30% of the hottest voxel within the ROI, FDG-avid tissue was automatically delineated within the tumor. We defined viable tissue as the FDG-avid fraction of the whole tumor. T2 images were used to analyze the amount of necrotic tissue within the tumors. A custom Matlab script (Mathworks v15.0, USA) was used to identify the necrotic proportionate from viable/undefined tissue based on SI per voxel. A ROI was manually drawn to delineate the tumor from other tissues. A second ROI was drawn in a region of tumor to annotate viable tissue SI (vSI) as a reference value for the necrotic tissue. The standard deviation of the vSI was calculated and used to determine the necrotic tissue, where voxel values of 3 standard deviations lower (outside the 99.7% confidence interval) than the vSI were deemed to be necrotic [41]. The amount of voxels containing necrotic tissue are represented as a percentage of the whole tumor.
Treatment regime
Mice were treated according to the following treatment schedule: 0.5 mg/kg bortezomib i.p. on days 0, 3, 7, and 10 in the first treatment cycle and 0.3 mg/kg bortezomib on days 21, 24, 28, and 31 in the second cycle. The combined treatment group received additional 30 MBq of Lu-177-DOTATOC i.v. on days 0 and 21. The PRRT only treatment group received 30 MBq of Lu-177- DOTATOC on days 0 and 21 as previously published [42]. Vehicle control was sterile saline. Animals were observed for maximum 15 weeks after start of treatment or euthanized at a tumor volume of 1400 mm3. MRI of tumor size was performed once per week.
Immunohistochemistry
Paraffin sections (1–2 µm) of formalin-fixed tissues were prepared from 2 different tumor areas, dewaxed and subjected to heat-induced epitope retrieval prior to incubation with anti-cleaved PARP1 (clone E51, Abcam), anti-53BP1 (polyclonal rabbit, Novusbio) or anti-Ki67 (clone MIB1, Agilent). For detection, Dako REAL EnVision Detection System or for detection of MIB1, alkaline phosphatase-labeled streptavidin and chromogen RED (both Agilent) were employed. Nuclei were counterstained with hematoxylin (Merck) and sections were coverslipped with glycerol gelatin (Merck).
Images were acquired using the AxioImager Z1 microscope (Carl Zeiss MicroImaging, Inc.). All evaluations were performed in a blinded manner.
HIF1-alpha, cPARP, and 53BP1 immunohistochemistry was quantitated by a 12-point immunoreactivity scoring (IRS) [43] integrating the product of the expression level (0 = no; 1 = low; 2 = intermediate; 3 = strong) with percentage of positive cells (0 = no; 1 = <10%; 2 = 10%–50%; 3 = 51%–80%; 4 = >80%). Necrosis and Ki-67 were displayed as percentage of cells.
Cell cycle analysis
For mitotic index flow cytometry, cells were treated with 50 nM bortezomib, 10 µM cisplatin or their combination for 24 h and stained following a previously published protocol [31]. For JC-1 staining, cells were harvested after treatment with 50 nM bortezomib, 10 µM cisplatin or 50 nM bortezomib + 10 µM cisplatin and suspended in PBS. Positive controls were incubated 1:500 with the generic mitochondrial membrane depolarizer Carbonylcyanide m-chlorophenylhydrazone (CCCP) for 5 min. All samples (negative controls excluded) were incubated with 1:500 JC-1 and incubated 30 min at 37 °C. Flow cytometry was conducted with FACSCalibur (Becton Dickinson) by BD Cell Quest Pro software and analyzed with FlowJo 8.7 software, Prism 7 and IBM SPSS Statistics 22.
nCounter multiplex gene expression analysis
BON cells were treated in triplets with 25 nM bortezomib, 10 µM cisplatin or their combination versus DMSO for 24 h or transfected with siRNA as explained below. Messenger RNA (mRNA) was isolated with RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA was measured using nanodrop (Thermo Fisher Scientific). 60 ng RNA was analyzed using the PanCancer pathway panel kit (NanoString Technologies, Seattle, WA, USA) according to the manufacturers instruction [44,45] and nSolver v2.5 (based on R v3.1.1). The differentially expressed (DE) genes of the gene expression analysis were analyzed with nSolver v2.5 software (NanoString Technologies) using first principal component analysis and regression analysis with and without Benjamini-Yekutieli procedure. Raw data was normalized to the following housekeeping genes: AGK, DDX50, EIF2B4, ZC3H14, CNOT10, MRPS5, PRPF38A, NUBP1, AMMECR1L, PIAS1, HDAC3, ACAD9, EDC3, RBM45, NOL7, USP39, COG7, ZNF384, SF3A3, VPS33B, SAP130, PIK3R4, TLK2, SLC4A1AP, ZKSCAN5, ZNF346, MTMR14, ERCC3, CNOT4, TMUB2, C10orf76.
Pathway visualization was done with the nSolver software based on the KEGG ontology pathway system [46], [47], [48].
To compare bortezomib treated cells with cells under FOXM1 knockdown, we performed RNA interference for 72 h as described below. Knockdown efficiency was determined as 93% to 94% FOXM1 reduction. Messenger-RNA of FOXM1 knockdown and control cells was prepared and analyzed as described above. Raw data was normalized to the following housekeepers: CNOT10, SF3A3, DDX50, USP39, PIAS1, SLC4A1AP, FCF1, NOL7, SAP130, HDAC3, EIF2B4, ZC3H14, ERCC3, PRPF38A, EDC3, ZNF384, TMUB2, ACAD9, AMMECR1L, VPS33B, TLK2, C10orf76, CNOT4, AGK, PIK3R4, MTMR14, ZNF143, MRPS5, TRIM39, ZKSCAN5, DNAJC14.
Matching differentially expressed genes were identified. A gene pathway overrepresentation analysis was performed with the http://pantherdb.org/ database using the PANTHER Overrepresentation Test (release 20,170,413, reference list: homo sapiens) based on the “panther pathway” annotation data set (PANTHER version 12.0 Released 2017–07–10) with Bonferroni correction for multiple testing [49]. Gene enrichment was estimated by use of the ENRICHR platform [50,51].
RNA interference
Cells were transfected with 40 pmol/mL siRNA in appropriate amounts of Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions for 72 h. An endoribonuclease-prepared heterogeneous siRNA pool was used in order to enhance specificity and reduce off-target effects: enhanced siRNA (esiRNA) against FOXM1 by Sigma (EHU124431: NCBI reference sequences: NM_021953, NM_202002, NM_202003) or negative control: Sigma Mission siRNA Universal negative control #1. Knockdown efficiency was measured by qPCR.
Real time qPCR
RNA was isolated by use of the Qiagen RNeasy RNA isolation kit, quantified with Nanodrop 2000 and DNA digesting was carried out with 0.1 µL/100 ng RNA DNase I peqGOLD (Peqlab, VWR International GmbH, Erlangen, Germany). 1000 ng RNA was transcribed into cDNA using the High-Capacity RNA-to-cDNA Kit according to the manufacturer's instructions (Thermo Fisher, Carlsbad CA, USA). Real time quantitative (q) PCR was performed with 25 ng cDNA with the StepOne Real time PCR System (Thermo Fisher) and analyzed with StepOne v2.3 Software and MS Excel using the ΔΔCt-Method. Figure was prepared with Prism 7 software. The PCR primers were used as follows: FOXM1 forward 5′GGA GCA GCG ACA GGT TAA GG 3′, FOXM1 reverse 5′GTT GAT GGC GAA TTG TAT CAT GG 3′; GUS (Housekeeper) forward 5′GAA AAT ATG TGG TTG GAG AGC TCA TT 3′, GUS reverse 5′ CCG AGT GAA GAT CCC CTT TTT A 3′ (all Metabion).
Statistical analyses
Data (excluding the Nanostring gene expression analysis) was analyzed using IBM SPSS Statistics v22.0.0.0 (based on Python 2.7), Prism 7 and 8 (v7.04 and v8.4.1; GraphPad Software, San Diego, USA). Unpaired Students t test was used for 2 groups and one-way analysis of variance (ANOVA) or 2-way ANOVA for multiple groups of normally distributed data, respectively. Kruskal-Wallis Test was applied for multiple group data with rejected assumption of normality. Dunn's or Sidak's correction was used for correction of multiple testing. The choice was dependent on the structure of the data (number of groups, comparison with one or multiple groups), low numbers of samples (<5) were always analyzed with non-parametric tests, parametric tests were chosen after testing for normality in data with n≥5. Normality was assumed after Shapiro-Wilk normality test. All differences were considered to be significant with alpha = 0.05.
Results
Bortezomib has antiproliferative effects GEP-NEN and sensitizes to cisplatin therapy by induction of G2/M arrest and death receptor-driven apoptosis in GEP-NEN cell lines
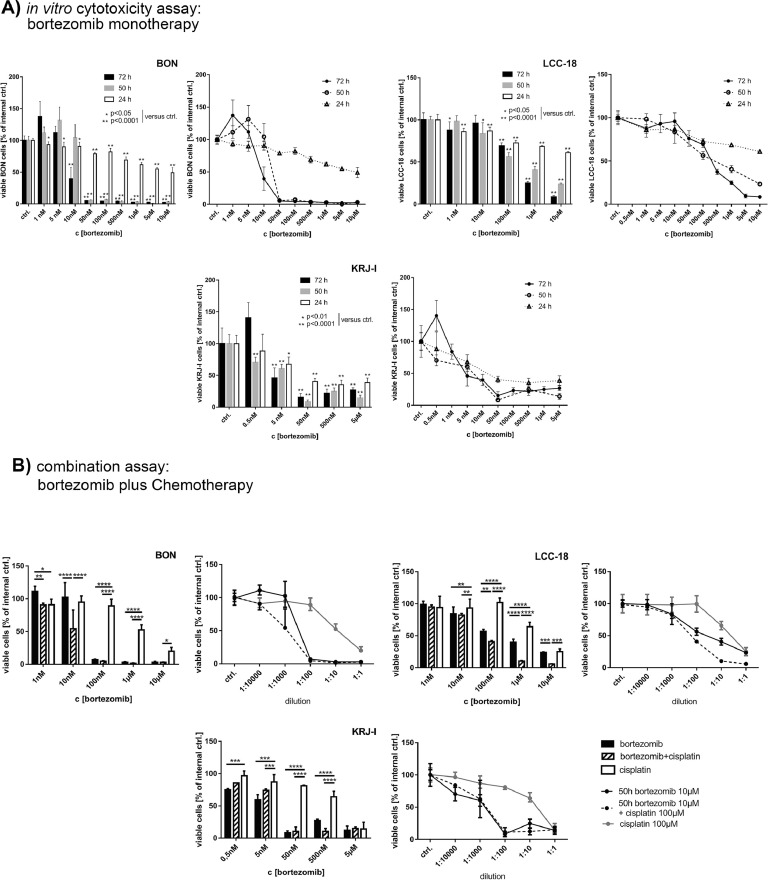
To analyze the effects of bortezomib monotherapy, we treated the cell lines BON, LCC-18 and KRJ-I with increasing concentrations of bortezomib for 24, 50, and 72 h and analyzed the cell viability. All cell lines responded significantly in a time- and concentration-dependent manner by the reduction of cell viability. The IC50 values for 72 h of treatment ranged between 4.7 nM in KRJ-I and 270 nM in LCC-18 cells (Figure 1).
Figure 1.
Therapeutic effect of bortezomib in vitro: (A) BON, LCC-18 and KRJ-I cell lines were treated with increasing concentrations of bortezomib for 24 h, 50 h, and 72 h, respectively, and cell viability was measured by WST-1 proliferation assay. Dose-response curve and histogram of the same data (representative data out of three independent experiments is shown, bar represents means and SD of n = 5 replicates) is shown for a more concise presentation. All cell lines showed a significant antiproliferative response to the treatment (one-way ANOVA: P< 0.0001; and Holm-Sidak's multiple comparisons test; normality was assumed after both, Shapiro-Wilk and Kolmogorov-Smirnov normality test). Dose of relative half-maximal inhibitory effect (IC50) was determined by non-linear regression using Prism 7. Captions: *P< 0.05; **P< 0.0001. (B) In vitro combinatory effects of cisplatin and bortezomib after 50 h of treatment: cell lines were treated with a combination of bortezomib and cisplatin 1:10 (starting from 100 or 50 µM cisplatin + 10 or 5 µM bortezomib, respectively, and subsequent dilutions) versus the corresponding single agents (representative experiment of three independent experiments, bar shows median and interquartile range of n = 5 replicates). Dose response curves were analyzed according to the method of Chou and Talalay. BON cells responded in all measured dilutions (excluding the starting concentrations) with synergistic effects. In LCC-18 cells, bortezomib induced synergism in doses >10 nM, which is lower than the respective IC50. KRJ-I cells, which showed very strong effects even at low concentrations of bortezomib alone, exhibited a very strong response, however, synergism only substantiated for intermediate doses (500 nM cisplatin + 50 nM bortezomib and 5 µM cisplatin + 500 nM bortezomib) (2-way ANOVA with Sidak's multiple comparisons test). *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
In order to study the chemosensitizing effects in neuroendocrine tumor cell lines, we chose cisplatin due to its low monotherapeutic effect in the cell lines. Subsequently, we performed viability studies after 24 h (not shown) and 50 h of combined treatment versus monotherapy control. Here, bortezomib exhibited additive and synergistic effects combined with cisplatin, leading to further decreases in IC50-values (Table 1). In KRJ-I, representing the only p53 wild type cell line in this study, bortezomib alone induced a very strong effect (including the lowest IC50 values of all tested cell lines), which was not further enhanced by cisplatin in the majority of the experimental settings. As demonstrated for BON cells, the antiproliferative effect was proven by HTCA. Here, bortezomib significantly reduced the cell's capacity to form new colonies and induced strong morphological changes (Supplementary Fig. 1).
Table 1.
IC50 values after cisplatin, bortezomib and combined treatment of cell lines in vitro: 95% confidence intervals (CI) of IC50 values after 50 h of treatment. For combined treatment with a fix ratio of bortezomib to cisplatin of 1:10, the respective IC50 values of the bortezomib content is shown.
| Cell Line | 95% CI (IC50 Cisplatin) | 95 % CI (IC50 Bortezomib) | 95% CI (IC50 Combination, Given c[Bor]) |
|---|---|---|---|
| BON | 4.960 to 11.54 µM | 0.01963 to 0.05009 µM | 0.01148 to 0.01921 µM |
| LCC-18 | 9.056 to 31.38 µM | 0.03125 to 0.1359 µM | 0.05334 to 0.07376 µM |
| KRJ-I | 2.386 to 6.931 µM | 0.003470 to 0.008452 µM | 0.001668 to 0.006319 µM |
While cisplatin induced S-phase arrest in accordance with the necessity to repair DNA crosslinks before entering the DNA replication step [52], bortezomib induced G2 and G2/M arrest respectively, indicating that crucial G2/M checkpoint mediators were affected by the treatment. The combination of both substances significantly induced apoptosis in both cell lines with an earlier induction in KRJ-I cells (Supplementary Figure 1).
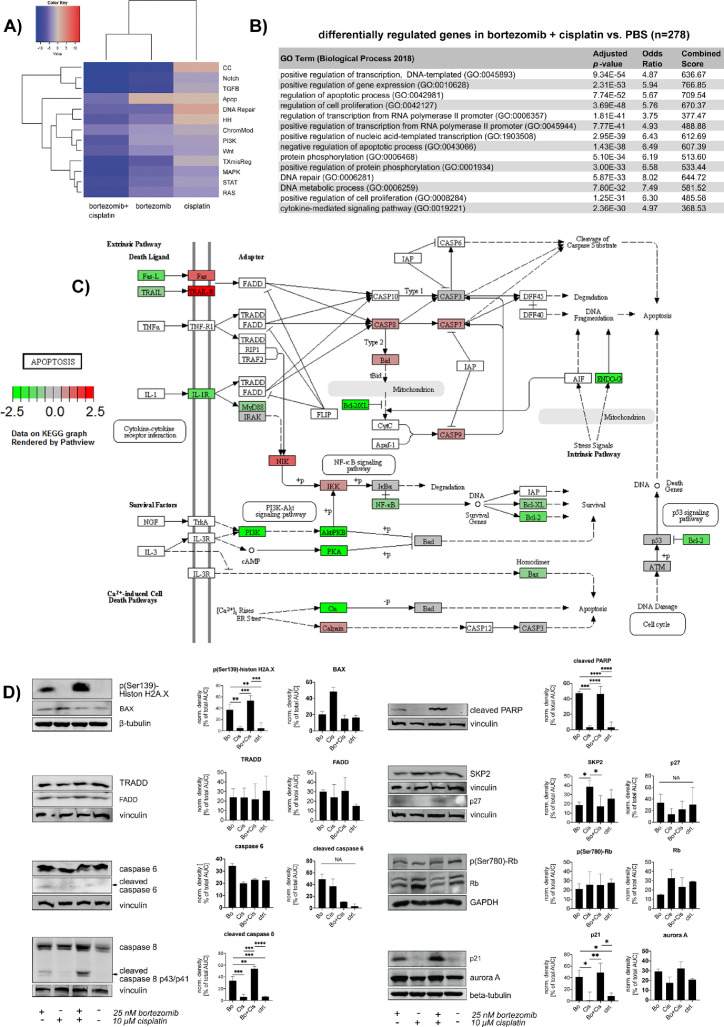
To clarify the underlying signal transduction, we analyzed BON cells after 24 h of combined treatment by targeted mRNA profiling on the Nanostring nCounter platform [45]. The affected signaling pathways (Figure 2) showed only slight changes upon cisplatin only treatment, including an upregulation of DDR and cell cycle-associated genes. In contrast, bortezomib alone and the combined treatment exhibited large effects, especially on cell cycle regulatory and transcription-associated genes (Figure 2A–C). The pathway-allocated gene groups include both, positive and negative regulatory genes. Here, the apoptotic signaling varied from an upregulation of primarily proapoptotic genes upon bortezomib monotherapy to overall downregulation of anti-apoptotic genes in the combined treatment arm, resulting in an overall proapoptotic signaling in either case. Furthermore, transcripts of the death receptor-related apoptotic pathways (encoding caspase 7 and 8, tumor necrosis factor receptors, I-kappaB kinase subunit gamma, calpain, BID and FAS) were overrepresented after combined treatment (Figure 2B). Here, as proven by western blot, the strongly increased DNA damage, indicated by histone H2AX phosphorylation, did not increase the expression of BAX as mediator of intrinsic apoptotic processes (Figure 2D). In contrast, cisplatin treatment alone induced a slight, but not significant, BAX upregulation. The combination of bortezomib plus cisplatin rather induced the expression of death receptors (Fas and TRAIL-receptors). The signals of Fas and TRAIL receptors are canonically transduced by Fas/TNF-receptor-associated proteins with death domain (FADD/TRADD) and cleavage of caspase 8, which in turn cleaves BID and activates effector caspases, which eventually results in apoptotic events such as in the cleavage of Poly- (ADP-ribose)- polymerase (PARP) in the nuclei [53]. The western blot only confirmed a slight upregulation of FADD (not significant), but verified the significantly increase in caspase and PARP cleavage. The additional cleavage of BID could be only demonstrated for KRJ-I, QGP-1 and LCC-18 cells (Supplementary Figure 2)
Figure 2.
Gene expression after bortezomib, cisplatin and combinatory therapy demonstrated induction of apoptosis: BON cells were treated with 25 nM bortezomib, 10 µM cisplatin or both versus saline for 24 h and analyzed by nCounter gene expression array. (A) Gene set analyses of affected pathways after cisplatin, bortezomib and combination related to control. The directed analysis heatmap displays each treatment's directed global significance scores. Directed global significance statistics measure the extent to which a pathway's genes are up- or downregulated after treatment. Red denotes pathways whose genes exhibit extensive over-expression upon treatment, blue denotes pathways with extensive under-expression. Cisplatin upregulated genes associated with positive regulation of DNA repair and cell cycle regulation. After combined treatment, genes of all analyzed pathways were found predominantly downregulated. The detailed analysis of apoptosis-related gene expression, color-coded as upregulated in the bortezomib arm and downregulated in the combination arm, revealed that the bortezomib treatment affected three genes (ENOG, BCL2, and NFKB1) in a negative way and 5 genes (CASP7, BID, IKBK, CAPN2, and MAP3K14) in a positive manner. The combination of bortezomib and cisplatin changed the expression of eleven genes (ENDOG, PRKAR1B, BCL2, NFKB1, FASLG, PIK3CB, PPP3CB, PIK3R2, IL1RAP, PPP3CA and AKT2) negatively and upregulated nine genes (CASP8, CAPN2, CASP7, BID, IKBKG, TNFRSF10A, MAP3K14, TNFRSF10D, und FAS). Although the resulting score would lead to the assumption that bortezomib regulates apoptosis in a more positive way than the combined treatment, the combination affected more antiapoptotic genes, including BCL2, AKT2, subunits of NF-kappaB, PI3K, PKA and Bcl-2 (to a lower extent), in a negative way. Therefore, the signaling changes result in a proapoptotic signaling. Significantly differentially expressed genes have been filtered by minimum fold change ≥1.5-fold before detailed analysis. Gene expression raw data can be found in Supplementary Tables 1-3. (B) Pathway analysis of apoptosis-related genes after bortezomib plus cisplatin treatment. Visualization by KEGG ontology pathway system. Although the overall pathway score for apoptosis-related genes was determined negative, the differential analysis of the distinct transcripts demonstrated pro-apoptotic mechanisms: Genes of pro-apoptotic death receptor-mediated pathways, such as of Fas, TRAIL-R, caspases 7 and 8, BID and NIK were upregulated (red), whereas predominantly crucial antiapoptotic genes, including those of Bcl-2, NF-kappaB and genes of the PI3K pathway, were downregulated (green). This resulted in an overall negative pathway score. (C) GO enrichment analysis using the ENRICHR data analysis tool verified a strong modulation of gene expression affecting apoptotic processes. (D) For verification of the gene expression profiling, BON cells were treated with 25 nM bortezomib, 10 µM cisplatin, combination of both or control for 24 h and analyzed by western blot. Bortezomib and combined therapy induced massive DNA damage (DNA double stand breaks indicated by H2AX phosphorylation), but did not induce internal apoptosis via BAX. Both treatment arms presented death receptor-dependent extrinsic apoptotic signaling as detected by FADD upregulation and cleavage of caspase 8, caspase 6 and finally PARP. The relative effects were stronger in the cells that were treated with the combination of both, cisplatin and bortezomib. Expression of the p27 regulator and DNA repair associated protein SKP2 was reduced after bortezomib and combined therapy in association with an upregulation of the p27 cell cycle regulator. Independent from Rb protein abundance (which is repressed upon bortezomib and combined treatment), p21 is upregulated following bortezomib or combined treatment. CK2-alpha and aurora A were also upregulated upon bortezomib and the combination. Western Blot data was densitometrically quantified by use of ImageJ and statistically evaluated with Prism 8. Normal distribution of the data was assumed after passing Shapiro-Wilk test and according to the results, data was tested by Kruskal-Wallis or one-way ANOVA analysis. Graphs show mean with SD. Apop, apoptosis; CC, cell cycle; ChromMod, chromatin modifications; HH, hedgehog; TXmisReg, transcriptional misregulation; Bo, bortezomib; Cis, cisplatin, ctrl, control; NA, not applicable (due to too little contrast of the signal and error-prone densitometric measurements); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (Color version of figure is available online.)
The cell cycle-related effects of bortezomib were further explored by western blot (Figure 2D). Here, we found a significant downregulation of S-phase Kinase Associated Protein 2 (SKP2). The FOXM1 target SKP2 is a subunit of the SCF E3 ubiquitin-protein ligase complex and a crucial cell cycle regulator, which recognizes phosphorylated p27 and is involved in regulation of G1/S transition [54]. Loss of p27 and overexpression of FOXM1 have been described as frequent events in GEP-NEN before [31,55]. Following slightly reduced SKP2 expression, we demonstrated an upregulation of p27 and p21, independent from Rb (which can transactivate p21 expression [56] and inhibit SKP2-mediated p27 ubiquitination [57]) and without effect on Rb Serin-780 phosphorylation. In particular, the phosphorylation of Rb did not change after bortezomib (and combined) treatment, whereas the total protein abundance showed a tendency to decrease (although not significantly), which could be due to the enrichment of G2/M arrested cells (as demonstrated in Supplementary Figure 1). Accordingly, the M-phase protein aurora kinase A (aurora A) was slightly more abundant in the bortezomib or combination treated cells without reaching significance. Comparable results can be shown for other GEP-NEN cell lines and upon bortezomib-combined radiation therapy (Supplementary Figs. 2 and 3).
Bortezomib shows sensitizing effects to PRRT in a chicken egg-based in vivo model
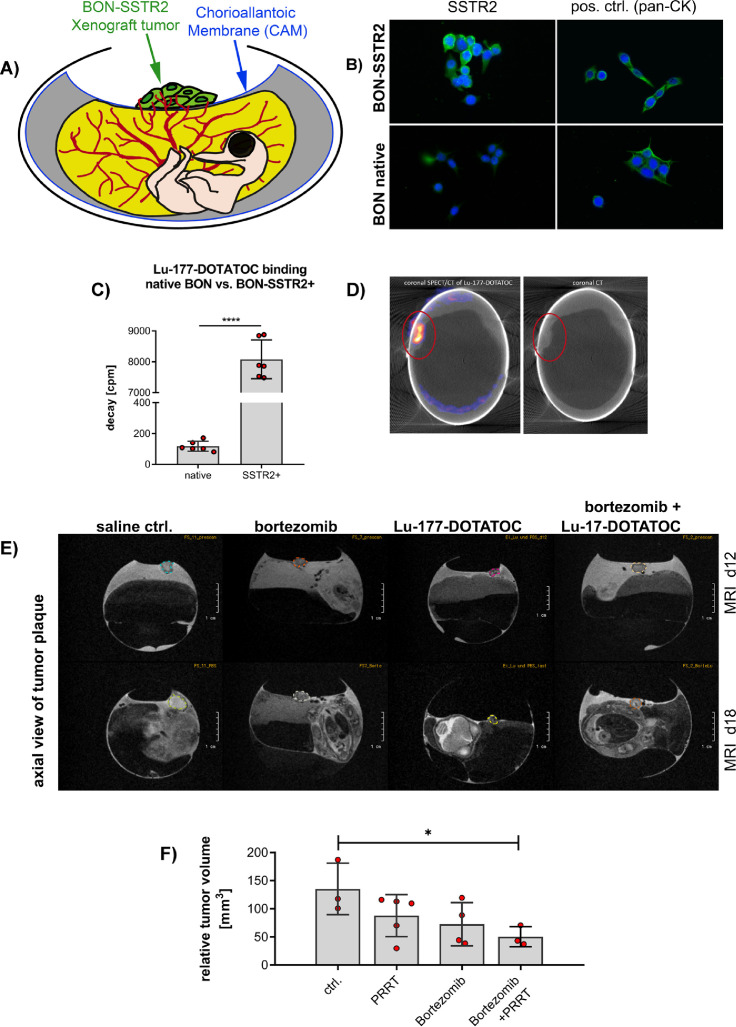
To adapt to a more clinically relevant pancreatic NEN model by exploiting PRRT-induced signaling changes as therapeutic targets, we established a short-term preclinical in ovo model by implanting a pancreatic, SSTR2-positive cell line-derived 3D spheroid, sensitive to Lu-177-DOTATOC PRRT, onto the chorioallantoic membrane (CAM) of fertilized chicken eggs (Figure 3A). Since native BON cells lack functional SSTR2 expression, we used a human SSTR2-transfected BON cell line [32]. Those BON-SSTR2+ were positive for anti-SSTR2 immunofluorescent staining and showed a ∼60-fold higher Lu-177-DOTATOC binding compared to native BONs (Figure 3B, C).
Figure 3.
In vivo analysis of protein expression after bortezomib-combined peptide receptor radionuclide therapy (PRRT) in the chicken chorioallantoic membrane (CAM) model: (A) Schematic depiction of the SSTR2+ chicken CAM model: cells were mixed with matrigel to form a tumor plaque and inoculated on the CAM of a fertilized, chicken egg on d6 of embryonal development. (B-D) Generation of SSTR2-positive NEN cell line and analysis for receptor functionality in vitro and in vivo: (B) BON cells were transfected with human somatostatin receptor 2 (SSTR2). Immunofluorescent staining of SSTR2 of native and SSTR2+- BON cells demonstrated a low native expression of SSTR2 in native BON cells und a positive membrane staining in the genetically modified cell line. Green: SSTR2 or pan-cytokeratin control; blue: DAPI. (C) Quantification of the functional SSTR2 expression using ligand binding assay: BON-SSTR2+ cells ∼60-fold higher than in native BON cells (P < 0.0001; unpaired t test; n = 6). BON and BON-SSTR2+ cells were incubated in vitro with 0.0074 MBq/mL Lu-177-DOTATOC. After washing and cell lysis, the radiopharmaceutical uptake was measured using a gamma counter. Graph (bars indicate mean with SD) shows representative results of 3 independent experiments. (D) SPECT/CT of CAM model: tumor plaque was imaged 3 h after i.v. injection of 20 MBq Lu-177-DOTATOC showing a 1.6% uptake of the total injected dose. Representative image, left side: coronal SPECT/CT of Lu-177-DOTATOC, right side: coronal CT. (E) T2w MR imaging of SSTR2+ BON tumors. Tumors (circle) were either treated once with saline, 25 nM bortezomib, 20 MBq Lu-177-DOTATOC or a combination of bortezomib and Lu-177-DOTATOC on day 12 and tumor size measured before (d12) and after treatment (d18). Representative images of n > 3 independent experiments. (F) Statistical evaluation of the tumor plaque growth in the four treatment groups on d18. PRRT alone resulted in an overall 13% relative mass reduction related to baseline volume, bortezomib inhibited tumor growth with average 28% mass reduction. The combined treatment reduced relative tumor mass significantly by 50% (P = 0.041; ANOVA with Dunnett's test). pos. ctrl., positive control; CK, cytokeratin; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; SSTR2+: SSTR2-transfected, d, day; MRI: magnetic resonance imaging, PET: positron emission tomography; T2w, T2 weighted; Bo, Bortezomib, Lu-177, Lu-177-DOTATOC; Bo+Lu-177, combination therapy bortezomib plus Lu-177-DOTATOC. (Color version of figure is available online.)
To visualize SSTR2 expression in an in vivo setting we inoculated the BON- SSTR2+ 3D spheroids onto the CAM of 7 d old fertilized chicken eggs. Following an incubation time of 3 to 5 d, during which BON-SSTR2+ spheroids connected to the embryonic vasculature while the matrigel was degraded, we verified functional SSTR2 expression in those vascularized microtumors by SPECT/CT imaging. In fact, we could demonstrate an increased uptake (0.8%–2.2% ID) of Lu-177-DOTATOC 3 h post inoculation (Figure 3D). Multiparametric in vivo imaging using PET/MRI allowed for monitoring individual tumor development of BON-SSTR2+ microtumors under treatment in a noninvasive manner. Five days after inoculation of the microtumors, treatment started with bortezomib, or Lu-177-DOTATOC, or a combination of both or saline (vehicle treatment). While the size of vehicle-treated tumors increased by ∼35.2% within 7 d, confirming significant growth of the microtumors in the in ovo setup, all treatment arms induced a shrinkage below the initial tumor volume at the beginning of the treatment (day 0). Lu-177-DOTATOC and bortezomib monotherapy showed approximately 13% and 28% tumor volume reduction, respectively. The strongest effect was seen after combined treatment, as BON-SSTR2+ microtumors not only stopped to grow, but decreased in volume by approximately 50% (Figure 3F). Treatment effects were analyzed on molecular level after tumor excision.
Bortezomib-dependent apoptosis is induced by DNA damage repair insufficiency
After demonstrating the induction of cell cycle arrest and apoptosis upon bortezomib-combined DNA damaging therapy of NEN cells in vitro and delay of tumor growth in vitro and when implanted “in ovo” (CAM model) we aimed to clarify what caused the induction of apoptosis in vitro and in the chicken CAM model. Therefore, we reanalyzed the in vitro mRNA profiling results and determined effects of bortezomib plus cisplatin on DNA damage response gene expression and subsequently analyzed the expression of major candidate target proteins in vitro and in the bortezomib plus PRRT treated chicken CAM model.
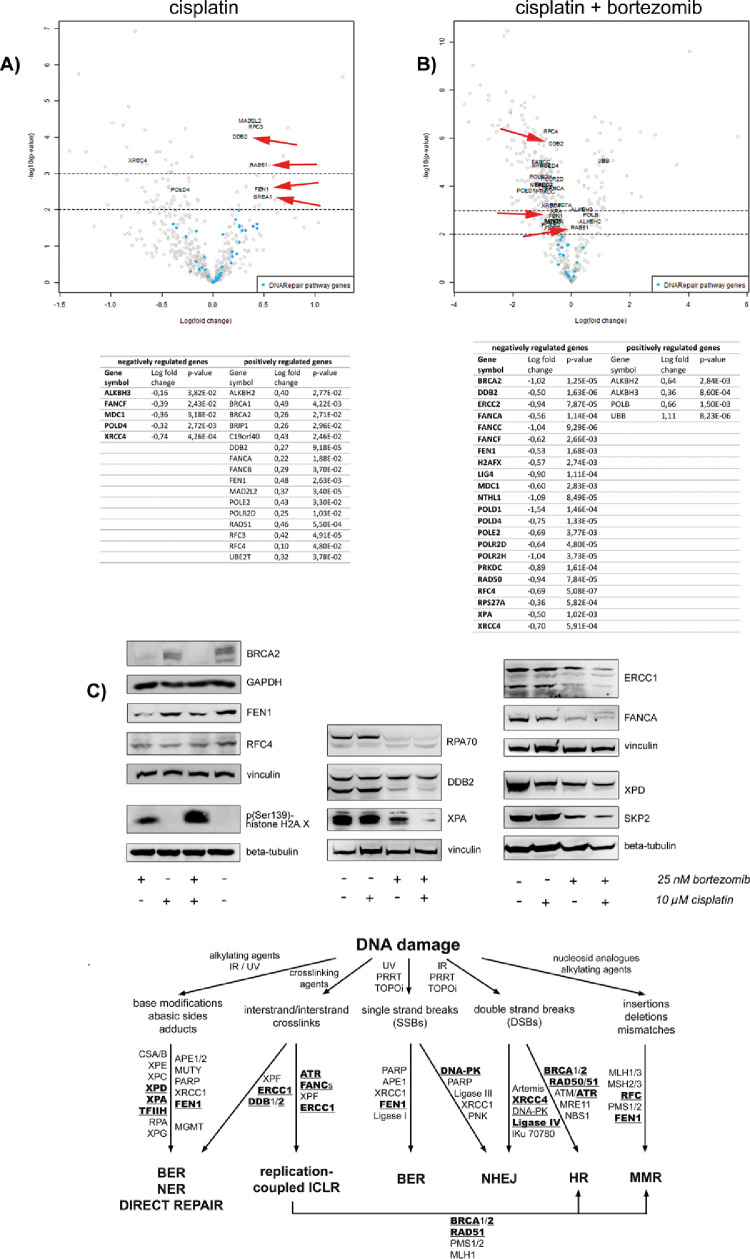
In vitro, cisplatin alone caused the upregulation of a notable number of DNA damage repair associated genes in BON cells (Figure 4; Supplementary Table 1). In contrast, bortezomib alone triggered the downregulation of identical or associated repair genes (Supplementary. Table 2). Under treatment with both substances, the cells retained the bortezomib-controlled pattern, resulting in a massive downregulation of DNA damage repair genes (including a large number of those upregulated by cisplatin) and a vast accumulation of DNA damage (Figure 4; Supplementary Table 3). Under the combined bortezomib/cisplatin treatment, all common DNA damage repair pathways were negatively affected in BON cells (Figure 4; related data from other cell lines refer to Supplementary Figure 2). This indicates a versatile DNA repair inhibitory effect of bortezomib treatment letting us to assume that the combination of PRRT with DNA damaging treatment could have synthetic lethal effects irrespective from the source of the DNA damage. To prove this theory, we analyzed the effect of combination with PRRT instead of cisplatin on the DNA repair protein expression in the tissues obtained from the chicken CAM model.
Figure 4.
Regulation of DNA damage repair related genes by bortezomib-combined chemotherapy in vitro: BON cells were either treated with 25 nM bortezomib, 10 µM cisplatin, combination of both or control for 24 h in vitro and analyzed by nCounter mRNA expression array. (A) Cisplatin induced the upregulation of several DNA repair genes whereas (B) Addition of bortezomib reversed the effect and triggered the downregulation of identical genes and genes associated with the same repair pathways, respectively. Arrows exemplarily mark DDB2, RAD51, FEN1, and BRCA1. (C) BON cells were treated with 25 nM bortezomib, 10 µM cisplatin, combination of both or control for 24 h and analyzed by western blot. Crucial DNA damage repair proteins were downregulated after bortezomib and combined treatment. These include proteins of the HR (RPA, SKP2), FA (FANCA), MMR (RPA, RFC), BER (FEN1) and primarily of the NER (RPA, DDB2, XPA, XPD, ERCC1) pathways of DNA repair. These effects strongly induced DNA damage stress, indicated by histone H2A.X phosphorylation. Representative data of three independent experiments are shown. Corresponding data obtained from KRJ-I and LCC-18 cells are presented in Supplementary Figure 2. (D) Bortezomib affected important molecules of the human response to DNA damage: Alkylating agents, ionizing radiation (IR), ultra violet (UV) radiation, crosslinking agents (e.g., platin-based chemotherapeutics), inhibitors of topoisomerases (TOPOi), PRRT or nucleoside analogs induce different DNA lesions (list is not exhaustive). Major pathways of DNA repair, which depend on the type of damage and the cell cycle phase, are direct repair (e.g., by methyltransferases), base excision repair (BER), nucleotide excision repair (NER), interstrand crosslink repair (ICLR), nonhomologous end joining (NHEJ), homologous recombination (HR) and mismatch repair (MMR). Bolded proteins have been shown to be downregulated by the proteasome inhibitor bortezomib. BER, Base Excision Repair; FA, Fanconi Anemia; HR, Homologous Recombination; MMR, Mismatch Repair; NER, Nucleotide Excision Repair.
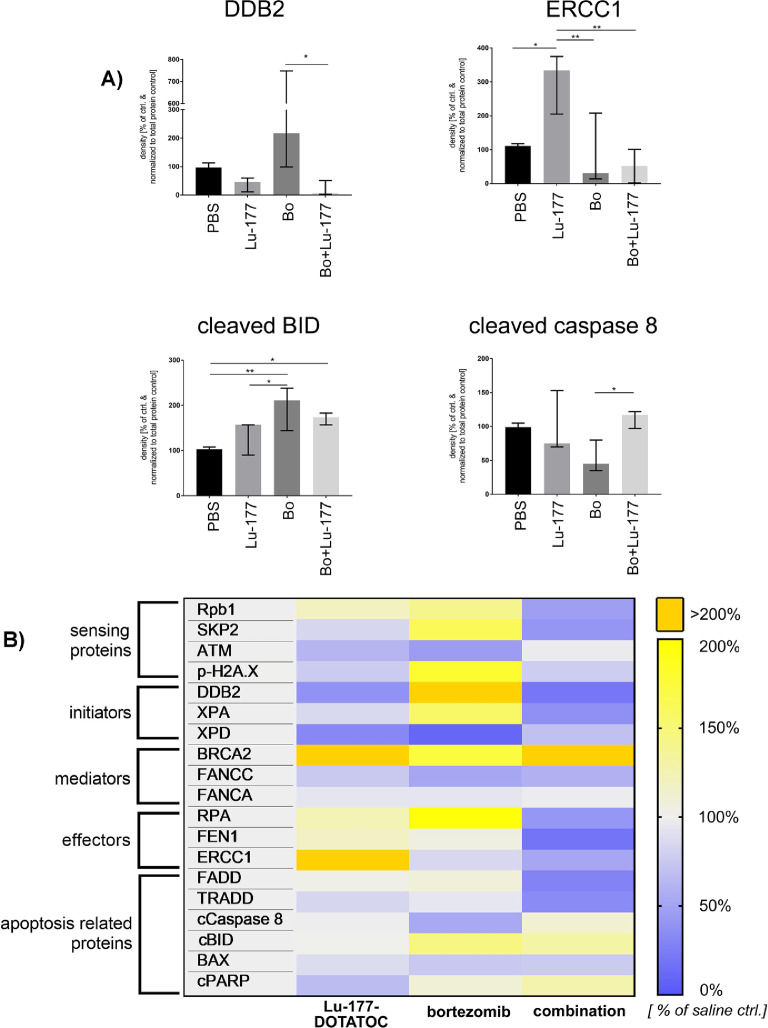
Quantitative analysis of the respective protein expression upon bortezomib-combined PRRT in the tissues derived from the chicken CAM in ovo model showed significant protein expression alterations for several DNA damage and apoptosis-related proteins according to western blot densitometry data (Figure 5 and raw data Supplementary Figure 4). Despite relatively high variances within the biological replicates of one treatment arm––which reduced the statistical power on a single protein level––the overall protein expression revealed a general downregulation of DNA repair related genes in the combined treatment group. Here, especially the candidate effector proteins of the BER pathway were affected.
Figure 5.
Regulation of DNA damage repair related proteins after bortezomib-combined PRRT in vivo: Chicken CAM xenografted tumors were treated i.v. with a single dose of ∼20 MBq 177-Lu-DOTATOC with and without 25 nM bortezomib versus saline control and explanted 6 d after treatment. Protein abundance of proteins involved in the sensing of DNA damage, initiation, signal transduction and execution of DNA repair and apoptosis was analyzed by western blot and densitometry was performed using Image J v1.53. Data was normalized to densitometric data of total protein stain (ponceau S). (A, B) We identified significant protein expression alterations in the combined treatment groups for DDB2, ERCC1, cleaved BID and p38, p41/p43 cleaved caspase 8 (Fisher's Least Significant Difference Test) and an overall downregulation of DNA repair-related genes, especially of the BER pathway (RNA polymerase II, DDB2, XPD, XPA, RPA, ERCC1, FEN1). (B) Heatmap shows mean of ≥3 independent experiments per treatment group normalized to total protein stain and relative to saline control. Raw data is shown in Supplementary Figure 4. BER, Base Excision Repair,; Bo, bortezomib; Lu-177, Lu-177-DOTATOC; Rpb1, RNA polymerase II subunit B1 c-terminal domain, *P < 0.05; **P < 0.01.
While bortezomib predominantly repressed the expression of proteins acting in late stages of DNA repair, leaving the sensing and initiating proteins unaffected or upregulated, PRRT reduced the expression of some early recognition proteins such as ATM. This effect might interfere with apoptosis induction since DNA lesion-sensing proteins are involved in apoptosis regulation. However, in contrast to the in vitro data, we could not clearly distinguish whether an extrinsic or intrinsic induction of apoptosis occurred after the combined treatment, since proteins of both pathways were affected. This variability is most likely caused by the fact that bortezomib was dropped onto the engrafted tumor plaques in ovo resulting in a heterogeneous distribution of the pharmaceutical.
The role of FOXM1
The forkheadbox transcription factor FOXM1 is a crucial regulator of G2/M transition and mitosis and can be suppressed by proteasome inhibition [58,59]. In neuroendocrine tumors, we recently clarified the role of FOXM1 as frequently expressed proliferation marker [21,31]. FOXM1 has further been attributed to the control of DNA damage response and resistance to chemotherapy [60], [61], [62], [63]. Taken together, we assumed that bortezomib might exert its cell cycle regulatory and DNA damage preserving effect by inhibition of FOXM1.
We therefore treated GEP-NEN cell lines with bortezomib and analyzed the FOXM1 expression by western blot and real time qPCR. All cell lines responded with a downregulation of FOXM1 mRNA and protein (Supplementary Figure 5). To prove that bortezomib exerts its anticancer effect by inhibition of FOXM1, we silenced the FOXM1 translation by RNA interference in BON cells and compared the differential expression upon bortezomib (25 nM for 24 h) with the expression upon FOXM1 knockdown (Supplementary table 4) by Nanostring nCounter pathway gene expression array. Knockdown efficacy was determined by qPCR (Supplementary Figure 5E). We found 39 corresponding genes. Interestingly, those genes were predominantly associated with cell cycle and apoptosis, according to the PANTHER pathway overrepresentation analysis [49]. There was no overrepresentation of DNA damage repair-related genes within the 39 corresponding genes indicating that FOXM1 is involved in the cell cycle regulation upon bortezomib treatment rather than in DNA damage response.
Addition of bortezomib to conventional lu-177-PPRT does not affect overall tumor volume or viable tumor cell mass according to in vivo FDG-PET/MRI analyses in an orthotopic mouse model
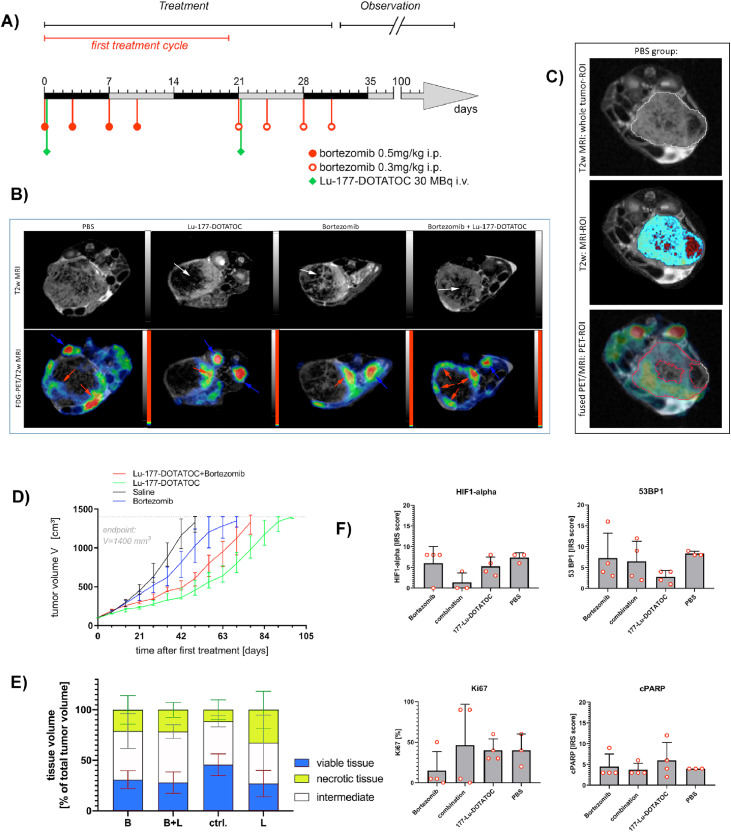
To confirm the preliminary results obtained from the chicken CAM model, we established an orthotopic xenograft mouse model bearing intrapancreatic BON-SSTR2+ tumors [39]. When the tumors reached 100 mm3, treatment was started according to the scheme presented in Figure 6 including 2 therapy cycles of 3 weeks. As preliminary in vivo studies had shown that a single injection (i.e., one cycle) of Lu-177-DOTATOC induced a growth plateau of about 2 weeks, we designed the study in such a way that the second cycle started once the tumor in the Lu-177 DOTATOC treated animals commenced to progress again. As seen in Figure 6, BON-SSTR2+ tumors responded to 177-Lu-DOTATOC monotherapy by showing a growth delay of about 6.5 weeks compared to saline-treated BON-SSTR2+ xenograft tumors, while bortezomib monotherapy delayed the growth by only 3 weeks. Interestingly, the combined treatment (bortezomib 4 hours before PRRT) delayed tumor growth by 4 weeks. Hence, the Kaplan-Maier survival analysis of cumulative time to endpoint (defined by a tumor volume of 1400 mm3) indicates that the combination of bortezomib and Lu-177-DOTATOC does not result in a longer overall time to end point compared to PRRT monotherapy (Supplementary Figure 6).
Figure 6.
Analysis of treatment efficacy of Lu-177-DOTATOC plus bortezomib therapy in orthotopic pancreatic mice xenograft tumors: Mice were xenotransplanted with BON-SSTR2+ cells and treated with either saline, Lu-177-DOTAOC, bortezomib or Lu-177-DOTATOC/bortezomib when tumor volume reached ≥100 mm3. (A) The therapeutic scheme was based on the human bortezomib treatment schedule, including 2 cycles of 3 weeks each (2 weeks treatment and one week pause). Mice received either a single dose of ∼30 MBq Lu-177-DOTATOC (i.v.), PBS (i.p.), i.p. injection of 0.5 mg/kg bortezomib or a combination of Lu-177-DTATOC and bortezomib on day 0 and d 21. For combined treatment, bortezomib was injected 4 h prior to Lu-177-DOTATOC to ensure full absorption. Due to its rapid clearance, bortezomib treatment was repeated on days 3, 7, and 10 in the first cycle and day 24, 28, and 31 during the second cycle. During week 3 of the first cycle, the treatment was paused, before the second analogous cycle started in week 4 with a reduced dosage of bortezomib. (B) Representative high-resolution T2-weighted images of all treatment groups (tumor center) 2 mo after the second cycle was finished, revealed morphologic heterogeneity due to treatment effects in the tumor tissue. Viable tumor tissue is represented as hyperintense signal intensity (SI) within the tumor, while degraded, necrotic tumor tissue is represented by hypointense SI (white arrows). Fused FDG-PET/T2w tumor images clearly show reduced FDG-avid fractions of a representative tumor per treatment group confirming the tumor tissue heterogeneity seen in corresponding T2w images. (C) MRI-and PET-based tumor texture analysis, representative images of tumor center (PBS group): Based on T2w images a whole tumor ROI was defined. T2w MRI-analysis of SI based on threshold settings: red areas represent true necrosis, light blue areas living and intermediate tumor tissue. Fused MRI/PET: FDG-PET analysis based on threshold settings: red ROI represents FDG-avid tumor tissue. (D) The tumor growth was monitored by MRI once a week until the tumors reached the endpoint of 1400 mm3 in tumor size. The Lu-177-DOTATOC monotherapy arm showed the strongest growth delay. (E) FDG-PET/T2w demonstrated a lower living tumor tissue content in the treatment groups related to the control group, without reaching significance in either the viable tumor or the necrotic tumor content comparison. One-Way ANOVA and Dunnett's multiple comparisons test (after assuming normality by Shapiro-Wilk test) demonstrated a tendency to differences in vital tumor volume in the Lu-177-DOTATOC versus ctrl. Group (p = 0.1042). (F) Immunohistochemical analyses of HIF1-alpha, Ki-67, 53BP1, and cleaved PARP in formalin-fixed mice tumor rim tissue resulted in no significant differences between the four treatment arms due to high intratumoral heterogeneity. Kaplan-Meier plot and corresponding raw data are shown in Supplementary Figs. 6 and 7. d, day; FDG, 18F-fluorodeoxyglucose; MRI, magnetic resonance imaging; PET, positron emission tomography; ROI, Region of Interest; T2w, T2 weighted; Captions: arrows: red = tumor tissue; white = necrotic/damaged tumor tissue; blue = kidneys/intestine; treatment arms: B, bortezomib monotherapy; B+L, combined treatment; ctrl., control; L, Lu-177-DOTATOC. (Color version of figure is available online.)
Notably, 5 to 6 weeks after the second cycle, the treatment-induced, reduced tumor growth terminated and a metabolically active tumor fraction started to develop in all treatment groups as compared to the saline group. This unexpected tumor outgrowth may be an indication that therapy-resistant tumor clones are able to develop throughout the treatment. In order to check whether the outgrowing tumor cells are actually vital, we performed combined FDG-PET/MRI scans once tumors reached a tumor volume of approximately 1000 mm3. The T2-weighted MRI sequence used here was established in such a way that viable tumor tissue results in hyperintense and necrotic/degrading tissue in hypointense signal intensity (SI). This allowed to perform a 3D-tumor texture analysis for this particular time point, in which FDG-PET identified viable tissue whereas T2w imaging identified true necrotic regions by analyzing SI in each voxel (Figure 6). The resulting third tumor section can be described as a mixture of different cellular states including cells in transition from viable to apoptotic and/or necrotic tissue under hypoxia or normoxia.
When PBS and Lu-177-DOTATOC treated animals were compared, PBS-treated animals showed far less tissue damage than Lu-177-DOTATOC treated tumors or those treated with bortezomib alone or in combination with Lu-177-DOTATOC. This confirmed induction of necrosis as a secondary treatment effect. The reduced nutrient perfusion, which typically occurs in the tumor center and thereby affects both treated and control groups, triggers increased necrosis in the DNA-damaged tumor core. Visual inspection of the T2w images clearly indicated that PRRT-treated tumors as well as tumor treated with bortezomib in mono- or combined therapy showed large areas of tissue damage. This tissue most likely consisted of necrosis, cell debris, and damaged of various intermediate states, varying from ∼21% to ∼33% of the tumor mass in the three treatment arms compared to 11% in the control arm (Figure 6).
As expected, the saline treated tumors exhibited a high percentage of viable tumor tissue and only small parts of necrotic tissue (mean of 45.6% and 11.4% respectively). The Lu-177-DOTATOC treatment group showed in 26.8% viable and 32.5% necrotic tumor tissue, respectively. The bortezomib and bortezomib/Lu-177-DOTATOC group showed almost identical distribution of the individual tumor tissue states (approximately 28%–31% living tumor tissue versus approximately 21% necrosis). Here, bortezomib monotherapy resulted in a slightly higher portion of viable tumor tissue. The combination treatment resulted in less vital tumor tissue, but a higher percentage of intermediate state tumor tissue at the time of investigation.
Immunohistochemical analysis of necrosis induction, proliferation, and apoptosis markers such as nuclear assembly of 53BP1 and PARP cleavage revealed no statistically significant differences between the treatment arms. However, the histology results are strongly determined by the analyzed tumor area and only reflect a snapshot. These variances indicate that biological heterogeneity resulting from responsive and non-responsive tumor areas is very high and that clonal selection after PRRT may cause disease relapse (Figure 6 and Supplementary Figure 7).
Discussion
Targeting DNA-damage repair is an emerging therapy approach in cancers that are characterized by a high mutational burden, impairing genes responsible for DNA repair. Whereas PARP inhibitors induce synthetic lethality in cancers with deleterious germline or somatic genetic defects in homologous recombination (HR) repair, clinical development of combination with DNA-damaging chemotherapy was limited by the more-than-additive toxicity, in particular dose-limiting myelosuppression. Multiple other DNA damage repair inhibitors have been developed and entered preclinical and clinical testing in the last two decades [64]. However, profound molecular knowledge of agent- or mutation-specific vulnerabilities in DDR is crucial to stratify patients for DDR inhibitory regimens.
Because of the specific molecular features of neuroendocrine tumors, which is dominated by somatostatin (SST) signaling and overactivation of mitogenic signal transduction (reviewed in [3]), DDR inhibition has not been a focus of basic research in NENs so far. Furthermore, the relatively low mutational burden decreases the likelihood of presence of synthetic lethal mutations.
However, DDR plays a clinically relevant role in neuroendocrine tumors. A study published in 2017 found DDR defect-related gene signatures (BRCA, MUTYH, and APOBEC) as well as biallelic mutations in the DDR genes MUTYH, CHEK2 or BRCA2 in 8% of pancreatic NETs [65]. Other studies demonstrated a considerably high prevalence of aberrant regulation of MMR genes by microsatellite instability, promoter hypermethylation and loss of heterozygosity in insulinomas and other pancreatic NENs [66,67], although contradictory studies exist [68]. Furthermore, PTEN, which is frequently lost in NENs due to epigenetic alterations or copy loss, is involved in DDR (reviewed in [69]). Finally, the expression of MGMT is predictive of the response to temozolomide in pancreatic neuroendocrine tumors [70].
In contrast, predictive markers or broad mechanistic knowledge of DDR upon PRRT is rare (reviewed in [71]). PRRT using Lu-177 as beta emitter has been shown to induce mainly SSBs and, to a lower extent, DSBs [11,72,73]. SSBs are primarily repaired by components of base excision repair (BER), nucleotide excision repair (NER) and mismatch repair (MMR) whereas DSB are cleared predominantly by non-homologous end joining (NHEJ) or homologous recombination (HR), depending on the availablilty of homologous DNA material and genetic background. Although both kinds of lesions induce different ways of damage repair, DSB repair generates intermediates, such as single-stranded DNA (ssDNA), which are present in MMR, BER, and NER pathways of DNA repair (but not in direct repair). Accordingly, common interacting proteins are recruited in specific steps of the repair pathways to protect ssDNA, polymerize second strands and control ligation. Those versatile proteins, including PARP1, RPA, FEN1 are crucial to more than one pathway of DNA repair.
Since there was no clear evidence, which of those components might be relevant to which extent in PRRT (and beyond that also targetable), we decided to use cisplatin as a model for our basic in vitro experiments. This decision was made on the fact that crosslink repair integrates a large number of components of DSB repair and thus, in contrast to alkylating agents and antimetabolites, platin-based chemotherapy might have considerable functional overlaps with DNA damage response to PRRT. Platinum-induced DNA crosslinks are repaired by NER or by interstrand crosslink repair, which makes use of components of NER, DSB repair pathways such as homologous recombination (HR), and translesion synthesis [74]. In contrast, TEM- and STZ-induced DNA adducts are primarily repaired through direct repair by methyltransferases such as O6-methylguanine DNA methyltransferase (MGMT), ALKB family demethylases and BER [75]. Since we wanted to focus specifically on PRRT-induced DNA repair mechanisms, we excluded the more clinically relevant alkylating agents (TEM, STZ) and antimetabolites (5-FU, CAP) from the study, despite the fact that our preliminary experiments have demonstrated enhancement effects when we combined them with bortezomib as well (unpublished).
Not surprisingly, transcripts of BAX, CDKN1A, DDB2, FDXR, GADD45A, MDM2, STAT5B and XPC showed time-dependent upregulation in peripheral blood cells upon radionuclide exposure in a recent study [76]. This indicates induction of apoptosis and cell cycle arrest along with NER induction. In our study, we verified an activation of NER components upon PRRT. Precisely, we demonstrated a significant upregulation of the endonuclease ERCC1 upon Lu-177-DOTATOC treatment in the chicken CAM model, along with postexcision features, including upregulation of the Flap Endonuclease 1 (FEN1), which is involved in the final ligation step of NER and BER, and of replication protein A (RPA), which recruits protein complexes to ssDNA in various processes where ssDNA is an intermediate structure [77].
In the last years, PARP inhibitors have been discussed as PPRT sensitizers. PARP family proteins are multifaced repair proteins, involved in BER, NER, NHEJ and HR pathways and thus play a role in both, SSB and DSB repair [78]. Accordingly, preclinical studies, combining PARP1 inhibition with PRRT, have shown promising results [72,79,80]. Concomitant inhibition of PARP1, leads to a PARP1 trapping, stalling of the replication fork and induction of DSBs [81]. However, in the context of HR deficiency (e.g., by BRCA1 or 2 mutations), which is present in a small fraction of NENs [65], this approach would induce synthetic letality [81]. In the case of HR proficiency, the cells could bypass fork stalling by induction of HR and therefore survive [81], [82], [83]. In our study we present further evidence that the DDR upon PRRT is very complex and involves aspects of several pathways. Hence, a versatile inhibiton of DDR might be necessary to reduce the induction of bypassing processes.
Therefore, we hypnotized that a major effect, which orchestrates the complex interplay of repair proteins, such as posttranslational regulation, could perturb DDR more efficiently than targeting specific components. So far, multiple studies have demonstrated that the ubiquitin-proteasome-system is involved in regulation of DNA-damage proteins [19,20]. Accordingly, proteasome inhibition has been shown to sensitize tumor cells to DNA-damaging therapy in vitro [22,[84], [85], [86], [87]. Although bortezomib monotherapy has not been effective in phase II studies of metastatic neuroendocrine tumors of different offspring [23] and for the treatment of neuroendocrine SCLC [88], the addition of a strong proapoptotic trigger, such as DNA damage, presented encouraging results concerning response and tolerability of bortezomib/chemotherapy combinations in a number of clinical studies [89], [90], [91], [92], [93], [94], [95], [96], [97], [98]. Recently, we have demonstrated a significant benefit from a bortezomib/cisplatin regimen in neuroendocrine lung cancer therapy in vivo models [21]. In the study presented herein, were able to to show that bortezomib affects gene expression of all common DNA repair mechanisms, including radiation/cisplatin-induced NER and SSB-repairing BER in vitro and in vivo. Both cellular stress and apoptosis are induced in reponse to unfolded protein and/or DNA damage accumulation. However, since the molecular mechanisms induced by bortezomib are not completely understood and presumably dependent on cellular context, we expected to detect evidence of altered protein decay due to proteasome inhibition rather than effect on gene transcription. Nevertheless, recent data suggests that bortezomib targets Sp family transcription factors in cancer cells [99], which in turn regulate the expression of DDR genes following genotoxic stress (reviewed in [100]).
We further hypothesized that bortezomib could be beneficial to overcome resistance to DNA damaging therapy in TP53 mutated and wildtype tumors, since bortezomib exerts its pro-apoptotic effect mainly by death receptor-dependent pathways and thus, is fairly independent from TP53 mutations [101], [102], [103]. Therefore it might have applicability in both, rarely TP53-mutated PRRT-treated G1/G2 NENs and frequently mutated G3 NECs which qualify for platinum-based chemotherapy [65,[104], [105], [106]. Accordingly, BON cells which are TP53 mutated [16,107] showed a relative low induction of primarily p53-driven intrinsic (mitochondrial) apoptosis and the major treatment effect of bortezomib in the analyzed NEN cell lines was induced by extrinsic apoptotic pathways.
Current data suggest that PRRT might be effective in the approximately 20% to 30% SSTR2-positive G3 NETs [108], which is currently discussed as therapeutic option for SSTR imaging-positive tumors [109,110]. However, there is still a great need for treatment options for G3 NETs (which proliferate fast and show better-differentiated morphology). As demonstrated in our preclinical study, bortezomib might be a novel approach to sensitize to cisplatin (which exploits high proliferation rates but with limited success in G3 NETs). It might also enhance the effect of PRRT (which targets SSTR expression, but requires more research in G3 NETs). In this preclinical study we give detailed insight into common resistance-associated effects and provide groundwork for subsequent translational considerations.
Several groups have described FOXM1 as a crucial regulator of the DNA damage reponse (reviewed in [60,111,112]). This suggests that inhibition of FOXM1 impairs DNA damage repair and that repression of FOXM1 upon bortezomib treatment mediates the DNA damage repair inhibition. For instance, FOXM1 was discussed as regulator of the HR proteins BRCA2, DNA polymerase delta, BRIP1 or Rad51 [60]. Surprisingly, we could demonstrate that FOXM1 knockdown was associated with apoptosis and cell cycle arrest, but we were not able to detect considerable effects on DNA repair-related gene expression. As we could confirm sufficient knockdown by verification of FOXM1 decrease by western blot and real time PCR of FOXM1, we assume that these DNA repair genes are not regulated by FOXM1 directly, but follow the downregulation of interposed regulatory events. Even FOXM1 knockdown in presence of cisplatin did not alter the expression of DNA damage-related genes compared to cisplatin plus mock siRNA, suggesting that the inhibition of FOXM1 alone is not sufficient to interfere with DNA damage response (data not shown).Thus, we assume that FOXM1 is not involed in the DNA damage control upon proteasome inhibition, but mediates some of it's effects via apoptosis induction and cell cycle arrest.
The strength of our study is the broad, multimodel-based assessment of detailed mechanistic data about therapies that have already entered clincial use in NENs, but are still not fully charaterized from a molecular point of view. We provide further information about weaknesses in cellular response to DNA damaging therapy that might be exploited for therapeutic purposes in the future.
However, our study also has some shortcomings. Most notably, the availability of proper preclinical models is limited in NEN research. In our study we used two different pancreatic cell line models: native BON and QGP-1 cells were generated from well-differentiated pancreatic NET, however, their mutational pattern, lack in SSTR2 and Chromogranin A expression, their reduced SSTR5 expression and their relatively high proliferation rates represent features, that are not completely representative of well-differentiated NETs [113], but might be exploited for preclinical evaluation of therapy approaches that are attributed to less differentiated tumors. However, after re-expression of SSTR2 in BON cells, the model was also applicable for PRRT, which requires specific epitopes such as SSTR2 and has limited applicability in poorly differentiated tumors [14].
Despite encouraging results in short-term in vitro experiments and in the 3D CAM model, we failed to prove a bortezomib-induced PRRT-sensitizing effect in the presented long-term mouse model. Two treatment cycles did not result in an overall reduction of tumor volume in any of the treatment arms, while tumor growth was delayed to some extent (depending on the type of treatment) when compared to the control group. In summary, the Kaplan-Meier survival curve revealed that the combination of bortezomib with Lu-177-DOTATOC was not beneficial concerning the overall time to the endpoint when compared to PRRT monotherapy.
Interestingly, FDG-PET/MRI, performed when tumors reached an average size of 1000 mm3, allowed a basic total tumor texture analysis separating the tumor into different metabolic states: the center of all tumors did not accumulate FDG, indicating necrotic or degraded tumor tissue, while the tumor margins were still metabolically active which could be best observed on fused FDG-PET/T2w images. However, as demonstrated by fused images, metabolically active tumor areas on PET images can considerably differ from the actual total tumor volume which is in line with the definition of metabolic tumor volume as a reliable parameter for tumor characterization and prognosis by FDG PET [114,115]. The approach of tumor texture analysis has been lately addressed in a few publications [116,117], but so far did not result in a generally suggested procedure. Nonetheless, simultaneous FDG-PET/MR imaging of the tumor allowed for clear identification of the viable tumor sections by FDG-PET and the nonvital/necrotic tumor areas by T2w imaging (MRI signal intensity analysis) resulting in hypointense SI versus hyperintense signals in viable or intermediate tumor tissue states.
As the analysis of the in ovo experiments clearly indicated an increase in apoptotic-related gene expression in the bortezomib/Lu177 DOTATOC treatment group, we assume that the identified third fraction by tumor texture analysis may consist of a higher percentage of cells already undergoing apoptotic stress compared to both monotherapies. This would result in higher percentages of dead cells at a later time point. CAM assay studies however are methodologically limited to a period of approximately 10 to 15 d after inoculation before the chicken hatch around day 21. However, as we did not use a marker for apoptosis at this timepoint of the longitudinal study, this hypothesis is rather speculative.
The scattering of the imaging results regarding the percentages of remaining viable tumor tissue or necrotic tumor volume was confirmed by immunohistochemistry. Here, no significant differences between treatment arms were shown. Especially the high variance of proliferation index, DNA damage accumulation and apoptosis induction were confirmed by anti-Ki67, anti-53BP1, and anticleaved PARP immunostaining. This effect was considerably dependent on the localization of the tissue specimen within the tumor. Since cell line-derived xenograft tumors usually represent rather clonal tumors even when obtained from a bulk, the morphologically and histologically variable patterns suggest a de novo acquired resistance. Presumably, the microvasculature of the xenograft tumors caused an unequal distribution of the treatment compounds throughout the tumors.
To further investigate why the in vivo study did not clearly confirm the beneficial effect of bortezomib plus PRRT that we showed in the short-term experiments, we immunohistochemically stained HIF1-alpha, a marker for hypoxia. High levels of hypoxia are known to trigger resistance to radiotherapy (reviewed in [118]). As bortezomib can increase hypoxia in some tumor entities [119], we assumed that bortezomib may induce hypoxia in the combination treatment compared to bortezomib monotherapy. However, no significant differences were identified in the treatment groups when compared to each other. As HIF1-alpha was analyzed only on one tissue section of each tumor, it does not reflect the whole tumor.
Some studies describe severe side effects of bortezomib in terms of delayed toxicity [120,121]. Nevertheless, in the study presented herein all animals tolerated the treatments according to the expected course. The group treated with either bortezomib alone or in combination with Lu-177-DOTATOC also displayed no different health states or showed unexpected deaths.
Concluding, the current study presents comprehensive data about the molecular effects of PRRT in neuroendocrine neoplasms. It gives also insight into NEN-specific resistance to cisplatin therapy. Understanding unintended effects of therapy is the basis of ascertaining how these might be overcome. Especially in NENs, broadening the applicability of both regimens might help to fill the therapeutic gap for NET G3, which often do not qualify perfectly for either therapy. However, while increased sensitivity to chemo-, radio-, and peptide radionuclide therapy was shown in bortezomib-combined regimens in short-term models, data generated in a long-term orthotopic mouse tumor model displayed highly variable effects on various analyzed levels including tumor size, tissue composition and protein expression. Some effects were limited to certain tumor areas, indicating clonal heterogeneity induced by development of resistant clones, which could not be encountered by combination therapy. Since detailed analysis of these resistant clones, for example by use of single cell techniques, might provide valuable information of how cancer cells bypass DNA repair defects, this observation should be addressed in an independent study.
Author contributions
Project was conceptualized by Franziska Briest and Eva J. Koziolek. Experiments were conducted by Franziska Briest, Eva J. Koziolek, Fränze Schmidt, Jakob Albrecht, Dagmar Sedding, Teresa Hartung, Anja A. Kühl, Samantha Exner and Martina Welzel. Data was curated, analyzed and visualized by Franziska Briest, Anja A. Kühl, and Eva J. Koziolek. Statistical analyses were conducted by Franziska Briest. Animal model was established by Jakob Albrecht. Imaging and imaging analysis were performed by Eva J. Koziolek, Jakob Albrecht, Joost Haeck, Monique Bernsen, and Fränze Schmidt. Manuscript was written by Franziska Briest and Eva J. Koziolek (original draft) and Franziska Briest (revision). The subprojects were administrated by Franziska Briest (in vitro and in vivo analyses, funding acquisition), Eva J. Koziolek (in vivo analyses and imaging), Christian Fischer (in vitro analyses), Carsten Grötzinger (in vitro analyses), Winfried Brenner (imaging analyses, lab leader, funding acquisition), Richard P. Baum (consulting clinical perspective of nuclear medicine, funding acquisition) and Patricia Grabowski (lab leader, funding acquisition).
Acknowledgments
We thank the Berlin Experimental Radionuclide Imaging Center (BERIC), Charité - Universitätsmedizin Berlin, Germany for performing in vivo experiments including PET/MR and SPECT/CT imaging. The authors thank Nicola Beindorff for coordination of the BERIC infrastructure.
Footnotes
Funding: This work was supported by the Theranostics Research Network. The project was further supported by a Young Investigators in Neuroendocrine Neoplasia for Germany Grant from Novartis Pharma GmbH. This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG) for PET/MRI use (INST 335/454-1FUGG) and in part by the Technologiestiftung Berlin (TSB) for SPECT/CT use. The funding sources had no involvement in the conduct of the research and/or preparation of the article, in study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication.
Conflicts of Interests: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
FB and PG were supported by the Theranostic Research Network Germany. The group of Dr. Patricia Grabowski receives financial support from Ipsen Pharma, Novartis and Pfizer. FB and PG received a travel grant and a lecture salary from Ipsen Pharma.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.11.004.
Appendix. Supplementary materials
References
- 1.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science (New York, NY) 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao JC, Fazio N, Singh S, Buzzoni R, Carnaghi C, Wolin E, Tomasek J, Raderer M, Lahner H, Voi M. Everolimus for the treatment of advanced, non-functional neuroendocrine tumours of the lung or gastrointestinal tract (RADIANT-4): a randomised, placebo-controlled, phase 3 study. Lancet. 2015 doi: 10.1016/S0140-6736(15)00817-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briest F, Grabowski P. PI3K-AKT-mTOR-signaling and beyond: the complex network in gastroenteropancreatic neuroendocrine neoplasms. Theranostics. 2014;4:336–365. doi: 10.7150/thno.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubomierski N, Kersting M, Bert T, Muench K, Wulbrand U, Schuermann M, Bartsch D, Simon B. Tumor suppressor genes in the 9p21 gene cluster are selective targets of inactivation in neuroendocrine gastroenteropancreatic tumors. Cancer research. 2001;61:5905–5910. [PubMed] [Google Scholar]
- 5.Lee J, Sung CO, Lee EJ, Do IG, Kim HC, Yoon SH, Lee WY, Chun HK, Kim KM, Park YS. Metastasis of neuroendocrine tumors are characterized by increased cell proliferation and reduced expression of the ATM gene. PloS one. 2012;7:e34456. doi: 10.1371/journal.pone.0034456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu W, Feng Z, Modica I, Klimstra DS, Song L, Allen PJ, Brennan MF, Levine AJ, Tang LH. Gene Amplifications in Well-Differentiated Pancreatic Neuroendocrine Tumors Inactivate the p53 Pathway. Genes & cancer. 2010;1:360–368. doi: 10.1177/1947601910371979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briest F, Grabowski P. The p53 network as therapeutic target in gastroenteropancreatic neuroendocrine neoplasms. Cancer Treat Rev. 2015;41(5):423–430. doi: 10.1016/j.ctrv.2015.03.006. Epub 2015 Mar 21. [DOI] [PubMed] [Google Scholar]
- 8.Brabander T, van der Zwan WA, Teunissen JJM, Kam BLR, Feelders RA, de Herder WW, van Eijck CHJ, Franssen GJH, Krenning EP, Kwekkeboom DJ. Long-Term Efficacy, Survival, and Safety of [(177)Lu-DOTA(0),Tyr(3)]octreotate in Patients with Gastroenteropancreatic and Bronchial Neuroendocrine Tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:4617–4624. doi: 10.1158/1078-0432.CCR-16-2743. [DOI] [PubMed] [Google Scholar]
- 9.Strosberg J, El-Haddad G, Wolin E, Hendifar A, Yao J, Chasen B, Mittra E, Kunz PL, Kulke MH, Jacene H. Phase 3 Trial of 177Lu-Dotatate for Midgut Neuroendocrine Tumors. The New England journal of medicine. 2017;376:125–135. doi: 10.1056/NEJMoa1607427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Al-Toubah T, El-Haddad G, Strosberg J. (177)Lu-DOTATATE for the treatment of gastroenteropancreatic neuroendocrine tumors. Expert Rev Gastroenterol Hepatol. 2019;13:1023–1031. doi: 10.1080/17474124.2019.1685381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graf F, Fahrer J, Maus S, Morgenstern A, Bruchertseifer F, Venkatachalam S, Fottner C, Weber MM, Huelsenbeck J, Schreckenberger M. DNA double strand breaks as predictor of efficacy of the alpha-particle emitter Ac-225 and the electron emitter Lu-177 for somatostatin receptor targeted radiotherapy. PloS one. 2014;9:e88239. doi: 10.1371/journal.pone.0088239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer letters. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Rinke A, B W, Auernhammer C, Bartenstein P, Bartsch D, Begum N, Faiss S, Fottner C, Gebauer B, Goretzki P. [Practice guideline neuroendocrine tumors - AWMF-Reg. 021-27] Z Gastroenterol. 2018;56:583–681. doi: 10.1055/a-0604-2924. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Carbonero R, Sorbye H, Baudin E, Raymond E, Wiedenmann B, Niederle B, Sedlackova E, Toumpanakis C, Anlauf M, Cwikla JB. ENETS Consensus Guidelines for High-Grade Gastroenteropancreatic Neuroendocrine Tumors and Neuroendocrine Carcinomas. Neuroendocrinology. 2016;103:186–194. doi: 10.1159/000443172. [DOI] [PubMed] [Google Scholar]
- 15.Pellat A, Coriat R. Well Differentiated Grade 3 Neuroendocrine Tumors of the Digestive Tract: A Narrative Review. J Clin Med. 2020;9(6) doi: 10.3390/jcm9061677. https://www.mdpi.com/about/announcements/784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briest F, Grass I, Sedding D, Möbs M, Christen F, Benecke J, Fuchs K, Kaemmerer D, Mende S, Sänger J. Mechanisms of targeting the MDM2-p53-FOXM1 axis in well-differentiated intestinal neuroendocrine tumors. Neuroendocrinology. 2018;107(1):1–23. doi: 10.1159/000481506. Epub 2017 Sep 14. [DOI] [PubMed] [Google Scholar]
- 17.Cipolla L, Maffia A, Bertoletti F, Sabbioneda S. The Regulation of DNA Damage Tolerance by Ubiquitin and Ubiquitin-Like Modifiers. Frontiers in genetics. 2016;7:105. doi: 10.3389/fgene.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daulny A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA repair. 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Falaschetti C, Mirkin E, Raha S, Paunesku T, Woloschak G. The Ubiquitin-Proteasome System and DNA Repair. In: Storici F, editor. DNA Repair - On the Pathways to Fixing DNA Damage and Errors. InTech; Rijeka, Croatia: 2011. pp. 255–286. editor. [Google Scholar]
- 20.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer research. 2007;67:7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 21.Taromi S, Lewens F, Arsenic R, Sedding D, Sänger J, Kunze A, Möbs M, Benecke J, Freitag H, Christen F. Proteasome inhibitor bortezomib enhances the effect of standard chemotherapy in Small Cell Lung Cancer. Oncotarget. 2017;8:97061–97078. doi: 10.18632/oncotarget.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao A, Wang TH. Molecular mechanisms for synergistic effect of proteasome inhibitors with platinum-based therapy in solid tumors. Taiwanese journal of obstetrics & gynecology. 2016;55:3–8. doi: 10.1016/j.tjog.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Shah MH, Young D, Kindler HL, Webb I, Kleiber B, Wright J, Grever M. Phase II study of the proteasome inhibitor bortezomib (PS-341) in patients with metastatic neuroendocrine tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:6111–6118. doi: 10.1158/1078-0432.CCR-04-0422. [DOI] [PubMed] [Google Scholar]
- 24.Evers BM, Townsend CM, Jr., Upp JR, Allen E, Hurlbut SC, Kim SW, Rajaraman S, Singh P, Reubi JC, Thompson JC. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology. 1991;101:303–311. doi: 10.1016/0016-5085(91)90004-5. [DOI] [PubMed] [Google Scholar]
- 25.Kaku M, Nishiyama T, Yagawa K, Abe M. Establishment of a carcinoembryonic antigen-producing cell line from human pancreatic carcinoma. Gan. 1980;71:596–601. [PubMed] [Google Scholar]
- 26.Pfragner R, Wirnsberger G, Niederle B, Behmel A, Rinner I, Mandl A, Wawrina F, Luo J, Adamiker D, Hoger H. Establishment of a continuous cell line from a human carcinoid of the small intestine (KRJ-I) International journal of oncology. 1996;8:513–520. doi: 10.3892/ijo.8.3.513. [DOI] [PubMed] [Google Scholar]
- 27.Lundqvist M, Mark J, Funa K, Heldin NE, Morstyn G, Wedell B, Layton J, Oberg K. Characterisation of a cell line (LCC-18) from a cultured human neuroendocrine-differentiated colonic carcinoma. European journal of cancer (Oxford, England: 1990) 1991;27:1663–1668. doi: 10.1016/0277-5379(91)90441-f. [DOI] [PubMed] [Google Scholar]
- 28.Hofving T, Arvidsson Y, Almobarak B, Inge L, Pfragner R, Persson M, Stenman G, Kristiansson E, Johanson V, Nilsson O. The neuroendocrine phenotype, genomic profile and therapeutic sensitivity of GEPNET cell lines. Endocrine-related cancer. 2018;25:367–380. doi: 10.1530/ERC-17-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez MJ, Subramaniam PS, Tang LH, Grunn A, Aburi M, Rieckhof G, Komissarova EV, Hagan EA, Bodei L, Clemons PA. A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors. Nature genetics. 2018;50:979–989. doi: 10.1038/s41588-018-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez MJ, Yan P, Alpaugh ML, Bowden M, Sicinska E, Zhou CW, Karan C, Realubit RB, Mundi PS, Grunn A. Reply to 'H-STS, L-STS and KRJ-I are not authentic GEPNET cell lines'. Nature genetics. 2019;51:1427–1428. doi: 10.1038/s41588-019-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briest F, Berg E, Grass I, Freitag H, Kaemmerer D, Lewens F, Christen F, Arsenic R, Altendorf-Hofmann A, Kunze A. FOXM1: A novel drug target in gastroenteropancreatic neuroendocrine tumors. Oncotarget. 2015;6:8185–8199. doi: 10.18632/oncotarget.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Exner S, Prasad V, Wiedenmann B, Grotzinger C. Octreotide Does Not Inhibit Proliferation in Five Neuroendocrine Tumor Cell Lines. Front Endocrinol (Lausanne) 2018;9:146. doi: 10.3389/fendo.2018.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 34.http://www.combosyn.com/. CompuSyn v1.0; access date 01.12.2014.
- 35.Nowak-Sliwinska P, Segura T, Iruela-Arispe ML. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis. 2014;17:779–804. doi: 10.1007/s10456-014-9440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigues J, Abramjuk C, Vásquez L, Gamboa N, Domínguez J, Nitzsche B, Höpfner M, Georgieva R, Bäumler H, Stephan C. New 4-maleamic acid and 4-maleamide peptidyl chalcones as potential multitarget drugs for human prostate cancer. Pharmaceutical research. 2011;28:907–919. doi: 10.1007/s11095-010-0347-8. [DOI] [PubMed] [Google Scholar]
- 37.Koziolek E, Briest F, Exner S, Sedding D, Grötzinger C, Grabowski P, Brenner W. A BON-SSTR2 chicken chorioallantoic membrane (CAM) model and imaging portfolio for the analysis of Lu-177-DOTATOC therapeutic effects and sensitizing agents (#461). European Molecular Imaging Meeting - EMIM 2018; San Sebastián, Spain; 2018. [Google Scholar]
- 38.Barnes AE, Jensen WN. Blood volume and red cell concentration in the normal chick embryo. The American journal of physiology. 1959;197:403–405. doi: 10.1152/ajplegacy.1959.197.2.403. [DOI] [PubMed] [Google Scholar]
- 39.Aristizabal Prada ET, Spottl G, Maurer J, Lauseker M, Koziolek EJ, Schrader J, Grossman A, Pacak K, Beuschlein F, Auernhammer CJ. The role of GSK3 and its reversal with GSK3 antagonism in everolimus resistance. Endocrine-related cancer. 2018;25:893–908. doi: 10.1530/ERC-18-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albrecht J, Polenz D, Kuhl AA, Rogasch JMM, Leder A, Sauer IM, Babos M, Mocsai G, Beindorff N, Steffen IG. Diffusion-weighted magnetic resonance imaging using a preclinical 1 T PET/MRI in healthy and tumor-bearing rats. EJNMMI Res. 2019;9:21. doi: 10.1186/s13550-019-0489-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelemen P, Alaoui J, Sieron D, Chan A, Kamm CP, Heldner MR, Gralla J, Wiest R, Verma RK. T1-weighted Grey Matter Signal Intensity Alterations After Multiple Administrations of Gadobutrol in Patients with Multiple Sclerosis, Referenced to White Matter. Scientific reports. 2018;8:16844. doi: 10.1038/s41598-018-35186-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albrecht J, Exner S, Groetzinger C, Prasad S, Konietschke F, Beindorff N, Kuehl AA, Prasad V, Brenner W, Koziolek EJ. Multimodal imaging of two-cycle PRRT with (177)Lu-DOTA-JR11 and (177)Lu-DOTATOC in an orthotopic neuroendocrine xenograft tumor mouse model. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2020 doi: 10.2967/jnumed.120.250274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaemmerer D, Peter L, Lupp A, Schulz S, Sanger J, Baum RP, Prasad V, Hommann M. Comparing of IRS and Her2 as immunohistochemical scoring schemes in gastroenteropancreatic neuroendocrine tumors. International journal of clinical and experimental pathology. 2012;5:187–194. [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Current protocols in molecular biology / edited by Frederick M Ausubel [et al]. 2011; Chapter 25: Unit25B 10. [DOI] [PubMed]
- 45.Fortina P, Surrey S. Digital mRNA profiling. Nature biotechnology. 2008;26:293–294. doi: 10.1038/nbt0308-293. [DOI] [PubMed] [Google Scholar]
- 46.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45:D353–Dd61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Research. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 53.Kaufmann T, Strasser A, Jost PJ. Fas death receptor signalling: roles of Bid and XIAP. Cell Death & Differentiation. 2012;19:42–50. doi: 10.1038/cdd.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Molecular and cellular biology. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grabowski P, Schrader J, Wagner J, Horsch D, Arnold R, Arnold CN, Georgieva I, Stein H, Zeitz M, Daniel PT. Loss of nuclear p27 expression and its prognostic role in relation to cyclin E and p53 mutation in gastroenteropancreatic neuroendocrine tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:7378–7384. doi: 10.1158/1078-0432.CCR-08-0698. [DOI] [PubMed] [Google Scholar]
- 56.Decesse JT, Medjkane S, Datto MB, Cremisi CE. RB regulates transcription of the p21/WAF1/CIP1 gene. Oncogene. 2001;20:962–971. doi: 10.1038/sj.onc.1204169. [DOI] [PubMed] [Google Scholar]
- 57.Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, Zhu L. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Molecular cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 58.Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PloS one. 2009;4:e6593. doi: 10.1371/journal.pone.0006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Advances in cancer research. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 60.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochimica et biophysica acta. 2014;1839:1316–1322. doi: 10.1016/j.bbagrm.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, Myatt SS, Lam EW. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Molecular cancer research: MCR. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Qiu W, Liu B, Yao R, Liu S, Yao Y, Liang J. Forkhead box transcription factor 1 expression in gastric cancer: FOXM1 is a poor prognostic factor and mediates resistance to docetaxel. Journal of translational medicine. 2013;11:204. doi: 10.1186/1479-5876-11-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Chen X, Gu Y, Zhu L, Qian Y, Pei D, Zhang W, Shu Y. FOXM1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and mediates sensitivity to cisplatin in A549 cells via the JNK/mitochondrial pathway. Neoplasma. 2015;62:61–71. doi: 10.4149/neo_2015_008. [DOI] [PubMed] [Google Scholar]
- 64.Yap TA, Plummer R, Azad NS, Helleday T. The DNA Damaging Revolution: PARP Inhibitors and Beyond. American Society of Clinical Oncology Educational Book. 2019:185–195. doi: 10.1200/EDBK_238473. [DOI] [PubMed] [Google Scholar]
- 65.Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 66.Mei M, Deng D, Liu T-H, Sang X-T, Lu X, Xiang H-D, Zhou J, Wu H, Yang Y, Chen J. Clinical Implications of Microsatellite Instability and MLH1 Gene Inactivation in Sporadic Insulinomas. The Journal of Clinical Endocrinology & Metabolism. 2009;94:3448–3457. doi: 10.1210/jc.2009-0173. [DOI] [PubMed] [Google Scholar]
- 67.House MG, Herman JG, Guo MZ, Hooker CM, Schulick RD, Cameron JL, Hruban RH, Maitra A, Yeo CJ. Prognostic value of hMLH1 methylation and microsatellite instability in pancreatic endocrine neoplasms. Surgery. 2003;134:902–908. doi: 10.1016/s0039-6060(03)00412-4. discussion 9. [DOI] [PubMed] [Google Scholar]
- 68.Arnold CN, Sosnowski A, Schmitt-Graff A, Arnold R, Blum HE. Analysis of molecular pathways in sporadic neuroendocrine tumors of the gastro-entero-pancreatic system. International journal of cancer Journal international du cancer. 2007;120:2157–2164. doi: 10.1002/ijc.22569. [DOI] [PubMed] [Google Scholar]
- 69.Liu IH, Ford JM, Kunz PL. DNA-repair defects in pancreatic neuroendocrine tumors and potential clinical applications. Cancer treatment reviews. 2016;44:1–9. doi: 10.1016/j.ctrv.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Cros J, Hentic O, Rebours V, Zappa M, Gille N, Theou-Anton N, Vernerey D, Maire F, Lévy P, Bedossa P. MGMT expression predicts response to temozolomide in pancreatic neuroendocrine tumors. Endocrine-related cancer. 2016;23:625–633. doi: 10.1530/ERC-16-0117. [DOI] [PubMed] [Google Scholar]
- 71.Bodei L, Schöder H, Baum RP, Herrmann K, Strosberg J, Caplin M, Öberg K, Modlin IM. Molecular profiling of neuroendocrine tumours to predict response and toxicity to peptide receptor radionuclide therapy. The lancet oncology. 2020;21:e431–ee43. doi: 10.1016/S1470-2045(20)30323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nonnekens J, van Kranenburg M, Beerens CE, Suker M, Doukas M, van Eijck CH, de Jong M, van Gent DC. Potentiation of Peptide Receptor Radionuclide Therapy by the PARP Inhibitor Olaparib. Theranostics. 2016;6:1821–1832. doi: 10.7150/thno.15311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Neill E, Kersemans V, Allen PD, Terry SYA, Torres JB, Mosley M, Smart S, Lee BQ, Falzone N, Vallis KA. Imaging DNA Damage Repair In Vivo After (177)Lu-DOTATATE Therapy. Journal of nuclear medicine: official publication. Society of Nuclear Medicine. 2020;61:743–750. doi: 10.2967/jnumed.119.232934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. The lancet oncology. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 75.Soll JM, Sobol RW, Mosammaparast N. Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities. Trends in biochemical sciences. 2017;42:206–218. doi: 10.1016/j.tibs.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edmondson DA, Karski EE, Kohlgruber A, Koneru H, Matthay KK, Allen S, Hartmann CL, Peterson LE, DuBois SG, Coleman MA. Transcript Analysis for Internal Biodosimetry Using Peripheral Blood from Neuroblastoma Patients Treated with (131)I-mIBG, a Targeted Radionuclide. Radiation research. 2016;186:235–244. doi: 10.1667/RR14263.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mocquet V, Lainé JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. The EMBO journal. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Experimental cell research. 2014;329:18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 79.Cullinane C, Waldeck K, Kirby L, Rogers BE, Eu P, Tothill RW, Hicks RJ. Enhancing the anti-tumour activity of 177Lu-DOTA-octreotate radionuclide therapy in somatostatin receptor-2 expressing tumour models by targeting PARP. Scientific reports. 2020;10:10196. doi: 10.1038/s41598-020-67199-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Purohit NK, Shah RG, Adant S, Hoepfner M, Shah GM, Beauregard JM. Potentiation of (177)Lu-octreotate peptide receptor radionuclide therapy of human neuroendocrine tumor cells by PARP inhibitor. Oncotarget. 2018;9:24693–24706. doi: 10.18632/oncotarget.25266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Molecular oncology. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arun B, Akar U, Gutierrez-Barrera AM, Hortobagyi GN, Ozpolat B. The PARP inhibitor AZD2281 (Olaparib) induces autophagy/mitophagy in BRCA1 and BRCA2 mutant breast cancer cells. International journal of oncology. 2015;47:262–268. doi: 10.3892/ijo.2015.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. Journal of molecular biology. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 84.Cron KR, Zhu K, Kushwaha DS, Hsieh G, Merzon D, Rameseder J, Chen CC, D'Andrea AD, Kozono D. Proteasome inhibitors block DNA repair and radiosensitize non-small cell lung cancer. PloS one. 2013;8:e73710. doi: 10.1371/journal.pone.0073710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karpov DS, Spasskaya DS, Tutyaeva VV, Mironov AS, Karpov VL. Proteasome inhibition enhances resistance to DNA damage via upregulation of Rpn4-dependent DNA repair genes. FEBS letters. 2013;587:3108–3114. doi: 10.1016/j.febslet.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 86.Denlinger CE, Rundall BK, Keller MD, Jones DR. Proteasome inhibition sensitizes non-small-cell lung cancer to gemcitabine-induced apoptosis. The Annals of thoracic surgery. 2004;78:1207–1214. doi: 10.1016/j.athoracsur.2004.04.029. discussion -14. [DOI] [PubMed] [Google Scholar]
- 87.Wang H, Yu Y, Jiang Z, Cao WM, Wang Z, Dou J, Zhao Y, Cui Y, Zhang H. Next-generation proteasome inhibitor MLN9708 sensitizes breast cancer cells to doxorubicin-induced apoptosis. Scientific reports. 2016;6:26456. doi: 10.1038/srep26456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lara PN, Jr., Chansky K, Davies AM, Franklin WA, Gumerlock PH, Guaglianone PP, Atkins JN, Farneth N, Mack PC, Crowley JJ. Bortezomib (PS-341) in relapsed or refractory extensive stage small cell lung cancer: a Southwest Oncology Group phase II trial (S0327) Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2006;1:996–1001. [PubMed] [Google Scholar]
- 89.Davies AM, Chansky K, Lara PN, Jr., Gumerlock PH, Crowley J, Albain KS, Vogel SJ, Gandara DR. Bortezomib plus gemcitabine/carboplatin as first-line treatment of advanced non-small cell lung cancer: a phase II Southwest Oncology Group Study (S0339) Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2009;4:87–92. doi: 10.1097/JTO.0b013e3181915052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davies AM, Ruel C, Lara PN, Lau DH, Gumerlock PH, Bold R, Shibata S, Lenz HJ, Schenkein DP, Gandara DR. The proteasome inhibitor bortezomib in combination with gemcitabine and carboplatin in advanced non-small cell lung cancer: a California Cancer Consortium Phase I study. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2008;3:68–74. doi: 10.1097/JTO.0b013e31815e8b88. [DOI] [PubMed] [Google Scholar]
- 91.Badros A, Goloubeva O, Fenton R, Rapoport AP, Akpek G, Harris C, Ruehle K, Westphal S, Meisenberg B. Phase I trial of first-line bortezomib/thalidomide plus chemotherapy for induction and stem cell mobilization in patients with multiple myeloma. Clinical lymphoma & myeloma. 2006;7:210–216. doi: 10.3816/CLM.2006.n.061. [DOI] [PubMed] [Google Scholar]
- 92.Aghajanian C, Dizon DS, Sabbatini P, Raizer JJ, Dupont J, Spriggs DR. Phase I trial of bortezomib and carboplatin in recurrent ovarian or primary peritoneal cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:5943–5949. doi: 10.1200/JCO.2005.16.006. [DOI] [PubMed] [Google Scholar]
- 93.Jandial DA, Brady WE, Howell SB, Lankes HA, Schilder RJ, Beumer JH, Christner SM, Strychor S, Powell MA, Hagemann AR. A phase I pharmacokinetic study of intraperitoneal bortezomib and carboplatin in patients with persistent or recurrent ovarian cancer: An NRG Oncology/Gynecologic Oncology Group study. Gynecologic oncology. 2017;145:236–242. doi: 10.1016/j.ygyno.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jatoi A, Dakhil SR, Foster NR, Ma C, Rowland KM, Jr., Moore DF, Jr., Jaslowski AJ, Thomas SP, Hauge MD, Flynn PJ. Bortezomib, paclitaxel, and carboplatin as a first-line regimen for patients with metastatic esophageal, gastric, and gastroesophageal cancer: phase II results from the North Central Cancer Treatment Group (N044B) Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2008;3:516–520. doi: 10.1097/JTO.0b013e31816de276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramirez PT, Landen CN, Jr., Coleman RL, Milam MR, Levenback C, Johnston TA, Gershenson DM. Phase I trial of the proteasome inhibitor bortezomib in combination with carboplatin in patients with platinum- and taxane-resistant ovarian cancer. Gynecologic oncology. 2008;108:68–71. doi: 10.1016/j.ygyno.2007.08.071. [DOI] [PubMed] [Google Scholar]
- 96.Turkington RC, Purcell C, James CR, Millar J, Napier E, Law D, Gallagher R, Morris M, Wilson RH, Eatock MM. A phase I trial of bortezomib in combination with epirubicin, carboplatin and capecitabine (ECarboX) in advanced oesophagogastric adenocarcinoma. Investigational new drugs. 2014;32:250–260. doi: 10.1007/s10637-013-9970-7. [DOI] [PubMed] [Google Scholar]
- 97.Zhao Y, Foster NR, Meyers JP, Thomas SP, Northfelt DW, Rowland KM, Jr., Mattar BI, Johnson DB, Molina JR, Mandrekar SJ. A phase I/II study of bortezomib in combination with paclitaxel, carboplatin, and concurrent thoracic radiation therapy for non-small-cell lung cancer: North Central Cancer Treatment Group (NCCTG)-N0321. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2015;10:172–180. doi: 10.1097/JTO.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma C, Mandrekar SJ, Alberts SR, Croghan GA, Jatoi A, Reid JM, Hanson LJ, Bruzek L, Tan AD, Pitot HC. A phase I and pharmacologic study of sequences of the proteasome inhibitor, bortezomib (PS-341, Velcade), in combination with paclitaxel and carboplatin in patients with advanced malignancies. Cancer chemotherapy and pharmacology. 2007;59:207–215. doi: 10.1007/s00280-006-0259-9. [DOI] [PubMed] [Google Scholar]
- 99.Karki K, Harishchandra S, Safe S. Bortezomib Targets Sp Transcription Factors in Cancer Cells. Molecular pharmacology. 2018;94:1187–1196. doi: 10.1124/mol.118.112797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Christmann M, Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Research. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yerlikaya A, Okur E, Ulukaya E. The p53-independent induction of apoptosis in breast cancer cells in response to proteasome inhibitor bortezomib. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:1385–1392. doi: 10.1007/s13277-012-0386-3. [DOI] [PubMed] [Google Scholar]
- 102.Saulle E, Petronelli A, Pasquini L, Petrucci E, Mariani G, Biffoni M, Ferretti G, Scambia G, Benedetti-Panici P, Cognetti F. Proteasome inhibitors sensitize ovarian cancer cells to TRAIL induced apoptosis. Apoptosis: an international journal on programmed cell death. 2007;12:635–655. doi: 10.1007/s10495-006-0025-9. [DOI] [PubMed] [Google Scholar]
- 103.Sayers TJ. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer immunology, immunotherapy: CII. 2011;60:1173–1180. doi: 10.1007/s00262-011-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vijayvergia N, Boland PM, Handorf E, Gustafson KS, Gong Y, Cooper HS, Sheriff F, Astsaturov I, Cohen SJ, Engstrom PF. Molecular profiling of neuroendocrine malignancies to identify prognostic and therapeutic markers: a Fox Chase Cancer Center Pilot Study. British journal of cancer. 2016;115:564–570. doi: 10.1038/bjc.2016.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scarpa A. The landscape of molecular alterations in pancreatic and small intestinal neuroendocrine tumours. Ann Endocrinol (Paris) 2019;80:153–158. doi: 10.1016/j.ando.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 106.Raj N, Valentino E, Capanu M, Tang LH, Basturk O, Untch BR, Allen PJ, Klimstra DS, Reidy-Lagunes D. Treatment Response and Outcomes of Grade 3 Pancreatic Neuroendocrine Neoplasms Based on Morphology: Well Differentiated Versus Poorly Differentiated. Pancreas. 2017;46:296–301. doi: 10.1097/MPA.0000000000000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vandamme T, Peeters M, Dogan F, Pauwels P, Van Assche E, Beyens M, Mortier G, Vandeweyer G, de Herder WW, Van Camp G. Whole exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. J Mol Endocrinol. 2015;54(2):137–147. doi: 10.1530/JME-14-0304. Epub 2015 Jan 22. [DOI] [PubMed] [Google Scholar]
- 108.Kaemmerer D, Träger T, Hoffmeister M, Sipos B, Hommann M, Sänger J, Schulz S, Lupp A. Inverse expression of somatostatin and CXCR4 chemokine receptors in gastroenteropancreatic neuroendocrine neoplasms of different malignancy. Oncotarget. 2015;6:27566–27579. doi: 10.18632/oncotarget.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carlsen EA, Fazio N, Granberg D, Grozinsky-Glasberg S, Ahmadzadehfar H, Grana CM, Zandee WT, Cwikla J, Walter MA, Oturai PS. Peptide receptor radionuclide therapy in gastroenteropancreatic NEN G3: a multicenter cohort study. Endocrine-related cancer. 2019;26:227–239. doi: 10.1530/ERC-18-0424. [DOI] [PubMed] [Google Scholar]
- 110.Nicolini S, Severi S, Ianniello A, Sansovini M, Ambrosetti A, Bongiovanni A, Scarpi E, Di Mauro F, Rossi A, Matteucci F. Investigation of receptor radionuclide therapy with (177)Lu-DOTATATE in patients with GEP-NEN and a high Ki-67 proliferation index. European journal of nuclear medicine and molecular imaging. 2018;45:923–930. doi: 10.1007/s00259-017-3925-8. [DOI] [PubMed] [Google Scholar]
- 111.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochimica et biophysica acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 112.Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Advances in cancer research. 2013;119:191–419. doi: 10.1016/B978-0-12-407190-2.00016-2. [DOI] [PubMed] [Google Scholar]
- 113.Luley KB, Biedermann SB, Künstner A, Busch H, Franzenburg S, Schrader J, Grabowski P, Wellner UF, Keck T, Brabant G. A Comprehensive Molecular Characterization of the Pancreatic Neuroendocrine Tumor Cell Lines BON-1 and QGP-1. Cancers (Basel) 2020;12(3):691. doi: 10.3390/cancers12030691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Apostolova I, Steffen IG, Wedel F, Lougovski A, Marnitz S, Derlin T, Amthauer H, Buchert R, Hofheinz F, Brenner W. Asphericity of pretherapeutic tumour FDG uptake provides independent prognostic value in head-and-neck cancer. Eur Radiol. 2014;24:2077–2087. doi: 10.1007/s00330-014-3269-8. [DOI] [PubMed] [Google Scholar]
- 115.O JH, Lodge MA, Wahl RL. Practical PERCIST: A Simplified Guide to PET Response Criteria in Solid Tumors 1.0. Radiology. 2016;280:576–584. doi: 10.1148/radiol.2016142043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Just N. Improving tumour heterogeneity MRI assessment with histograms. British journal of cancer. 2014;111:2205–2213. doi: 10.1038/bjc.2014.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jardim-Perassi BV, Huang S, Dominguez-Viqueira W, Poleszczuk J, Budzevich MM, Abdalah MA, Pillai SR, Ruiz E, Bui MM, Zuccari D. Multiparametric MRI and Coregistered Histology Identify Tumor Habitats in Breast Cancer Mouse Models. Cancer research. 2019;79:3952–3964. doi: 10.1158/0008-5472.CAN-19-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Horsman MR, Overgaard J. The impact of hypoxia and its modification of the outcome of radiotherapy. J Radiat Res. 2016;57(Suppl 1):i90–ii8. doi: 10.1093/jrr/rrw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun X, Ackerstaff E, He F, Xing L, Hsiao HT, Koutcher JA, Ling CC, Li GC. Visualizing the antivascular effect of bortezomib on the hypoxic tumor microenvironment. Oncotarget. 2015;6:34732–34744. doi: 10.18632/oncotarget.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Croghan GA, Suman VJ, Maples WJ, Albertini M, Linette G, Flaherty L, Eckardt J, Ma C, Markovic SN, Erlichman C. A study of paclitaxel, carboplatin, and bortezomib in the treatment of metastatic malignant melanoma: a phase 2 consortium study. Cancer. 2010;116:3463–3468. doi: 10.1002/cncr.25191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Edelman MJ, Burrows W, Krasna MJ, Bedor M, Smith R, Suntharalingam M. Phase I trial of carboplatin/paclitaxel/bortezomib and concurrent radiotherapy followed by surgical resection in Stage III non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands) 2010;68:84–88. doi: 10.1016/j.lungcan.2009.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.