Abstract
Intramolecular motions in proteins are one of the important factors that determine their biological activity and interactions with molecules of biological importance. Magnetic relaxation of 15N amide nuclei allows one to monitor motions of protein backbone over a wide range of time scales. 15N{1H} nuclear Overhauser effect is essential for the identification of fast backbone motions in proteins. Therefore, exact measurements of NOE values and their accuracies are critical for determining the picosecond time scale of protein backbone. Measurement of dynamic NOE allows for the determination of NOE values and their probable errors defined by any sound criterion of nonlinear regression methods. The dynamic NOE measurements can be readily applied for non-deuterated or deuterated proteins in both HSQC and TROSY-type experiments. Comparison of the dynamic NOE method with commonly implied steady-state NOE is presented in measurements performed at three magnetic field strengths. It is also shown that improperly set NOE measurement cannot be restored with correction factors reported in the literature.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-020-00346-6) contains supplementary material, which is available to authorized users.
Keywords: NMR, Protein dynamics, Heteronuclear NOE, Dynamic NOE, Errors of NOE measurements
Introduction
Since its first use of magnetic relaxation measurements of 15N nuclei applied to the protein, the staphylococcal nuclease (Kay et al. 1989), this method has become indispensable in the determination of molecular motions in biopolymers (Jarymowycz and Stone 2006; Kempf and Loria 2003; Palmer, III 2004; Reddy and Rayney 2010; Stetz et al. 2019). The canonical triad of relaxation parameters—longitudinal (R1) and transverse (R2) relaxation rates accompanied by the 15N{1H} nuclear Overhauser effect (NOE)—have been most often used in studies investigating the mobility of backbone in proteins. It is a common opinion that 15N{1H} NOE is unique among the mentioned three relaxation parameters because it is regarded as essential for the accurate estimation of the spectral density function at high frequencies (ωH ± ωN), and it is crucial for the identification of fast backbone motions. (Idiyatullin et al. 2001; Gong and Ishima 2007; Ferrage et al. 2009).
The most common method for the determination of X{1H} NOE is a steady-state approach. It requires measurements of the longitudinal polarization at the thermal equilibrium of spin X system, S0, and the steady-state longitudinal X polarization under 1H irradiation, Ssat (Noggle and Schirmer 1971). Note that the nuclear Overhauser effect, defined as , should not be mistaken with nuclear Overhauser enhancement, (Harris et al. 1997).
It has to be pointed out that NOE measurements appear to be very demanding and artifact prone observations. One of severe obstacles in these experiments is their ca. tenfold lower sensitivity in comparison to R1N and R2N which is due to the fact that the NOE experiments with 1H detection start with the equilibrium 15N magnetization rather than 1H. The steady-state 15N{1H} NOEs (ssNOE) are normally determined as a ratio of cross-peak intensities in two experiments—with and without saturation of HN resonances. Such arrangement creates problems with computing statistically validated assessment of experimental errors. 15N{1H} NOE pulse sequence requires a very careful design as well. Properly chosen recycle delays between subsequent scans and saturation time of HN protons have to take into account the time needed to reach the equilibrium or stationary values of 15 N and HN magnetizations (Harris and Newman 1976; Canet 1976; Renner et al. 2002). Exchange of HN protons with the bulk water combined with the long longitudinal relaxation time of water protons leads to prolonged recycle delay in the spectrum acquired without saturation of HN resonances. Unintentional irradiation of the water resonance suppresses HN and other exchangeable signals owing to the saturation transfer and many non-exchangeable 1H resonances via direct or indirect NOE with water (Grzesiek and Bax 1993) while interference of DD/CSA relaxation mechanisms of 15N amide nuclei disturbs the steady-state 15N polarization during 1H irradiation (Ferrage et al. 2009). All aforementioned processes depend directly or indirectly on the longitudinal relaxation rates of amide 1H and 15N nuclei R1H and R1N as well as the longitudinal relaxation rate of water protons, R1W, and the exchange rate between water and amide protons, k.
In this study, the dynamic NMR experiment (DNOE), a forgotten method of the NOE determination in proteins, was experimentally tested, and the results were compared with independently performed steady-state NOE measurements at several magnetic fields for widely studied, small, globular protein ubiquitin. Additionally, several difficulties inherent in 15N{1H} NOEs and methods for overcoming or minimizing these difficulties are cautiously discussed.
Experimental
The uniformly labeled U-[15N] human ubiquitin was obtained from Cambridge Isotope Laboratories, Inc in lyophilized powder form and dissolved to 0.8 mM protein concentration in buffer containing 10 mM sodium phosphate at pH 6.6 and 0.01% (m/v) NaN3. DSS-d6 of 0.1% (m/v) in 99.9% D2O was placed in a sealed capillary inserted into the 5 mm NMR tube.
Amide resonance assignments of ubiquitin were taken from BioMagResBank (BMRB) using the accession code 6457 (Cornilescu et al. 1998).
NMR experiments were performed on three Bruker Avance NEO spectrometers operating at 1H frequencies of 700, 800 and 950 MHz equipped with cryogenic TCI probes. The temperature was controlled before and after each measurement with an ethylene glycol reference sample (Rainford et al. 1979) and was set to 25 °C. The temperature was stable with maximum detected deviation of ± 0.3 °C. Chemical shifts in the1H NMR spectra were reported with respect to external DSS-d6 while chemical shifts of the 15N signals were referenced indirectly using frequency ratio of 0.101329118 (Wishart et al. 1995). The spectral widths were set to 12 ppm and 22 ppm for 1H and 15N, respectively. The number of complex data points collected for 1H and 15N dimensions 2048 and 200, respectively. In each experiment, 8 scans were accumulated per FID. Double zero filling and a 90°-shifted squared sine-bell filter were applied prior to Fourier transformation. Data were processed using the program nmrPipe (Delaglio et al. 1995) and analyzed with the program SPARKY (Goddard and Kneller). Resonance intensities were used in calculating relaxation times and NOE values obtained from a nonlinear least-squares analysis performed using Fortran routines written in-house, based on the Newton–Raphson algorithm (Press et al. 2007).
The pulse programs used in this work were based on the HSQC-type R1(15N) and 15N{1H} NOE experiments (Lakomek et al. 2012). The carrier frequency during 1H saturation with 22 ms spaced 180° hard pulses on 1H was moved from water frequency to the centre of amide region (8.5 ppm). Evolution times in R1(15N) and dynamic NOE experiments were collected in random order. Reproducibility of experiments was excellent. Therefore, the interleaved mode was not used since it could introduce instabilities of water magnetization (Renner et al. 2002). The list of delays applied in the experiments used in this work is given in Table S3.
Results and discussion
Dynamic NOE measurement—introduction
It can be concluded from the Solomon equations (Solomon 1955) that in the heteronuclear spin system X–H, the heteronuclear Overhauser effect is built up with the rate R1(X) under the condition of proton saturation as shown for the 13C-1H spin system (Kuhlmann et al. 1970; Kuhlmann and Grant 1971). As a consequence of this observation a dynamic NOE was employed for the simultaneous determination of R1(13C) and 13C{1H} NOE using Eq. (1)
| 1 |
Measurements of time dependent changes of signal intensities S(t) allow for the determination of ε, R1, and their probable errors, as defined by any standard criterion of nonlinear regression methods. The DNOE can be especially beneficial in studying nuclei with negative magnetogyric ratios since in unfavorable circumstances, nulling of the resonance in a proton saturated spectrum can occur. Therefore, the DNOE has been successfully used in relaxation studies of 29Si (Kimber and Harris 1974; Ejchart et al. 1992) and 15N (Levy et al. 1976) nuclei in organic molecules. The 15N-DNOE has been also investigated in small protein (Zhukov and Ejchart 1999). This approach can be especially profitable in studies of medium to large size proteins displaying highly dynamic fragments.
Time schedule of NOE measurement
Both nitrogen polarizations, Ssat and S0, depend on a number of physical processes in the vicinity of amide nitrogen nuclei. Dipolar interaction between 15N and 1HN brings about the nuclear Overhauser effect. Additional processes as chemical shift anisotropy relaxation mechanism of 15N and its interference with 15N/1HN dipolar interaction, direct NOE and saturation transfer from water to 1HN protons due to chemical exchange influence both nitrogen polarizations, especially if the pulse sequence itself will result in non equilibrium state of water protons. Presaturation of the water resonance resulting in partial saturation of water magnetization attenuates 1HN signal intensities mostly through the chemical exchange or through homonuclear NOE with water protons. (Grzesiek and Bax 1993; Lakomek et al. 2012). Therefore, evolution of the spin system towards Ssat or S0 nitrogen polarizations depends on the rates of the processes mentioned above, the longitudinal relaxation rates of 15N, 1HN, and water protons, R1N, R1H, and R1W, and the chemical exchange rate, k, between amide and water protons. These rates strongly determine the time schedule of NOE measurements, which is schematically shown in Fig. 1. Hence, their knowledge is a prerequisite for choice of optimal delays. The numerical data of R1H and R1W for the sample studied here are given in Table 1. Nevertheless, one should be aware that the R1W depends on temperature, pH, and protein concentration. Residue specific R1N values for the ubiquitin sample will be discussed further.
Fig. 1.
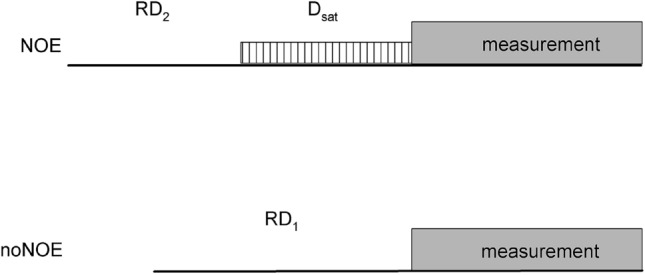
Steady-state NOE measurement is composed of two sequences: NOE and noNOE with saturated and unperturbed HN protons, respectively. Dynamic NOE measurement comprises several NOE type sequences with a set of different Dsat values
Table 1.
Longitudinal relaxation rates of water protons R1W and averaged rates of amide protons R1H for ubiquitin sample at 25 °C
| B0 [T] | R1W [s−1] | dR1W [s−1] | R1H [s−1] | dR1H [s−1] |
|---|---|---|---|---|
| 16.4 | 0.389 | 0.003 | 0.92 | 0.06 |
| 18.8 | 0.404 | 0.003 | Not determined | |
| 22.3 | 0.412 | 0.005 | 0.85 | 0.07 |
Detailed information on the sample composition are given in the experimental results
In the noNOE reference measurement, 15N nuclei have to reach the thermal equilibrium at the end of delay RD1. During the block denoted as measurement in Fig. 1, the pulse sequence resulting in the 2D 15N/1H spectrum with the desired cross peak intensities is executed. At the start of acquisition, several coupled relaxation processes take place, resulting in multi-exponential decay of 15N, 1HN, and water protons (Ferrage et al. 2008). Keeping in mind that R1W is much smaller than the rates of other processes, it can be reasonably assumed that R1W rate mainly defines RD1. Fulfillment of the condition
| 2 |
where factor 0.02 has been chosen to some extent arbitrarily, should properly determine RD1 values in most of the cases. Still one has to be aware that the smallest decay rate resulting from the exact solution of full relaxation matrix can be smaller than R1W.
In NOE measurement, the buildup of 15N magnetization takes place with the rate R1N. 15N relaxation rates can be, however, broadly dispersed if mobility of N–H vectors in a studied molecule differ significantly. Therefore, to meet the condition
| 3 |
a compromise may be required (c.f. Table S1). Experiments of steady-state and dynamic NOE measurements differ in the RD2 setting. In the case of steady-state NOE, the value RD2 = 0 is adequate. Even if the nitrogen polarization displays a nonzero value at the beginning of the Dsat period, it will still have enough time to reach the steady-state condition. In dynamic NOE, however, the nitrogen polarization has to start from closely controlled thermal equilibrium. Therefore, condition (2) with RD1 replaced with RD2 has to be fulfilled. The description (RD1–RD2–Dsat)/B0 will be further adopted to characterize particular NOE experiments used in this work.
Analysis of systematic errors resulting from an incorrect delay setting in NOE values, ε = Ssat/S0, for nuclei with γ < 0 should take into account that these errors can be caused by false S0 values and/or Ssat values. The apparent S0,app value in not fully relaxed spectrum is always smaller than the S0 of true equilibrium value. On the other hand, the non-equilibrium apparent Ssat,app value is always larger than the Ssat, equilibrium value, i.e. more positive for ε > 0 or less negative for ε < 0. The joint effect of erroneous Ssat and S0, however, does not always result in the relation εapp > ε as could be hastily concluded. An attenuated S0 value in conjunction with properly determined, negative Ssat results in εapp < ε, and this is experimentally confirmed by ε values observed for the C-terminal, mobile residue G76. Its values obtained in the measurements free of systematic errors (10-10-8)/16.4 T and (10-10-5)/22.3 T are equal to − 0.812 and − 0.246, respectively. Herein, both, S0 and Ssat values are expected to be error free. In the measurements (10-10-4)/16.4 T and (10-10-1.3)/22.3 T with proper S0 value and Ssat,app > Ssat owing to too short Dsat, εapp are equal to − 0.738 and 0.162, respectively, while in (3-0-3)/22.3 T with too short RD1 and Dsat delays, S0,app < S0 and εapp = − 0.379 (cf. Figure 8). Such misleading behavior could be expected for mobile residues in flexible loops, unstructured termini, or intrinsically disordered proteins.
Fig. 8.
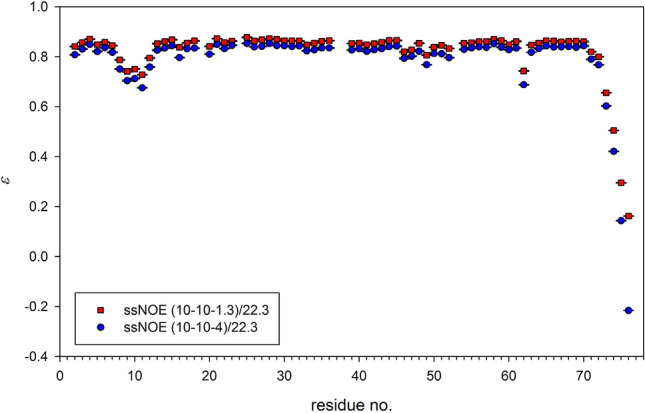
Nuclear Overhauser effect values obtained in steady-state NOE experiments with the saturation period Dsat set to 1.3 s (red squares) or 4 s (blue circles)
Setup and data processing of DNOE measurement
Relation between signal intensities and evolution times in a dynamic NOE experiment, Dsat, depend on three parameters: nuclear Overhauser effect, ε, nitrogen longitudinal relaxation rate, R1N, and signal intensity at the thermal equilibrium, S0 (Eq. 1). Provided that the longitudinal relaxation rates have been previously obtained in a separate experiment, their values can be entered in Eq. 1, reducing the number of determined parameters in a computational task further denoted as a sequential one. Influence of the propagation of R1N errors on the ε values is usually negligible; variation of R1N values within the range ± σ (standard deviation) typically results in dε changes smaller than 10–5 except for residues exhibiting ε < 0.4 (Figs. S1, S2). In ubiquitin, such residues are located at the C-terminus. This behavior is attributed to the stronger correlation between ε and R1N parameters owing to the increased range of signal intensities for smaller ε values (Fig. 2). Another possibility of data processing, simultaneous use of dynamic NOE and relaxation rate data in one computational task, brings about results (ε and dε values) practically identical to those obtained in the sequential task.
Fig. 2.
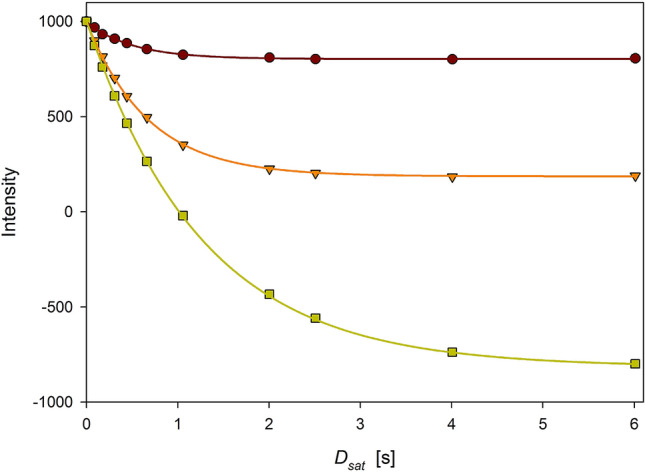
Experimental data obtained in DNOE measurement at 16.4 T for D58 residue (brown circles), R74 (orange triangles), and G76 (light green squares). NOE values determined in the sequential task are: (D58) = 0.805, ε (R74) = 0.186, and ε (G76) = − 0.813. Color-coded lines correspond to the nonlinear least-square fit of the Eq. (1) to the experimental data. Correlations between ε and R1, c(ε,R1), in the simultaneous task are: c(D58) = − 0.003, c(R74) = 0.013, and c(G76) = 0.099. The larger range of intensities results in larger correlation c(ε,R1) between fitted parameters
The dynamic NOE data can also be used without support from separate R1N data. Such data processing delivers the ε values and their errors close to those resulted from the sequential or simultaneous approach (Figs. S3, S5). On the other hand, derived R1 relaxation rates are less accurate with errors an order of magnitude larger than those obtained in the dedicated R1 experiment (Figs. S4, S6). Therefore, a dynamic NOE measurement cannot be regarded as a complete equivalence of a separate R1 experiment. Numerical data for three different data processing methods of dynamic NOE at 22.3 T are given in the Table S2, and a comparison of the discussed numerical methods are presented in Table 2 using data acquired for ubiquitin at 16.4 and 22.3 T. The pairwise root-mean-square deviations (RMSD) for ε values are extremely small in all cases, while those for R1 values are larger. Their values, together with average standard deviations, are given in Table 3. Recently, an experimentally demanding TROSY-based pulse sequence dedicated to deuterated proteins has been invented for simultaneous measurement of R1N relaxation rates and ε values. The accuracy of the proposed technique has been verified by comparison to the results of both relaxation parameters measured conventionally (O'Brien and Palmer III 2018).
Table 2.
Values of standard error ratios averaged over 70 amino acid residues of ubiquitin available from our experiments
| B0 [T] | dR1/R1 [s−1] | dε/ε | ||
|---|---|---|---|---|
| B/A | C/A | B/A | C/A | |
| 16.4 | 1.11 | 7.52 | 1.02 | 0.99 |
| 22.3 | 0.97 | 7.59 | 1.30 | 0.98 |
Comparison of the data reduction methods in dynamic NOE experiments: (A) sequential determination of R1 from the dedicated R1 measurement followed by the ε determination from DNOE measurement using previously determined R1 values, (B) simultaneous use of DNOE and R1 measurements in a single computational task, (C) DNOE measurement data alone used for the determination of R1 and ε values
Table 3.
The pairwise RMSDs and the mean values of standard deviations determined for three data reduction methods for dynamic NOE experiments
| Pairwise RMSD | The mean of standard deviations | ||||
|---|---|---|---|---|---|
| B0 = 22.3 T | ε | R1 [s−1] | σ (ε) | σ (R1) [s−1] | |
| A/B | 0.0002 | 0.0082 | A | 0.0029 | 0.0081 |
| A/C | 0.0016 | 0.0862 | B | 0.0036 | 0.0077 |
| B/C | 0.0017 | 0.0852 | C | 0.0028 | 0.0611 |
Labels A, B, and C are defined in the caption to Table 2
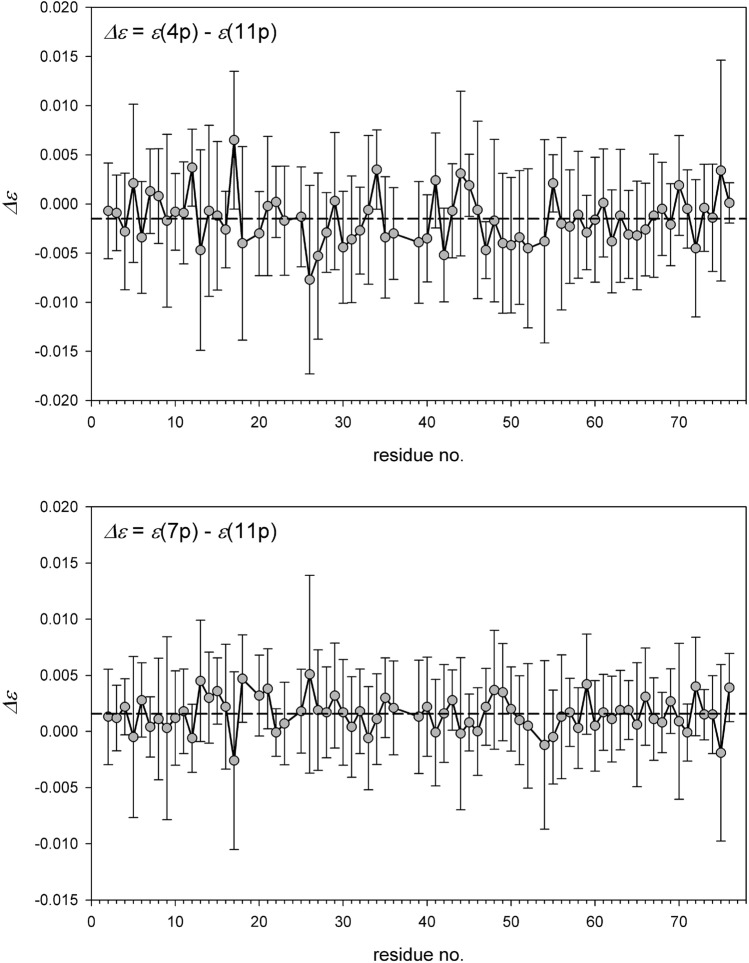
Dynamic NOE measurements, as with relaxation rate experiments, require optimization of a number and length of saturation periods, Dsat. One important assumption in the selection of Dsat values is to sample a broad range of intensities I(t) ~ S(t) in a uniform manner. The shortest Dsat equal to zero delivers I0 ~ S0. The longest Dsat should be as close to a value fulfilling the condition (2) as is practically feasible (c.f. Table S1). These assumptions were checked on the DNOE measurement comprising 11 delays. Next the number of delays was reduced to seven and then to 4 selected delays, and results were compared. Apparent NOE values and their standard deviations changed only slightly. Residue specific differences in ε values between the full experiment and each of the reduced ones were smaller than appropriate dε values. They are compared in Fig. 3, and the presented data assure that four correctly chosen Dsat values do not deteriorate ε values and their accuracies. This conclusion allows us to state that DNOE measurement can require an acceptable amount of spectrometer time.
Fig. 3.
Residue specific differences with error bars between DNOE measurement at 22.3 T comprising 11 Dsat values and curtailed DNOE measurements composed of four or seven Dsat values (upper part and lower part, respectively). Horizontal, dashed lines represent averages of Δε values given in plots. Full set of Dsat values [0.0, 0.11, 0.22, 0.35, 0.55, 0.66, 0.79, 1.10, 1.30, 3.00, 4.00]. Four values: 0.22, 0.66, 1.10, and 3.00 were rejected to get seven Dsat value measurement. Further rejection of 0.11, 0.55, and 1.30 Dsat values resulted in four-value set
Error determination of NOE measurements
The NOE errors are equally important to NOE values themselves. They are used to weigh the NOE data in the relaxation-based backbone protein dynamics calculation (Palmer et al. 1991; d’Auvergne 2008; Jaremko et al. 2015). Inaccurate values of NOE errors can result in the erroneous estimation of protein backbone dynamics. Particularly, the overestimation of NOE leads to significant errors in the local dynamics parameters as evidenced by appropriate simulations (Ferrage et al. 2008). Occasionally, the average values of the NOE and standard errors in the mean have been determined from several separate NOE data sets (Stone et al. 1992; Renner et al. 2002). Nonetheless, it has been most often accepted to use signal-to-noise ratios (SNR) in the determination of steady-state NOE errors (Farrow et al. 1994; Tjandra et al. 1995; Fushman 2003).
| 4 |
The Eq. (4) is an approximation of exact formulation of experimental error determination since it takes into account only this part of experimental errors which arises from the thermal noise. It can be safely used if the thermal noise dominates other contributions to the total experimental error. A weak point in Eq. (4) arises also from the fact that amino acid residues located in flexible parts of macromolecules often display NOE values close to zero, which results in the underestimation of dε, owing to the factor as shown in Eq. (4).
Justification of an SNR-based approach should comprise two issues: checking of the reliability of SNR determination delivered by commonly used processing tools and comparison of the SNR-determined errors with those obtained from the statistical analysis of a series of independent NOE measurements. To the best of authors' knowledge, such study has not been yet undertaken for 15N nuclei in proteins and has only be performed once for 13C nuclei (Bernatowicz et al. 2010). In our study, we found that SNR values automatically derived in the peak intensity determination differed from those obtained semi-manually; their larger part was overestimated. Therefore, automatically delivered SNR values concomitant cross peak intensities cannot be taken for granted. Description of the SNR issue is given in the Supporting Material (section: Determination of signal-to-noise ratio). In order to closely analyze the relevance of SNR-based NOE errors, a series of 10 NOE measurements was performed at 22.3 T using identical spectrometer setup. A comparison of standard deviations (σ) calculated for each of 70 residues of ubiquitin with corresponding means of SNR-based NOE errors is presented in Fig. 4. It can be concluded from Fig. 4 that values of two presented sets of NOE errors are very similar, and their means are close to one another with a difference of 8⋅10−5. Individual ε values for the residue A46 showing the largest NOE data dispersion are compared with the mean and the standard deviation in Fig. 5. Examination of Figs. 4 and 5 allows us to conclude that properly determined SNR-based NOE errors are reliable and can be safely used in further applications.
Fig. 4.
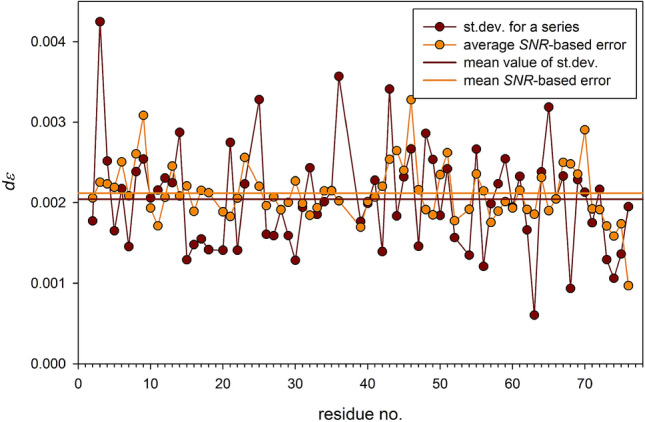
Standard deviations (σ) calculated for 70 residues of ubiquitin (brown circles) and their mean (solid horizontal brown line) determined for series of ten measurements. Means of ten SNR-based errors calculated for each residue (orange circles) and their mean (solid horizontal orange line)
Fig. 5.
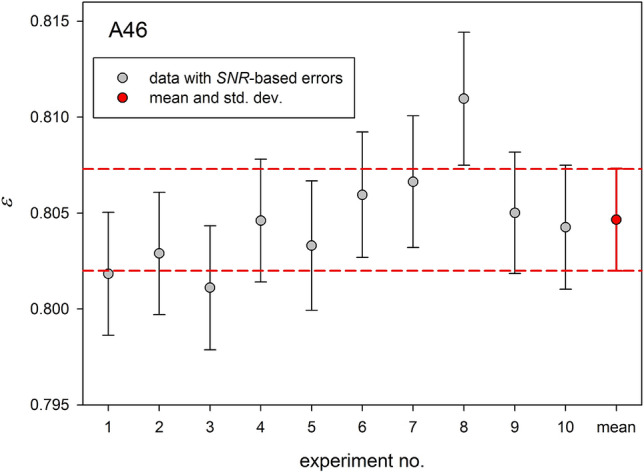
The NOE values of A46 residue obtained in a series of 10 measurements with appropriate SNR-based errors (gray circles with SNR-based error bars) and their mean with standard deviation (red circle). Dashed red lines correspond to the mean ± σ
Saturation of HN protons
Originally, saturation of proton resonances was achieved by a train of 250° pulses at 10 ms intervals (Markley et al. 1971). In protein relaxation studies, however, a train of 120° pulses spaced 20 ms apart was commonly used for this purpose (Kay et al. 1989). In search of the optimal 1H saturation scheme, different pulse lengths (120°, 180°, 250°) and different pulse spacings (5 ms, 10 ms, 20 ms) were employed (Renner et al. 2002). Finally, it was concluded that pulses of approximately 180° at l0 ms intervals performed slightly better than other settings.
Extensive experimental survey of HN proton saturation accompanied by theoretical calculations based on averaged Liouvillian theory was carried out on all components of saturation sequence (Ferrage et al. 2009, 2010). It was concluded that the best results were obtained using the symmetric 180° pulse train (τ/2 − 180° − τ/2)n with τ = k/JNH, where n—the integer determining length of saturation time (Dsat = n⋅ τ) and k—a small integer, usually k = 2, giving τ about 22 ms. It was also suggested to move the proton carrier frequency from water resonance to the center of the amide region and reduce the power of the 180° pulses to minimize sample heating.
Analysis of NOE experiments
NOE experiments performed to analyze the influence of a particular sequence of parameters on the apparent nuclear Overhauser effects values, εapp, are presented in Table S3. Experiments, ssNOE(10-10-8)/16.4, DNOE/16.4, ssNOE(14-0-14)/18.8, ssNOE(13-0-3)/22.3, and DNOE/22.3 can be expected to deliver the most accurate results. They are regarded as a kind of reference point for a selected magnetic field.
The importance of using appropriate Dsat values in steady-state NOE measurements is demonstrated by comparing NOEs in the experiments (14-0-4)/18.8 and (14-0-14)/18.8. The first displays a systematic increase of εapp owing to incomplete HN saturation during Dsat. Residue specific differences between the mentioned experiments are shown in Fig. 6. Residues G75 and G76 with negative ε values display decreased εapp as discussed earlier (section: Time schedule of NOE measurement).
Fig. 6.
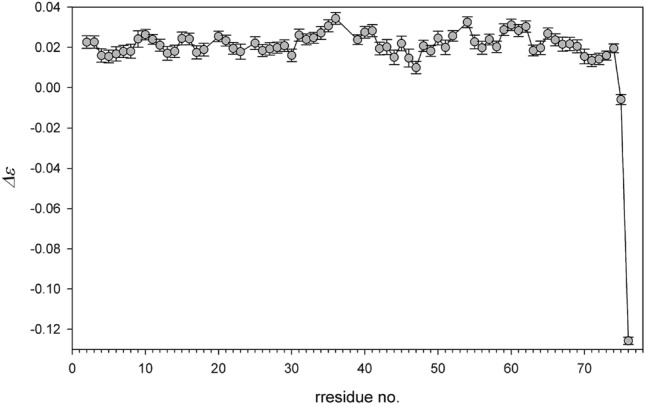
NOE differences Δε = εapp − ε obtained in measurements performed at 18.8 T with Dsat = 4 s (εapp) and Dsat = 14 s (ε). Average difference after rejection of G75 and G76 with ε < 0 is equal to 0.022
Calculation of factors using residue specific R1N data is presented in Fig. 7 for Dsat values utilized in the measurements performed at 22.3 T as listed in Table S3. The Dsat = 3 s is sufficiently long for all residues except the last two C-terminal glycines, G75 and G76. In fact, even Dsat = 4 s is not long enough for the observation of unperturbed G76. Therefore, it is not surprising that Dsat = 1.3 s is much too short, and εapp values derived from the experiment (10-10-1.3)/22.3 are significantly larger than those obtained at the longer period of Dsat = 4 s (Fig. 8), on average, 0.0348.
Fig. 7.
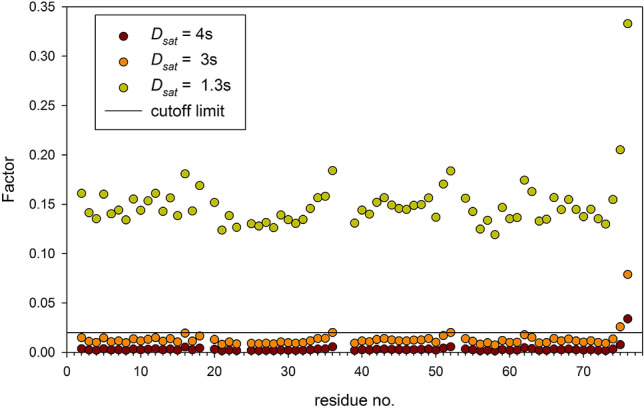
Factor characterizing efficiencies of the saturation of nitrogen magnetization for different Dsat values were calculated using residue specific R1N values determined at 22.3 T in a separate measurement. A common sense but arbitrary limit 0.02 is marked with a horizontal line
The effect of a very short RD1 delay can be demonstrated by comparing experiments ssNOE(13-0-3)/22.3, ssNOE(10-10-3)/22.3, ssNOE(6-0-3)/22.3, and ssNOE(3-0-3)/22.3 (Fig. 9). The RD1 = 3 s and RD1 = 6 s result in the increase of ε magnitudes relative to the RD1 = 13 s on average, 0.0544 and 0.0042, respectively. On the other hand, average difference between measurements with RD1 = 13 s and RD1 = 10 s is negligible − 0.0007. This result gives evidence that RD1 delay equal to 10 s allows to reach the equilibrium state of HN protons in the studies system.
Fig. 9.
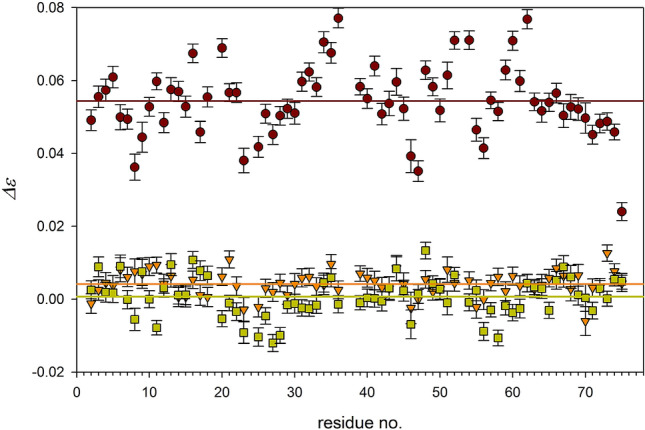
NOE differences Δε = εapp − ε obtained for measurements performed at 22.3 T: ssNOE(13-0-3)/22.3, ssNOE(10-10-3)/22.3 (extracted from DNOE), ssNOE(6-0-3)/22.3, and ssNOE(3-0-3)/22.3. Δ for the RD1 pair: 3 s and 13 s (brown circles), the RD1 pair: 6 s and 13 s (orange triangles), the RD1 pair: 10 s and 13 s(light green squares). Color coded average differences after rejection of G76 with ε < 0 are equal to 0.0544, 0.0042, and 0.0007
Concluding, comparison of the NOE values obtained at different settings of Dsat or RD1 highlights the importance of the correct choice of delays in the determination of accurate ε values.
Correction factors
As has been shown above, the effect of slow spin-lattice relaxation of water protons and the chemical exchange of amide protons with water combined with too short relaxation delays in the steady-state NOE experiments usually results in substantial systematic NOE errors owing to the incomplete relaxation towards the steady-state or equilibrium 15N polarization. Therefore, several correction factors were introduced to compensate such errors using the following equation
| 5 |
where ε and εapp are exact and apparent NOE values, respectively.
It has been claimed that the effect of incomplete R1W recovery can be corrected by substituting the factor
| 5A |
into Eq. 5 (Skelton et al. 1993). It has been also suggested that factor
| 5B |
allows for the correction of the not sufficiently long relaxation delay RD with respect to R1H (Grzesiek and Bax 1993). Another correction that takes into consideration the inconsistency of both R1N and R1H with relaxation delays has also been recommended (Freedberg et al. 2002):
| 5C |
Efficiencies of all three corrections were checked on the NOE measurement with the intentionally too short delays: RD1 = 3 s, RD2 = 0, and Dsat = 3 s, (3-0-3)/22.3. As shown earlier (Fig. 9), all εapp in (3-0-3)/22.3 measurement were larger than corresponding ε values in the correctly performed measurement (13-0-3)/22.3. The mean of differences was equal to 0.054. None of these above-listed corrections was able to fully compensate the effect of wrong adjustment of RD1 delay. Three corrections allowing for R1W (Eq. 5), R1H (Eq. 5B), and R1H and R1N (Eq. 5C) resulted in the means of absolute differences equal to 0.019, 0.048, and 0.036, respectively (Fig. 10). Therefore, these corrections have compensated for the delay missetting by 67%, 17%, and 38%, respectively. Obviously, the R1W effect is the most important factor for compensation.
Fig. 10.
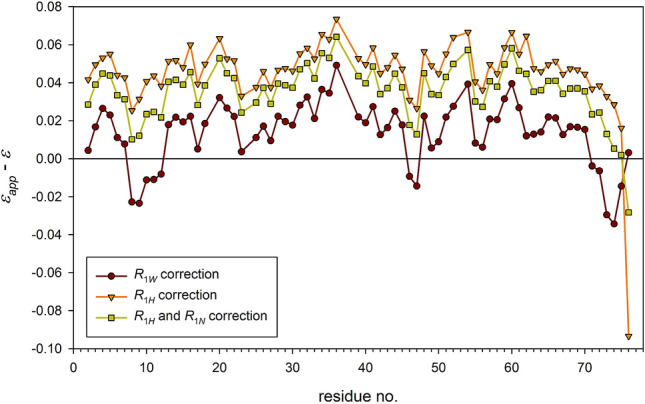
Residue specific differences between corrected εapp and ε values obtained in (13-0-3)/22.3 measurement. The εapp values were obtained from (3-0-3)/22.3 experiment after compensation for R1W (Eq. 5A, brown circles), R1H (Eq. 5B, orange triangles), and R1H, R1N (Eq. 6, light green squares). Horizontal color-coded lines correspond to appropriate means of difference magnitudes
Compensation for a not long enough Dsat period with properly chosen RD1 is an easier task. The experiment (10-10-1.3)/22.3 was discussed earlier, and its results were shown in Fig. 8. Use of another correction,
| 6 |
results in the corrected εapp values, which differ from the DNOE experiment by an average of 0.003 (Fig. S7). Nevertheless, in view of the above-mentioned results, it is obvious that none of the existing correction terms should be used as a substitute for a properly designed experiment.
Conclusions
In this study, it has been shown that dynamic NOE measurement is an efficient and accurate method for NOE determination. In particular, it presents its usefulness in cases of NOE values that are close to zero. This method provides a robust and more accurate alternative to widely used steady-state NOE measurement. The DNOE measurement allows for the determination of NOE values and their accuracies with standard nonlinear regression methods. If high accuracy longitudinal relaxation rates R1 are not of great importance, they can be simultaneously obtained with a reduced accuracy as a "by-product" in the DNOE data processing without any significant reduction of the accuracy and precision of determined NOE values.
It has been proven that commonly used methods of NOE accuracy based on the signal-to-noise ratio accompanying steady-state NOE measurements are reliable provided that root-mean-square noise has been determined correctly.
It has to be stressed that in view of the results presented in this work, none of the existing correction terms are able to restore accurate NOE values in cases where measurements are improperly set up and performed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplementary material 1 (PDF 1100 kb)
Acknowledgements
The authors would like to thank the Imaging and Characterization Core Lab at the King Abdullah University of Science and Technology (KAUST) for the access to the NMR facilities. This publication is based upon work supported by KAUST Office of Sponsored Research (OSR) under Award No. OSR-CRG2018-3792 (LJ) and through baseline-funds (LJ and MJ).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- d’Auvergne E (2008) Protein Dynamics A Study of the Model-free Analysis of NMR Relaxation Data. VDM Verlag Dr. Müller, Saarbrücken, Germany
- Bernatowicz P, Ruszczynska-Bartnik K, Ejchart A, Dodziuk H, Kaczorowska E, Ueda H. Carbon-13 NMR relaxation study of the internal dynamics in cyclodextrins in isotropic solution. J Phys Chem B. 2010;114:59–65. doi: 10.1021/jp9084734. [DOI] [PubMed] [Google Scholar]
- Canet D. Systematic errors due to improper waiting times in heteronuclear overhauser effect measurements by the gated decoupling technique. J Magn Reson. 1976;23:361–364. [Google Scholar]
- Cornilescu G, Marquardt J, Ottiger M, Bax A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ejchart A, Gryff-Keller A, Szczeciński P. Silicon-29 and carbon-13 longitudinal nuclear spin relaxation study on molecular dynamics of 1,4-Bis(trimethylsilyl)-1,3-butadiyne in solution. J Magn Reson. 1992;97:313–321. [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Ferrage F, Piserchio A, Cowburn D, Ghose R. On the measurements of 15H-{1H} nuclear Overhauser effects. J Magn Reson. 2008;192:302–313. doi: 10.1016/j.jmr.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrage F, Cowburn D, Ghose R. Accurate sampling of high-frequency motions in proteins by steady-state 15N-{1H} nuclear Overhauser effect measurements in the presence of cross-correlated relaxation. J Am Chem Soc. 2009;131:6048–6049. doi: 10.1021/ja809526q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrage F, Reichel A, Battacharya S, Cowburn D, Ghose R. On the measurements of 15H-{1H} nuclear Overhauser effects. 2. Effects of the saturation scheme and water signal suppression. J Magn Reson. 2010;207:294–303. doi: 10.1016/j.jmr.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D. In: BioNMR in drug research. Zerbe O, editor. Weinheim: Wiley; 2003. pp. 283–308. [Google Scholar]
- Goddard TD, Kneller DG, SPARKY 3: University of California, San Francisco. https://www.cgl.ucsf.edu/home/sparky
- Gong Q, Ishima R. 15N–{1H} NOE experiment at high magnetic field strengths. J Biomol NMR. 2007;37:147–157. doi: 10.1007/s10858-006-9125-7. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- Harris RK, Newman RH. Choice of pulse spacings for accurate T1, and NOE measurements in NMR spectroscopy. J Magn Reson. 1976;24:449–455. [Google Scholar]
- Harris RK, Kowalewski J, Cabral de Menezes S. Parameters and symbols for use in nuclear magnetic resonance. Pure Appl Chem. 1997;69:2489–2495. doi: 10.1016/s0926-2040(97)00079-9. [DOI] [PubMed] [Google Scholar]
- Idiyatullin D, Daragan VA, Mayo KM. Improved measurement of 15N–{1H} NOEs in the presence of H(N)-water proton chemical exchange. J Magn Reson. 2001;153:138–143. doi: 10.1006/jmre.2001.2412. [DOI] [PubMed] [Google Scholar]
- Jaremko Ł, Jaremko M, Nowakowski M, Ejchart A. The quest for simplicity: remarks on the free-approach models. J Phys Chem B. 2015;119:11978–11987. doi: 10.1021/acs.jpcb.5b07181. [DOI] [PubMed] [Google Scholar]
- Jarymowycz VA, Stone MJ. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kempf JG, Loria JP. Protein dynamics from solution NMR. Cell Biochem Biophys. 2003;37:187–211. doi: 10.1385/cbb:37:3:187. [DOI] [PubMed] [Google Scholar]
- Kimber BJ, Harris RK. Measurement of spin-lattice relaxation times in the null-signal case. J Magn Reson. 1974;16:354–356. [Google Scholar]
- Kuhlmann KF, Grant DM, Harris RK. Nuclear Overhauser effects and 13C relaxation times in 13C-{H} double resonance spectra. J Chem Phys. 1970;52:3439–3448. [Google Scholar]
- Kuhlmann KF, Grant DM. Carbon-13 relaxation and internal rotation in mesitylene and o-xylene. J Chem Phys. 1971;53:2998–3007. [Google Scholar]
- Levy GC, Holloway CE, Rosanske RC, Hewitt JM, Bradley CH. Natural abundance nitrogen-15 n.m.r. spectroscopy. Spin-lattice relaxation in organic compounds. Org Magn Reson. 1976;8:643–647. [Google Scholar]
- Lakomek NA, Ying J, Bax A. Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Horsley WJ, Klein M. Spin-lattice relaxation measurements in slowly relaxing complex spectra. J Chem Phys. 1971;55:3604–3605. [Google Scholar]
- Noggle JH, Schirmer RE. The nuclear Overhauser effect. Chemical application. New York: Academic Press; 1971. [DOI] [PubMed] [Google Scholar]
- O'Brien PA, Palmer AG., III TROSY pulse sequence for simultaneous measurement of the 15N R1 and {1H}–15N NOE in deuterated proteins. J Biomol NMR. 2018;70:205–209. doi: 10.1007/s10858-018-0181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AG, III, Rance M, Wright PE. Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance 13C heteronuclear NMR spectroscopy. J Am Chem Soc. 1991;113:4371–4380. [Google Scholar]
- Palmer AG., III NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes: the art of scientific computing. New York: Cambridge University Press; 2007. [Google Scholar]
- Raiford DS, Fisk CL, Becker ED. Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Anal Chem. 1979;51:2050–2051. [Google Scholar]
- Reddy T, Rayney JK. Interpretation of biomolecular NMR spin relaxation parameters. Biochem Cell Biol. 2010;88:131–142. doi: 10.1139/o09-152. [DOI] [PubMed] [Google Scholar]
- Renner C, Schleicher M, Moroder R, Holak TA. Practical aspects of the 2D 15N-{1H}-NOE experiment. J Biomol NMR. 2002;23:23–33. doi: 10.1023/a:1015385910220. [DOI] [PubMed] [Google Scholar]
- Skelton NJ, Palmer AG, III, Akke M, Kördel J, Rance M, Chazin WJ. Practical aspects of two-dimensional proton-detected 15N spin relaxation measurements. J Magn Reson B. 1993;102:253–264. [Google Scholar]
- Solomon I. Relaxation processes in a system of two spins. Phys Rev. 1955;99:559–565. [Google Scholar]
- Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur J Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- Stone MJ, Fairbrother WJ, Palmer AG, III, Reizer J, Saier MH, Jr, Wright PE. Backbone dynamics of the Bacillus subtilis glucose permease IIA domain determined from 15N NMR relaxation measurements. Biochemistry. 1992;31:4394–4406. doi: 10.1021/bi00133a003. [DOI] [PubMed] [Google Scholar]
- Stetz MA, Caro JA, Kotaru S, Yao X, Marques BS, Valentine KG, Wand AJ. Characterization of internal protein dynamics and conformational entropy by NMR relaxation. Method Enzymol. 2019;615:237–284. doi: 10.1016/bs.mie.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao CG, Abildgaad F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Zhukov I, Ejchart A. Factors improving the accuracy of determination of 15N relaxation parameters in proteins. Acta Biochim Pol. 1999;46:665–671. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic supplementary material 1 (PDF 1100 kb)