Abstract
The human zinc finger (C2H2-type) protein ZNF750 is a transcription factor regulated by p63 that plays a critical role in epithelial tissues homoeostasis, as well as being involved in the pathogenesis of cancer. Indeed, missense mutations, truncation and genomic deletion have been found in oesophageal squamous cell carcinoma. In keeping, we showed that ZNF750 negatively regulates cell migration and invasion in breast cancer cells; in particular, ZNF750 binds and recruits KDM1A and HDAC1 on the LAMB3 and CTNNAL1 promoters. This interaction, in turn, represses the transcription of LAMB3 and CTNNAL1 genes, which are involved in cell migration and invasion. Given that ZNF750 is emerging as a crucial transcription factor that acts as tumour suppressor gene, here, we show that ZNF750 represses the expression of the small GTPase, Ras-related C3 botulinum toxin substrate 1 (RAC1) in breast cancer cell lines, by directly binding its promoter region. In keeping with ZNF750 controlling RAC1 expression, we found an inverse correlation between ZNF750 and RAC1 in human breast cancer datasets. More importantly, we found a significant upregulation of RAC1 in human breast cancer datasets and we identified a direct correlation between RAC1 expression and the survival rate of breast cancer patient. Overall, our findings provide a novel molecular mechanism by which ZNF750 acts as tumour suppressor gene. Hence, we report a potential clinical relevance of ZNF750/RAC1 axis in breast cancer.
Subject terms: Breast cancer, Tumour biomarkers
Introduction
ZNF750 is a zinc finger transcription factor that plays an important role in controlling the homoeostasis of epithelial tissue1,2. Indeed, ZNF750 deregulation results in neoplastic transformation3. ZNF750 consists of an atypical C2H2 zinc finger motif in the N-terminal domain, which is required for ZNF750 transcriptional function. In addition, two highly conserved PLNLS motifs, that are required for protein–protein interaction, are present in the C-terminal domain4. ZNF750 regulates epidermal differentiation by activating differentiation gene expression cooperatively with KLF4 and by inhibiting the expression of progenitor genes in association with KDM1A. In addition, ZNF750 has been described as a tumour suppressor protein in squamous cell carcinomas (SCCs) of the oesophagus and lung5,6. In SCCs, ZNF750 results frequently mutated, and truncation and missense mutations represent the most frequent genetic alterations. Notably, overexpression of ZNF750 inhibits cell proliferation and migration of SCC cells lines by regulating the expression of terminal differentiation-induced ncRNA (TINCR)7, through which it controls cancer cell proliferation and inhibits the expression of Laminin Subunit Gamma 2 (LAMC2), a component of Laminin-332 (ref. 5). Accordingly, low expression of ZNF750 has been associated with poor prognosis. We have recently shown that ZNF750 negatively regulates cell migration, invasion and proliferation in breast cancer cells. Specifically, ZNF750 binds and recruits KDM1A and HDAC1 on the Laminin Subunit Beta 3 (LAMB3) and Catenin Alpha Like 1 (CTNNAL1) promoters. This interaction in turn represses the transcription of LAMB3 and CTNNAL1 genes, two well-known proteins involved in cell migration and invasion8.
Breast cancer is the most common invasive cancer diagnosed in women. It is a complex disease that shows both inter- and intratumoral heterogeneity. Gene expression profiling studies have identified five distinct subtypes of breast cancers: luminal A, luminal B, HER2-enriched, basal-like and normal-like. Each of these subtypes is characterized by distinct disease progression, therapeutic response and clinical outcome. Therefore, a stratification of the patients is necessary to achieve better clinical outcome and for predicting the clinical course of the disease9,10. This is a general problem in clinical cancer research11–13, and current research is very active in this respect14, including for example on neuroblastoma9,15–17. To achieve this, novel molecular pathways that are implicated in the pathogenesis of cancer should be uncovered and identification of novel biomarkers is needed for predicting patient clinical outcome15,16,18.
Although in the last few years several studies have highlighted the role of ZNF750 as tumour suppressor gene, the molecular mechanisms underlying the inhibition of cancer onset and progression are not well characterized. In this study, we add a layer of complexity by showing that ZNF750 acts as tumour suppressor protein by transcriptional inhibition of Ras-related C3 botulinum toxin substrate 1 (RAC1), a member of the Rho/Rac GTPase family19.
Results
RAC1 is upregulated in breast cancer cells and its expression is controlled by ZNF750
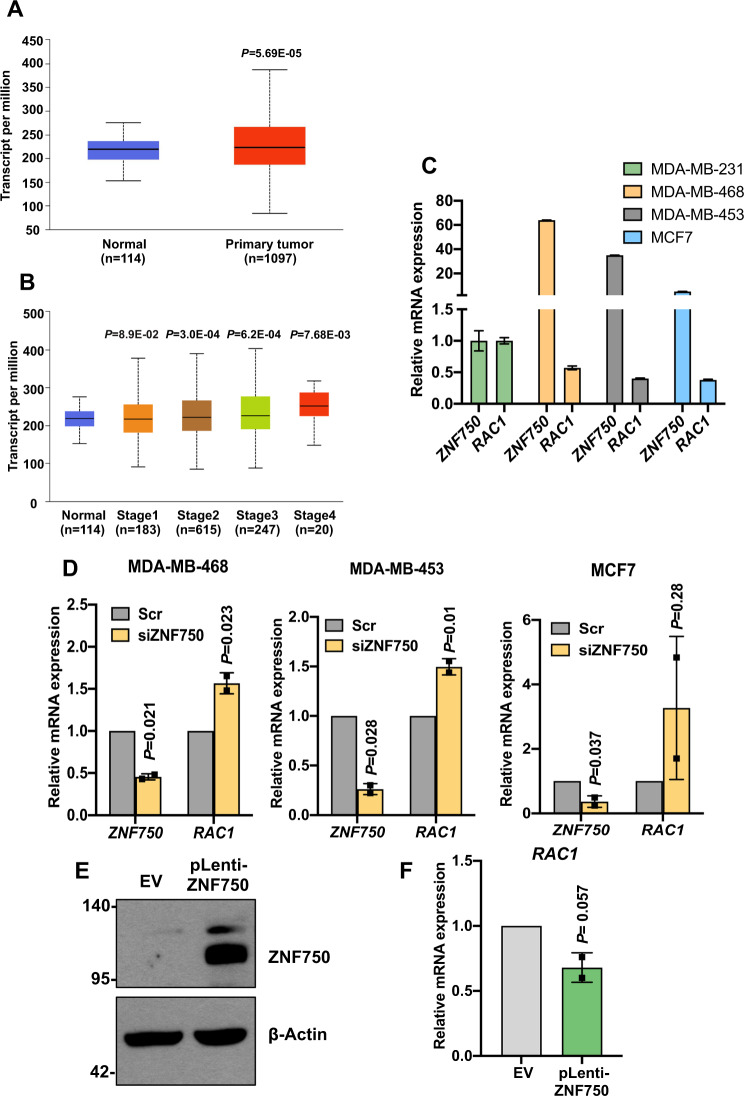
We have previously demonstrated that ZNF750 acts as a tumour suppressor in breast cancer8. In particular, ZNF750 inhibits migration and invasion of the breast cancer cell lines by downregulating Wnt signalling. Among the several players involved in the Wnt signalling, RAC1 mediates the transduction of the non-canonical Wnt pathway and therefore is implicated in the establishment of cell polarity and cell migration20. We have recently found that RAC1 was upregulated after ZNF750 depletion in breast cancer cell lines. To gain insight into the role of RAC1 in breast cancer, we first performed a comprehensive analysis of the expression of RAC1 in breast cancer tissue samples. As shown in Fig. 1A, high levels of RAC1 mRNA are found in primary tumour when compared with the normal tissue. Moreover, RAC1 expression positively correlates with tumour stage (Fig. 1B). To further explore the relationship between ZNF750 and RAC1, we tested RAC1 expression by qPCR in several breast cancer cell lines characterized by different ZNF750 expression levels: MDA-MB-468 (high expression), MDA-MB-453 (high expression), MCF7 (moderate expression) and MDA-MB 231 (low expression). Interestingly, we observed that RAC1 expression inversely correlated with ZNF750. In particular, as shown in Fig. 1C, in the high-ZNF750-expressing MDA-MB-468 and MDA-MB-453 cell lines, RAC1 was expressed at lower levels as compared to the more aggressive MDA-MB-231. Conversely, MDA-MB-231 cell line, which was previously characterized by the lowest ZNF750 levels8, showed the highest levels of RAC1 (Fig. 1C). In order to confirm that the RAC1 expression was ZNF750-dependent, we depleted ZNF750 using a specific siRNA in MDA-MB468, MDA-MB-453 and MCF7. Interestingly, ZNF750 depletion resulted in a significant RAC1 upregulation in all the three breast cancer lines (Fig. 1D). On the contrary, overexpression of ZNF750 in MDA-MB-231 (Fig. 1E) cells resulted in a reduction of RAC1 mRNA levels (Fig. 1F). Collectively, these data suggest that ZNF750 controls the expression of RAC1 in breast cancer cell lines.
Fig. 1. RAC1 expression is controlled by ZNF750 in breast cancer cells.
A, B Bioinformatic analysis showing the increased expression of RAC1 in breast cancer and in the different cancer stages. The graphs were directly downloaded from the UALCAN website (http://ualcan.path.uab.edu/index.html). The datasets analysed were from TCGA. C Expression of ZNF750 and RAC1 across several breast cancer cell lines. Data represent the mean of three technical replicates (N = 3, PCR runs) ± SD and are representative of one experiment. D Depletion of ZNF750 caused RAC1 upregulation at the mRNA level. Bars represent the mean ± SD of two independent experiments. E Immunoblot showing the overexpression of ZNF750 in transduced MDA-MB-231. F Overexpression of ZNF750 represses RAC1 mRNA expression. Bars represent the mean ± SD of two independent experiments (N = 2).
ZNF750 directly binds RAC1 promoter region
To gain insight into the molecular mechanism by which ZNF750 regulates the expression of RAC1, we performed a careful analysis of RAC1 proximal promoter using the JASPAR21 and MEME suite websites22. By scanning the 1 kbp-upstream region of RAC1 gene, we found two putative binding sites for ZNF750 located at 882 bp and 897 bp, respectively (Fig. 2A). To investigate whether ZNF750 directly binds the promoter region of RAC1, we performed chromatin immunoprecipitation (ChIP) in MDA-MB-468 cells. As shown in Fig. 2B, endogenous ZNF750 directly binds the RAC1 promoter region suggesting a potential direct transcriptional control. Overall these data indicate that ZNF750 negatively modulates RAC1 expression through a direct binding to its promoter.
Fig. 2. ZNF750 binds the promoter region of RAC1.
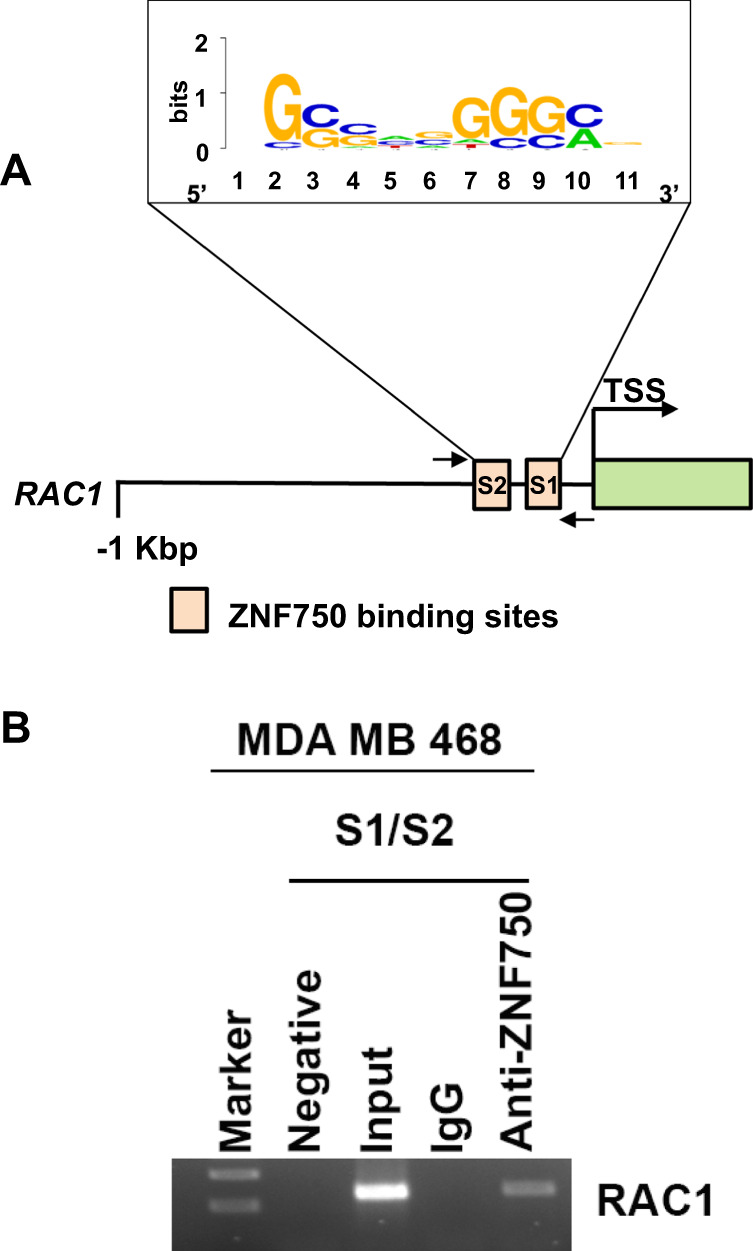
A Schematic representation of the scanned RAC1 promoter region with two putative binding sites for ZNF750 containing the consensus sequence reported. B Chromatin immunoprecipitation assay showing the binding of ZNF750 to RAC1 promoter region. Arrows indicate the primer position for PCR amplification.
ZNF750 and RAC1 negatively correlate in human breast cancer
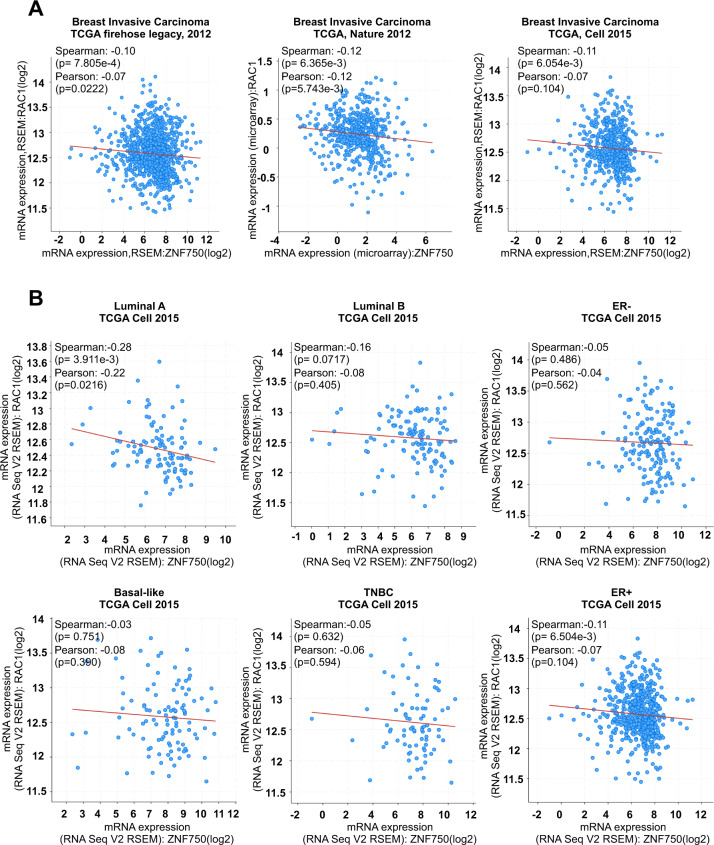
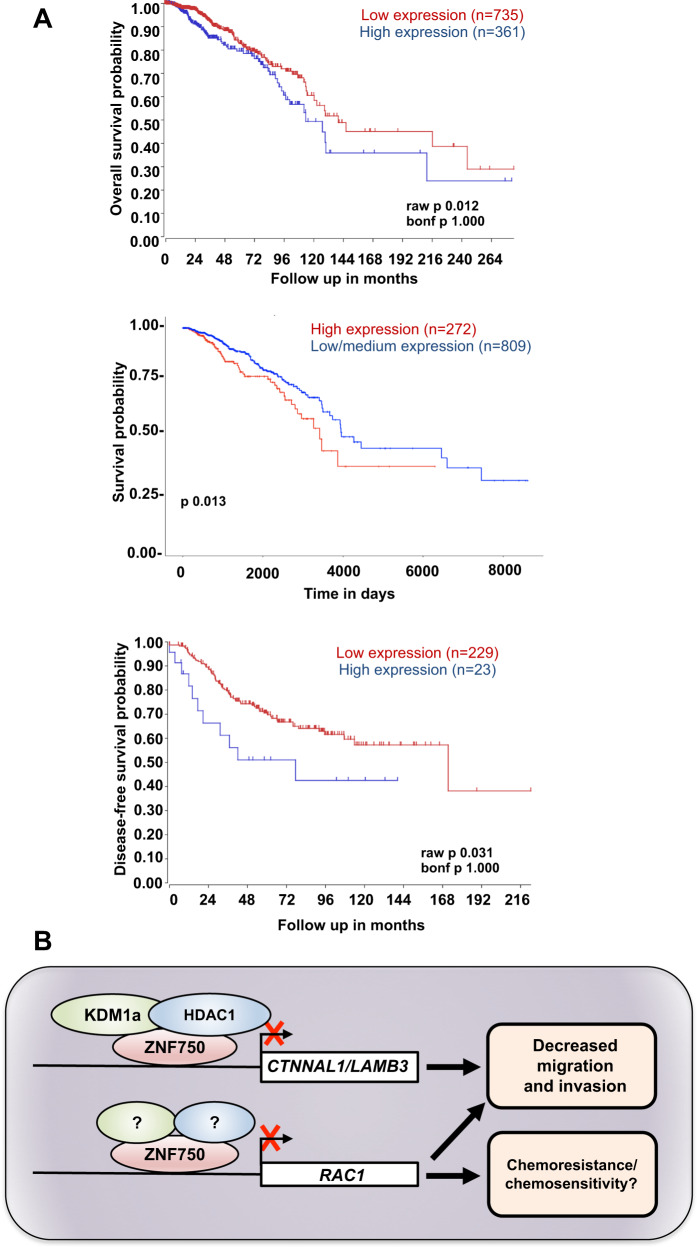
RAC1 expression increases with the tumour progression, and hyperactivation and/or overexpression are associated with a very poor prognosis23, while, on the contrary, ZNF750 levels significantly decrease with the breast cancer stage and negatively correlate with cancer aggressiveness. Then, we asked whether the negative correlation between ZNF750 and RAC1 may have a clinical relevance. Thus, using the cBioPortal database24,25, we asked whether the negative correlation between ZNF750 and RAC1 observed in breast cancer cell lines was also conserved in human breast cancer by using publicly available breast cancer datasets. As shown in Fig. 3A, we found a significant negative correlation between ZNF750 and RAC1 in all the breast invasive carcinoma datasets analysed. Given that breast cancer is characterized by high molecular and cellular heterogeneity26, we analysed the correlation between ZNF750 and RAC1 among the different breast cancer subtypes. The bioinformatic analysis reported a significant correlation (p < 0.05) only for the luminal A subtype (Fig. 3B). Remarkably, RAC1 expression was discriminatory of good and bad prognosis in breast cancer patients. Indeed, as shown in Fig. 4, high levels of RAC1 were associated with a worse survival in all the cancer datasets assessed. Overall our data indicate that ZNF750 acts as tumour suppressor gene in breast cancer by negatively regulating the expression of RAC1.
Fig. 3. ZNF750 and RAC1 expressions negatively correlate in breast cancer patients.
A Bioinformatic analysis showing a negative correlation between ZNF750 and RAC1 in breast invasive carcinoma. The datasets analysed were TCGA, Cell 2015; TCGA Nature 2012; TCGA Firehose Legacy, 2012. B ZNF750 and RAC1 inversely correlated in different breast cancer subtypes. The dataset analysed was TCGA, Cell 2015. For A and B, the analysis was carried out using the cBioPortal for Cancer Genomics database (http://www.cbioportal.org).
Fig. 4. High level of RAC1 is associated with reduced patient survival.
A Kaplan–Meier survival analysis showing that higher expression of RAC1 associates with a poor patient survival. The analysis was performed across several datasets of breast invasive carcinoma using cBioPortal for Cancer Genomics database, the R2 Genomic Platform (https://r2.amc.nl) and UALCAN website (http://ualcan.path.uab.edu/index.html). B Schematic representation of ZNF750 role in breast cancer. ZNF750 represses breast cancer migration and invasion by recruiting the epigenetic modifiers KDM1a and HDAC1 on CTNNAL1 and LAMB3 promoters (ref. 8). Furthermore, ZNF750 represses RAC1 expression by binding to its promoter, therefore modulating the cytoskeleton remodelling and possibly affecting migration and chemoresistance in breast cancer.
Discussion
In the last decade, OMICS technologies have contributed to our understanding of the pathogenesis of cancer27–30 leading to the development of precision oncology31,32, where selection of the patients for treatment is fundamental for better therapy outcome33. Therefore, identification of novel biomarkers for guiding treatment selection is a key requirement14,34–36.
We have previously identified the involvement of p63 in the progression of breast cancer through regulation of Sonic Hedgehog signalling37 in epithelial stem cells38 that is able to interact with p5339 or IKK40 to keep epithelial homoeostasis41,42 or to kill43. Although p63 belongs to the p53 family44–46, this is a unique property of p6347.
In the current study, we show that the axis ZNF750/RAC1 may function as a potential biomarker for predicting patient survival in breast cancer. In particular, ZNF750 transcriptionally represses the expression of RAC1, a member of the Rho/Rac GTPase family48 by binding its promoter region. In agreement, ZNF750 expression negatively correlates with RAC1 expression in human breast cancer and in particular in luminal A subtypes. RAC1 is a molecular switch that can be found either in an active state, when complexed to GTP, and in an inactive state when bound to GDP. When activated, RAC1 is implied in the cell migration, through a cytoskeleton rearrangement, and in cell survival. In the latter case, a large number of studies have demonstrated that the cell survival potential occurs via activation of the MAPK kinase pathway, through the formation of the Raf/MEK/ERK complex49. In addition, RAC1 directly interacts with PI3K, stimulating the PI3K/AKT signalling and survival50–52. Of relevance, a somatic mutation of RAC1 has been found as an oncogenic driver in melanoma, head and neck and prostate cancers53,54. Moreover, high expression of RAC1 was shown to be associated with poor outcome in several human cancers, such as colorectal cancers, and leukaemia19,55,56. In agreement with this scenario, our findings provide evidences for a parallel and/or alternative molecular mechanism by which ZNF750 functions as tumour repressor in breast cancer. In fact, we would like to speculate that besides repressing LAMB3 and CTNNAL1 expression, ZNF750 might also inhibit migration and invasion in breast cancer by repressing the expression of RAC1 (Fig. 4B). Despite its role in migration and survival, ZNF750 is also involved in resistance to chemoradiotherapy. Indeed, high levels of ZNF750 are associated with a good response to chemoradiotherapy in oesophageal SCC57. Interestingly, recently it has been shown that overexpression of RAC1 confers resistance to neoadjuvant chemotherapy in triple negative breast cancer23,58. Therefore, it is tempting to hypothesize that ZNF750 sensitizes cancer cells to chemotherapy by repressing the expression of RAC1. Although this should be tested in clinical setting, our data may also suggest that the ZNF750/RAC1 axis may serve as a novel candidate biomarker for chemoradiotherapy sensitivity.
In cancer, treatment is becoming crucial to identify biomarkers that will allow the stratification of the patients in order to select the most appropriate treatment for a subgroup of patients. Here, we identify the axis ZNF750/RAC1 as a potential novel prognostic biomarker for predicting clinical outcome in breast cancer. Moreover, our findings highlight a possible alternative molecular mechanism by which ZNF750 functions as tumour suppressor gene in breast cancer.
Methods
Cell culture and transfection
All cell lines used were obtained from American Type Culture Collection (ATCC) and maintained at 37 °C in 5% CO2 in culture medium. MCF7 (adenocarcinoma), MDA-MB-468 (ductal carcinoma, basal like-1), MDA-MB-453 (carcinoma, LAR) and MDAMB-231 (adenocarcinoma, mesenchymal-stem-like) were grown in Dulbecco’s Modified Eagle’s medium with 4.5 g/l glucose supplemented with 250 μM l-glutamine (Gibco), penicillin/streptomycin 1 U/ml (Gibco) and 10% FBS (Invitrogen). MDA-MB-231 stably overexpressing ZNF750 were previously generated and used for the study. Silencing was performed using 50 nM of ZNF750-specific siRNAs with Lipofectamine RNAimax according to the manufacturer’s protocol. Forty-eight hours after transfection, cells were harvested for experimental procedures.
RNA isolation and quantitative real-time PCR
Total RNA from cells was isolated using an RNeasy minikit (Qiagen) according to the manufacturer’s instructions. RNA samples were treated with RNase-free DNase I (Qiagen), and RNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific). Total RNA was reverse transcribed using the SensiFAST cDNA Synthesis kit (Bioline) according to the manufacturer’s protocol. qRT-PCR was performed using GoTaq qPCR Mastermix (Promega) with SYBR Green ready mix. The expression of each gene was defined according to the threshold cycle (Ct), and relative expression levels were calculated by using the 2−ΔΔCt method after normalization to the expression of the housekeeping gene β-actin. All the primer sequences are listed as follows: ZNF750 for: 5′-AGCTCGCCTGAGTGTGAC-3′; ZNF750 rev: 5′-TGCAGACTCTGGCCTGTA-3′; RAC1 for: 5′-GCTGACTCCCATCACCTATCC-3′; RAC1 rev: 5′-CGAGGGGCTGAGACATTTACAA-3′; β-ACTIN for: 5′-GTTGCTATCCAGGCTGTG-3′; β-ACTIN rev: 5′-AATGTCACGCACGATTTCCCG-3′.
Western blot analysis
For immunoblot analysis, proteins were extracted with RIPA buffer containing cocktail inhibitors (Roche), separated on SDS polyacrylamide gels and then transferred onto nitrocellulose membranes (GE Healthcare) by a wet-transfer system. Membranes were blocked with TBS-0.1% Tween and 10% milk and incubated overnight with primary antibodies. The following day, membranes were washed and then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody. Proteins were visualized with the Super Signal chemiluminescence kit (Thermo Scientific). The following antibodies were used: anti-ZNF750 (1:1000; Sigma HPA023012) and anti-β-actin (1:50,000; Sigma A5441).
ChIP assay
MDA-MB-468 were cross-linked for 10 min in a solution containing 1% formaldehyde. After fixation, ChIP assay was performed using a MAGnify ChIP system (Invitrogen) according to the manufacturer’s instructions. In brief, cells were lysed and then sonicated to obtain chromatin fragments of ~250–400 bp. The lysate was immunoprecipitated using an anti-ZNF750 specific antibody (Sigma HPA023012), and nonspecific IgG as a negative control. Collected DNA fragments were tested by canonical PCR. In order to confirm the specificity of the antibody, a negative control region belonging to the β-actin promoter was chosen. The primer sequences used for the amplification reaction are reported as follows: RAC1 promoter for: 5′- TCCGAGCATTCCCGAAGTCC-3′; RAC1 promoter rev: 5′-AAATGGCCGCTCCACTCAC-3′.
Bioinformatics analysis
For bioinformatic analysis, the following websites were used: cBioPortal for Cancer Genomics, http://www.cbioportal.org24,25, UALCAN, http://ualcan.path.uab.edu59 and R2 (R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl)).
The UALCAN website was interrogated to analyse the expression levels of RAC1 between breast cancer and the normal tissue counterpart. Expression levels were obtained by RNA sequencing of normal and tumoral tissues and results were represented as transcripts per million (TPM). Even more, the UALCAN website reported the Kaplan–Meier survival curve of patients stratified with high (with TPM values above upper quartile) and low (with TPM values below upper quartile) RAC1 levels. The cBioPortal was interrogated to analyse the ZNF750–RAC1 correlation in TCGA Breast Cancer Invasive Carcinoma Datasets. The following datasets were analysed: TCGA, firehose legacy, 2012; TCGA Nature 2012 (ref. 60); TCGA, Cell 2015 (ref. 61). The latter was further used to study the correlation among the different breast cancer subtypes. The R2 platform was interrogated to enlarge the number of datasets for the Kaplan–Meier analysis. The interrogated datasets were: Breast Tumour GSE21653 (ref. 62) and Tumour Breast Invasive Carcinoma-TCGA.
Statistical analysis
All results are expressed as the mean ± SDs. p-Values < 0.05 were considered statistically significant.
Acknowledgements
We would like to thank Richard A. Knight for helpful comments and suggestions. This research was funded by Associazione Italiana per la Ricerca contro il Cancro (AIRC) to G.M. (IG#20473; 2018–2022), Ministry of Health and MAECI Italy-China Science and Technology Cooperation (#PGR00961) to G.M. This work has been also supported by Regione Lazio through LazioInnova Progetto Gruppo di Ricerca no. 85-2017-14986.
Author contributions
G.M. and M.A. designed the research; A.B., M.C. and F.R. performed the research; M.A. and A.B. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Massimiliano Agostini, Email: m.agostini@med.uniroma2.it.
Gerry Melino, Email: melino@uniroma2.it.
References
- 1.Sen GL, et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassandri M, et al. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3:17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin DC, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boxer LD, Barajas B, Tao SY, Zhang JJ, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Gene Dev. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazawa M, et al. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36:2243–2254. doi: 10.1038/onc.2016.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nambara S, et al. Clinical significance of ZNF750 gene expression, a novel tumor suppressor gene, in esophageal squamous cell carcinoma. Oncol. Lett. 2017;14:1795–1801. doi: 10.3892/ol.2017.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun BK, et al. CALML5 is a ZNF750-and TINCR-induced protein that binds stratifin to regulate epidermal differentiation. Gene Dev. 2015;29:2225–2230. doi: 10.1101/gad.267708.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassandri M, et al. ZNF750 represses breast cancer invasion via epigenetic control of prometastatic genes. Oncogene. 2020;39:4331–4343. doi: 10.1038/s41388-020-1277-5. [DOI] [PubMed] [Google Scholar]
- 9.Baali I, Acar DAE, Aderinwale TW, HafezQorani S, Kazan H. Predicting clinical outcomes in neuroblastoma with genomic data integration. Biol. Direct. 2018;13:20. doi: 10.1186/s13062-018-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu QY, et al. Differentially expressed tRFs in CD5 positive relapsed & refractory diffuse large B cell lymphoma and the bioinformatic analysis for their potential clinical use. Biol. Direct. 2019;14:23. doi: 10.1186/s13062-019-0255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polewko-Klim A, Lesinski W, Mnich K, Piliszek R, Rudnicki WR. Integration of multiple types of genetic markers for neuroblastoma may contribute to improved prediction of the overall survival. Biol. Direct. 2018;13:17. doi: 10.1186/s13062-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suo C, et al. Accumulation of potential driver genes with genomic alterations predicts survival of high-risk neuroblastoma patients. Biol. Direct. 2018;13:14. doi: 10.1186/s13062-018-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihaylov I, Kandula M, Krachunov M, Vassilev D. A novel framework for horizontal and vertical data integration in cancer studies with application to survival time prediction models. Biol. Direct. 2019;14:22. doi: 10.1186/s13062-019-0249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han YT, et al. Integrative analysis based on survival associated co-expression gene modules for predicting Neuroblastoma patients' survival time. Biol. Direct. 2019;14:4. doi: 10.1186/s13062-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han YT, et al. Integration of molecular features with clinical information for predicting outcomes for neuroblastoma patients. Biol. Direct. 2019;14:16. doi: 10.1186/s13062-019-0244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hidalgo MR, Amadoz A, Cubuk C, Carbonell-Caballero J, Dopazo J. Models of cell signaling uncover molecular mechanisms of high-risk neuroblastoma and predict disease outcome. Biol. Direct. 2018;13:16. doi: 10.1186/s13062-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris ZN, Dhungel E, Mosior M, Ahn TH. Massive metagenomic data analysis using abundance-based machine learning. Biol. Direct. 2019;14:12. doi: 10.1186/s13062-019-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazanietz MG, Caloca MJ. The Rac GTPase in cancer: from old concepts to new paradigms. Cancer Res. 2017;77:5445–5451. doi: 10.1158/0008-5472.CAN-17-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hein AL, et al. RAC1 GTPase promotes the survival of breast cancer cells in response to hyper-fractionated radiation treatment. Oncogene. 2016;35:6319–6329. doi: 10.1038/onc.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, J. J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal.6, pl1 (2013). [DOI] [PMC free article] [PubMed]
- 25.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data (vol 2, pg 401, 2012) Cancer Discov. 2012;2:960. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeo SK, Guan JL. Breast cancer: multiple subtypes within a tumor? Trends Cancer. 2017;3:753–760. doi: 10.1016/j.trecan.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SY, Jeong HH, Kim J, Moon JH, Sohn KA. Robust pathway-based multi-omics data integration using directed random walks for survival prediction in multiple cancer studies. Biol. Direct. 2019;14:8. doi: 10.1186/s13062-019-0239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tranchevent LC, et al. Predicting clinical outcome of neuroblastoma patients using an integrative network-based approach. Biol. Direct. 2018;13:12. doi: 10.1186/s13062-018-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons, J. & Francavilla, C. ‘Omics approaches to explore the breast cancer landscape. Front. Cell Dev. Biol.7, 395 (2020). [DOI] [PMC free article] [PubMed]
- 31.Blucher AS, Choonoo G, Kulesz-Martin M, Wu GM, McWeeney SK. Evidence-based precision oncology with the cancer targetome. Trends Pharm. Sci. 2017;38:1085–1099. doi: 10.1016/j.tips.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimes T, Walker AR, Datta S, Datta S. Predicting survival times for neuroblastoma patients using RNA-seq expression profiles. Biol. Direct. 2018;13:11. doi: 10.1186/s13062-018-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francescatto M, et al. Multi-omics integration for neuroblastoma clinical endpoint prediction. Biol. Direct. 2018;13:5. doi: 10.1186/s13062-018-0207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int. J. Mol. Sci. 2019;20:4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobon B, Montanucci L, Pereto J, Bertranpetit J, Laayouni H. Gene connectivity and enzyme evolution in the human metabolic network. Biol. Direct. 2019;14:17. doi: 10.1186/s13062-019-0248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larmuseau M, Verbeke LPC, Marchal K. Associating expression and genomic data using co-occurrence measures. Biol. Direct. 2019;14:10. doi: 10.1186/s13062-019-0240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Memmi EM, et al. p63 sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc. Natl Acad. Sci. USA. 2015;112:3499–3504. doi: 10.1073/pnas.1500762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melino, G., Memmi, E. M., Pelicci, P. G. & Bernassola, F. Maintaining epithelial stemness with p63. Sci. Signal.8, re9 (2015). [DOI] [PubMed]
- 39.Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Candi E, et al. p63 is upstream of IKK alpha in epidermal development. J. Cell Sci. 2006;119:4617–4622. doi: 10.1242/jcs.03265. [DOI] [PubMed] [Google Scholar]
- 41.Shalom-Feuerstein R, et al. Delta Np63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Candi E, et al. p63 in epithelial development. Cell. Mol. Life Sci. 2008;65:3126–3133. doi: 10.1007/s00018-008-8119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melino G, Knight RA, Nicotera P. How many ways to die? How many different models of cell death? Cell Death Differ. 2005;12:1457–1462. doi: 10.1038/sj.cdd.4401781. [DOI] [PubMed] [Google Scholar]
- 44.Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Nemajerova A, et al. Non-oncogenic roles of TAp73: from multiciliogenesis to metabolism. Cell Death Differ. 2018;25:144–153. doi: 10.1038/cdd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marini A, et al. TAp73 contributes to the oxidative stress response by regulating protein synthesis. Proc. Natl Acad. Sci. USA. 2018;115:6219–6224. doi: 10.1073/pnas.1718531115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Compagnone M, et al. Delta Np63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl Acad. Sci. USA. 2017;114:13254–13259. doi: 10.1073/pnas.1711777114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bokoch GM. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 49.Slack-Davis JK, et al. PAKI phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J. Cell Biol. 2003;162:281–291. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du J, et al. PI3K and ERK-induced Rac1 activation mediates hypoxia-induced HIF-1 alpha expression in MCF-7 breast cancer cells. PLoS ONE. 2011;6:e25213. doi: 10.1371/journal.pone.0025213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murga C, Zohar M, Teramoto H, Gutkind JS. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappa B. Oncogene. 2002;21:207–216. doi: 10.1038/sj.onc.1205036. [DOI] [PubMed] [Google Scholar]
- 52.Rao M, Chen DF, Zhan P, Jiang JQ. MDA19, a novel CB2 agonist, inhibits hepatocellular carcinoma partly through inactivation of AKT signaling pathway. Biol. Direct. 2019;14:9. doi: 10.1186/s13062-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis MJ, et al. RAC1(P29S) is a spontaneously activating cancer-associated GTPase. Proc. Natl Acad. Sci. USA. 2013;110:912–917. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espina C, et al. Critical role for Rac1 in tumor progression of human colorectal adenocarcinoma cells. Am. J. Pathol. 2008;172:1153–1153. doi: 10.2353/ajpath.2008.070561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofbauer SW, et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood. 2014;123:2181–2188. doi: 10.1182/blood-2013-08-523563. [DOI] [PubMed] [Google Scholar]
- 57.Otsuka R, et al. ZNF750 expression as a novel candidate biomarker of chemoradiosensitivity in esophageal squamous cell carcinoma. Oncology. 2017;93:197–203. doi: 10.1159/000476068. [DOI] [PubMed] [Google Scholar]
- 58.Li QJ, et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat. Commun. 2020;11:1456. doi: 10.1038/s41467-020-15308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandrashekar DS, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciriello G, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabatier R, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res. Treat. 2011;126:407–420. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]