Abstract
Babesia bovis is a hemoprotozoan parasite of cattle that has a complex life cycle within vertebrate and invertebrate hosts. In the mammalian host, B. bovis undergoes asexual reproduction while in the tick midgut, gametes are induced, fuse, and form zygotes. The zygote infects tick gut epithelial cells and transform into kinetes that are released into the hemolymph and invade other tick tissues such as the ovaries, resulting in transovarial transmission to tick offspring. To compare gene regulation between different B. bovis life stages, we collected parasites infecting bovine erythrocytes and tick hemolymph. Total RNA samples were isolated, and multiplexed libraries sequenced using paired-end 100 cycle reads of a HiSeq 2500. The data was normalized using the TMM method and analysed for significant differential expression using the generalized linear model likelihood ratio test (GLM LRT) in edgeR. To validate our datasets, ten genes were selected using NormFinder. Genes that had no significant fold change between the blood and tick stages in the RNA-Seq datasets were tested by quantitative PCR to determine their suitability as “housekeeping” genes. The normalized RNA-Seq data revealed genes upregulated during infection of the mammalian host or tick vector and six upregulated genes were validated by quantitative PCR. These datasets can help identify useful targets for controlling bovine babesiosis.
Keywords: Bovine babesiosis, Babesia, Bovine, Gene expression, Kinetes, Rhipicephalus microplus
Specifications Table
| Subject | Parasitology |
| Specific subject area | Transcriptome analysis of Babesia bovis life stages. |
| Type of data | Tables Graphic Excel file |
| How data were acquired | Illumina HiSeq™ 2500 Bio-RAD CFX96™ Real-Time PCR |
| Data format | Raw Analysed |
| Parameters for data collection | Babesia bovis blood stages were collected from bovines with acute parasitemia and suspended in TRIzol (Thermo Fisher Scientific, Waltham, MA). Babesia bovis kinetes were collected from infected replete female ticks by extraction via pressurized capillary tubing, pooled, concentrated, and suspended in TRIzol. Total RNA was extracted, and library construction performed according to Illumina TruSeq mRNA library protocols. |
| Description of data collection | Counts were generated from alignments for each gene using the Subread feature of Counts v1.6.0. Genes without at least 1 read per million mapped reads across all three samples within a group were removed, data was normalized using the TMM method, and analysed for differential expression significance testing using the generalized linear model likelihood ratio test (GLM LRT) method in edgeR v3.20.9. The false discovery rate (FDR) method was employed to correct for multiple testing and genes were termed significantly differentially expressed if their logFold Change (logFC) value was greater than or equal to 1 and the FDR set to 5%. Triplicate RNA samples from B. bovis blood or kinete stages were used for qPCR. Quantitative PCR was performed in triplicate using a Bio-RAD CFX96™ Real-Time PCR Detection System. The transcript level of six genes of interest was normalized by dividing the transcript level of the gene of interest with that of the housekeeping genes. To identify suitable “housekeeping” genes, Excel (Microsoft Office 2013) with NormFinder and the comparative delta-Ct method from the RNA-Seq dataset were used to select ten genes that had no significant fold changes between the blood and kinete stages. The top five housekeeping genes were then used to normalize transcript levels in the qPCR data. |
| Data source location | The U.S. Department of Agriculture-ARS-Animal Disease Research Unit. Pullman/Washington State United States of America 46.730873 ° N, -117.163475 ° E |
| Data accessibility | Raw data were deposited at the NCBI Gene Expression Omnibus under accession number GSE144066. Direct URL to data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144066. |
| Related research article | M.W. Ueti, W.C. Johnson, L.S. Kappmeyer, D.R. Herndon, M.R. Mousel, K.E. Reif, N.S. Taus, O.O. Ifeonu, J.C. Silva, C.E. Suarez, K.A. Brayton. Comparative analysis of gene expression between Babesia bovis blood stages and kinetes allowed by improved genome annotation. Int. J. Parasitol. doi: 10.1016/j.ijpara.2020.08.006. |
Value of the Data
-
•
Comparison between life stages can help identify useful targets for therapeutic intervention and vaccine development to control bovine babesiosis or block parasite infection of the tick vector.
-
•
These data can benefit scientists at universities, federal agencies, and international institutions working towards prevention of B. bovis.
-
•
These datasets may be the foundation to define potential targets for understanding transmission mechanisms of tick-borne protozoan parasites of humans and animals.
-
•
The kinete dataset contains reads for R. microplus hemocytes that could be used to elucidate tick responses to Babesia infection.
1. Data Description
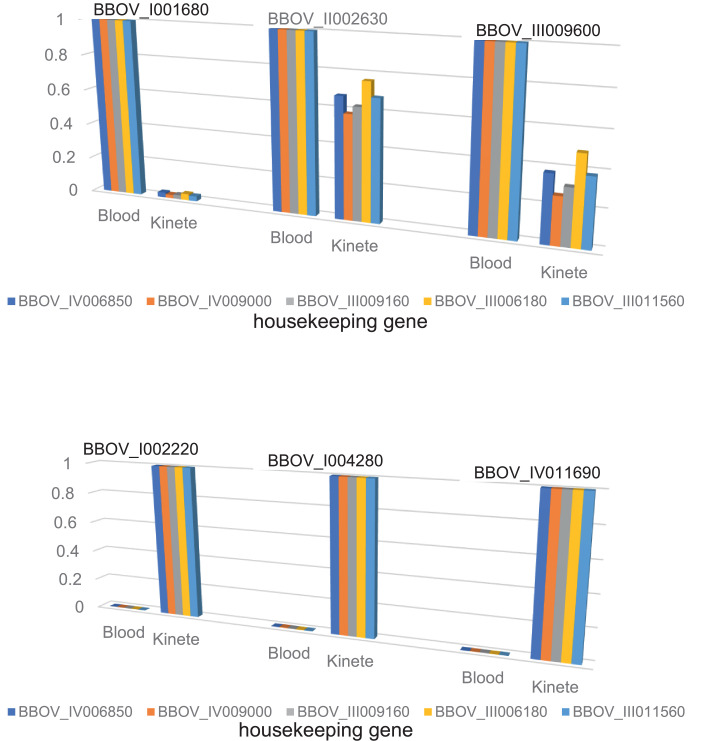
Tables 1–4 show differential expression of gene families by B. bovis blood stages or kinetes, including SMORFs, SBPs, GCC2 and GCC3 domain-containing proteins, and VESAs. Normalized data were used to determined fold increase by dividing normalized counts of one stage with the other. Table 5 indicates differential gene expression associated with elements of the B. bovis glideosome between blood stages and kinetes. To validate the RNA-Seq, three test genes from either kinete stage (BBOV_I002220, BBOV_I004280, BBOV_IV011690) or blood stages (BBOV_I001680, BBOV_II002630, BBOV_III009600) were selected based on their magnitude of transcription. A panel of housekeeping genes (BBOV_IV006850, BBOV_IV009000, BBOV_III009160, BBOV_III006180, BBOV_III011560) demonstrates a consistent pattern of gene expression differences between B. bovis blood stages and kinetes. Fig. 1 illustrates the consistent ratios of housekeeping gene relative expression as compared to the test genes. Supplementary Table S1 is an Excel Spreadsheet with normalized RNA-Seq data and analyzed in the context of differential gene expression between B. bovis blood stages and kinetes. Table S1 contains B. bovis gene identifiers, Log Fold Change, Log Count per Million Reads, False Discovery Rate, triplicate reads for B. bovis blood stages and kinetes (normalized and raw counts).
Table 2.
Differential expression of the spherical body protein gene family between B. bovis blood stages and kinetes.
| GeneID | norm blood stages | norm kinetes | Fold increase in blood stages | Fold increase in kinetes |
|---|---|---|---|---|
| BBOV_I004210 | 9564.525657 | 716.5825508 | 13.34741635 | 0.074920867 |
| BBOV_IV005390 | 4059.333716 | 31.38524799 | 129.3389084 | 0.007731625 |
| BBOV_II000680 | 3355.546841 | 21.10031815 | 159.0282581 | 0.006288191 |
| BBOV_II000740 | 1058.592079 | 23.79627366 | 44.48562387 | 0.022479172 |
| BBOV_III005860 | 679.0452333 | 5.850819611 | 116.0598478 | 0.008616244 |
| BBOV_III005600 | 384.621592 | 4.203584114 | 91.49848831 | 0.010929142 |
| BBOV_III006540 | 289.7373304 | 8.541515569 | 33.92106799 | 0.029480204 |
| BBOV_III005830 | 252.4609668 | 4.173006004 | 60.498587 | 0.016529312 |
| BBOV_III006460 | 239.8983689 | 413.0159404 | 0.580845303 | 1.721628798 |
| BBOV_III005840 | 162.7218894 | 12.47769378 | 13.04102282 | 0.076681102 |
| BBOV_III006520 | 50.57652699 | 0.628796749 | 80.43382394 | 0.012432581 |
| BBOV_III005630 | 36.93756706 | 1.347535007 | 27.41121148 | 0.036481423 |
| BBOV_III005790 | 19.96872969 | 3.655263286 | 5.463007211 | 0.183049365 |
| BBOV_III006480 | 18.77035174 | 0.901596442 | 20.81901711 | 0.048033007 |
| BBOV_III006500 | 4.383303317 | 0.20015859 | 21.89915163 | 0.045663869 |
Table 3.
Differential expression of the GCC2 and GCC3 domain containing protein gene family between B. bovis blood stages and kinetes.
| GeneID | norm blood stages | norm kinetes | Fold increase in blood stages | Fold increase in kinetes |
|---|---|---|---|---|
| BBOV_IV006260 | 319.5826078 | 22.37882551 | 14.28057999 | 0.070025167 |
| BBOV_III011740 | 197.335074 | 35.38007157 | 5.577577016 | 0.179289322 |
| BBOV_IV006250 | 4.042298771 | 4387.570389 | 0.000921307 | 1085.414671 |
| BBOV_III011730 | 3.208575878 | 2445.955909 | 0.001311788 | 762.3182379 |
Table 1.
Differential expression of the small open reading frame (smorf) gene family between B. bovis blood stages and kinetes.
| GeneID | norm blood stages | norm kinetes | Fold increase in blood stages | Fold increase in kinetes |
|---|---|---|---|---|
| BBOV_III011930 | 2113.913468 | 14.36620151 | 147.1449127 | 0.006796022 |
| BBOV_III001320 | 536.3841036 | 2.982448672 | 179.8468851 | 0.005560285 |
| BBOV_I003880 | 518.6261427 | 7.285320965 | 71.18782345 | 0.014047346 |
| BBOV_III007740 | 398.7738069 | 110.4001374 | 3.612077088 | 0.276849019 |
| BBOV_II004160 | 380.0745053 | 2.545392465 | 149.3186259 | 0.006697088 |
| BBOV_II004220 | 369.9142849 | 2.877928198 | 128.534925 | 0.007779987 |
| BBOV_I003890 | 288.1595966 | 2.645425925 | 108.9274864 | 0.009180419 |
| BBOV_IV007930 | 286.4893288 | 1.926687667 | 148.6952627 | 0.006725164 |
| BBOV_II000060 | 280.9586624 | 1.984377164 | 141.5853133 | 0.007062879 |
| BBOV_III000050 | 240.2349071 | 1.13933101 | 210.8561122 | 0.004742571 |
| BBOV_I001120 | 222.2547257 | 36.4316058 | 6.10060196 | 0.16391825 |
| BBOV_II006810 | 208.0321558 | 5.086740939 | 40.89694331 | 0.024451705 |
| BBOV_IV006430 | 207.1816437 | 0.937498517 | 220.9941028 | 0.004525008 |
| BBOV_I001180 | 202.7519971 | 1.332583402 | 152.1495741 | 0.00657248 |
| BBOV_II002275 | 194.4475113 | 11.84410832 | 16.41723515 | 0.060911596 |
| BBOV_IV007970 | 187.8401442 | 0.724434865 | 259.2919714 | 0.003856656 |
| BBOV_II001390 | 169.3409791 | 10.7185897 | 15.79881158 | 0.0632959 |
| BBOV_IV006390 | 126.4761588 | 12.76775186 | 9.905906709 | 0.100949871 |
| BBOV_II002280 | 109.3296517 | 3.105479146 | 35.20540521 | 0.028404729 |
| BBOV_I001370 | 108.4269976 | 0.615054735 | 176.2883713 | 0.005672524 |
| BBOV_IV000090 | 99.97305676 | 0.97946984 | 102.0685402 | 0.009797338 |
| BBOV_III011960 | 97.89184227 | 1.043228584 | 93.8354679 | 0.010656951 |
| BBOV_II007780 | 80.91934755 | 0.623937094 | 129.6915159 | 0.007710605 |
| BBOV_III000020 | 77.72837628 | 0.441915862 | 175.8895368 | 0.005685387 |
| BBOV_IV012140 | 69.65059029 | 2.152564713 | 32.35702503 | 0.03090519 |
| BBOV_III007710 | 66.79064133 | 32.61366171 | 2.047934449 | 0.488296879 |
| BBOV_IV000040 | 66.34182859 | 3.766155266 | 17.61526647 | 0.05676894 |
| BBOV_III002350 | 63.3051779 | 3.209907951 | 19.72180476 | 0.050705299 |
| BBOV_I003860 | 61.03939933 | 1.178418838 | 51.79771178 | 0.019305872 |
| BBOV_I001160 | 56.8016851 | 2.876162626 | 19.74912148 | 0.050635164 |
| BBOV_III002340 | 51.26993531 | 0.410873441 | 124.7827924 | 0.008013925 |
| BBOV_I001420 | 45.33252697 | 0.838674649 | 54.05257813 | 0.018500505 |
| BBOV_I001170 | 43.73955786 | 0.743409174 | 58.83645158 | 0.016996266 |
| BBOV_II007820 | 32.65136192 | 0.406850737 | 80.25390869 | 0.012460452 |
| BBOV_II004150 | 31.64665067 | 0.486700297 | 65.02287115 | 0.015379204 |
| BBOV_IV007960 | 29.20461566 | 0.486700297 | 60.00533771 | 0.016665184 |
| BBOV_II002290 | 25.70031072 | 11.88324468 | 2.162735129 | 0.462377471 |
| BBOV_II006800 | 16.08857604 | 607.0183122 | 0.026504268 | 37.72977241 |
| BBOV_II001380 | 13.69017871 | 2.146657517 | 6.377439625 | 0.156802739 |
| BBOV_II000400 | 8.75589443 | 406.4511482 | 0.021542305 | 46.42028881 |
| BBOV_I005150 | 4.467630153 | 3.39732353 | 1.315044067 | 0.760430791 |
| BBOV_I003850 | 3.639805192 | 0.281682051 | 12.92167951 | 0.077389321 |
| BBOV_IV006420 | 1.559208382 | 4.05065043 | 0.384927905 | 2.59788908 |
| BBOV_III000690 | 0.288793277 | 92.75983833 | 0.003113344 | 321.198053 |
Table 4.
Differential expression of the variant erythrocyte surface antigen (ves1) gene family between B. bovis blood stages and kinetes.
| GeneID | norm blood stages | norm kinetes | Fold increase in blood stages | Fold increase in kinetes |
|---|---|---|---|---|
| BBOV_III000015 | 1.728967521 | 0.601312722 | 2.875321706 | 0.34778717 |
| BBOV_III001300 | 1.02316527 | 0.401991082 | 2.545243701 | 0.392889687 |
| BBOV_II002310 | 11.46011732 | 4.90734948 | 2.335296756 | 0.428211103 |
| BBOV_I001190 | 1.921214622 | 0.906828737 | 2.118608006 | 0.472008034 |
| BBOV_IV007950 | 1.919285603 | 0.94926437 | 2.021866261 | 0.494592555 |
| BBOV_I005825 | 156.5772135 | 79.50478851 | 1.969406075 | 0.507767297 |
| BBOV_II002320 | 423.6835178 | 220.0983315 | 1.924973783 | 0.519487594 |
| BBOV_IV005670 | 331.7736784 | 174.558981 | 1.900639408 | 0.526138728 |
| BBOV_II000380 | 41.83335358 | 22.80774353 | 1.834173272 | 0.545204761 |
| BBOV_IV005660 | 140.5719566 | 82.52085286 | 1.703471932 | 0.587036382 |
| BBOV_II000040 | 16.7797131 | 10.55649865 | 1.589515014 | 0.629122714 |
| BBOV_III001270 | 24.07677147 | 15.19922865 | 1.58407851 | 0.631281842 |
| BBOV_II007810 | 7.620384667 | 4.871933002 | 1.564139873 | 0.639329012 |
| BBOV_II007830 | 3.44796873 | 2.208278049 | 1.561383419 | 0.640457679 |
| BBOV_III001150 | 18.99083241 | 12.39033947 | 1.532712841 | 0.652437934 |
| BBOV_I000060 | 3.967674309 | 2.651030862 | 1.496653383 | 0.668157378 |
| BBOV_I000070 | 10.11356796 | 7.128294751 | 1.418792055 | 0.704824922 |
| BBOV_IV002830 | 23.37186412 | 16.55999767 | 1.41134465 | 0.708544153 |
| BBOV_III002370 | 12.47428832 | 8.974695799 | 1.389939959 | 0.719455537 |
| BBOV_II000900 | 24.01138202 | 22.98856169 | 1.044492576 | 0.957402688 |
| BBOV_I001410 | 10.98244456 | 12.27491138 | 0.894706627 | 1.117684803 |
| BBOV_II000030 | 6.160960597 | 7.564935187 | 0.814410229 | 1.227882417 |
| BBOV_III011940 | 26.85581707 | 36.03944587 | 0.745178413 | 1.341960506 |
| BBOV_I000050 | 34.99932496 | 48.47120721 | 0.722064231 | 1.384918345 |
| BBOV_II000945 | 8.125332914 | 12.97149013 | 0.626399344 | 1.596425681 |
| BBOV_III007700 | 19.30319549 | 31.04248606 | 0.621831494 | 1.608152706 |
| BBOV_II002260 | 0.539728944 | 0.905991786 | 0.595732712 | 1.678605154 |
| BBOV_IV000030 | 14.53940728 | 24.77284468 | 0.586909072 | 1.703841443 |
| BBOV_II006820 | 14.94653581 | 29.50277234 | 0.50661462 | 1.973886975 |
| BBOV_III007110 | 420.2763021 | 50.88074443 | 8.260026594 | 0.121064986 |
| BBOV_III007140 | 563.2564757 | 77.88629427 | 7.231779108 | 0.13827856 |
| BBOV_III006920 | 478.708575 | 140.9323883 | 3.396725059 | 0.29440122 |
| BBOV_III003060 | 250.1297463 | 78.89379732 | 3.170461491 | 0.315411495 |
| BBOV_III006080 | 798.0562832 | 5.063209234 | 157.6186656 | 0.006344426 |
| BBOV_IV001490 | 499.5313164 | 5.215861947 | 95.77157554 | 0.010441511 |
| BBOV_IV007980 | 128.4011235 | 2.044485845 | 62.80362559 | 0.015922648 |
| BBOV_III011950 | 53.41646004 | 1.155794466 | 46.21622756 | 0.021637422 |
| BBOV_I001320 | 4.013872157 | 0.104520474 | 38.40273571 | 0.026039812 |
| BBOV_I005865 | 193.6617495 | 5.222233452 | 37.08408505 | 0.026965746 |
| BBOV_III002320 | 44.98541218 | 1.343047993 | 33.49501463 | 0.029855189 |
| BBOV_I000010 | 32.39996069 | 1.048088238 | 30.91339021 | 0.032348442 |
| BBOV_III003090 | 4982.64518 | 189.2864097 | 26.32331179 | 0.037989141 |
| BBOV_III000090 | 8.739195077 | 0.401154131 | 21.78513044 | 0.04590287 |
| BBOV_I005120 | 165.8736178 | 9.685269174 | 17.12638181 | 0.058389449 |
| BBOV_I005835 | 90.24457916 | 5.64889545 | 15.97561505 | 0.062595399 |
| BBOV_IV000350 | 201.2939525 | 17.46459112 | 11.52583253 | 0.086761628 |
| BBOV_I001340 | 6.804356674 | 0.614217784 | 11.07808476 | 0.090268311 |
| BBOV_IV002840 | 1937.126989 | 201.8771272 | 9.595574376 | 0.10421471 |
| BBOV_II004130 | 16.85364966 | 2.155539876 | 7.818760323 | 0.127897513 |
| BBOV_I005945 | 68.75377453 | 9.111370654 | 7.545931028 | 0.132521752 |
| BBOV_I005905 | 96.32245181 | 13.5837803 | 7.090990116 | 0.14102403 |
| BBOV_II000100 | 22.9740958 | 3.478312299 | 6.604954881 | 0.151401488 |
| BBOV_III000700 | 20.52508246 | 3.406670199 | 6.024969034 | 0.165975957 |
| BBOV_II001370 | 117.6823372 | 21.98983005 | 5.351671067 | 0.186857523 |
| BBOV_I005160 | 58.84307326 | 14.5715978 | 4.038203227 | 0.247634887 |
| BBOV_IV005640 | 16.05860815 | 5.164916596 | 3.109170856 | 0.321629157 |
| BBOV_I005885 | 58.78805667 | 19.15570372 | 3.068958339 | 0.325843459 |
| BBOV_III001295 | 2.354492652 | 0.778101658 | 3.025944783 | 0.330475297 |
| BBOV_I005925 | 323.1792347 | 120.5558705 | 2.680742409 | 0.373030992 |
| BBOV_IV000200 | 1221.015774 | 33.55945 | 36.38366462 | 0.027484862 |
| BBOV_II004170 | 25.25477425 | 0.100497771 | 251.2968606 | 0.003979357 |
| BBOV_III006070 | 971.8226855 | 4.00958644 | 242.3747935 | 0.004125842 |
| BBOV_III003100 | 4179.375093 | 21.0944595 | 198.1266737 | 0.005047276 |
| BBOV_IV001500 | 736.2579763 | 6.232070813 | 118.1401814 | 0.008464521 |
| BBOV_I005955 | 156.0360625 | 2.111219752 | 73.90801565 | 0.013530332 |
| BBOV_I005140 | 122.6569861 | 2.272128462 | 53.98329723 | 0.018524248 |
| BBOV_IV007990 | 399.8551031 | 8.592072955 | 46.5376755 | 0.021487966 |
| BBOV_II000370 | 203.6019731 | 4.45954771 | 45.65529653 | 0.021903264 |
| BBOV_III002330 | 82.99101801 | 2.13898475 | 38.79925651 | 0.02577369 |
| BBOV_III002310 | 47.30825938 | 1.381833563 | 34.23585926 | 0.02920914 |
| BBOV_I005935 | 204.0221091 | 6.004055555 | 33.98071641 | 0.029428455 |
| BBOV_III007690 | 56.20382698 | 1.679979071 | 33.45507569 | 0.029890831 |
| BBOV_II004190 | 31.41995041 | 1.005279965 | 31.25492549 | 0.031994957 |
| BBOV_I005895 | 36.94778066 | 1.235341766 | 29.90895449 | 0.033434803 |
| BBOV_I005915 | 69.19411686 | 2.391600542 | 28.93213798 | 0.03456364 |
| BBOV_III000040 | 22.04538552 | 0.841860402 | 26.18650963 | 0.038187602 |
| BBOV_I005845 | 85.75392604 | 3.346863777 | 25.622174 | 0.039028694 |
| BBOV_III000710 | 24.83032387 | 0.978632889 | 25.37246004 | 0.039412812 |
| BBOV_II004120 | 62.60907076 | 2.697046176 | 23.21394098 | 0.043077563 |
| BBOV_III007730 | 100.4983749 | 4.340448271 | 23.15391604 | 0.043189238 |
| BBOV_III007720 | 52.02941561 | 2.278964278 | 22.83029011 | 0.043801458 |
| BBOV_IV006410 | 37.49204361 | 1.667538318 | 22.48346752 | 0.044477125 |
| BBOV_I005875 | 56.76625474 | 3.27019997 | 17.3586494 | 0.057608169 |
| BBOV_II001400 | 133.9557957 | 7.782021543 | 17.21349587 | 0.058093952 |
| BBOV_I005815 | 72.0948074 | 4.198351819 | 17.17216911 | 0.058233761 |
| BBOV_IV006400 | 39.62063076 | 2.351978021 | 16.84566369 | 0.059362458 |
| BBOV_IV000060 | 20.97555289 | 1.258803089 | 16.66309295 | 0.060012868 |
| BBOV_IV007910 | 38.04169291 | 2.348792269 | 16.19627815 | 0.06174258 |
| BBOV_I003870 | 19.33495425 | 1.212344753 | 15.94839604 | 0.06270223 |
| BBOV_I001330 | 9.327555329 | 0.692555493 | 13.46831471 | 0.074248339 |
| BBOV_II002270 | 9.706371156 | 0.770428891 | 12.59865936 | 0.079373525 |
| BBOV_III000100 | 25.84479748 | 2.285406166 | 11.30862333 | 0.088428093 |
| BBOV_I000020 | 110.8671163 | 9.939986566 | 11.1536485 | 0.089656761 |
| BBOV_II001410 | 55.15815454 | 6.181125463 | 8.923642607 | 0.112061861 |
| BBOV_IV005680 | 278.7178629 | 33.63615638 | 8.286257791 | 0.120681739 |
| BBOV_I004520 | 17.03969061 | 2.704022755 | 6.301607696 | 0.158689663 |
| BBOV_II006780 | 54.01289193 | 8.834899055 | 6.113583369 | 0.163570191 |
| BBOV_IV002850 | 2371.499271 | 427.4573271 | 5.547920507 | 0.180247716 |
| BBOV_I001140 | 10.81090535 | 2.000911002 | 5.402991609 | 0.185082649 |
| BBOV_II002300 | 2.672564091 | 0.513720014 | 5.202374875 | 0.192219904 |
| BBOV_IV007920 | 25.37602988 | 5.163242694 | 4.914746679 | 0.203469286 |
| BBOV_II000940 | 125.7918935 | 25.9247027 | 4.852201968 | 0.206091998 |
| BBOV_I005180 | 101.9253517 | 21.35619549 | 4.772636203 | 0.209527808 |
| BBOV_I003840 | 9.184526328 | 2.050646761 | 4.478843701 | 0.223271913 |
| BBOV_II004140 | 15.36332175 | 3.952571843 | 3.886917773 | 0.257273258 |
| BBOV_I001430 | 17.06123265 | 4.433667203 | 3.848108545 | 0.259867929 |
| BBOV_II006830 | 23.9118131 | 6.226676466 | 3.8402209 | 0.260401687 |
| BBOV_I003830 | 5.60933158 | 1.611452341 | 3.480916832 | 0.287280636 |
| BBOV_II000920 | 35.70692278 | 11.62363102 | 3.071925004 | 0.32552878 |
| BBOV_I003900 | 16.74032752 | 6.142248224 | 2.72543976 | 0.366913265 |
| BBOV_I001440 | 6.681265191 | 2.610360801 | 2.559517898 | 0.390698577 |
| BBOV_II000110 | 24.00063777 | 9.819882162 | 2.444086128 | 0.409150884 |
| BBOV_II000390 | 11.18485422 | 4.944482435 | 2.262087966 | 0.442069457 |
| BBOV_IV002860 | 18.92967276 | 37.88463844 | 0.499666185 | 2.001336153 |
| BBOV_III003110 | 89.20183852 | 188.362042 | 0.473565892 | 2.111638562 |
| BBOV_III001220 | 1.72155826 | 4.18135367 | 0.411722709 | 2.42881915 |
| BBOV_I003910 | 10.52155624 | 26.5396865 | 0.396446139 | 2.522410743 |
| BBOV_IV001510 | 2.698503508 | 7.92944196 | 0.340314428 | 2.938459015 |
| BBOV_II000910 | 25.91710922 | 93.57840916 | 0.276956078 | 3.610680819 |
| BBOV_I005805 | 8.085385626 | 43.16715254 | 0.187304122 | 5.338910788 |
| BBOV_II006790 | 28.98624709 | 163.5550893 | 0.177226201 | 5.642506559 |
| BBOV_I000030 | 45.94475751 | 345.461503 | 0.132995304 | 7.519062496 |
| BBOV_III001280 | 55.72201753 | 624.213682 | 0.089267536 | 11.20228071 |
| BBOV_I005110 | 5.925071676 | 74.15213411 | 0.079904264 | 12.51497672 |
| BBOV_IV006380 | 8.428348062 | 113.9017072 | 0.073996679 | 13.51412001 |
| BBOV_III001160 | 42.67923143 | 686.2582539 | 0.06219121 | 16.07944264 |
| BBOV_I004510 | 17.47781955 | 597.9515367 | 0.029229492 | 34.21202141 |
| BBOV_I005190 | 9.67496017 | 562.8547934 | 0.017189087 | 58.17644554 |
| BBOV_III000670 | 4.364457017 | 969.2748263 | 0.004502807 | 222.0837146 |
Table 5.
Differential gene expression of glideosome elements between B. bovis blood stages and kinetes
| GeneID | norm blood stages | norm kinetes | Fold increase in blood stages | Fold increase in kinetes |
|---|---|---|---|---|
| BBOV_II006100 | 5.338814 | 70978.19 | 7.52177E-05 | 13294.74914 |
| BBOV_II006070 | 1.414999 | 4259.571 | 0.000332193 | 3010.298479 |
| BBOV_II002650 | 96.21683 | 164058.8 | 0.000586478 | 1705.095019 |
| BBOV_II006080 | 8.861071 | 13495.36 | 0.000656601 | 1522.994638 |
| BBOV_IV003210 | 352.6652 | 26835.99 | 0.013141504 | 76.09479332 |
| BBOV_IV008510 | 325.7201 | 6390.77 | 0.050967269 | 19.62043538 |
| BBOV_IV009790 | 93.05779 | 1545.97 | 0.060193791 | 16.61300904 |
| BBOV_I000300 | 1768.456 | 22238.81 | 0.079521131 | 12.57527387 |
| BBOV_II004420 | 118.6838 | 1229.741 | 0.096511253 | 10.36148606 |
| BBOV_I003490 | 637.5578 | 6256.353 | 0.101905671 | 9.812996615 |
| BBOV_II006000 | 509.7894 | 3351.442 | 0.152110493 | 6.574168436 |
| BBOV_II005470 | 286.0339 | 670.0373 | 0.426892467 | 2.342510298 |
| BBOV_II005940 | 32.09359 | 0.249803 | 128.4757525 | 0.00778357 |
| BBOV_IV011430 | 584.8345 | 5.444342 | 107.4206191 | 0.0093092 |
| BBOV_IV011230 | 537.6968 | 24.69678 | 21.77194044 | 0.045930679 |
| BBOV_II003100 | 132.8903 | 6.408557 | 20.73637991 | 0.048224425 |
| BBOV_I001630 | 500.6437 | 28.21142 | 17.7461369 | 0.056350292 |
| BBOV_II005930 | 150.9749 | 10.4024 | 14.51346609 | 0.068901529 |
| BBOV_II002890 | 679.6739 | 61.55152 | 11.04235702 | 0.090560376 |
| BBOV_II002630 | 824.4089 | 197.8921 | 4.165952089 | 0.240041167 |
Fig. 1.
Ratios of housekeeping gene relative expression as compared to the test genes. The top panel are genes upregulated in B. bovis blood stages and the bottom panel are genes upregulated in kinetes.
2. Experimental Design, Materials and Methods
Three splenectomized Holstein calves approximately four months of age and determined to be Babesia-free by competitive enzyme-linked immunosorbent assay and PCR [1,2,3] were used for acquisition of B. bovis Texas strain by R. microplus, La Minita strain, as previously described [1,4]. Approximately 40,000 R. microplus larvae were placed under a cloth patch on calves. When ∼1% of the ticks had molted to the adult stage, calves were intravenously inoculated with B. bovis stabilate containing 107 infected erythrocytes to synchronize female tick repletion with an ascending parasitemia. Replete female ticks were collected and incubated at 26 °C in 96% relative humidity to allow B. bovis development [1,2].
To increase the percent parasitized erythrocytes, blood samples from an acute parasitemia were cultured for 5 days at 3% oxygen and 5% carbon dioxide [5,6]. These in vitro cultured B. bovis blood stages were centrifuged, media removed, cells suspended in TRIzol (Thermo Fisher Scientific, Waltham, MA) and stored at -80 °C. Babesia bovis kinetes were collected from replete female ticks by extraction using pressurized capillary tubing, pooled, concentrated by centrifugation at 3,000 × g for 2 min, suspended in TRIzol and stored at -80 °C. To isolate RNA, the samples were thawed, transferred into Lysing Matrix H tubes and homogenized three times for 30 s at a speed setting of 6.0 m/s (MP Biomedicals, Solon, OH) with cooling of samples on ice for 5 min between runs. Homogenized samples were transferred into microfuge tubes and centrifuged for 1 min at 16,000 x g at 4 °C to remove large particulates. Sample supernatants were transferred to microfuge tubes to which 200 µl of chloroform per ml of TRIzol supernatant was added. The samples were vortexed for 15 s and incubated at room temperature for 3 min. For phase separation, samples were centrifuged at 16,000 x g for 15 min at 4 °C and the upper aqueous phase transferred to a new microfuge tube. Subsequent RNA isolation was accomplished using the RNA Cleanup and Concentrator kit (Zymo Research, Irvine, CA) according to the manufacturer's instructions. In brief, two volumes of RNA binding buffer were added to the aqueous phase, mixed, an equal volume of 100% ethanol added, and mixed again. A volume of 800 µl of each sample was transferred to RNA-25 concentrator column assemblies and the RNA bound to the column matrix by passing the sample through the column by centrifuging at 13,000 x g for 30 s. To the sample-bound column, 400 µl of RNA prep buffer was added and spun at 13,000 x g for 30 s. Samples were washed by adding 800 µl of RNA wash buffer to each column, centrifuged at 13,000 x g for 30 s, washed again with 400 µl of RNA wash buffer, and centrifuged for 2 min at 13,000 x g after the final wash. Columns were transferred to new 1.5 ml tubes and samples eluted by adding 60 µl of nuclease-free water. Samples were treated with TURBO DNase (ThermoFisher Scientific) according to the manufacturer's instructions. Reactions were terminated and samples concentrated using RNA Cleanup and Concentrator RNA-5 or RNA-25 columns as described above. Sample RNA concentrations were determined using a NanoDrop 1000 (ThermoFisher Scientific) and tested for residual DNA by Real-Time SYBR Green PCR targeting of BBOV_II006950. If DNA contamination was observed, samples were re-treated with TURBO DNase as described above until samples were confirmed to be free of DNA. RNA samples were stored at -80 °C. Total RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip (Agilent Technologies, Santa Clara, CA) and NanoDrop absorbance ratios for 260/280nm and 260/230nm. Library construction was performed according to the Illumina TruSeq mRNA (Illumina, San Diego, CA) stranded protocol. Using an input quantity for total RNA within the recommended range, mRNA was enriched using oligo dT magnetic beads. The enriched mRNA was chemically fragmented. First strand synthesis used random primers and reverse transcriptase to make cDNA. After second strand synthesis the ds cDNA was cleaned using AMPure XP beads (Beckman Coulter, Brea, CA) and the cDNA was end repaired and the 3’ ends adenylated. Illumina barcoded adapters were ligated to the ends and the adapter ligated fragments were enriched by nine cycles of PCR. The resulting libraries were validated by qPCR and sized using an Agilent Bioanalyzer DNA high sensitivity chip. The library concentrations were normalized and then multiplexed. The multiplexed libraries were sequenced using paired end 100 cycle chemistry for the HiSeq 2500 (Illumina). The version of HiSeq control software was HCS 2.2.58 with real time analysis software, RTA 1.18.64. Low quality reads were filtered before alignment to the new reference genome using STAR v2.5.2a (2-pass mapping). Counts were generated from alignments for each gene using the Subread feature of Counts v1.6.0. Genes without at least 1 read per million mapped reads across all three samples within a group were removed, data were normalized using the TMM method, and analysed for differential expression significance testing using the generalized linear model likelihood ratio test (GLM LRT) method in edgeR v3.20.9. The false discovery rate (FDR) method was employed to correct for multiple testing and genes were termed differentially expressed if their log Fold Change (logFC) value was greater than or equal to 1 with the FDR set to 5%. Triplicate RNA samples from B. bovis blood or kinete stages were used for qPCR. Approximately 100 ng of total RNA from each preparation of parasites was reverse transcribed using a Superscript III™ cDNA Synthesis Kit (ThermoFisher Scientific) following the manufacturer's protocol. Quantitative PCR was performed in triplicate using a Bio-RAD CFX96™ Real-Time PCR Detection System. Reaction volumes were 20 μl using 10 μl SsoFast™ EvaGreen® Supermix (Bio-Rad, Hercules, CA, USA), 1 μl of 500 nM of each primer set, 6 μl of nuclease-free water and 2 μl of a 1:10 dilution of cDNA as template. The conditions consisted of an enzyme activation step of 98 °C for 2 min followed by 40 cycles of 98 °C for 5 s and 55 °C for 5 s. The transcript level of six genes of interest was normalized by dividing the transcript level of the gene of interest with that of the housekeeping genes. To identify suitable “housekeeping” genes, Excel (Microsoft Office 2013) with NormFinder and the comparative delta-Ct method from the RNA-Seq dataset were used to select five genes that had no significant fold changes between the blood and kinete stages [7]. The selected housekeeping genes were then used to normalize transcript levels in the qPCR data.
3. Ethics Statement
Animal experiments were conducted with the approval of the Institutional Animal Care and Use Committee of the University of Idaho, Moscow, Idaho, in accordance with institutional guidelines based on the U.S. National Institutes of Health Guide for the Care and Use of Laboratory Animals. Splenectomies were performed under sedation with xylazine and isoflurane inhalation, and all efforts were made to minimize suffering. All animals exposed to an exotic pathogen and ticks were euthanized. These animals were sedated with xylazine, brought to a recumbent position and euthanized by intravenous injection of sodium pentobarbitone. IACUC #2018-16.
CRediT Author Statement
Massaro Ueti, Wendell Johnson, and Kelly Brayton: Conceptualization. Massaro Ueti, Wendell Johnson, Lowell Kappmeyer, David Herndon, Kathryn Reif: Methodology. Massaro Ueti, Wendell Johnson, Lowell Kappmeyer, David Herndon, Michelle Mousel, Carlos Suarez and Kelly Brayton: Data curation, Writing-Original draft preparation. Massaro Ueti, Wendell Johnson, and Kelly Brayton: Visualization, Investigation. Massaro Ueti: Supervision. Massaro Ueti, Wendell Johnson and Kelly Brayton: Validation of data. Massaro Ueti, Wendell Johnson, Naomi Taus, Olukemi Ifeonu, Joana Silva, Carlos Suarez, and Kelly Brayton: Writing- Reviewing and Editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
We would like to thank Nicholas Durfee, Paul Lacy, and Jacob Laughery for their laboratory support and Ralph Horn, Kathy Mason, James Allison, and Megan Jacks for their excellent assistance with animal care. This work was supported by the United States Department of Agriculture-Agricultural Research Service (Project # 2090-32000-039-00D), U.S. Agency for International Development (Grant # AID-BFS-P-13-00002) and National Institute of Food and Agriculture-Agriculture and Food Research Initiative Competitive Grant (Project # 2016-67015-24968 to MWU).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2020.106533.
Contributor Information
Massaro W. Ueti, Email: massaro.ueti@usda.gov.
Kelly A. Brayton, Email: kbrayton@wsu.edu.
Appendix. Supplementary materials
References
- 1.Howell J.M., Ueti M.W., Palmer G.H., Scoles G.A., Knowles D.P. Transovarial transmission efficiency of Babesia bovis tick stages acquired by Rhipicephalus (Boophilus) microplus during acute infection. J. Clin. Microbiol. 2007;45:426–431. doi: 10.1128/JCM.01757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson W.C., Taus N.S., Reif K.E., Bohaliga G.A.R., Kappmeyer L.S., Ueti M.W. Analysis of stage-specific protein expression during Babesia bovis development within female Rhipicephalus microplus. J. Proteome Res. 2017;16:1327–1338. doi: 10.1021/acs.jproteome.6b00947. [DOI] [PubMed] [Google Scholar]
- 3.Chung C.J., Suarez C.E., Bandaranayaka-Mudiyanselage C.L., Bandaranayaka-Mudiyanselage C.B., Rzepka J., Heiniger T.J., Chung G., Lee S.S., Adams E., Yun G., Waldron S.J. A novel modified-indirect ELISA based on spherical body protein 4 for detecting antibody during acute and long-term infections with diverse Babesia bovis strains. Parasit.Vectors. 2017;10:77. doi: 10.1186/s13071-017-2016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell J.M., Ueti M.W., Palmer G.H., Scoles G.A. D.P. Knowles. Persistently infected calves as reservoirs for acquisition and transovarial transmission of Babesia bovis by Rhipicephalus (Boophilus) microplus. J. Clin. Microbiol. 2007;45:3155–3159. doi: 10.1128/JCM.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy M.G., Ristic M. Babesia bovis: continuous cultivation in a microaerophilous stationary phase culture. Science. 1980;207:1218–1220. doi: 10.1126/science.7355284. [DOI] [PubMed] [Google Scholar]
- 6.J.A. Alvarez, C. Rojas, J.V. Figueroa. Diagnostic tools for the identification of Babesia sp. in persistently infected cattle. Pathogens, 8(2019):143. doi:10.3390/pathogens8030143. [DOI] [PMC free article] [PubMed]
- 7.Andersen C.L., Jensen J.L., Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer datasets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.