Abstract
Objectives:
The purpose of this study was to examine parental preferences for researchers accessing their child’s electronic health record across 3 groups: those with a child with (1) a known genetic condition (fragile X syndrome FXS), (2) a suspected genetic condition (autism spectrum disorder [ASD]), and (3) no known genetic condition (typically developing).
Methods:
After extensive formative work, a discrete choice experiment was designed consisting of 5 attributes, each with 2 or 3 levels, including (1) type of researcher, (2) the use of personally identifiable information, (3) the use of sensitive information, (4) personal importance of research, and (5) return of results. Stratified mixed logit and latent class conditional logit models were examined.
Results:
Parents of children with FXS or ASD had relatively higher preferences for research conducted by nonprofits than parents of typically developing children. Parents of children with ASD also preferred research using non-identifiable and nonsensitive information. Parents of children with FXS or ASD also had preferences for research that was personally important and returned either summary or individual results. Although a few child and family characteristics were related to preferences, they did not overall define the subgroups of parents.
Conclusions:
Although electronic health record preference research has been conducted with the general public, this is the first study to examine the opinions of parents who have a child with a known or suspected genetic condition. These parents were open to studies using their child’s electronic health record because they may have more to gain from this type of research.
Keywords: discrete choice experiment, electronic health records, genetic conditions, fragile X syndrome, autism spectrum disorder
Introduction
Electronic health records (EHRs) are a valuable source in obtaining real-world evidence for researchers, clinicians, regulators, and policy makers.1,2 The volume of clinical data contained within EHRs as well as the large numbers of patients within EHR systems enable researchers to mine vast amounts of information on a scale that was previously unattainable. In addition to patient demographics, diagnosis and procedure codes, laboratory tests, patient history, and prescribed medications, EHRs contain clinical notes that can be analyzed with natural language processing.3 A broad range of studies have used EHRs, including retrospective observational, epidemiological, descriptive, and comparative effectiveness studies, among others.4–6 There are noted challenges, however, with the quality of EHR data,7–9 which has led some to call for and develop national standards or frameworks for their use in research.10,11 Despite these issues, EHRs show promise as a useful health technology tool for clinical research.12
EHRs can be especially useful in genomic and rare disease research. If the disease or condition of interest has a low prevalence rate, EHRs are a practical, cost-effective method for gathering data on patients who are geographically dispersed. One way in which EHRs have been used in genomic or rare disease research has been to explore phenotypic variability. Using International Classification of Diseases, Tenth Revision codes, researchers can identify individuals with a specific genetic condition, then examine other data in the EHR to better understand healthcare utilization patterns or comorbid medical conditions.13,14 A second application has been combining data from EHRs with genetic information gathered from biobank samples. For example, the Electronic Medical Records and Genomics Network has successfully used algorithms to identify patterns of patient phenotypes using EHRs and then conducted exploratory genome-wide association studies to diagnose diseases.15,16 Finally, efforts are under way to combine information from EHRs with biospecimens as well as patient-reported data to further understand the burden of both common and rare diseases and their impact on quality of life.17–20 In the United States, this is exemplified in the All of Us Research Program.21
Nevertheless, there are ethical considerations for using EHRs when conducting research in these special populations.22 Even if a study is exempt from federal guidelines that protect the rights of human participants in research, and thus does not require informed consent, patients and their caregivers from rare disease communities want to know how their or their child’s data are being used.23 Some of this is owing to the risk of re-identification when analyzing data from small or unique samples.24 To guide future EHR research on those with genetic or rare diseases and to aid in the development of responsive policies in this area, it is important to understand patient preferences and, in particular, whether they perceive that the benefits outweigh the risks.
Some preference studies on access to EHRs for research purposes have been conducted, but mainly among those in the general population and some groups with special healthcare needs.25–27 Although a few have been quantitative, most studies have used focus groups or interviews. Overall, most individuals in these studies support EHR research, but many voiced concerns about the privacy and security of their information.28–30 This was of particular concern when sensitive data, such as mental or sexual health information, were being accessed, or if pharmaceutical or insurance companies were seeking access, because patients worried about possible discrimination and stigmatization.31,32 Participants in genetic biobank research have expressed similar privacy concerns.33,34 Notably, the risk of re-identification was specifically mentioned.35
No work, however, has been conducted to date to examine preferences of using EHR data to conduct research among those with, or who have a child with, a known or suspected genetic condition. This group has much to gain from EHR research, because it provides a method for further understanding genotype-phenotype associations and, ultimately, leading to personalized medicine.36,37 Nevertheless, individuals, especially children, with known or suspected genetic conditions may need special protections given their genetic or disability status to ensure there are no negative consequences of using EHRs for research.23,38
In this study, we sought to gather preference data from parents of a child with a likely or established genetic condition regarding research use of their child’s EHR using a discrete choice experiment. Three groups of parents were included: (1) parents of children with fragile X syndrome (FXS), (2) parents of children with autism spectrum disorder (ASD), and (3) a comparison group of parents with children who are typically developing (TD). In addition to intellectual disability, children with FXS often present with a range of co-occurring conditions such as attention problems, anxiety, aggression or self-injurious behavior, and seizures.39 Children with ASD may also have intellectual disability, but core diagnostic criteria focus on communication challenges, social impairment, and restricted behaviors or interests.40 Although ASD is not linked to a single gene, it has been associated with several genetic variants.41 Although parents of children with FXS or ASD do not represent the entire spectrum of parents who have a child with a known or suspected genetic condition, they are excellent prototypes for understanding patient preferences given their prevalence rate (1:58 for ASD, ~1:4000 for FXS) and heritability patterns. Understanding the preferences of parents who have a child with an established or likely genetic condition will lay the groundwork for ethical research practices and policies that take into account their perspectives on both risks and benefits of this type of research, regardless of whether consent is needed. We addressed 3 research questions:
Which factors drive parental decision making about research use of their child’s EHR?
Do parents’ preferences differ between those who have a child with a known or suspected genetic condition, and are either different than preferences of those who have a TD child?
What contributing factors, such as parental health literacy or severity of the child’s condition, are related to parents’ preferences regarding EHR research?
Methods
Participant Recruitment
We conducted an online discrete choice survey with 3 groups of parents. Parents of children aged 14 to 17 with FXS or ASD were eligible, as were legal guardians of an adult with FXS or ASD aged 18 to 40. Parents of TD children were eligible if they had a child aged 14 to 17 who did not have an intellectual or developmental disability or genetic condition, and also did not have a moderate or severe chronic health condition (eg, asthma, diabetes, epilepsy) or a psychiatric diagnosis (eg, anxiety, attention-deficit disorder). We targeted parents of adolescent children to not confound any potential ethical issues related to conducting research on younger children. Parents of children with FXS and ASD were a convenience sample recruited through research registries (eg, Our Fragile X World, Interactive Autism Network), parent advocacy organizations (eg, National Fragile X Foundation, Autism Society), and university partners (TEACCH Autism Program at University of North Carolina at Chapel Hill). Parents of TD children were recruited from an online survey panel (Qualtrics) with an effort to obtain a nationally representative sample. All participants resided in the United States.
Instrument
Discrete choice experiment
The design, analysis, and administration of the DCE followed current recommended guidelines in healthcare research.42 We conducted 2 steps of formative work to identify potential attributes and levels. First, we conducted a scoping review of research use of electronic health records and potential implications for those with genetic conditions.43 Next, we held a series of focus groups with parents of individuals with ASD or FXS.44 Based on these activities and discussions with expert advisers, we included 5 attributes, or characteristics, in the DCE. Each attribute took on 2 or 3 different levels, also informed by the focus groups. Combined, our final list was (1) who is conducting the research (for-profit, nonprofit, or government health researchers), (2) whether the research includes identifiable information (identifiable or not identifiable), (3) whether the research includes sensitive information (sensitive or not sensitive), (4) importance of the research topic (important to you or not important to you), and (5) how research results will be shared (individual, summary, or no results shared). A short description of each attribute and level was provided, including examples. For instance, for-profit companies, included pharmaceutical companies whereas nonprofits included universities and foundations. Sensitive information included mental health, substance abuse, or sexual health information, whereas not sensitive examples were data on height, weight, blood pressure, and allergies. A full description of the attributes and levels is available upon request. We used a paired comparison format in which respondents were asked to compare a series of hypothetical research studies, study A and study B, described by the attributes and levels. An example DCE comparison question is shown in Table 1.
Table 1.
Sample DCE item.
| Study A | Study B | |
|---|---|---|
| Who is conducting research | For-profit researcher | Nonprofit researcher |
| Whether the research includes identifiable information | Identifiable | Not identifiable |
| Whether the research includes sensitive information | Sensitive | Not sensitive |
| Importance of research topic | Not important to you | Important to you |
| How research results shared | No results | No results |
| I would prefer to give permission for: | Study A | Study B |
DCE indicates discrete choice experiment.
Before the DCE items were presented to participants, we provided an introduction to EHRs that included a definition, examples of what is typically included in an EHR, and how EHRs can be used in research. Next, we described each attribute and level. Participants were given 2 sample DCE questions; the first allowed participants to practice comparing only the first 3 attributes and the second included all 5 attributes and served as a validity test. The sample DCE item shown in Table 1 describes the choice options presented to participants in the validity test, where we assumed most participants would prefer study B. Approximately 90% showed preference for the expected choice in the second item.
Experiences with the healthcare system
After the DCE questions, participants were asked additional questions about their experiences with the healthcare system. Three questions, taken from the revised Health Care System Distrust scale,45 asked participants about their level of trust of their healthcare provider, the healthcare system, and medical researchers (5-point Likert-type scale, 1 = strongly disagree [low trust] to 5 = strongly agree [high trust]). We created a trust in the healthcare system index (range 3–15) by summing these items (see Table 2).
Table 2.
Demographics of survey participants across 3 parent groups.
| Parent subgroups |
Total (n = 1503) | |||||||
|---|---|---|---|---|---|---|---|---|
| FXS (n = 397) |
ASD (n = 611) |
TD (n = 495) |
||||||
| n | % | n | % | n | % | n | % | |
| Sex | ||||||||
| Male | 28 | 7 | 62 | 10 | 205 | 41 | 295 | 20 |
| Female | 369 | 93 | 549 | 90 | 290 | 59 | 1208 | 80 |
| Race/ethnicity | ||||||||
| Non-Hispanic white alone | 348 | 88 | 515 | 84 | 360 | 73 | 1223 | 81 |
| Non-Hispanic black alone | 11 | 3 | 39 | 6 | 55 | 11 | 105 | 7 |
| Non-Hispanic other race | 10 | 3 | 24 | 4 | 31 | 6 | 65 | 4 |
| Hispanic | 18 | 5 | 33 | 5 | 49 | 10 | 100 | 7 |
| Missing | 10 | 3 | 0 | 0.0 | 0 | 0.0 | 10 | 1 |
| Education | ||||||||
| High school or less | 150 | 38 | 217 | 36 | 245 | 49 | 612 | 41 |
| 4-year college degree | 128 | 32 | 198 | 32 | 152 | 31 | 478 | 32 |
| Graduate degree | 119 | 30 | 196 | 32 | 98 | 20 | 413 | 27 |
| Income | ||||||||
| Less than $50 000 | 73 | 18 | 153 | 25 | 185 | 37 | 411 | 27 |
| $50 001 to $100 000 | 141 | 36 | 216 | 35 | 174 | 35 | 531 | 35 |
| More than $100 000 | 169 | 43 | 215 | 35 | 133 | 27 | 517 | 34 |
| Missing | 14 | 4 | 27 | 4 | 3 | 1 | 44 | 3 |
| Child sex | ||||||||
| Male | 332 | 84 | 486 | 80 | 262 | 53 | 1080 | 72 |
| Female | 65 | 16 | 125 | 20 | 233 | 47 | 423 | 28 |
| Child age, M (SD) | 23.22 | (7.08) | 17.10 | (3.95) | 15.57 | (1.08) | 18.21 | (5.42) |
| Trust in healthcare system | 11.28 | (1.78) | 11.09 | (1.89) | 11.45 | (2.08) | 11.25 | (1.93) |
| Satisfaction with HCP | 4.03 | (0.74) | 4.00 | (0.77) | 4.02 | (0.80) | 4.01 | (0.77) |
| Health literacy | 13.43 | (1.83) | 13.66 | (1.75) | 12.48 | (2.38) | 13.21 | (2.06) |
| Healthcare decision making | 4.14 | (0.80) | 4.07 | (0.74) | 4.07 | (0.77) | 4.09 | 0.77 |
| Child’s co-occurring health conditions | 4.39 | (1.85) | 3.71 | (2.12) | 0.45 | (1.03) | 2.82 | 2.43 |
| W-ADL | 20.67 | (6.93) | 22.27 | (7.33) | 29.41 | (4.54) | 24.20 | 7.42 |
HCP indicates healthcare provider; M, mean; W-ADL, Waisman Activities of Daily Living scale.
The next 2 questions, which were developed by the study team, asked participants to rate how well their child’s healthcare provider explains things so they can understand them and whether the provider considered and respected the healthcare choices the parent thinks are best for their child (4-point Likert-type scale, 1 = never to 4 = always). The final question asked participants if, overall, they were satisfied with the care their child receives from their healthcare provider (5-point Likert-type scale, 1 = strongly disagree to 5 = strongly agree). We rescaled the 2 4-point items to match the response range of the third question and created a satisfaction with the child’s healthcare provider scale by taking their average (see Table 2).
Health literacy
Three questions assessed a participant’s level of health literacy:46 (1) how often they needed help reading health materials (5-point Likert-type scale, 1 = always to 5 = never), (2) how confident they are in completing medical forms (5-point Likert-type scale, 1 = extremely to 5 = not at all), and (3) how often they have problems learning about their child’s health because of difficulty understanding written information (5-point Likert-type scale, 1 = always to 5 = never). We summed these items to create a summary health literacy score (see Table 2).
Healthcare decision making
A single item developed by the study team assessed the participant’s views about how confident they were that their healthcare decisions aligned with their child’s preferences (5-point Likert-type scale, 1 = extremely to 5 = not at all) (see Table 2).
Child’s co-occurring health conditions
We also asked parents whether their child had ever been diagnosed or treated by a medical professional for 9 commonly co-occurring health conditions associated with ASD and FXS: attention problems, hyperactivity, aggressiveness toward others, self-injurious behavior, seizures, anxiety, depression, general developmental delay or mental retardation, and specific learning disability. The response options for each condition were 0 = no or 1 = yes. We created a summary score for these items representing the total number of co-occurring health conditions for each child (see Table 2).
Waisman Activities of Daily Living Scale (W-ADL)
We assessed the child’s level of independence using the 17-item W-ADL.47 All items had a common question stem (ie, Rate your child’s level of independence in …), which was followed by a description of an activity varying in difficulty (eg, making his/her own bed; banking and managing daily finances). Each question had a 3-level response scale: 0 = does not do at all, 1 = does with help, and 2 = independent/does on own. We calculated a composite scale by taking the sum of all 17 items (mean = 24.20, SD = 7.42, range 0–51). Higher values on the W-ADL indicate greater independence (see Table 2).
Experimental Design
The attributes and levels describe 72 possible hypothetical research studies (32·23), or 2556 unique study pairs ([72·71] / 2). Nevertheless, only a small subset of well-chosen pairs needed to be shown to participants to achieve robust statistical identification.48 We used NGene 1.2.0 software (Choice Metrics, Sydney, Australia) to select a D-efficient experimental design subset for the final study. Specifically, the design was optimized for main effects (no attribute interactions) multinomial logistic model with effects coded attributes and priors for the preference parameters based on study pretests. A constraint prohibiting a nonsensical combination of “not identifiable” with “individual” or “summary” results was also imposed. Candidate designs were compared for statistical efficiency, orthogonality (low correlation across attributes and levels), and level balance (near-evenly distributed combinations of attribute and levels). The final design consisted of 120 profiles or pairs, distributed in sets of 10 across 12 blocks, or versions, of the survey. In fielding, each respondent was asked to compare and indicate their preference over a set of 10 different study pairs. We randomly assigned each respondent to 1 of the 12 different versions (blocks). To eliminate any ordering effects, we randomized the order in which the 10 pairs were shown within a block and also randomized the left/right order of A and B.
Procedures
Prior to collecting data, we conducted cognitive interviews with participants (n = 10; 4 FXS, 3 ASD, 3 TD) to ensure that the survey instrument was understandable and that the practice items were helpful. Minor edits were made to the instrument based on participant feedback. Participants in the main study were invited to complete the survey online. The study team invited parents of children with FXS or ASD to participate and sent up to 2 reminders. These parents were given a $20 Amazon gift card as a thank-you for their participation. Our survey vendor managed recruitment, reminders, and incentives for the sample of parents with a TD child. Data collection took approximately 12 weeks. Institutional review board approval was received before any human participant research was conducted.
Statistical Analyses
We developed several models to compare preferences across 3 participant subgroups and to examine associations between individual characteristics and preferences. In all models, a binary-dependent variable indicated which alternative each participant selected in the 10 choice sets. We used effects coding for the attribute levels, which were entered as explanatory variables in the models.
Stratified mixed logit model
Discrete choice models are based on random utility theory, which assumes that the utility or net benefit an individual derives from choosing one alternative in a choice set consists of a systematic component and a random component.49–51 The systematic component is made up of individual-specific parameters that account for differences in preferences and the observable attributes that differentiate choice alternatives. The random component is an error term summarizing all unexplainable factors that affect choice. We estimated stratified mixed logit models that allow for observed and unobserved heterogeneity in preferences.51 Equation 1 shows the utility function for a mixed logit model predicting the utility for individual n associated with alternative j in choice set s:
| (1) |
where χ′njs is a vector of observed explanatory variables (eg, attribute levels), b is a vector of unknown parameters to be estimated representing the relative contribution of attribute levels to the utility respondents assign to an alternative (eg, preference weights), ηn is a variance term, and εnjs is a random error term. Individuals are assumed to choose alternatives that maximize utility, and when aggregated over many choices, we are able to use choice data to estimate relative preferences for various attribute levels. Mixed logit models allow preferences to differ among participants and produce estimates of this distribution, or heterogeneity, for each attribute level.50 The model output includes a set of coefficients representing the mean preference weight for each attribute level and another set for the SD of preference weights of these attribute levels across participants. We used the mixlogit command in Stata 15.0 to estimate the stratified mixed logit model.52,53
A large number of model parameters makes testing for subgroup differences in preference weights using interaction terms intractable, so we instead conducted a stratified analysis by estimating separate models for each of the 3 subgroups (TD, FXS, ASD). To determine whether differences across subgroups were owing to differences in the preference weights and the scale of utility functions, we applied the Swait-Louviere test procedure.54 We used a likelihood ratio test55 to compare the combined fit of these stratified models against an aggregate model that did not take the subgroups into account, to determine whether the stratified model fit the data better than the aggregate model despite a loss of parsimony. The formula for the likelihood ratio test statistic is
| (2) |
where LLAggregate is the log-likelihood of the aggregate model, and (LLTD + LLASD + LLFXS) is the sum of the log-likelihoods from the separate models for the 3 subgroups. The degrees of freedom for this test are equal to the number of attribute levels in the models (ie, 7 in our case).56 We used Wald tests to test equivalence of individual preference weights across subgroups.57
Latent-class conditional logit model
An alternative to the continuous heterogeneity specified by mixed logit models is latent class analysis, which models preference heterogeneity as varying by discrete clusters (classes) of respondents with similar response patterns. We used latent class analysis to identify classes of participants within each respondent subgroup (TD, FXS, ASD) and tested associations of class membership with demographic factors. The utility function for these models allow for heterogeneity at the participant level (n):
| (3) |
where βn is a vector of preference weights, λ′n is a vector of individual characteristics that remain constant across alternatives and choice sets, χ′j is a vector of attribute levels, and εnj is a random error term. The latent-class conditional logit model clusters together participants by common preference weights that are fixed within a class but heterogeneous across the classes. We estimated models with up to 10 classes and selected the optimum number of classes in each subgroup by comparing the Bayesian information criterion (BIC) and the consistent Akaike information criteria (CAIC) of these models. Both the BIC and CAIC apply penalty terms to more complex models with a large number of parameters, and lower values indicate a better fitting model.58,59 Class membership was estimated with categorical demographic variables entered as binary dummy variables, including race/ethnicity (non-Hispanic white only, non-Hispanic black only, non-Hispanic other race, or Hispanic), annual household income ($50 000 or less, $50 001 to $100 000, or more than $100 000), and child’s sex (male or female). Interval-level variables included a trust in the healthcare system index (range 3 [low trust] to 15 [high trust]), health literacy (range 3 [low literacy] to 15 [high literacy]), parental confidence that healthcare decisions align with the child’s wishes (range 1 [not at all confident] to 5 [extremely confident]), and the number of health conditions the child has been diagnosed with other than FXS or autism (range 0–9).
We reasoned that parents’ preferences related to sharing their child’s EHR for research purposes may differ as a function of these variables. For example, perceived alignment between parents and their children regarding healthcare decision making was included because of the age of the children in the study. In some studies, parents can provide consent when their child is under 18, yet the study could last past the age when the child could consent himself or herself. Conceivably, research studies that use identifiable or sensitive information may be less appealing to parents who are not confident that their decision making aligns with their child’s wishes. We wanted to determine if preferences were related to perceived accordance of beliefs. We estimated the latent-class conditional logit model using the lclogit command in Stata 15.0.60
Results
Description of Participants
A total of 1531 parents completed the survey. The final analysis sample (N = 1503; see Table 2) excludes participants who skipped 1 or more choice tasks (n = 21) or who completed the DCE in an amount of time that was ±3 SDs from the average time that participants took to complete it (n = 7). Most participants were female (80%) and non-Hispanic white (81%). Overall, most participants had a 4-year degree or more education (59%) and an income of $50 001 or more (69%). Most parents had a male child (72%) whose average age was 18.21 years (SD = 5.42).
Stratified Mixed Logit Model
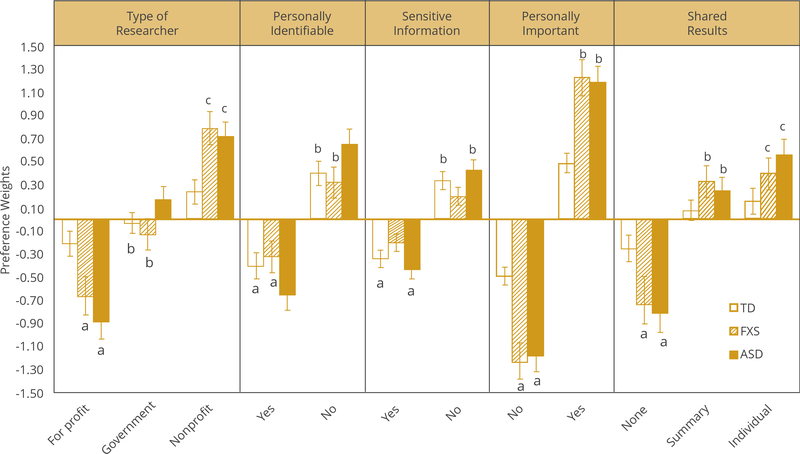
Results from the first step of the Swait-Louviere test procedure revealed that preference weights for attribute levels differed significantly across parent subgroups, χ2S-L (2) = 240.72, P < .001. Preference estimates from the final stratified mixed logit model are shown in Table 3 and also depicted in Figure 1. Preference weights within attribute levels with the same letter (eg, ‘a’) are not significantly different across subgroups at α = 0.05. The SDs of the preference weights revealed significant preference heterogeneity in each parent subgroup for most attribute levels; the only exception are preferences for summary results among parents of typically developing children (BSD = −0.18, SE = 0.15, 95% CI [–0.48 to 0.13], z = −1.14, P = .252).
Table 3.
Mixed logistic regression analysis predicting parental choice for research use of their child’s EHR, stratified by group.
| Factor | FXS |
ASD |
TD |
|||
|---|---|---|---|---|---|---|
| Coef | SE | Coef | SE | Coef | SE | |
| Mean preference weights | ||||||
| Type of researcher | ||||||
| Nonprofit | 0.79* | 0.07 | 0.72* | 0.06 | 0.24* | 0.05 |
| Government | −0.13 | 0.07 | 0.17† | 0.06 | −0.03 | 0.05 |
| For-profit‡ | −0.66* | 0.08 | −0.88* | 0.08 | −0.21* | 0.05 |
| Personally identifiable information | ||||||
| Not identifiable | 0.32* | 0.07 | 0.65* | 0.07 | 0.40* | 0.06 |
| Identifiable‡ | −0.32* | 0.07 | −0.65* | 0.07 | −0.40* | 0.06 |
| Sensitive information about child | ||||||
| Not sensitive | 0.20* | 0.04 | 0.43* | 0.04 | 0.34* | 0.04 |
| Sensitive‡ | −0.20* | 0.04 | −0.43* | 0.04 | −0.34* | 0.04 |
| Personal importance of research topic | ||||||
| Important | 1.23* | 0.08 | 1.18* | 0.07 | 0.49* | 0.04 |
| Not important‡ | −1.23* | 0.08 | −1.18* | 0.07 | −0.49* | 0.04 |
| Shared results | ||||||
| Individual results | 0.40* | 0.07 | 0.56* | 0.07 | 0.16† | 0.06 |
| Summary results | 0.33* | 0.07 | 0.25* | 0.06 | 0.08 | 0.05 |
| No results‡ | −0.74* | 0.09 | −0.81* | 0.08 | −0.25* | 0.06 |
| Standard deviation of preference weights | ||||||
| Type of researcher | ||||||
| Nonprofit | 0.60* | 0.08 | 0.81* | 0.08 | 0.55v | 0.06 |
| Government | 0.91* | 0.09 | 0.78* | 0.07 | 0.59* | 0.06 |
| For-profit§ | – | – | – | – | – | – |
| Personally identifiable information | ||||||
| Not identifiable | 0.70* | 0.09 | 0.92* | 0.08 | 0.74* | 0.07 |
| Identifiable§ | – | – | – | – | – | – |
| Sensitive information about child | ||||||
| Not sensitive | 0.38* | 0.06 | 0.61* | 0.05 | 0.62* | 0.05 |
| Sensitive§ | – | – | – | – | – | – |
| Personal importance of research topic | ||||||
| Important | 0.93* | 0.08 | 1.11* | 0.07 | 0.59* | 0.04 |
| Not important§ | – | – | – | – | – | – |
| Shared results | ||||||
| Individual results | 0.57* | 0.09 | 0.88* | 0.09 | 0.64* | 0.07 |
| Summary results | 0.50* | 0.12 | 0.55* | 0.10 | −0.18 | 0.15 |
| No results§ | – | – | – | – | – | – |
| Model | ||||||
| Log likelihood | −1997.25 | −2996.68 | −2899.90 | |||
| AIC | 3802.50 | 6021.36 | 5827.80 | |||
| BIC | 3900.21 | 6125.11 | 5928.60 | |||
| χ2 7∥ | 588.04* | 1098.04* | 630.50* | |||
| N | 397 | 611 | 495 | |||
| Observations | 7940 | 12 220 | 9900 | |||
Note. Likelihood ratio test of equivalence for stratified models: χ2 (14) = 273.88, P < .001.
Coefficients in the means section of the table are the average preference weights for each attribute level in the designated subgroups. Significant coefficients in the SDs section indicate that there is preference heterogeneity for that attribute level and subgroup.
AIC indicates Akaike information criterion; ASD, parents of children with autism spectrum disorder; BIC, Bayesian information criterion; FXS, parents of children with fragile X syndrome; SE, standard error; TD, parents of typically developing children.
P < .01.
P < .001.
Omitted reference level of attribute, computed from estimated coefficients.
Omitted reference level of attribute.
Likelihood ratio test for the joint significance of the standard deviations, testing the null hypothesis that all standard deviations in the model are equal to 0.
Figure 1.
Average preference weights by attribute level and study population. Error bars denote 95% confidence intervals. Preference weights within attribute levels sharing a letter in common (eg, ‘a’) are not significantly different across subgroups at α = 0.05. Comparisons across subgroups assume no covariance between estimates.
Average preference weights for levels within attributes and subgroups are significantly different from one another (see Fig. 1), except that there are no pairwise differences between preferences for individual results versus summary results among parents of TD children (z = 0.89, P = .371) or among parents of children with FXS (z = 0.60, P = .546). Whereas parents of TD children or children with FXS appear to have made little distinction in their preferences between individual and summary results, parents of children with ASD were the only group that had a clear preference hierarchy on this attribute: no results was the level with the lowest preference weight, studies promising individual results had the highest preference weight, and preferences for summary results fell between the other 2 levels.
Preferences for several attribute levels also differed across subgroups. Compared to parents of TD children, parents of children with FXS (Wald χ2 = 42.64, P = .000) and parents of children with ASD (Wald χ2 = 37.26, P = .000) had higher preferences for studies conducted by nonprofit researchers; in line with this, parents of children with FXS had relatively lower preference for studies by for-profit researchers (Wald χ2 = 20.23, P = .000) or ASD (Wald χ2 = 51.56, P = .000). Parents of children with ASD also had higher preference for research conducted by government researchers, compared to parents of TD children (Wald χ2 = 6.96, P = .008) or those with FXS (Wald χ2 = 10.23, P = .001).
Parents of children with ASD had higher preferences for research using information that is not personally identifiable, compared to both parents of TD children (Wald χ2 = 7.74, P = .005) or those with FXS (Wald χ2 = 11.34, P = .001); however, this is only a difference of magnitude. On average, parents in all 3 groups preferred studies that rely on non-identifiable information to studies that use identifiable information.
Parents of children with FXS had lower preference for studies involving sensitive information about their child than did parents of TD children (Wald χ2 = 6.50, P = .011); however, they still preferred studies that use non-sensitive information over sensitive. Parents of children with ASD showed higher preferences for studies involving nonsensitive information compared to parents of children with FXS (Wald χ2 = 16.01 P = .000), but there was no difference compared to parents of TD children.
Compared to parents of TD children, parents of those with FXS (Wald χ2 = 67.96, P = .000) or ASD (Wald χ2 = 70.96 P = .000) had higher preference for studies with a research topic that was personally important to them or their family. Parents of children with FXS (Wald χ2 = 22.09, P = .000) or ASD (Wald χ2 = 31.89, P = .000) also had lower preference for studies that would not return results than did parents of TD children. Parents of children with FXS or ASD also had higher preferences for studies that would return summary results (FXS: Wald χ2 = 8.56, P = .003; ASD: Wald χ2 = 4.89, P = .027) or individual results (FXS: Wald χ2 = 7.16, P = 07; ASD: Wald χ2 = 19.54, P = .000) compared to parents of TD children.
Latent-Class Conditional Logit Model
Based on the BIC and CAIC criteria, models with 3 and 4 classes fit the data indicating significant preference heterogeneity, which is consistent with preference variation seen in the mixed logit results above (see Appendix Table in Supplemental Materials found at http://dx.doi.org/10.1016/j.jval.2020.06.016). The 3-class models were appropriate for parents with TD children (BIC = 6043.65, CAIC = 6086.65) and parents of children with FXS (BIC = 3830.11, CAIC = 3873.11). The 4-class model was best for parents of children with ASD (BIC = 6001.65, CAIC = 6062.65). The results of the latent-class conditional logit model for each parent subgroup are presented in Tables 4 to 6.
Table 4.
Estimated parameters for the latent-class conditional logit model among parents with a child with FXS (n = 373).
| Factor | Class 1 |
Class 2 |
Class 3 |
|||
|---|---|---|---|---|---|---|
| Coef | SE | Coef | SE | Coef | SE | |
| DCE attributes | ||||||
| Type of researcher | ||||||
| Nonprofit | 0.39‡ | 0.07 | 1.01‡ | 0.13 | 0.37‡ | 0.09 |
| Government | 0.28‡ | 0.07 | −0.77‡ | 0.12 | 0.00 | 0.10 |
| For-profit§ | −0.67‡ | 0.09 | −0.24* | 0.11 | −0.37‡ | 0.11 |
| Personally identifiable information | ||||||
| Not identifiable | −0.03 | 0.07 | 0.57‡ | 0.11 | 0.37‡ | 0.10 |
| Identifiable§ | 0.03 | 0.07 | −0.57‡ | 0.11 | −0.37‡ | 0.10 |
| Sensitive information about child | ||||||
| Not sensitive | −0.03 | 0.04 | 0.38‡ | 0.07 | 0.22† | 0.07 |
| Sensitive§ | 0.03 | 0.04 | −0.38‡ | 0.07 | −0.22† | 0.07 |
| Personal importance of research topic | ||||||
| Important | 0.28‡ | 0.05 | 0.11 | 0.06 | 1.45‡ | 0.10 |
| Not important§ | −0.28‡ | 0.05 | −0.11 | 0.06 | −1.45‡ | 0.10 |
| Shared results | ||||||
| Individual results | 0.39‡ | 0.08 | −0.05 | 0.11 | 0.40‡ | 0.10 |
| Summary results | 0.24† | 0.08 | 0.13 | 0.11 | 0.29† | 0.11 |
| No results§ | −0.64‡ | 0.09 | −0.08 | 0.12 | −0.69v | 0.12 |
| Participant characteristics | ||||||
| Race/ethnicity | ||||||
| Non-Hispanic white alone∥ | – | – | – | – | – | – |
| Non-Hispanic black alone | 22.49 | 727.24 | 21.70 | 727.24 | – | – |
| Non-Hispanic other alone | 0.84 | 0.86 | −0.01 | 1.42 | – | – |
| Hispanic | −0.13 | 0.69 | 0.33 | 0.76 | – | – |
| Income | ||||||
| Less than $50 000∥ | – | – | – | – | – | – |
| $50 001 to $100 000 | 0.33 | 0.42 | −0.27 | 0.46 | – | – |
| More than $100 000 | 0.14 | 0.42 | −0.21 | 0.44 | – | |
| Child sex∥ | ||||||
| Male | – | – | – | – | – | – |
| Female | 0.18 | 0.40 | 0.19 | 0.44 | – | – |
| Co-occurring conditions | 0.07 | 0.08 | −0.15 | 0.09 | – | – |
| Trust in healthcare system | 0.05 | 0.09 | −0.28† | 0.09 | – | – |
| Health literacy | −0.15 | 0.08 | −0.10 | 0.09 | – | – |
| Healthcare decision making | 0.21 | 0.19 | 0.09 | 0.20 | – | – |
| Intercept | −0.45 | 1.65 | 3.92* | 1.71 | – | – |
| FXS parents per class | 32.2% | 19.3% | 48.5% | |||
Note. Coefficients in the DCE attributes section of the table are the preference weights for each attribute level in the class. Significant coefficients in the participant characteristics section indicate variables that differentiate membership in the groups, where class 3 is the reference category.
DCE indicates discrete choice experiment; FXS, parents of children with fragile X syndrome; SE, standard error.
P < .05.
P < .01.
P < .001.
Omitted reference level of attribute, computed from estimated coefficients.
Omitted reference level of factor.
Table 6.
Estimated parameters for the latent-class conditional logit model among parents with a TD child (n = 492).
| Factor | Class 1 |
Class 2 |
Class 3 |
|||
|---|---|---|---|---|---|---|
| Coef | SE | Coef | SE | Coef | SE | |
| DCE attributes | ||||||
| Type of researcher | ||||||
| Nonprofit | 0.19 | 0.12 | 0.07 | 0.08 | 0.21‡ | 0.04 |
| Government | 0.19 | 0.11 | −0.07 | 0.08 | −0.02 | 0.05 |
| For-profit§ | −0.38† | 0.13 | 0.00 | 0.08 | −0.19‡ | 0.05 |
| Personally identifiable information | ||||||
| Not identifiable | 0.10 | 0.13 | 1.14‡ | 0.14 | 0.08 | 0.05 |
| Identifiable§ | −0.10 | 0.13 | −1.14‡ | 0.14 | −0.08 | 0.05 |
| Sensitive information about child | ||||||
| Not sensitive | 0.02 | 0.07 | 1.02‡ | 0.11 | 0.02 | 0.03 |
| Sensitive§ | −0.02 | 0.07 | −1.02‡ | 0.11 | −0.02 | 0.03 |
| Personal importance of research topic | ||||||
| Important | 1.14‡ | 0.14 | 0.22‡ | 0.05 | 0.08* | 0.04 |
| Not important§ | −1.14‡ | 0.14 | −0.22‡ | 0.05 | −0.08* | 0.04 |
| Shared results | ||||||
| Individual results | 0.43† | 0.12 | −0.02 | 0.09 | 0.12* | 0.05 |
| Summary results | 0.24 | 0.12 | 0.02 | 0.09 | 0.10* | 0.05 |
| No results§ | −0.67‡ | 0.15 | 0.00 | 0.10 | −0.22‡ | 0.06 |
| Participant characteristics | ||||||
| Race/ethnicity | ||||||
| Non-Hispanic white alone∥ | – | – | – | – | – | – |
| Non-Hispanic black alone | 0.07 | 0.44 | −0.65 | 0.50 | – | – |
| Non-Hispanic other alone | 0.00 | 0.76 | 0.65 | 0.60 | – | – |
| Hispanic | −0.35 | 0.52 | 0.21 | 0.43 | – | – |
| Income | ||||||
| Less than $50 000∥ | – | – | – | – | – | – |
| $50 001 to $100 000 | −0.11 | 0.36 | −0.09 | 0.32 | – | – |
| More than $100 000 | 0.39 | 0.36 | 0.22 | 0.34 | – | |
| Child sex∥ | ||||||
| Male | – | – | – | – | – | – |
| Female | 0.12 | 0.31 | 0.22 | 0.28 | – | – |
| Co-occurring conditions | −0.19 | 0.16 | −0.23 | 0.18 | – | – |
| Trust in healthcare system | 0.09 | 0.07 | 0.03 | 0.07 | – | – |
| Health literacy | 0.16* | 0.07 | 0.26‡ | 0.07 | – | – |
| Healthcare decision making | 0.08 | 0.20 | −0.06 | 0.18 | – | – |
| Intercept | −4.09† | 1.41 | −3.93† | 1.22 | – | – |
| TD parents per class | 24.1% | 28.8% | 47.1% | |||
Note. Coefficients in the DCE attributes section of the table are the preference weights for each attribute level in the class. Significant coefficients in the participant characteristics section indicate variables that differentiate membership in the groups, where class 3 is the reference category.
DCE indicates discrete choice experiment; SE, standard error; TD, parents of typically developing children.
P < .05.
P < .01.
P < .001.
Omitted reference level of attribute, computed from estimated coefficients.
Omitted reference level of factor.
Almost a third of parents of children with FXS were clustered into class 1. Parents in this class preferred studies conducted by nonprofit or governmental researchers and were dissuaded from participating in studies conducted by for-profit researchers (see Table 4). Members of this group were also drawn to studies that would share individual or summary results—with an aversion toward those that would not return any results—and preferred studies dealing with a personally important research topic. There were no differences in this group’s preferences for identifiability or sensitivity of information accessed. No defining family or child characteristics set this group apart from the comparison group (class 3). In the next subgroup, about a fifth of FXS parents (class 2) showed an especially high preference for participating in studies conducted by nonprofit researchers with corresponding low preference for government and for-profit studies. Participants who were clustered into this group also preferred studies that did not include personally identifiable information or sensitive information about the child (relative to those that did). Compared with participants in class 3, those who had lower trust in the healthcare system were more likely to be clustered into class 2 (B = −0.28, SE = 0.09, 95% CI [−0.46 to −0.10], z = −3.12, P = .002). The preferences of the remaining FXS parents (class 3), comprising nearly half the sample, were influenced to some degree by all 5 of the DCE attributes.
Among parents of children with ASD (see Table 5), there were 4 distinct subgroups. Approximately 17% were grouped together into class 1. These parents displayed an especially strong preference for participating in studies conducted by nonprofit and, to a lesser extent, governmental researchers. Members of this group also preferred studies that did not include personally identifiable information or sensitive information about their child, were on a personally important topic, and shared summary results. No defining family or child characteristics set this group apart from the comparison group (class 4). Members of class 2 make up a little more than a third of parents who have a child with ASD and are distinguished from the other groups by their particularly strong preference for research on a personally important topic. In other respects, the preferences of class 2 align most to those of class 1, with the exception of the shared results attribute; members of class 2 had a higher preference for individual results, were indifferent toward summary results, and had lower preference for studies that do not share any results. Compared to parents in class 4, parents of children with a greater number of co-occurring conditions (B = 0.12, SE = 0.06, 95% CI [0.00–0.24], z = 1.97, P = .048) or higher trust in the healthcare system (B = 0.19, SE = 0.07, 95% CI [0.05–0.32], z = 2.76, P = .006) were more likely to be grouped into class 2. A quarter of parents who have a child with ASD were clustered into class 3 and showed especially high preferences for participating in studies that shared individual or summary results and a corresponding aversion toward studies that would not return any results. Parents in class 3 were indifferent to the identifiability or sensitivity of information used in a study, and they showed a preference for studies conducted by nonprofit researchers over for-profit studies. Parents of children with ASD who had higher trust in the healthcare system were also more likely to be grouped into class 3 (B = 0.21, SE = 0.08, 95% CI [0.06–0.37], z = 2.66, P = .008) than class 4. The remaining parents of children with ASD who were grouped together in class 4 had especially high preferences for studies that did not include personally identifiable or sensitive information about their child (versus studies that relied on personally identifiable or sensitive information), preferred studies that dealt with a personally important research topic, and—similar to class 3—preferred studies by nonprofit over for-profit researchers. Members of class 4 were indifferent to whether results would be shared.
Table 5.
Estimated parameters for the latent-class conditional logit model among parents of children with ASD (n = 584).
| Factor | Class 1 |
Class 2 |
Class 3 |
Class 4 |
||||
|---|---|---|---|---|---|---|---|---|
| Coef | SE | Coef | SE | Coef | SE | Coef | SE | |
| DCE attributes | ||||||||
| Type of researcher | ||||||||
| Nonprofit | 1.28‡ | 0.14 | 0.38† | 0.12 | 0.21† | 0.07 | 0.25† | 0.08 |
| Government | 0.27* | 0.12 | 0.30* | 0.12 | 0.02 | 0.09 | 0.02 | 0.09 |
| For-profit§ | −1.55‡ | 0.16 | −0.68‡ | 0.16 | −0.23* | 0.09 | −0.26† | 0.10 |
| Personally identifiable information | ||||||||
| Not identifiable | 0.25* | 0.11 | 0.37† | 0.13 | 0.08 | 0.07 | 1.23‡ | 0.15 |
| Identifiable§ | −0.25* | 0.11 | −0.37† | 0.13 | −0.08 | 0.07 | −1.23‡ | 0.15 |
| Sensitive information about child | ||||||||
| Not sensitive | 0.17‡ | 0.06 | 0.15* | 0.07 | 0.03 | 0.04 | 0.82‡ | 0.09 |
| Sensitive§ | −0.17‡ | 0.06 | −0.15* | 0.07 | −0.03 | 0.04 | −0.82‡ | 0.09 |
| Personal importance of research topic | ||||||||
| Important | 0.28‡ | 0.07 | 1.64‡ | 0.13 | 0.31‡ | 0.05 | 0.33‡ | 0.06 |
| Not important§ | −0.28‡ | 0.07 | −1.64‡ | 0.13 | −0.31‡ | 0.05 | −0.33‡ | 0.06 |
| Shared results | ||||||||
| Individual results | −0.04 | 0.13 | 0.36‡ | 0.13 | 0.67‡ | 0.09 | 0.18 | 0.09 |
| Summary results | 0.26* | 0.11 | 0.14 | 0.12 | 0.32‡ | 0.08 | −0.01 | 0.08 |
| No results§ | −0.22 | 0.13 | −0.50‡ | 0.15 | −0.99‡ | 0.12 | −0.17 | 0.10 |
| Participant characteristics | ||||||||
| Race/ethnicity | ||||||||
| Non-Hispanic white alone∥ | – | – | – | – | – | – | – | – |
| Non-Hispanic black alone | 0.29 | 0.60 | −0.13 | 0.56 | 0.18 | 0.60 | – | – |
| Non-Hispanic other alone | 0.30 | 0.80 | 0.05 | 0.71 | −0.30 | 1.08 | – | – |
| Hispanic | 0.17 | 0.76 | 0.26 | 0.62 | 0.37 | 0.69 | – | – |
| Income | ||||||||
| Less than $50 000∥ | – | – | – | – | – | – | – | – |
| $50 001 to $100 000 | −0.08 | 0.46 | −0.69 | 0.38 | −0.88* | 0.41 | – | – |
| More than $100 000 | −0.67 | 0.48 | −0.82* | 0.38 | −1.22† | 0.42 | – | |
| Child sex∥ | ||||||||
| Male | – | – | – | – | – | – | – | – |
| Female | 0.33 | 0.39 | 0.30 | 0.32 | −0.04 | 0.40 | – | – |
| Co-occurring conditions | 0.07 | 0.08 | 0.12* | 0.06 | 0.10 | 0.07 | – | – |
| Trust in healthcare system | 0.01 | 0.08 | 0.19† | 0.07 | 0.21† | 0.08 | – | – |
| Health literacy | −0.08 | 0.09 | 0.02 | 0.08 | −0.10 | 0.09 | – | – |
| Healthcare decision making | −0.09 | 0.22 | −0.21 | 0.18 | −0.12 | 0.20 | – | – |
| Intercept | 1.02 | 1.63 | −1.05 | 1.44 | 0.10 | 1.64 | – | – |
| FXS parents per class | 16.8% | 34.5% | 25.8% | 22.9% | ||||
Note. Coefficients in the DCE attributes section of the table are the preference weights for each attribute level in the class. Significant coefficients in the participant characteristics section indicate variables that differentiate membership in the groups, where class 4 is the reference category.
ASD indicates parents of children with autism spectrum disorder; DCE, discrete choice experiment; FXS, parents of children with fragile X syndrome; SE, standard error.
P < .05.
P < .01.
P < .001.
Omitted reference level of attribute, computed from estimated coefficients.
Omitted reference level of factor.
Among parents of TD children (see Table 6), roughly a quarter of participants (class 1) had an aversion toward studies conducted by for-profit researchers, showed an especially high preference for studies on research topics that were personally important to them, and preferred studies that would return individual research results—with a corresponding indifference toward participating in studies that would only return summary results and a disinclination to participate in studies where no results would be returned. Type of researcher, personal importance of the research topic, and the shared results attributes were also important to class 3, which made up nearly half of TD participants. Parents who were grouped into this class had higher preferences for studies conducted by nonprofit researchers and lower preferences for studies by for-profit researchers, preferred personally important research topics, and had low preference for studies that would not return any research results, favoring those that would return either summary or individual results. The remaining participants who were grouped into class 2 (almost 30%) were most concerned about privacy, data sensitivity, and the personal importance of research topics, showing higher preferences for studies that did not include personally identifiable or sensitive information about the child and dealt with a personally important research topic (compared to studies that used personally identifiable data, sensitive information, and did not address a personally important topic, respectively). Among the participant characteristic variables included in the model, participants with higher health literacy scores were more likely to be in class 1 (B = 0.16, SE = 0.07, 95% CI [0.02–0.30], z = 2.26, P = .024) or class 2 (B = 0.26, SE = 0.07, 95% CI [0.13–0.40], z = 3.90, P < .001) than in class 3. Race/ethnicity, income, child’s sex, co-occurring conditions, trust in the healthcare system, and perceptions of healthcare decision making were not associated with group membership.
Discussion
This study provides much-needed quantitative information about the views of parents who have a child with a known or suspected genetic condition about research use of their child’s EHR. Several factors have pushed the need to understand the opinions of this unique group of stakeholders, such as the precision medicine research initiative, which uses EHR data along with other health information to individualize treatment.21 Preference studies are essential to understanding the perspectives of these unique stakeholders, which can help to guide future research and inform policy decisions.
In keeping with our prior research findings,44 parents had strong and clear preferences about trust. Parents of children with FXS or ASD favored nonprofit researchers to conduct studies using their child’s EHR. This was also the case for parents of TD children, but to a lesser degree. Nevertheless, 2 subgroups of parents had exceptionally strong preferences for nonprofit researchers: about 19% of parents with a child with FXS (class 2) and 17% of parents of a child with ASD (class 1). In contrast, just over one-half of parents of a TD child (class 1 and 2) were indifferent to who was conducting the research. The subgroup of parents of children with FXS had less trust in the healthcare system, which may have influenced their desire to have more trust in the researcher accessing their child’s EHR data. These results are consistent with earlier studies that found higher levels of trust for nonprofit research institutions or universities and lower levels of trust for pharmaceutical or insurance companies.61–63 Thus, future EHR research sponsored or conducted by for-profit companies, such as pharmaceutical companies, may consider partnering with nonprofit organizations or academic research institutes.
A second key finding was that most parents of children with FXS or ASD and a subgroup of parents with a TD child preferred research use of EHRs that used non-identifiable and nonsensitive data. Nevertheless, this was especially true for a subgroup of parents of a child with ASD (class 1). This is an interesting twist on genetic exceptionalism (ie, the belief that genetic information is unique when compared with other types of medical or health information and as such deserves special protection).64 It would not have been surprising if parents of children with FXS, who have a known genetic condition, espoused genetic exceptionalism and thus were more in favor of research using non-identifiable and nonsensitive data than parents of children with ASD, a suspected genetic condition. Nevertheless, the opposite was true. In fact, preferences of the subgroup of parents of a child with ASD were twice as high for non-identifiable and nonsensitive information than the preferences of parents of a child with FXS. Perhaps this finding is related to parents’ fear of possible discrimination against individuals with ASD who, despite having the same diagnosis, have quite variable functioning. The subgroup of parents with a TD child had similarly strong preference (class 2). Previous studies conducted with the general public have found mixed evidence for the idea of genetic exceptionalism as well.65,66
It is unclear what factors are driving the difference between the groups with strong preferences for the privacy of their child’s data. Only a handful of the child and family characteristics (parent’s income, amount of trust in the healthcare system, level of health literacy, and severity of the child’s condition as reflected in the number of co-occurring health conditions) were related to the groups who preferred the use of non-identifiable and nonsensitive data, but there was no consistent pattern across the subgroups of parents. All of the subgroups of parents with a child with FXS or ASD, though, who prioritized the use of non-identifiable and nonsensitive information also had preferences for nonprofits to conduct the research. This may reflect their fear of privacy breaches and possible negative consequences for their child, such as stigmatization or discrimination. Other researchers have noted possible misuses of EHR data in their work as well.35,67,68 For the subgroup of parents with a TD child, their concerns about privacy align with other research that has highlighted the need for the security of EHR data.61,67,69
Not surprisingly, most parents of children with FXS or ASD had strong preferences for research that was personally relevant and returned results. Our previous research44 indicated that desire to participate in personally relevant research that returns results is related to altruism, specifically a desire to help their child or others in their community. Altruism has been found to be a motivating factor for participating in research in earlier studies as well.28,62,70 Those who have been diagnosed with a particular condition often want to pay it forward by participating in research that hopefully will lead to better treatment for those with the same condition.71,72 Return of results in genetic research has similarly been supported by both those in the general public as well as individuals with genetic conditions.71,72 Return of results is also seen as a personal benefit of research participation.72–74 In studies of parents of individuals with ASD, some stated the return of results helps to alleviate guilt75 or prepare for the future.76 Finally, return of results in genetic studies often is seen as a benefit for other family members to help increase awareness and make more informed treatment or reproductive choices.72,75
Implications
Taken together, these findings have implications for policy and practice. As echoed by others, there is a need to balance the privacy of individuals and the benefit that comes from mining EHRs.37,77 The views of the general public as well as those expressed here by parents of individuals with known or suspected genetic conditions demonstrate a clear preference for maintaining trust, transparency, and relevance of research being conducted with data from EHRs. These beliefs are aligned with the ethical principles outlined in the Belmont Report, including respect for persons, beneficence, and justice. In practice, these principles are embodied in the informed consent process. Nevertheless, research studies that use deidentified data from EHRs are not classified as human participant research and therefore do not require informed consent. Despite this, a growing body of work78,79 shows that there is a need for researchers to apply a social license framework, one that goes beyond the regulations for conducting research and embodies a broader set of ethical standards including reciprocity, nonexploitation, and service to the public good.80 Models of informed consent that exemplify these ideals are opt-out, tiered, or dynamic consent models.81,82 For European countries, these ethical principles have been incorporated into policy, specifically the General Data Protection Regulation, which governs the use of EHR data.83 In the United States, the research use of EHRs falls under the policy umbrella of the Final Rule and the Health Insurance Portability and Accountability Act. These rules, though, are not as comprehensive as General Data Protection Regulation.84 The results of this study highlight the need to use social license principles to guide the refinement of policies and practices for individuals with genetic conditions in the United States.
Limitations
The interpretation of results from this study should be tempered with the following limitations. First, although we had a large sample of parents across each of the 3 groups, they were not necessarily representative of the broader population. For instance, all participants were recruited through research registries or convenience sampling. In addition, most participants were non-Hispanic white and, in the FXS and ASD groups, more educated and had higher incomes. Given that these parents had participated in other research studies, their preferences for research use of their child’s EHR may differ from parents who have not participated. Additionally, the views of parents of children with FXS or ASD likely do not represent the views of parents whose children have other known or suspected genetic conditions. Finally, most participants had similar levels of trust in the healthcare system, health literacy, and satisfaction with their healthcare providers thus making it difficult to tease out differences between the subgroups of parents based on these characteristics. It is possible other unmeasured factors, such as the degree to which EHR research will make an impact on treatment options for their child, could be related to parents’ preferences.
Future Research
This study helps to shape the knowledge base of research use of EHR data from a unique population: parents of children with known or suspected genetic conditions. The preferences of these individuals have not been investigated previously, yet they are critical to understanding the motivations of parents to participate in the growing number of large-scale genetic research studies. Knowing the factors that affect parental decision making can help to shape not only the type of research being conducted but also how to better inform parents so they can make appropriate decisions for their child and family. Additional research should examine which types of studies parents are more inclined to participate in and whether they are willing to combine their child’s EHR data with other epidemiological (eg, lifestyle) or biological (blood or tissue samples) data to share with researchers.
Supplementary Material
Acknowledgement:
The authors wish to thank Drs. Don Bailey, Liz Berry-Kravis, Rob Christian, Megan Lewis, Marsha Mailick, Rud Turnbull, Jonathon Wald, Sue West, and Deb Zuver for their input to the study. We also wish to thank all the parents who participated in the study.
Conflict of Interest Disclosures: Dr. Raspa, Dr. Paquin, Ms. Andrews, Ms. Edwards, Ms. Moultrie, Ms. Wagner, and Dr. Wheeler report receiving grants from the Eunice Kennedy Shrive National Institute of Child Health and Human Development during the conduct of the study. Dr. Brown reports receiving personal, consulting fees from RTI International during the conduct of the study. Ms. Frisch and Dr. Turner-Brown report receiving a subcontract/grant from RTI International during the conduct of the study. No other disclosures were reported.
Funding/Support: The research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number 5R01HD086702 (PI: Raspa).
Footnotes
Supplemental Materials
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.jval.2020.06.016.
Role of the Funder/Sponsor: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Melissa Raspa, RTI International, Research Triangle Park, NC, USA.
Ryan S. Paquin, RTI International, Research Triangle Park, NC, USA.
Derek S. Brown, Brown School, Washington University, St. Louis, MO, USA.
Sara Andrews, RTI International, Research Triangle Park, NC, USA.
Anne Edwards, RTI International, Research Triangle Park, NC, USA.
Rebecca Moultrie, RTI International, Research Triangle Park, NC, USA.
Laura Wagner, RTI International, Research Triangle Park, NC, USA.
MaryKate Frisch, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Lauren Turner-Brown, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Anne C. Wheeler, RTI International, Research Triangle Park, NC, USA.
REFERENCES
- 1.Sherman RE, Anderson SA, Dal Pan GJ, et al. Real-world evidence—what is it and what can it tell us. N Engl J Med. 2016;375(23):2293–2297. [DOI] [PubMed] [Google Scholar]
- 2.Garrison LP Jr, Neumann PJ, Erickson P, Marshall D, Mullins CD. Using real-world data for coverage and payment decisions: the ISPOR real-world data task force report. Value Health. 2007;10(5):326–335. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, Weng C, Yu H. Natural language processing, electronic health records, and clinical research In: Clinical Research Informatics. London, England: Springer; 2012:293–310. [Google Scholar]
- 4.Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform. 2016;4(3):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hope C, Estrada N, Weir C, Teng CC, Damal K, Sauer BC. Documentation of delirium in the VA electronic health record. BMC Res Notes. 2014;7(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrinton LJ, Shorstein NH, Paschal JF, et al. Comparative effectiveness of antibiotic prophylaxis in cataract surgery. Ophthalmology. 2016;123(2):287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20(1):144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hersh WR, Weiner MG, Embi PJ, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013;51(8 Suppl 3):S30–S37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2012;20(1):117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safran C, Bloomrosen M, Hammond WE, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc. 2007;4(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS (Wash DC). 2016;4(1):1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casey JA, Schwartz BS, Stewart WF, Adler NE. Using electronic health records for population health research: a review of methods and applications. Ann Rev Public Health. 2016;37:61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC. Mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8(12):e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyler CV Jr, Schramm S, Karafa M, Tang AS, Jain A. Electronic health record analysis of the primary care of adults with intellectual and other developmental disabilities. J Policy Pract Intellect Disabil. 2010;7(3):204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilbert JE, Kissel JT, Luebbe EA, et al. If you build a rare disease registry, will they enroll and will they use it? Methods and data from the National Registry of Myotonic Dystrophy (DM) and Facioscapulohumeral Muscular Dystrophy (FSHD). Contemp Clin Trials. 2012;33(2):302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson R, Johnston L, Taruscio D, et al. RD-Connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med. 2014;29(3):780–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knapp EA, Fink AK, Goss CH, et al. The Cystic Fibrosis Foundation Patient Registry. Design and methods of a national observational disease registry. Ann Am Thorac Soc 2016;13(7):1173–1179. [DOI] [PubMed] [Google Scholar]
- 20.Nelson EC, Dixon-Woods M, Batalden PB, et al. Patient focused registries can improve health, care, and science. BMJ. 2016;354:i3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins S F, Varmus H. A new initiative on precision medicine. New England Journal of Medicine. 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayton EW, Smith M, Fullerton SM, et al. Confronting real time ethical, legal, and social issues in the Electronic Medical Records and Genomics (eMERGE) Consortium. Genetics in Medicine. 2010;12(10):616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormack P, Kole A, Gainotti S, et al. ‘You should at least ask’. The expectations, hopes and fears of rare disease patients on large-scale data and biomaterial sharing for genomics research. Eur J Hum Genet. 2016;24(10):1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulynych J, Greely HT. Clinical genomics, big data, and electronic medical records: reconciling patient rights with research when privacy and science collide. Journal of Law and the Biosciences. 2017;4(1):94–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willison DJ, Steeves V, Charles C, et al. Consent for use of personal information for health research: do people with potentially stigmatizing health conditions and the general public differ in their opinions? BMC Med Ethics. 2009;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill EM, Turner EL, Martin RM, Donovan JL. “Let’s get the best quality research we can”: Public awareness and acceptance of consent to use existing data in health research: A systematic review and qualitative study. BMC Med Res Methodol. 2013;13:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aitken M, de St Jorre J, Pagliari C, Jepson R, Cunningham-Burley S . Public responses to the sharing and linkage of health data for research purposes: A systematic review and thematic synthesis of qualitative studies. BMC Medical Ethics. 2016;17(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perera G, Holbrook A, Thabane L, Foster G, Willison DJ. Views on health information sharing and privacy from primary care practices using electronic medical records. Int J Med Inform. 2011;80(2):94–101. [DOI] [PubMed] [Google Scholar]
- 29.Hazin R, Brothers KB, Malin BA, et al. Ethical, legal, and social implications of incorporating genomic information into electronic health records. Genet Med. 2013;15(10):810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damschroder LJ, Pritts JL, Neblo MA, Kalarickal RJ, Creswell JM, Hayward RA. Patients, privacy and trust: Patients’ willingness to allow researchers to access their medical records. Social Science & Medicine. 2007;64(1):223–235. [DOI] [PubMed] [Google Scholar]
- 31.Caine K, Hanania R. Patients want granular privacy control over health information in electronic medical records. J Am Med Inform Assoc. 2012;20(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willison DJ, Keshavjee K, Nair K, Goldsmith C, Holbrook AM. Patients’ consent preferences for research uses of information in electronic medical records: interview and survey data. BMJ. 2003;326(7385):373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hull SC, Sharp RR, Botkin JR, et al. Patients’ views on identifiability of samples and informed consent for genetic research. Am J Bioethics. 2008;8(10):62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trinidad SB, Fullerton SM, Bares JM, Jarvik GP, Larson EB, Burke W. Genomic research and wide data sharing: views of prospective participants. Genet Med. 2010;12(8):486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow-White PA, MacAulay M, Charters A, Chow P. From the bench to the bedside in the big data age: ethics and practices of consent and privacy for clinical genomics and personalized medicine. Ethics Inf Technol. 2015;17(3):189–200. [Google Scholar]
- 36.Roden DM, Denny JC. Integrating electronic health record genotype and phenotype datasets to transform patient care. Clin Pharmacol Ther. 2016;99(3):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. [DOI] [PubMed] [Google Scholar]
- 38.Griggs RC, Batshaw M, Dunkle M, et al. Clinical research for rare disease: opportunities, challenges, and solutions. Mol Genet Metab. 2009;96(1):20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey DB Jr, Raspa M, Olmsted M, Holiday DB. Co-occurring conditions associated with FMR1 gene variations: findings from a national parent survey. Am J Med Genet A. 2008;146A(16):2060–2069. [DOI] [PubMed] [Google Scholar]
- 40.Nicholas JS, Charles JM, Carpenter LA, King LB, Jenner W, Spratt EG. Prevalence and characteristics of children with autism-spectrum disorders. Ann Epidemiol. 2008;18(2):130–136. [DOI] [PubMed] [Google Scholar]
- 41.Geschwind DH. Genetics of autism spectrum disorders. Trends Cognitive Sci. 2011;15(9):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bridges JF, Hauber AB, Marshall D, et al. Conjoint analysis applications in health—a checklist: a report of the ISPOR Good Research Practices for Conjoint Analysis Task Force. Value Health. 2011;14(4):403–413. [DOI] [PubMed] [Google Scholar]
- 43.Raspa M, Moultrie R, Wagner L, et al. Ethical, legal, and social issues related to the inclusion of individuals with intellectual disabilities in electronic health record research. J Med Internet Res. 2020;22(5):e16734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews SA, Raspa M, Edwards A, et al. “Just tell me what’s going on”: The views of parents of children with genetic conditions regarding the research use of their child’s EHR. J Am Med Inform Assoc. 2020;27(3):429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shea JA, Micco E, Dean LT, McMurphy S, Schwartz JS, Armstrong K. Development of a revised health care system distrust scale. J Gen Intern Med. 2008;23(6):727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chew LD, Bradley KA, Boyko EJ. Brief questions to identify patients with inadequate health literacy. Fam Med. 2004;36(8):588–594. [PubMed] [Google Scholar]
- 47.Maenner MJ, Smith LE, Hong J, Makuch R, Greenberg JS, Mailick MR. Evaluation of an activities of daily living scale for adolescents and adults with developmental disabilities. Disabil Health J 2013;6(1):8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson FR, Lancsar E, Marshall D, et al. Constructing experimental designs for discrete-choice experiments: report of the ISPOR Conjoint Analysis Experimental Design Good Research Practices Task Force. Value Health. 2013;16(1):3–13. [DOI] [PubMed] [Google Scholar]
- 49.Thurstone LL. A law of comparative judgment. Psychol Rev. 1927;34(4):273. [Google Scholar]
- 50.Louviere JJ, Flynn TN, Carson RT. Discrete choice experiments are not conjoint analysis. J Choice Model. 2010;3(3):57–72. [Google Scholar]
- 51.Hensher DA, Greene WH. The mixed logit model: the state of practice and warnings for the unwary. Inst Transport Stud. 2002;28:1–39. [Google Scholar]
- 52.Hauber AB, González JM, Groothuis-Oudshoorn CG, et al. Statistical methods for the analysis of discrete choice experiments: a report of the ISPOR Conjoint Analysis Good Research Practices Task Force. Value Health. 2016;19(4):300–315. [DOI] [PubMed] [Google Scholar]
- 53.Hole AR. Fitting mixed logit models by using maximum simulated likelihood. Stata J. 2007;7(3):388–401. [Google Scholar]
- 54.Swait J, Louviere J. The role of the scale parameter in the estimation and comparison of multinomial logit models. J Mark Res 1993;30(3): 305–314. [Google Scholar]
- 55.Viney R, Lancsar E, Louviere J. Discrete choice experiments to measure consumer preferences for health and healthcare. Exp Rev Pharmacoecon Outcomes Res. 2002;2(4):319–326. [DOI] [PubMed] [Google Scholar]
- 56.Janssen EM. Measuring treatment preferences of people with type 2 diabetes, an application of stated-preference methods [dissertation]. Baltimore, MD: Johns Hopkins University; 2017. [Google Scholar]
- 57.Clogg CC, Petkova E, Haritou A. Statistical methods for comparing regression coefficients between models. Am J Sociol. 1995;100(5):1261–1293. [Google Scholar]
- 58.Train KE. Discrete Choice Methods With Simulation. Cambridge: University Press; 2009. [Google Scholar]
- 59.Nylund KL, Asparouhov T, Muthén BO. Deciding on the number of classes in latent class analysis and growth mixture modeling: a Monte Carlo simulation study. Struct Equ Model. 2007;14(4):535–569. [Google Scholar]
- 60.Pacifico D, Yoo HI. lclogit: a Stata command for fitting latent-class conditional logit models via the expectation-maximization algorithm. Stata J. 2013;13(3):625–639. [Google Scholar]
- 61.Grande D, Mitra N, Shah A, Wan F, Asch DA. Public preferences about secondary uses of electronic health information. JAMA Intern Med. 2013;173(19):1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim KK, Joseph JG, Ohno-Machado L. Comparison of consumers’ views on electronic data sharing for healthcare and research. J Am Med Inform Assoc. 2015;22(4):821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harle CA, Golembiewski EH, Rahmanian KP, et al. Patient preferences toward an interactive e-consent application for research using electronic health records. J Am Med Inform Assoc. 2017;25(3):360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murray TH. Genetic exceptionalism and ‘future diaries’: is genetic information different from other medical information? In: Rothstein MA, ed. Genetic Secrets: Protecting Privacy and Confidentiality in the Genetic Era. New Haven, CT: Yale University Press; 1997. [Google Scholar]
- 65.Ruiz-Canela M, Valle-Mansilla JI, Sulmasy DP. What research participants want to know about genetic research results: the impact of “genetic exceptionalism”. J Empir Res Hum Res Ethics. 2011;6(3):39–46. [DOI] [PubMed] [Google Scholar]
- 66.Diergaarde B, Bowen DJ, Ludman EJ, Culver JO, Press N, Burke W. Genetic information: special or not? Responses from focus groups with members of a health maintenance organization. Am J Med Genet A. 2007;143A:564–569. [DOI] [PubMed] [Google Scholar]
- 67.Aitken M, Jorre JD, Pagliari C, Jepson R, Cunningham-Burley S. Public responses to the sharing and linkage of health data for research purposes: a systematic review and thematic synthesis of qualitative studies. BMC Med Ethics. 2016;17(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rogith D, Yusuf RA, Hovick SR, et al. Attitudes regarding privacy of genomic information in personalized cancer therapy. J Am Med Inform Assoc. 2014;21(e2):e320–e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stockdale J, Cassell J, Ford E. “Giving something back”: a systematic review and ethical enquiry into public views on the use of patient data for research in the United Kingdom and the Republic of Ireland. Wellcome Open Res. 2019;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Platt J, Kardia S. Public trust in health information sharing: implications for biobanking and electronic health record systems. J Pers Med. 2015;5(1):3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Treloar SA, Morley KI, Taylor SD, Hall WD. Why do they do it? A pilot study towards understanding participant motivation and experience in a large genetic epidemiological study of endometriosis. Public Health Genomics. 2007;10(2):61–71. [DOI] [PubMed] [Google Scholar]
- 72.Hallowell N, Cooke S, Crawford G, Lucassen A, Parker M, Snowdon C. An investigation of patients’ motivations for their participation in genetics-related research. J Med Ethics. 2010;36(1):37–45. [DOI] [PubMed] [Google Scholar]
- 73.Sabatello M, Chen Y, Zhang Y, Appelbaum PS. Disability inclusion in precision medicine research: a first national survey. Genet Med. 2019;21(10):2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoop JG, Roberts LW, Hammond KA. Genetic testing of stored biological samples: views of 570 US workers. Genet Testing Mol Biomarkers. 2009;13(3):331–337. [DOI] [PubMed] [Google Scholar]
- 75.Trottier M, Roberts W, Drmic I, et al. Parents’ perspectives on participating in genetic research in autism. J Autism Dev Disord. 2013;43(3):556–568. [DOI] [PubMed] [Google Scholar]
- 76.Baret L, Godard B. Opinions and intentions of parents of an autistic child toward genetic research results: two typical profiles. Eur J Hum Genet. 2011;19(11):1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Layman EJ. Ethical issues and the electronic health record. Health Care Manag (Frederick). 2008;27(2):165–176. [DOI] [PubMed] [Google Scholar]
- 78.Vayena E, Haeusermann T, Adjekum A, Blasimme A. Digital health: meeting the ethical and policy challenges. Swiss Med Wkly. 2018;148:w14571. [DOI] [PubMed] [Google Scholar]
- 79.Ballantyne A Where is the human in the data? A guide to ethical data use. Gigascience. 2018;7(7):giy076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carter P, Laurie GT, Dixon-Woods M. The social licence for research: why care. data ran into trouble. J Med Ethics. 2015;41(5):404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Botkin JR. Informed consent for genetic research. Curr Protoc Hum Genet. 2010;66(1). [DOI] [PubMed] [Google Scholar]
- 82.Kaye J, Whitley EA, Lund D, Morrison M, Teare H, Melham K. Dynamic consent: a patient interface for twenty-first century research networks. Eur J Hum Genet. 2015;23(2):141–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rumbold JM, Pierscionek B. The effect of the general data protection regulation on medical research. J Med Internet Res. 2017;19(2):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Terry N Existential challenges for healthcare data protection in the United States. Ethics Med Public Health. 2017;3(1):19–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.