Abstract
Background
Long non-coding RNAs (lncRNAs), a class of endogenous non-coding RNAs, play an important role in the development and metastasis of non-small cell lung cancer (NSCLC). However, the function and mechanism of action of long intergenic non-protein coding RNA 1089 (LINC01089) in NSCLC remains unclear. This study aimed to identify the role of LINC01089 in cell proliferation, migration, and invasion of NSCLC.
Methods
Expression of LINC01089 and the relationship between LINC01089 and overall survival (OS) in NSCLC were determined using GEPIA 2.0. Similarly, microRNAs (miRNAs) that showed increased expression in NSCLC and correlated with OS were identified using the online OncomiR cancer database. Target miRNAs of LINC01089 were predicted using starBase. Cell models of LINC01089 and miR-3187-3p overexpression were constructed using transfection. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to analyze the expression of LINC01089 and miR-3187-3p. MTS assay was used to assess cell proliferation. Transwell was used for migration and invasion assays.
Results
LINC01089 expression was significantly reduced in NSCLC tissues and cells. Gain-of-function studies further demonstrated that LINC01089 overexpression inhibited proliferation, migration, and invasion of lung cancer cell lines, A549 and SK-MES-1. Based on starBase prediction and subsequent verification, we revealed that miR-3187-3p is a target miRNA of LINC01089. Additionally, miR-3187-3p expression was significantly increased in NSCLC tissues and cells. Overexpression of miR-3187-3p promoted proliferation, migration, and invasion of A549 and SK-MES-1 cells, thereby reversing the effect of LINC01089.
Conclusion
LINC01089 attenuates tumor proliferation, migration, and invasion by sponging miR-3187-3p in NSCLC. LINC01089 acts as a tumor suppressor and represents a potential therapeutic target in NSCLC.
Keywords: lncRNA, lung cancer, miRNA, prognosis
Introduction
Lung cancer is one of the most common cancers worldwide with high morbidity and mortality.1 In China, a total of 781,500 new lung cancer cases were diagnosed in 2014, and the morbidity was 57.13 per 100 000 cases.2 Due to increased industrialization, accelerated urbanization, and changes in lifestyle, the incidence and mortality of lung cancer is on the rise in China and poses a major burden on public health.3 Air pollution and smoking are the main causes of lung cancer.4–6 According to the world health organization classification, lung cancer can be grouped into two main histological groups: small cell lung cancer and non-small cell lung cancer (NSCLC, 90% of all lung cancers). At present, the treatment modalities for NSCLC mainly consist of surgical resection combined with chemoradiotherapy. However, NSCLC has a high recurrence rate following surgery.7 Therefore, there is an urgent need to identify new molecular mechanisms to provide novel therapeutic targets for the treatment of NSCLC.
In recent years, an increasing number of studies have focused on the correlation between long non-coding RNAs (lncRNAs) and tumorigenesis. LncRNAs are endogenous non-coding RNA molecules >200 nucleotides in length. They have important functions in gene silencing, transcriptional activation, chromosome modification, and intranuclear transport.8,9 Abnormal lncRNA expression is closely associated with various steps in the development of cancer including regulation of tumor proliferation and formation, invasion, and metastasis, and lncRNA also participates in the relapse process.10,11 LncRNA plays a key role in various biological processes in NSCLC.12 LncRNAs such as MALAT1, GAS5, HIF1A-AS1, and XIST have been shown to play significant regulatory roles in tumor biology and may serve as noninvasive diagnostic and prognostic biomarkers and potential therapeutic targets in NSCLC.13 Long intergenic non-protein coding RNA 1089 (LINC01089) localizes to chromosome 12: 121,795,267–121,803,906 reverse strand. A previous study found that the expression of LINC01089 is downregulated in breast cancer and inhibited extracellular matrix invasion and tumor metastasis in breast cancer.14 M2-like tumor-associated macrophages promote epithelial ovarian cancer metastasis via suppressing LIMT expression.15 Additionally, LINC01089 inhibited cell proliferation and invasion through the Wnt/β-catenin signaling pathway and improved clinical prognosis in breast cancer.16 These results suggest that LINC01089 is a tumor suppressor gene in breast cancer. However, the expression and biological function of LINC01089 in NSCLC remains unclear.
In this study, we investigated the expression of LINC01089 in NSCLC including lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). We also evaluated the roles of LINC01089 and explored the molecular mechanisms underlying the regulatory effects of LINC01089 in NSCLC.
Materials and Methods
Database Analyses
GEPIA 2.0 is a publicly accessible online database based on tumor and normal samples from the cancer genome atlas and the genotype-tissue expression databases (http://gepia2.cancer-pku.cn/).17 Expression of LINC01089 was analyzed in normal tissues and NSCLC tissues, including LUSC and LUAD, using GEPIA 2.0. Additionally, the relationship between LINC01089, disease-free survival (DFS) and overall survival (OS) of the patients with LUSC and LUAD was also analyzed using GEPIA 2.0. OncomiR (http://www.oncomir.org/), an online resource for exploring pan-cancer microRNA (miRNA) dysregulation, was used to analyze miRNA dysregulation in LUSC and LUAD.18
Cell Culture
Normal lung epithelial cells BEAS-2B, lung adenocarcinoma cell line, A549, and squamous cell lung carcinoma cell line, SK-MES-1 were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). BEAS-2B cells were cultured in bronchial epithelial growth medium (BEGM, Walkersville, MD, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA); A549 cells were cultured in F-12K (Gibco) supplemented with 10% FBS (Gibco); and SK-MES-1 cells were cultured in Eagle’s minimum essential medium (Gibco) supplemented with 10% FBS (Gibco). All cells were maintained in a humidified incubator with 5% CO2 at 37°C.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
QRT-PCR was used to measure the expression of LINC01089 and miR-3187-3p, and the expression was normalized to β-actin or U6, respectively. The relative expression was calculated using the 2−ΔΔCt method. The sequence of the primers designed using primer-blast and synthesized by Sangon (Shanghai, China) is shown in Table 1. Briefly, RNA was isolated from the cells using TRIzol (Invitrogen, Carlsbad, CA, USA). RNA was reverse-transcribed using RT Master Mix Kit (Promega, Madison, WI, USA), and qRT-PCR was performed using SYBR Green qPCR SuperMix (Invitrogen).
Table 1.
Sequences
| Primers | Sequences (5ʹ–3ʹ) |
|---|---|
| LINC01089-F | GCAGTAAACAGTCCTCAGCGAAG |
| LINC01089-R | CGGTGCCATGGAGTCTAGAAGAT |
| β-actin-F | GCATGGGTCAGAAGGATTCCT |
| β-actin-R | TCGTCCCAGTTGGTGACGAT |
| miR-3187-3p-F | ACACTCCAGCTGGGTTGGCCATGGGGCT |
| miR-3187-3p-R | CTCAACTGGTGTCGTGGA |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| siRNAs | |
| si-NC | UUCUCCGAACGUGUCACGUTT |
| si- LINC01089-1 | GCCUACCCAACCCUGGAAATT |
| si- LINC01089-2 | GCCUGGCAGGCAACUCCAUTT |
| si- LINC01089-3 | GCGCUCAGCCUUCAGUAAATT |
Abbreviations: F, forward primer; R, reverse primer; NC, negative control.
Transfection
LINC01089 sequence (NR_002809.3) was synthesized by Genewiz (Suzhou, China) and inserted into pcDNA3.1 (ov-LINC01089). Empty pcDNA3.1 vector served as a negative control (ov-NC). Three LINC01089 siRNAs (si-LINC01089), negative control (si-NC), miR-3187-3p mimic and NC mimic were synthesized by Genepharma (Shanghai, China). ov- LINC01089, ov-NC, miR-3187-3p mimic, and NC mimic were transfected into cells using Lipofectamine 3000 (Life Technologies, Gaithersburg, MD, USA).
MTS Assay
Cell proliferation was measured using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS). After culturing for 24, 48, and 72 h, 10 μL of MTS reagent was added to each well and incubated for 4 h at 37°C. Then, the absorbance was measured at 490 nm using a Vmax microplate spectrophotometer (Molecular Devices, Sunnyvale, CA). Each sample was assayed thrice.
Transwell Assay
Migration and invasion of A549 and SK-MES-1 cells were measured using the transwell assay without or with Matrigel (BD Biosciences). For the transwell assay, 5 × 104 cells in 200 µL of medium containing 0.1% FBS was placed in the upper chamber of an insert (pore size, 8 µm) while medium (600 µL) containing 10% FBS was added to the lower chamber. After 48 h of incubation, the cells were removed from the upper chamber of the filter, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet in 20% ethanol. Cells that migrated and invaded the chamber were counted by photographing at 200× magnification using a LEICA microscope. Counts were performed in five independent fields per well.
Luciferase Reporter Assay
The sponged miRNA of LINC01089 was analyzed by starBase 3.0 which mainly focuses on miRNA–target interactions.19 Wild type LINC01089 (WT-LINC01089) and mutated LINC01089 (MUT-LINC01089) were inserted into psi-check2. Then, the WT-LINC01089 or MUT-LINC01089 and miR-3187-3p mimic (or NC mimic) were co-transfected into 293T using Lipofectamine 3000. At 48 h after transfection, luciferase assays were performed using the dual-luciferase reporter assay system (Promega), and ratio of Renilla to firefly luciferase activity was calculated. The targeted gene of miR-3187-3p was analyzed by starBase 3.0, TarBase v.8, and Targetscan 7.2.19–21
Statistical Analysis
Statistical analysis was performed using SPSS 19.0 software package (SPSS Inc, Chicago, IL, USA). All data are shown as mean ± standard deviation (SD). Differences between the two groups were analyzed using t-tests. Differences between more than three groups were evaluated using one-way analysis of variance, followed by a Dunnett post hoc test. All tests performed were two-sided. P < 0.05 was considered to be statistically significant.
Results
LINC01089 Expression is Reduced in LUSC and LUAD
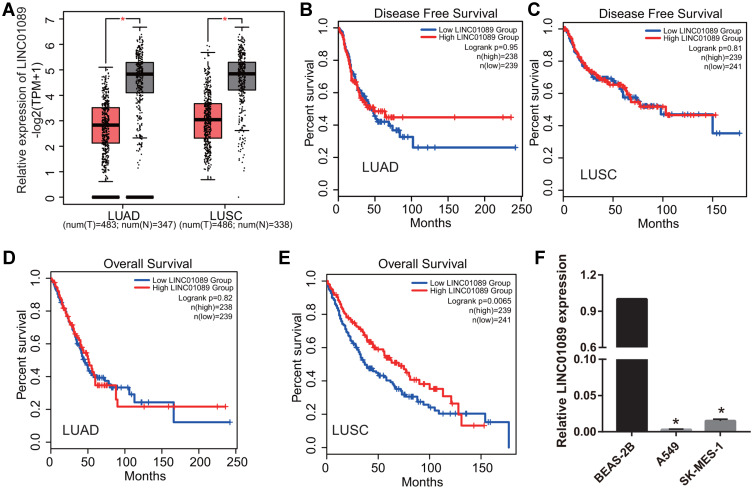
Based on the results of GEPIA 2.0 analysis, we found that LINC01089 expression was significantly reduced in LUAD and LUSC (Figure 1A). Additionally, LINC01089 expression was not significantly correlated with DFS in LUAD and LUSC (Figure 1B and C). Furthermore, LINC01089 expression had no significant correlation with OS in LUAD whereas it was significantly correlated with OS in LUSC (Figure 1D and E). Further, we found that LINC01089 expression was significantly lower in A549 and SK-MES-1 compared to BEAS-2B (Figure 1F).
Figure 1.
LINC01089 expression was analyzed in LUAD and LUSC tissues and cells. (A) LINC01089 expression was analyzed using the GEPIA 2.0 database. *P < 0.05. (B and C) The relationship between LINC01089 expression and the disease free survival of LUSC and LUAD was analyzed using GEPIA 2.0. (D and E) The relationship between LINC01089 expression and the overall survival of LUSC and LUAD was analyzed using GEPIA 2.0. (F) LINC01089 expression was measured by quantitative reverse transcription-polymerase chain reaction in normal lung epithelial cells, BEAS-2B, lung adenocarcinoma cell line, A549, and squamous cell lung carcinoma cell line, SK-MES-1. *P < 0.05 vs BEAS-2B.
Abbreviations: LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma; GEPIA, gene expression profiling interactive analysis; T, tumor; N, normal control.
LINC01089 Inhibits Proliferation, Migration, and Invasion in NSCLC
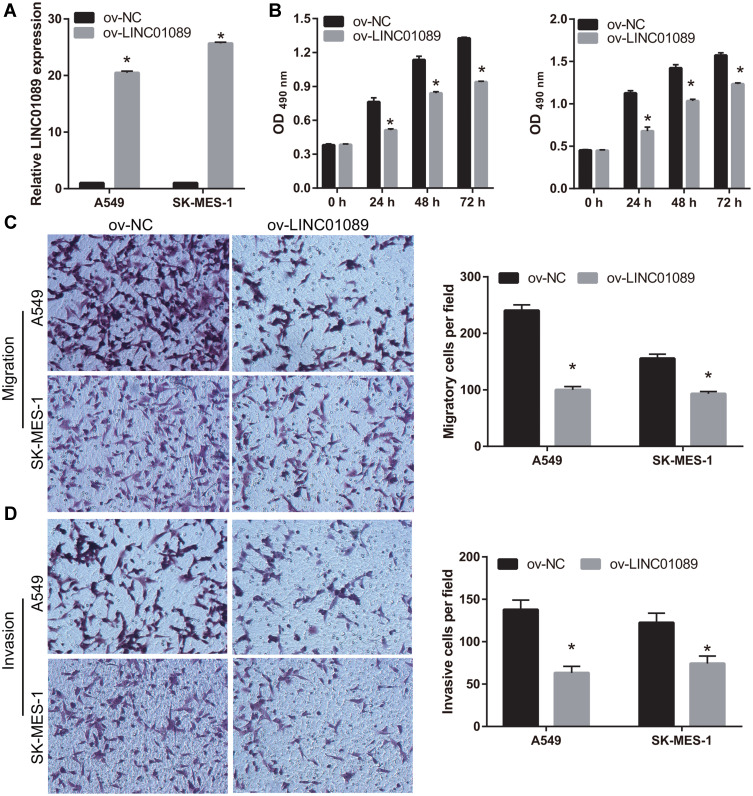
To examine the role of LINC01089 in NSCLC, LINC01089 was overexpressed in A549 and SK-MES-1 cells (Figure 2A). Compared to the ov-NC group, cell proliferation was markedly inhibited in the ov-LINC01089 group at 24, 48, and 72 h following transfection (Figure 2B). In addition, the migration and invasion of cells in the ov-LINC01089 group were also markedly inhibited at 48 h following transfection compared to the cells in the ov-NC group (Figure 2C and D).
Figure 2.
Overexpression of LINC01089 inhibited proliferation, migration, and invasion. (A) LINC01089 was overexpressed following transfection with LINC01089-pcDNA3.1. LINC01089 expression was measured by quantitative reverse transcription-polymerase chain reaction in A549 and SK-MES-1 following transfection with pcDNA3.1 (ov-NC) and LINC01089-pcDNA3.1 (ov- LINC01089) at 48 h. *P < 0.05. (B) Proliferation of A549 and SK-MES-1 was measured using MTS assay 48 h post-transfection. *P < 0.05. (C and D) Migration and invasion of A549 and SK-MES-1 were measured using transwell assay 48 h post-transfection. *P < 0.05.
Abbreviations: ov, overexpression; NC, negative control.
LINC01089 Sponges miR-3187-3p
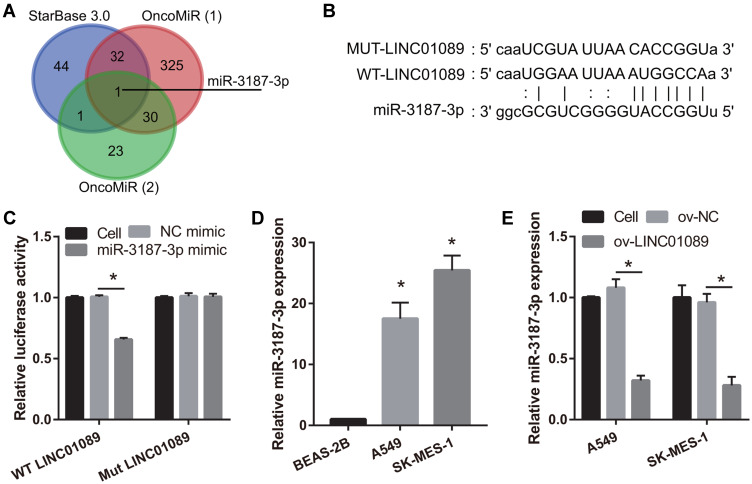
Using the bioinformatics tool, starBase 3.0, we identified 78 miRNAs that were potential target genes of LINC01089. Additionally, using the OncomiR cancer database, we found that 338 miRNAs were significantly increased in LUSC and LUAD [Figure 3A, OncomiR (1)]. Additionally, using the OncomiR cancer database, we also found that 55 miRNAs were associated with OS in LUSC and LUAD [Figure 3A, OncomiR (2)]. Based on these results, we identified miR-3187-3p as a common miRNA (Figure 3A). The potential binding sites between LINC01089 and miR-3187-3p are shown in Figure 3B. Dual-luciferase reporter assay was used to confirm the interaction between LINC01089 and miR-3187-3p in 293T cells. The results showed that the cells transfected with WT-LINC01089 and miR-3187-3p displayed lower luciferase activity than the cells transfected with MUT-LINC01089 and miR-3187-3p, suggesting an interaction between LINC01089 and miR-3187-3p (Figure 3C). Further, we found that miR-3187-3p expression was significantly inhibited in A549 and SK-MES-1, compared to BEAS-2B (Figure 3D). Compared with ov-NC group, miR-3187-3p expression in ov-LINC01089 group was significantly reduced (Figure 3E).
Figure 3.
MiR-3187-3p was sponged by LINC01089. (A) Potential target miRNAs of LINC01089 were identified using starBase 3.0. OncomiR (1): MiRNAs which were increased in NSCLC were identified using the bioinformatics tool, OncomiR. OncomiR (2): MiRNAs associated with overall survival in NSCLC were identified using OncomiR. (B) Potential binding sites between LINC01089 and miR-3187-3p are shown. (C) The interaction between LINC01089 and miR-3187-3p was verified using dual-luciferase reporter assay. *P < 0.05. (D) miR-3187-3p expression in BEAS-2B, A549, and SK-MES-1 was measured using quantitative reverse transcription-polymerase chain reaction (*P < 0.05 vs BEAS-2B). (E) The expression of miR-3187-3p after transfection with LINC01089 was measured using quantitative reverse transcription-polymerase chain reaction (*P < 0.05 vs ov-NC).
Abbreviations: NSCLC, non-small cell lung cancer; WT, wild type; MUT, mutation; ov, overexpression; NC, negative control.
miR-3187-3p Reverses the Effect of LINC01089 on Proliferation, Migration, and Invasion of NSCLC Cells
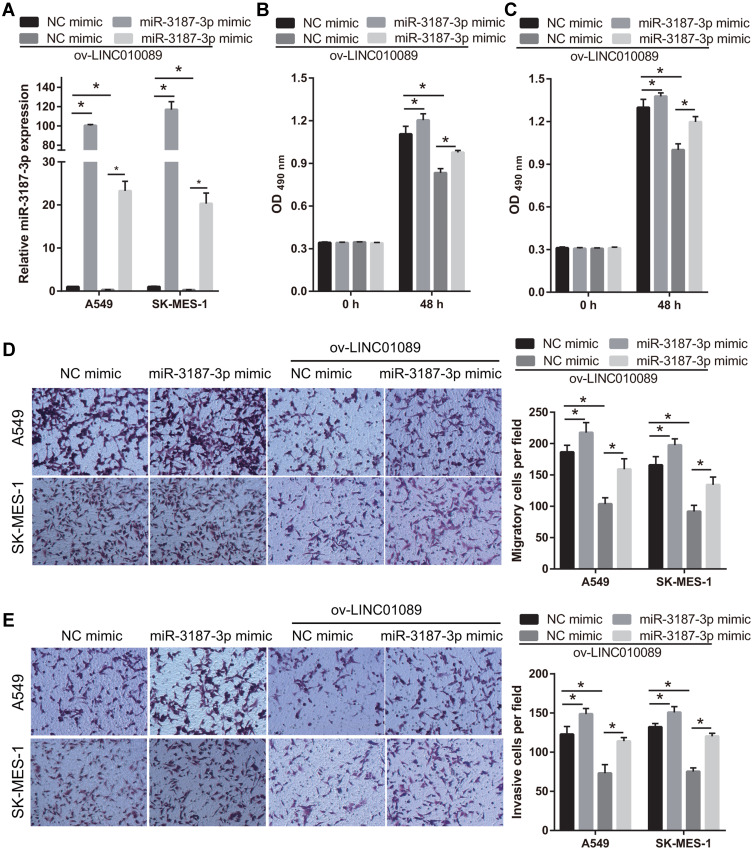
To further analyze the relationship between LINC01089 and miR-3187-3p, miR-3187-3p expression was enhanced in LINC01089 overexpressing-A549 and SK-MES-1 cells following transfection with miR-3187-3p mimic (Figure 4A). Compared to the NC mimic group, the proliferation, migration, and invasion of A549 and SK-MES-1 were markedly increased in the miR-3187-3p mimic group at 48 h following transfection (Figure 4B-E). Additionally, the proliferation, migration, and invasion of LINC01089 overexpressing-A549 and SK-MES-1 were markedly increased in the miR-3187-3p mimic group at 48 h following transfection, compared to the NC mimic group, suggesting the miR-3187-3p reverses the effect of LINC01089 in A549 and SK-MES-1 (Figure 4B-E).
Figure 4.
MiR-3187-3p reverses the effect of LINC01089 in A549 and SK-MES-1. (A) miR-3187-3p expression in A549 and SK-MES-1 was analyzed using quantitative reverse transcription-polymerase chain reaction at 48 h after transfection NC mimic, miR-3187-3p mimic, ov-LINC01089+ NC mimic, and ov-LINC01089+miR-3187-3p mimic. *P < 0.05. (B and C) Proliferation of A549 (B) and SK-MES-1 (C) was measured using MTS at 48 h after transfection. *P < 0.05. (D and E) Migration and invasion of A549 and SK-MES-1 was measured using transwell assay without or with Matrigel at 48 h after transfection. *P < 0.05.
Analysis of the Potentially Targeted Gene of miR-3187-3p
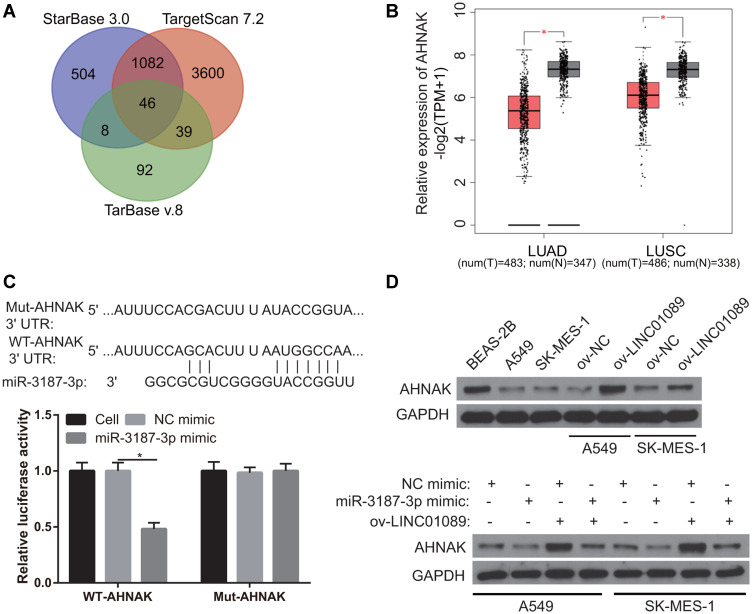
The targeted-gene of miR-3187-3p was analyzed by starBase 3.0, TarBase v.8, and Targetscan 7.2. The same targeted-gene was analyzed by Venn and 46 same targeted-genes were found (Figure 5A). Additionally, the expression of the 46 targeted-genes in LUSC and LUAD were analyzed by GEPIA 2.0. The result showed that only the AHNAK expression was significantly lower in the LUSC and LUAD tissues than that in normal control (Figure 5B), suggesting AHNAK may be the potential targeted-gene of miR-3187-3p because miR-3187-3p expression was significantly high in NSCLC. Potential binding sites between AHNAK and miR-3187-3p are shown in Figure 5C. The interaction between AHNAK 3ʹ-untranslated region and miR-3187-3p was verified using dual-luciferase reporter assay (Figure 5C). The results showed that the cells transfected with WT-AHNAK and miR-3187-3p displayed lower luciferase activity than the cells transfected with MUT-AHNAK and miR-3187-3p, suggesting an interaction between AHNAK and miR-3187-3p. Additionally, we found that AHNAK expression was remarkably lower in A549 and SK-MES-1 compared to BEAS-2B, and AHNAK expression was obviously higher in ov-LINC01089 group than that in ov-NC group. Furthermore, AHNAK expression was obviously inhibited after transfected miR-3187-3p mimic (Figure 5D).
Figure 5.
Analysis of the potentially targeted gene of miR-3187-3p. (A) The targeted-gene of miR-3187-3p was analyzed by starBase 3.0, TarBase v.8, and Targetscan 7.2 and the same targeted-gene was analyzed by Venn. (B) AHNAK expression was analyzed by GEPIA 2.0. *P < 0.05. (C) Potential binding sites between AHNAK 3ʹ-untranslated region and miR-3187-3p are shown. The interaction between AHNAK and miR-3187-3p was verified using dual-luciferase reporter assay. *P < 0.05. (D) AHNAK expression was measured by Western blot.
Abbreviations: LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma; GEPIA, gene expression profiling interactive analysis; T, tumor; N, normal control, ov, overexpression; NC, negative control.
Silenced LINC01089 or Overexpression miR-3187-3p Promotes Proliferation, Migration, and Invasion in BEAS-2B
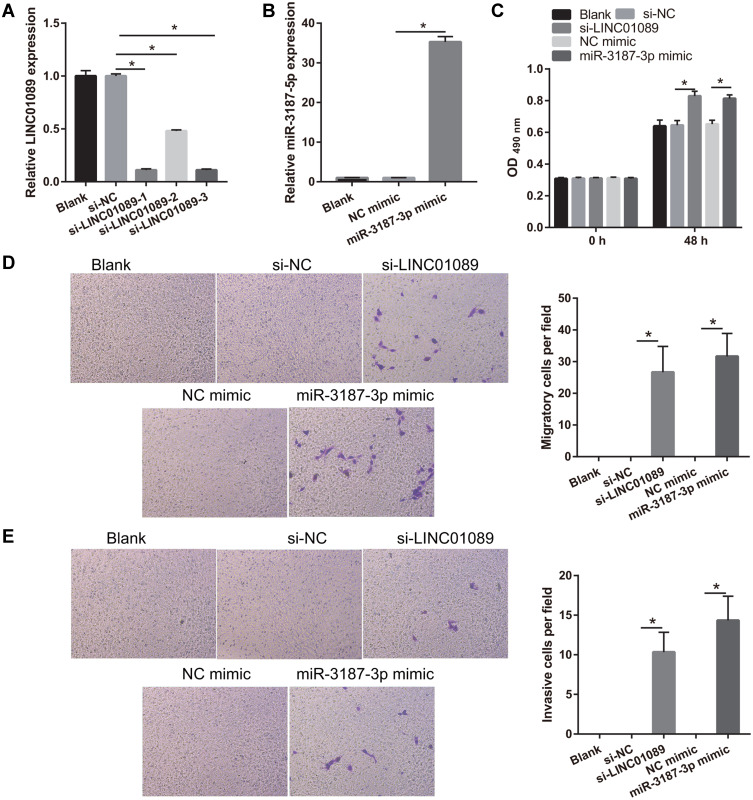
To examine the role of LINC01089 and miR-3187-3p in NSCLC development, LINC01089 was silenced or miR-3187-3p was overexpressed in BEAS-2B cells (Figure 6A and B). Compared to the si-NC or NC mimic group, cell proliferation, migration, and invasion were markedly promoted in the si-LINC01089 or miR-3187-3p mimic group at 48 h following transfection, respectively (Figure 6C-E).
Figure 6.
Silenced LINC01089 or overexpression miR-3187-3p promotes proliferation, migration, and invasion in BEAS-2B. (A) LINC01089 was down-expressed following transfection with si-LINC01089. LINC01089 expression was measured by quantitative reverse transcription-polymerase chain reaction in BEAS-2B following transfection with si-NC and three si-LINC01089 at 48 h. And si-LINC01089-1 was used for further study. *P < 0.05. (B) miR-3187-3p was overexpressed following transfection with miR-3187-3p mimic. miR-3187-3p expression was measured by quantitative reverse transcription-polymerase chain reaction in BEAS-2B following transfection with NC mimic and miR-3187-3p mimic at 48 h. *P < 0.05. (C) Proliferation of BEAS-2B was measured using MTS assay 48 h post-transfection. *P < 0.05. (D and E) Migration and invasion of BEAS-2B were measured using transwell assay 48 h post-transfection. *P < 0.05.
Abbreviations: si, siRNA; NC, negative control.
Discussion
LncRNA expression is significantly dysregulated in NSCLC and can thus be used as a diagnostic and prognostic marker for NSCLC. MALAT1 and GAS5 are diagnostic biomarkers for NSCLC.22,23 LINC00504 and NANCI are also associated with NSCLC prognosis.24,25 In this study, we found that LINC01089 expression was significantly reduced in LUAD, LUSC, and NSCLC cell lines. Additionally, LINC01089 expression was significantly associated with OS in LUSC. These results suggest that the downregulation of LINC01089 is associated with poor prognosis in LUSC. Surprisingly, although the expression of LINC01089 was significantly reduced in LUAD, it was not significantly associated with prognosis in LUAD.
Infinite proliferation and metastasis are important characteristics of malignant cells, and also the main processes that affect tumor treatment and relapse. LncRNAs that act as oncogenes and anti-oncogenes have a significant effect on the development, metastasis, and chemotherapeutic resistance of NSCLC. LINC01234 acts as an oncogene to promote cell proliferation and tumor growth in NSCLC.26 LncRNA LIMD1-AS1 acts as an anti-oncogene to suppress NSCLC progression.27 LncRNAs, HAS2-AS1 and XIST, accelerate resistance to chemotherapy in NSCLC.28,29 In this study, we found that LINC01089 overexpression inhibited the proliferation, migration, and invasion of A549 and SK-MES-1, suggesting that LINC01089 acts as an anti-oncogene to inhibit tumorigenesis and metastasis in NSCLC. A previous study found that LINC01089 inhibits cell invasion in vitro and tumor metastasis in vivo and acts as a tumor suppressor gene and prognostic biomarker in breast cancer and ovarian cancer.14–16 LINC01089 overexpression inhibited the β-catenin expression and blocked the Wnt/β-catenin pathway to inhibit breast cancer proliferation, migration, and invasion.16 According to our data, LINC01089 has an anti-cancer role in NSCLC, similar to its role in breast cancer. Additionally, cell proliferation, migration, and invasion were markedly promoted after silenced LINC01089 in normal lung epithelial cells, which also suggested LINC01089 has an anti-cancer role in NSCLC.
MiRNAs are single-stranded small non-coding RNA molecules, usually composed of 21–25 nt, which play a key role in several physiological and pathological processes in NSCLC. Abnormal expression of miRNAs in tissue, blood, and blood-exosomes can be used as a potential diagnostic, therapeutic, and prognostic biomarker in NSCLC patients.30,31 LncRNAs can serve as competing endogenous RNAs (ceRNAs) to sponge miRNAs and regulate tumorigenesis and metastasis of NSCLC. LncRNA, XIST, sponges miR-744 and miR-137 to inhibit cell proliferation, metastasis, and epithelial–mesenchymal transition in NSCLC.32,33 Another important finding of this study is the identification of miR-3187-3p as the downstream target of LINC01089. miR-3187-3p is associated with inflammation.34 The miR-3187-3p expression is increased in melanoma and drives immune escape.35 The miR-3187-3p expression is also increased in colorectal cancer and promotes colorectal cancer proliferation through suppressing ASXL1 expression.36 According to these results, miR-3187-3p has an oncogenic effect in melanoma and colorectal cancer. In this study, based on OncomiR analysis, we found that miR-3187-3p was significantly upregulated in NSCLC and higher miR-3187-3p expression was associated with lower OS in NSCLC. Additionally, miR-3187-3p overexpression promoted proliferation, migration, and invasion of A549 and SK-MES-1, reversing the effect of LINC01089. These results suggest that miR-3187-3p acts as an oncogene in NSCLC, similar to its role in melanoma and colorectal cancer. These results also suggest that LINC01089 and miR-3187-3p constitute a regulatory axis in NSCLC. Additionally, cell proliferation, migration, and invasion were markedly promoted after overexpressed miR-3187-3p in normal lung epithelial cells, which also suggested miR-3187-3p acts as an oncogene in NSCLC.
AHNAK is a scaffolding protein that implicated blood-brain barrier, cell migration, the regulation of cardiac calcium channels and muscle membrane repair, and the development of tumor metastasis.37 A previous study found that AHNAK expression was significantly reduced in human lung adenocarcinomas compared with matched normal lung tissues, and reduced AHNAK expression enhanced Type II Pneumocyte Hyperplasia and expedited the lung tumor development.38 AHNAK degradation could maintain the stemness of lung cancer stem-like cells.39 Consistently, this study showed that AHNAK expression was significantly decreased in NSCLC tissues and cells. Additionally, we found that AHNAK 3ʹ-untranslated region was bound to miR-3187-3p and AHNAK expression was significantly increased by LINC01089 overexpression while it was significantly decreased by miR-3187-3p overexpression. These results suggested that AHNAK, the target gene of miR-3187-3p, can regulate by LINC01089. However, the study has two limitations. First, LINC01089 expression was only obtained in GEPIA 2.0 online analysis and did not show in its own clinical sample. Second, the effect of LINC01089/miR-3187-3p/AHNAK axis on NSCLC needs to be elucidated through animal studies in the future.
Conclusions
In conclusion, LINC01089 attenuates tumor proliferation, migration, and invasion by sponging miR-3187-3p in NSCLC. LINC01089 acts as a tumor suppressor and represents a potential therapeutic target for NSCLC.
Abbreviations
lncRNA, long non-coding RNA; NSCLC, non-small cell lung cancer; LINC010089, long intergenic non-protein coding RNA 01089; OS, overall survival; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; miRNAs, microRNAs; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma; DFS, disease-free survival; ceRNAs, competing endogenous RNAs.
Disclosure
The authors have no conflicts of interest pertaining to this work.
References
- 1.Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun KX, Zheng RS, Zeng HM, et al. [The incidence and mortality of lung cancer in China, 2014]. Zhonghua Zhong Liu Za Zhi. 2018;40(11):805–811. Chinese. [DOI] [PubMed] [Google Scholar]
- 3.Zheng RS, Sun KX, Zhang SW, et al. [Report of cancer epidemiology in China, 2015]. Zhonghua Zhong Liu Za Zhi. 2019;41(1):19–28. Chinese. [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Chang Z, Wu J, Li W. Air pollution and lung cancer incidence in China: who are faced with a greater effect? Environ Int. 2019;132:105077. doi: 10.1016/j.envint.2019.105077 [DOI] [PubMed] [Google Scholar]
- 5.Parascandola M, Xiao L. Tobacco and the lung cancer epidemic in China. Transl Lung Cancer Res. 2019;8(Suppl 1):S21–s30. doi: 10.21037/tlcr.2019.03.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xing DF, Xu CD, Liao XY, et al. Spatial association between outdoor air pollution and lung cancer incidence in China. BMC Public Health. 2019;19(1):1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94(8):1623–1640. doi: 10.1016/j.mayocp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- 8.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 10.Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217. doi: 10.3727/096504016X14549667334007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9(7):1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 12.Ghafouri-Fard S, Shoorei H, Branicki W, Taheri M. Non-coding RNA profile in lung cancer. Exp Mol Pathol. 2020;114:104411. doi: 10.1016/j.yexmp.2020.104411 [DOI] [PubMed] [Google Scholar]
- 13.Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sas-Chen A, Aure MR, Leibovich L, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med. 2016;8(9):1052–1064. doi: 10.15252/emmm.201606198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng XY, Xie H, Yuan J, et al. M2-like tumor-associated macrophages-secreted EGF promotes epithelial ovarian cancer metastasis via activating EGFR-ERK signaling and suppressing lncRNA LIMT expression. Cancer Biol Ther. 2019;20(7):956–966. doi: 10.1080/15384047.2018.1564567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan H, Qin Y, Zeng B, et al. Long noncoding RNA LINC01089 predicts clinical prognosis and inhibits cell proliferation and invasion through the Wnt/beta-catenin signaling pathway in breast cancer. Onco Targets Ther. 2019;12:4883–4895. doi: 10.2147/OTT.S208830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–w560. doi: 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong NW, Chen Y, Chen S, Wang X. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics. 2018;34(4):713–715. doi: 10.1093/bioinformatics/btx627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Databaseissue):D92–97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1):D239–d245. doi: 10.1093/nar/gkx1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234(11):20721–20727. doi: 10.1002/jcp.28678 [DOI] [PubMed] [Google Scholar]
- 23.Pan J, Bian Y, Cao Z, et al. Long noncoding RNA MALAT1 as a candidate serological biomarker for the diagnosis of non-small cell lung cancer: a meta-analysis. Thorac Cancer. 2020;11(2):329–335. doi: 10.1111/1759-7714.13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moises J, Navarro A, Castellano JJ, et al. Long Non-Coding RNA NANCI/NKX2-1 duplex impacts prognosis in Stage I non-small-cell lung cancer. Arch Bronconeumol. 2020. [DOI] [PubMed] [Google Scholar]
- 25.Ma HP, Wang LX, Li W, Guo HH, Wu Y, Li XY. Upregulation of LINC00504 is associated with aggressive progression and poor prognosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2020;24(2):699–703. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Chen X, Lei T, et al. Integrative analysis of NSCLC Identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther. 2020;28(6):1479–1493. doi: 10.1016/j.ymthe.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan J, Tang Y, Liu S, et al. LIMD1-AS1 suppressed non-small cell lung cancer progression through stabilizing LIMD1 mRNA via hnRNP U. Cancer Med. 2020;9(11):3829–3839. doi: 10.1002/cam4.2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun P, Sun L, Cui J, Liu L, He Q. Long noncoding RNA HAS2-AS1 accelerates non-small cell lung cancer chemotherapy resistance by targeting LSD1/EphB3 pathway. Am J Transl Res. 2020;12(3):950–958. [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Zhou X, Chen Z, Gao C, Zhao L, Cui Y. Silencing of lncRNA XIST inhibits non-small cell lung cancer growth and promotes chemosensitivity to cisplatin. Aging (Albany NY). 2020;12(6):2726–4711. doi: 10.18632/aging.102673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Huang SX, Zhang F, et al. MicroRNAs: a novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017;50:6. doi: 10.1111/cpr.12394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Switlik WZ, Szemraj J. Circulating miRNAs as non-invasive biomarkers for non-small cell lung cancer diagnosis, prognosis and prediction of treatment response. Postepy Hig Med Dosw (Online). 2017;71:649–662. doi: 10.5604/01.3001.0010.3845 [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Zhang G, Cheng Z, et al. Knockdown of LncRNA-XIST suppresses proliferation and TGF-beta1-Induced EMT in NSCLC through the Notch-1 pathway by regulation of miR-137. Genet Test Mol Biomarkers. 2018;22(6):333–342. doi: 10.1089/gtmb.2018.0026 [DOI] [PubMed] [Google Scholar]
- 33.Li C, Wan L, Liu Z, et al. Long non-coding RNA XIST promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036 [DOI] [PubMed] [Google Scholar]
- 34.Agrawal M, Rastogi M, Dogra S, Pandey N, Basu A, Singh SK. Chandipura virus changes cellular miRNome in human microglial cells. J Med Virol. 2019. doi: 10.1002/jmv.25491 [DOI] [PubMed] [Google Scholar]
- 35.Vignard V, Labbe M, Marec N, et al. MicroRNAs in tumor exosomes drive immune escape in melanoma. Cancer Immunol Res. 2020;8(2):255–267. doi: 10.1158/2326-6066.CIR-19-0522 [DOI] [PubMed] [Google Scholar]
- 36.Yang G, Zhang T, Ye J, et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manag Res. 2019;11:6499–6509. doi: 10.2147/CMAR.S203137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis TA, Loos B, Engelbrecht AM. AHNAK: the giant jack of all trades. Cell Signal. 2014;26(12):2683–2693. doi: 10.1016/j.cellsig.2014.08.017 [DOI] [PubMed] [Google Scholar]
- 38.Park JW, Kim IY, Choi JW, et al. AHNAK loss in mice promotes Type II pneumocyte hyperplasia and lung tumor development. Mol Cancer Res. 2018;16(8):1287–1298. doi: 10.1158/1541-7786.MCR-17-0726 [DOI] [PubMed] [Google Scholar]
- 39.Gu J, Mao W, Ren W, et al. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 2019;443:125–134. doi: 10.1016/j.canlet.2018.11.029 [DOI] [PubMed] [Google Scholar]