Abstract
Free-text problem descriptions are brief explanations of patient diagnoses and issues, commonly found in problem lists and other prominent areas of the medical record. These compact representations often express complex and nuanced medical conditions, making their semantics challenging to fully capture and standardize. In this study, we describe a framework for transforming free-text problem descriptions into standardized Health Level 7 (HL7) Fast Healthcare Interoperability Resources (FHIR) models.
This approach leverages a combination of domain-specific dependency parsers, Bidirectional Encoder Representations from Transformers (BERT) natural language models, and cui2vec Unified Medical Language System (UMLS) concept vectors to align extracted concepts from free-text problem descriptions into structured FHIR models. A neural network classification model is used to classify thirteen relationship types between concepts, facilitating mapping to the FHIR Condition resource. We use data programming, a weak supervision approach, to eliminate the need for a manually annotated training corpus. Shapley values, a mechanism to quantify contribution, are used to interpret the impact of model features.
We found that our methods identified the focus concept, or primary clinical concern of the problem description, with an F1 score of 0.95. Relationships from the focus to other modifying concepts were extracted with an F1 score of 0.90. When classifying relationships, our model achieved a 0.89 weighted average F1 score, enabling accurate mapping of attributes into HL7 FHIR models. We also found that the BERT input representation predominantly contributed to the classifier decision as shown by the Shapley values analysis.
Keywords: Health Information Interoperability (D000073892), Deep Learning (D000077321), Systematized Nomenclature of Medicine (D039061), Semantics (D012660), Natural Language Processing (D009323)
Graphical Abstract
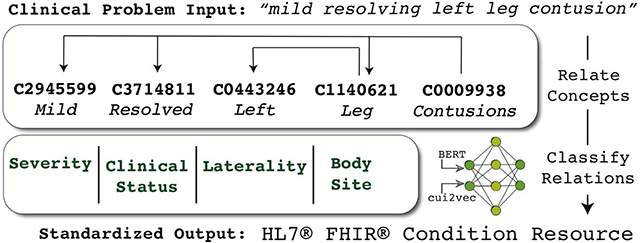
1. Introduction
The problem-oriented medical record (POMR) was a significant change in how the clinical patient record was structured.[1] Introduced in 1968, this strategy involves using concise descriptions of a patient’s current health concerns to serve as indexed headings into the larger medical chart.[2] These summary level “problem descriptions” describe complex clinical conditions with important supporting context such as severity/stage, body location, related or contributing conditions, and so on, and are an integral part of the POMR as a whole.
By orienting the record around clinical problems, the POMR is by definition predicated on the ability to accurately and succinctly describe a patient’s pertinent issues.[3] Furthermore, it also places a greater burden on ensuring that problems are described comprehensively and in a standardized way.[4] Although these challenges pre-dated the widespread implementation of the electronic health record (EHR), the structure inherent in EHRs did not alleviate issues regarding how clinical problems are represented.[5] Specifically, while the expressiveness of free-text is required by clinicians to convey their impressions and reasoning regarding a patient’s problems,[6] structured representation and standardization are beneficial for processing and analytics.[7]
Codification, or the assignment of codes or terms from a controlled terminology, is a common strategy for capturing and standardizing the semantics of a clinical problem. This can be done by the clinician directly, but requires significant time and effort[8] and adds to an already full clinical workload.[9] Alternatively, this coding may be accomplished using automated or semi-automated Natural Language Processing (NLP) techniques. Even if automated, codification often fails to capture the entirety of the clinician’s intent, a situation known as the “content completeness problem.”[10,11] This issue is rooted in the fact that natural language descriptions of medical problems are often too expressive to be fully represented via a finite set of terms.[12]
The content completeness problem is of particular importance to clinical problem descriptions, as it has been shown empirically that clinical problems often cannot be sufficiently described by a single concept, but instead require a set of concepts to capture modifiers and other related context.[13–15] To account for this, logical models can be paired with codification to create a more robust standard for data representation and exchange.[16] Health Level 7 (HL7) Fast Healthcare Interoperability Resources (FHIR), an emerging specification for representing clinical data, is a prominent example of this type of standardization.[17] FHIR specifies several models (or “Resources”) for many types of healthcare data, including representations specifically suited for clinical problems.
The goal of this study is to introduce a framework for encoding free-text clinical problem descriptions using HL7 FHIR. Our methods focus on combining machine learning techniques with rule-based methods and domain-specific knowledge bases to map free-text problem descriptions to FHIR-based structured representations.
2. Background and Significance
The standardization of free-text clinical problems has long been a focus of research. Concept extraction, or mapping text mentions to standardized terminologies or ontologies, is a fundamental clinical NLP task and an important step towards a standardized problem representation. Prominent implementations such as MetaMap[18] and Clinical Text Analysis and Knowledge Extraction System (cTAKES)[19] have been widely adopted and used for a variety of standardization applications.[20]
While concept extraction is an essential first step, further standardization may be applied by organizing the extracted concepts into logical structures that better capture their full context and semantics. The Medical Language Extraction and Encoding System (MedLEE)[21] accomplishes this using frames,[22] or structures that link concepts to their modifiers and related terms. Similar notions of combining, or “post-coordinating” concepts are natively built into the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT) ontology via the SNOMED CT compositional grammar.[23]
In their most structured form, clinical problems may be represented via common information models.[24] Efforts such as the Clinical Element Model (CEM)[25] and the Clinical Information Modeling Initiative (CIMI)[26] aim to define a standard set of attributes and modifiers for clinical data exchange. HL7 FHIR is the latest of these efforts, and for this study the Condition resource of the FHIR specification is the chosen target for clinical problem representation.
The use of NLP to extract information from clinical text as FHIR resources is a growing field of study,[27–29] driven in part by the increasing prominence of FHIR in the healthcare information landscape.[30] The NLP2FHIR project, based on several Unstructured Information Management Architecture (UIMA)[31] tools, extracts a broad range of FHIR resources from unstructured clinical notes.[32] In contrast to the broader scope of NLP2FHIR, our study exclusively focuses on encoding summary level problem descriptions into FHIR Condition resources. While care was taken to ensure our techniques could generally be applied to other free-text clinical data (such as procedures, labs, medications, and so on), the nuances of those transformations are beyond the scope of this work. While a focus on clinical conditions narrows our purview, several new challenges are introduced:
Although free-text problem descriptions are generally terse, they are surprisingly expressive, and routinely encompass semantics outside the bounds of single concepts from a controlled terminology or vocabulary.[14,15]
They are often phrased as collections of medical terms, which generally have quite different grammatical and linguistic characteristics as compared to the larger clinical note narrative.[33] Specifically, these problems are generally represented as noun phrases as opposed to full sentences.
It is known that non-standard grammar, sentence structure, and word usage, or non-canonical text, poses significant problems for NLP model reuse.[34] As stated, these problem descriptions do not follow a canonical notion of grammar or structure – and complex noun phrases have proven to be especially difficult for many NLP parsing tasks.[35,36] Given this, existing NLP models may perform poorly when applied to these problems.
Our main contribution in this study is a standardization framework for clinical problem descriptions using HL7 FHIR, an expansion of our previous work on codifying problems using the SNOMED CT compositional grammar.[37] We extend this previous study in the following areas: First, we update our target representation to the FHIR Condition resource to take full advantage of the growing FHIR healthcare ecosystem. Next, we increase the performance of our previous methods through the addition of training techniques based on incorporating rule-based methods, weak labeling, and distant supervision. This was necessary to make our previous methods more resilient to the linguistic heterogeneity seen in real-world clinical text – a main limitation of our previous work. Finally, we add a thorough analysis of our model features using the latest neural network explainability methods, giving us important insight into what model features are important and why.
3. Methods and Materials
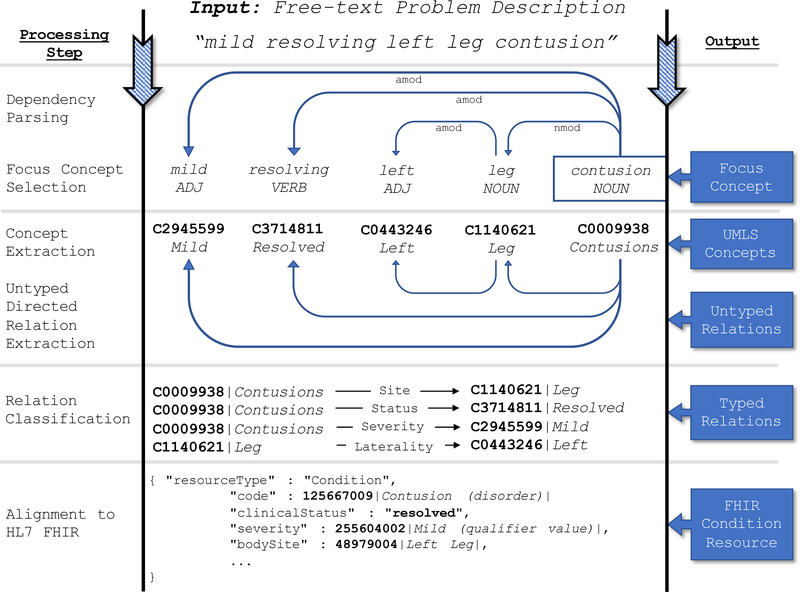
We define our clinical problem description standardization task as such: Given a free-text description of a patient’s clinical problem, output an HL7 FHIR Condition resource representing the codified problem and all relevant modifiers and context. We account for the specific challenges of processing problem descriptions using three general methodological foci: (1) an emphasis on leveraging existing pre-trained models to maximize transfer learning, using fine-tuning where necessary, (2) the incorporation of rule-based methods with neural network models to avoid manual training data annotation, and (3) the usage of recent advances in neural network explainability to examine the importance of the features in our model. Our methods are broadly segmented into five subtasks that gradually build an increasingly structured and standardized representation of the clinical problem. Figure 1 highlights the high-level steps of the standardization framework, the details of which are explained further below.
Figure 1:
The high-level processing steps for encoding a free-text clinical problem description into an HL7 FHIR Condition Resource.
3.1. Preprocessing: Dependency Parsing
Dependency parsing is the formalization of text into a graph of words and their syntactic relationships. It is an important input into many clinical NLP tasks such as concept extraction,[38] semantic parsing,[39] and negation detection,[40] and contributes prominently to several of the subtasks described in our methods below.
For all clinical problem dependency parsing we used the spaCy NLP platform with a custom parsing model fine-tuned from the pre-trained ScispaCy Biomedical model.[41] To fine-tune the model, we manually annotated 141 problem descriptions with their correct dependency parses. To keep the amount of manual annotation as low as possible, we used data augmentation, a strategy to increase the size and heterogeneity of training data. While many NLP data augmentation algorithms focus on expanding synonyms,[42,43] we used SNOMED CT to expand SNOMED CT Qualifier Value terms matched in the text, similar in concept to what Kobayashi describes as “contextual augmentation.”[44] For example, given a problem “severe contusion”, we recognize that the term severe is a child of the SNOMED CT concept 272141005|Severities|. Given this, we can expand this training example with other Severities, yielding “mild contusion”, “moderate contusion”, and so on. With data augmentation, we expanded our training set to 349 entries.
3.2. Subtask: Focus Concept Selection
We define the “focus concept” of a problem description as the semantic root, or primary concept from which the remaining concepts are either directly or indirectly connected. As summary level problem descriptions are primarily noun phrases, we hypothesize that the ROOT word of the dependency parse will align with the focus concept, a hypothesis based primarily on the work of Spasić et al.[45] Representing our text as a set of tokens S, this step aims to learn a function that inputs the problem description tokens S and outputs the root token r such that r ∈ S. This technique closely aligns with methods used in our previous work to select the focus concept for a SNOMED CT expression.[37] Note that this approach assumes each problem description primarily describes one and only one clinical problem. While a single problem description may contain several mentions of different conditions, signs, or symptoms, it is assumed that all serve to modify or add context to a single focus problem. Other formats of problem descriptions, such as concatenations of multiple, unrelated problems (for example: “Tonsillitis; fracture of the femur”) are beyond the scope of this subtask.
3.3. Subtask: Concept Extraction
We extracted concepts from the text using MetaMap,[18] a tool based on the Unified Medical Language System (UMLS)[46] used to link free-text mentions of biomedical concepts to their corresponding UMLS concepts via Concept Unique Identifiers (CUIs). Given problem description text S composed of s1,…,s|s| tokens, we used MetaMap to implement the mapping S↦E where E is a set of UMLS concepts.
3.4. Subtask: Untyped Directed Relation Extraction
It has been shown by Reichartz et al.[47] that using a dependency parse tree can be an effective way to extract semantic relationships between entities in text. Several biomedical relation extraction systems leverage the dependency parse tree, incorporating it into a wide variety of model architectures including rule-based approaches,[48,49] kernel-based methods,[50,51] and recently deep learning models.[52,53] The goal of this subtask is similarly to extract a set of untyped, directed entity relationships between a source and target entity, or (e1,e2). We implemented this by connecting pairs of entities via their shortest path in the dependency parse. Entities e1 and e2 are considered connected if (1) a path exists from e1 to e2, and (2) no other entity exists on the path between them.
3.5. Subtask: Relation Classification
Classifying relationships between biomedical concepts is an important task with wide-reaching applications,[54] with use cases including chemical-disease relations,[55,56] disease-symptom relations,[57] and protein–protein relations.[58] In this subtask we aim to classify the untyped relationships extracted via the Untyped Directed Relation Extraction subtask, or r(e1,e2), where r represents the relationship type, and e1 and e2 represent the source and target entity, respectively. Further details regarding the methodology for this subtask are detailed below.
3.5.1. Relation Type Selection
We selected twenty-one relation types for inclusion in this study. Relation types were chosen via satisfaction of one or more of the following criteria: (1) they can be directly mapped to an attribute of the FHIR v4.0.1 Condition resource, (2) they are associated with a standard FHIR Condition extension, or (3) they are included in prominent clinical data models other than the FHIR specification. This was done to give our classifier a broader semantic range given that the FHIR specification allows for extensibility. Relationship types that did not come directly from the FHIR specification were obtained via a survey of the following models: the Clinical Element Model (CEM) - ClinicalAssert model,[25] the openEHR - Problem/Diagnosis archetype,[59] and the Clinical Information Modeling Initiative (CIMI) FindingSiteConditionTopic logical model.[26]
Table 1 shows these relationship types and their mapping to the FHIR model. Note that as shown in the table, some map directly to attributes in the FHIR Condition resource, some map to standard FHIR extensions, and some have no direct mapping to FHIR at all. For those with no FHIR mapping, the standardization framework(s) from which they were selected are listed.
Table 1:
The set of twenty-one relation types considered in the Relation Classification subtask with their mappings to the FHIR Condition resource.
| Relation Type | FHIR Mappings |
|---|---|
| Base FHIR Condition | |
| clinicalStatus | Condition.clinicalStatus |
| verificationStatus | Condition.verificationStatus |
| severity | Condition.severity |
| bodySite | Condition.bodySite |
| stage | Condition.stage |
| Standard FHIR Extensions | |
| dueTo | condition-dueTo |
| ruledOut | condition-ruledOut |
| occurredFollowing | condition-occurredFollowing |
| associatedSignAndSymptom | condition-related |
| laterality | BodyStructure |
| anatomicalDirection | BodyStructure |
| Non-FHIR Attributes | |
| course | CIMI:FindingSiteAssertion - clinicalCourse |
| OpenEHR:Problem/Diagnosis Archetype - Course label | |
| periodicity | CIMI:FindingSiteAssertion - periodicity |
| exacerbatingFactor | CIMI:FindingSiteAssertion - exacerbatingFactor |
| interpretation | CIMI:FindingSiteAssertion - interpretation |
| findingMethod | CIMI:FindingSiteAssertion - method |
| historicalIndicator | CEM:ClinicalAssert - historicalInd |
| OpenEHR:Problem/Diagnosis Archetype - Current/Past? | |
| certainty | OpenEHR:Problem/Diagnosis Archetype - Diagnostic certainty |
| CEM:ClinicalAssert - likelihood | |
| risk | CEM:ClinicalAssert - riskForInd |
| negatedIndicator | generic negation modifier |
| otherwiseRelated | any other non-specified relationship |
3.5.2. Relation Classification Model Architecture
An artificial neural network model was chosen for the relation classification task. The architecture consists of a fully-connected neural network with one hidden layer containing 256 nodes using ReLU activation functions. The output layer of this network contains one node for each relation class (see Table 1) using softmax activation functions. Dropout rates of 0.5 were used to prevent overfitting. The ultimate output of the model is a probability for each relationship class. We used the Keras framework[60] to implement our model.
A variety of input representations ranging from text embeddings to facets of the extracted UMLS concepts were selected as input features. Special emphasis was placed on incorporating features based on pre-trained, domain-specific models where transfer learning could be leveraged. The full feature set is described below.
Source/Target Entity Text Embedding (BERT)
We transformed text into a suitable input format via Bidirectional Encoder Representations from Transformers (BERT),[61] a deep learning-based language representation aimed at capturing the semantic intent of words in context. We specifically used the pre-trained ClinicalBERT model, a BERT-based model trained on clinical notes.[62] For each of the source and target entities we obtained a 768-dimensional vector using mean pooling of the second-to-last BERT layer. BERT embeddings were incorporated into our pipeline via the bert-as-service project.[63]
Source/Target Entity Concept Embedding (cui2vec)
We incorporated vectors from the extracted source and target entity concepts using cui2vec, a UMLS concept embedding model.[64] The concept embedding of cui2vec was used specifically for transfer learning of UMLS semantics into our model. The UMLS concepts of the source and target entities were mapped to 500-dimensional vectors from a pre-trained cui2vec model.[64]
Dependency Parse Shortest Path
The shortest path through the dependency parse between the source and target entities is hypothesized to be helpful in determining their semantic relationship.[50] Furthermore, it has been shown that incorporating this as a feature in a machine learning model can improve relation extraction performance.[65] A BERT vector of the path was used to represent this feature.
Source/Target Entity Semantic Type
For both the source and target concepts extracted during the Concept Extraction step, MetaMap also assigns concepts one or more of the 127 UMLS semantic categories called semantic types.[66] These categories were used as features to represent the high-level semantics of the concepts.
Source/Target Entity Semantic Type Group
The source and target entity semantic types are additionally grouped into fifteen even broader categories called semantic groups,[67] representing the coarsest level of semantics in our feature set.
3.5.3. Data Programming
We merged rule-based and neural network NLP approaches by using data programming,[68] a technique for creating a weakly-labeled training data set given a set of “labeling functions,” or domain-specific rules crafted by subject matter experts or other domain-specific oracles. We used the Snorkel framework to train a generative model from our labeling functions, and used that model to generate training data for the downstream neural network model.[69] Data programming allows us to address two main challenges: (1) we avoid the cost and time of using specially trained clinical informaticians to hand-annotate training data,[70] and (2) we incorporate domain knowledge via symbolic approaches and distant supervision,[71,72] highlighting the importance of leveraging domain expertise via rule-based approaches.[73,74] For our data programming implementation, we created approximately thirty labeling functions using both hand-crafted rules and distant supervision using SNOMED CT. Our neural network training data set was generated by applying the generative data programming model to 100,000 problem descriptions extracted from a large clinical corpus.[14]
3.5.4. Interpretability & Feature Attribution
Shapley values are a mechanism to quantify the contributions (in terms of gains or losses) of members of a coalition cooperating toward a common goal.[75,76] Shapley values have been applied to determining feature importance of machine learning models,[77] and are used here to gain insight into our relationship classification model.
Feature attribution can be cast as a game theory problem as such: First, given a set of model features F, assume we wanted to determine the contribution of some feature j where j ∈F. Next, we generate a subset of F as S such that does not include the feature of interest j. We test the contribution of feature j by measuring its contribution v(S ∪ {j}) ― v(S) where v denotes the characteristic function, or the total contribution of a set of features toward the end goal. In our case, the characteristic function input is a set of features, and the output is the probability of the chosen relationship label. By repeating this for all subsets of F such that S ⊆ F\{j}, we compute the Shapley value ϕj as:
We then explain the approximate contribution of a feature by averaging all Shapley values over a random sampling of n training samples.[78]
3.6. Subtask: Alignment to HL7 FHIR
Table 1 shows the basic mappings of our chosen relation types to the FHIR Condition resource. Any relationship that does not map directly to a FHIR Condition attribute will be added as a FHIR extension, and the Condition.code attribute of the FHIR Condition resource will be set to the focus concept extracted via the Focus Concept Selection subtask. FHIR alignment also involves mapping each extracted UMLS concept to SNOMED CT. To do this, we used the UMLS Metathesaurus to find the SNOMED CT concept associated with the given UMLS concept. If the UMLS concept maps to more than one SNOMED CT concept, each SNOMED CT concept will be added as a FHIR Coding for the particular FHIR attribute.
3.7. Evaluation & Experiments
Six hundred problem descriptions extracted from a large clinical notes corpus were manually annotated by three annotators. Annotation consisted of finding the focus concept of the problem (i.e. the primary disease or finding), all related diseases, findings, or modifiers, and the relationship types that connect them (see Table 1). BRAT,[79] a freely-available annotation tool, was used to conduct the annotation. Inter-annotator agreement was measured via Krippendorff’s alpha score.[80] We conducted the following experiments to analyze the performance of our framework.
3.7.1. Untyped Directed Relation Extraction & Focus Concept Selection
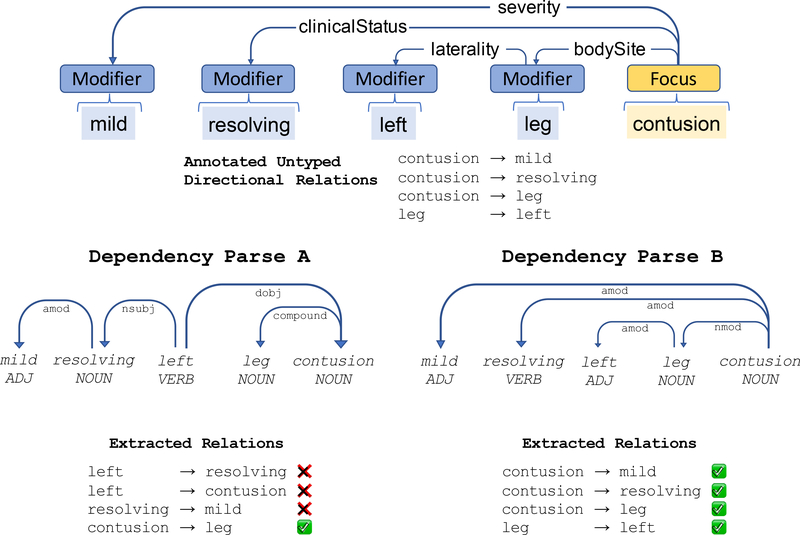
Because both Untyped Directed Relation Extraction and Focus Concept Selection subtasks are based on dependency parsing, we evaluated them in tandem. For Untyped Directed Relation Extraction, we evaluated the effectiveness of using dependency parsing to determine related concepts (regardless of relationship type). We used the evaluation corpus detailed above with the following experiment: First, we extracted from the evaluation corpus all annotated relationships and retained a list of all source/target tuples. Then, we compared the annotated relationships with those asserted from the dependency parse. Figure 2 illustrates this test. In this example, Dependency Parse A produces three incorrect relationships, while Dependency Parse B fully corresponds to the human-annotated example.
Figure 2:
An example of the evaluation of an annotated problem description. We evaluate the ability of our dependency parsing model to learn the correct (untyped) relationships. When compared to the human-annotated example (top), Dependency Parse A reflects poor alignment, while Dependency Parse B corresponds completely.
In our Focus Concept Selection subtask, we hypothesize that the ROOT dependency of the dependency parse will correspond to what a human annotator would specify as the focus concept of the problem description. We tested this by running our dependency parse model on each problem description in the evaluation set and measuring the accuracy with which the dependency parse ROOT dependency corresponds to the human-annotated focus.
We further hypothesize that performance for both tasks above will increase as the dependency parse model is increasingly tuned to the domain. To test this, we ran the above evaluations using four dependency parse models. First, we evaluated two unmodified pre-trained models: (1) the Default spaCy English model, and (2) ScispaCy Biomedical, a spaCy model specifically trained on biomedical data sets.[41] Next, we evaluated two fine-tuned models as described in the Dependency Parsing step: (1) ScispaCy Biomedical fine-tuned with 141 annotated dependency parses from a random set of problem descriptions, and (2) that same fine-tuned model plus data augmentation.
3.7.2. Relation Classification & Data Programming
To test the ability of our framework to determine the correct semantic relationship type between entities, we next evaluated the performance of our relation classification model. We specifically tested whether or not the data programming approach can effectively be used to train a neural network model. First, we evaluated the performance of our data programming rule-based model on the test set. Next, we trained the neural network classifier via data generated from the data programming model. Finally, we compared the performance of the two models. We hypothesize that the neural network model will have better performance than the rule-based model.
4. Results
The gold standard annotation of the evaluation set of six hundred problem descriptions by the three annotators resulted in 1553 relationship annotations and 2057 focal concept/modifier entity annotations. We recorded Krippendorff’s alpha inter-annotator agreement scores of 0.79 for the relationships and 0.94 for the focal concepts. In the case of annotator disagreement, simple majority vote was used to adjudicate. As a result of class imbalance in the evaluation set, results from any relationships with less than fifteen supporting evaluation annotations are not reported in this study.
Tables 2 & 3 show the results of the Focus Concept Selection and Untyped Directed Relation Extraction subtasks. Both focus concept and untyped directed relation F1 scores are the highest when using the domain-specific fine-tuned dependency parse model. Note that while fine-tuning resulted in a large performance boost, data augmentation had little if any positive performance impact.
Table 2:
Evaluation results from the Focus Concept Selection subtask.
| model | precision | recall | f1-score |
|---|---|---|---|
| ScispaCy Biomedical + fine-tuning + data augmentation | 0.95 | 0.94 | 0.94 |
| ScispaCy Biomedical + fine-tuning | 0.96 | 0.94 | 0.95 |
| ScispaCy Biomedical | 0.70 | 0.68 | 0.69 |
| Default spaCy English (baseline) | 0.68 | 0.66 | 0.67 |
Table 3:
Evaluation results from the Untyped Directed Relation Extraction subtask.
| model | precision | recall | f1-score |
|---|---|---|---|
| ScispaCy Biomedical + fine-tuning + data augmentation | 0.89 | 0.90 | 0.90 |
| ScispaCy Biomedical + fine-tuning | 0.88 | 0.9l | 0.89 |
| ScispaCy Biomedical | 0.84 | 0.70 | 0.76 |
| Default spaCy English (baseline) | 0.84 | 0.65 | 0.73 |
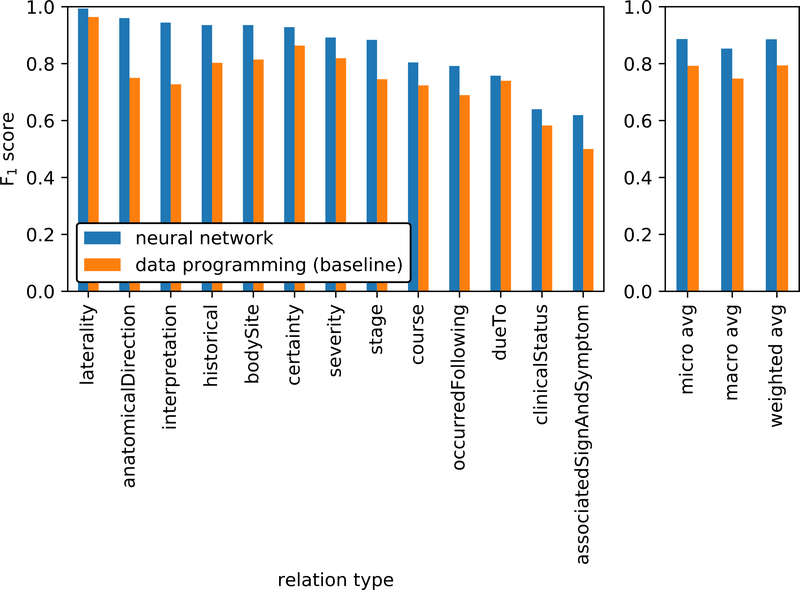
Table 4 shows the F1 scores for the neural network relationship classification model for all relationship types supported by more than fifteen annotated relationships in the test set.
Table 4:
Relation classification results of the neural network model trained via data programming.
| relation type | precision | recall | f1-score | # annotations |
|---|---|---|---|---|
| laterality | 0.99 | 0.99 | 0.99 | 167 |
| anatomicalDirection | 0.96 | 0.96 | 0.96 | 25 |
| interpretation | 0.94 | 0.94 | 0.94 | 18 |
| historical | 0.97 | 0.90 | 0.94 | 81 |
| bodySite | 0.98 | 0.90 | 0.94 | 235 |
| certainty | 1.00 | 0.87 | 0.93 | 45 |
| severity | 1.00 | 0.80 | 0.89 | 41 |
| stage | 0.95 | 0.83 | 0.88 | 23 |
| course | 0.76 | 0.85 | 0.80 | 41 |
| occurredFollowing | 1.00 | 0.66 | 0.79 | 29 |
| dueTo | 0.89 | 0.66 | 0.76 | 38 |
| clinicalStatus | 1.00 | 0.47 | 0.64 | 68 |
| associatedSignAndSymptom | 0.50 | 0.81 | 0.62 | 43 |
| — | — | — | — | — |
| micro avg | 0.92 | 0.85 | 0.89 | 854 |
| macro avg | 0.92 | 0.82 | 0.85 | 854 |
| weighted avg | 0.94 | 0.85 | 0.89 | 854 |
Figure 3 contrasts the F1 scores of the trained neural network model as compared to the data programming rule-based model used to create the training data. This figure highlights the amount of improvement gained via data programming when using the rule-based model as a baseline.
Figure 3:
Relation classification results compared to the rule-based data programming baseline model.
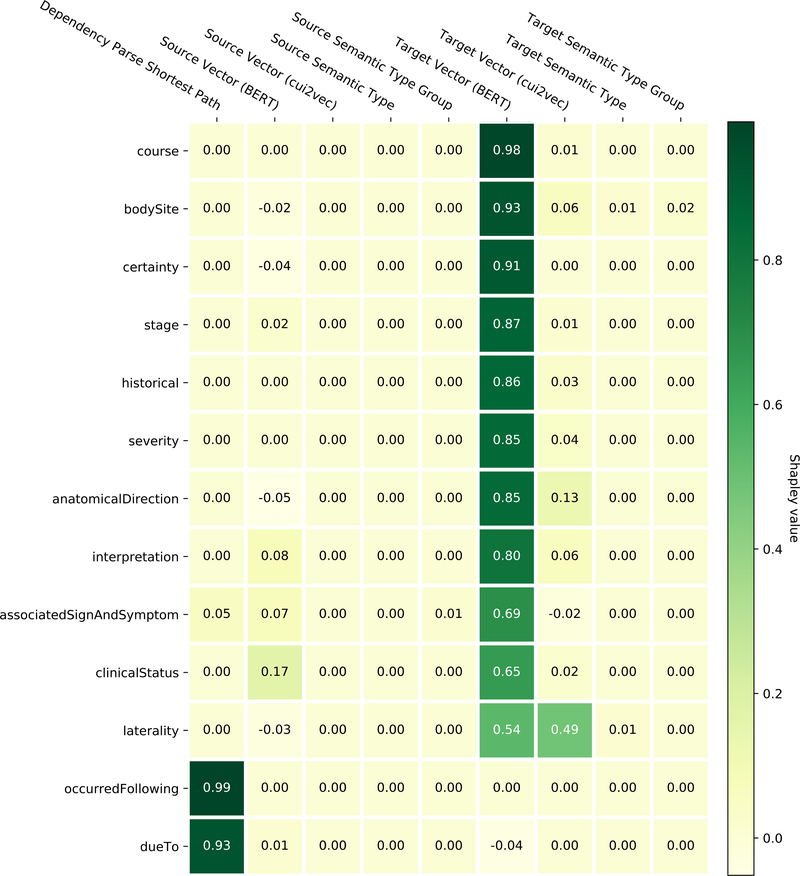
Table 5 shows the Shapley values for the nine relation classification model features. BERT vectors are shown to have the most impact, and features of the target entity contribute more to the model than the source entity. Shapley values for each of the individual relationships under test are shown in Figure 4. While BERT vectors are prominent, there are some differences to be noted in feature importance across classes – notably, that the dependency parse shortest path contributes almost exclusively to two relationship classes and relatively little to others.
Table 5:
Shapley values for the nine features input into the relation classifier.
| Shapley value | % total contribution | |
|---|---|---|
| Target Vector (BERT) | 0.715 | 78.71 |
| Dependency Parse Shortest Path | 0.1057 | 11.64 |
| Target Vector (cui2vec) | 0.0634 | 6.98 |
| Source Vector (BERT) | 0.0163 | 1.8 |
| Target Semantic Type | 0.0041 | 0.45 |
| Target Semantic Type Group | 0.0013 | 0.14 |
| Source Vector (cui2vec) | 0.0012 | 0.13 |
| Source Semantic Type | 0.0009 | 0.1 |
| Source Semantic Type Group | 0.0004 | 0.05 |
Figure 4:
Contrasting the Shapley values for the nine source and target entity features of the relation classifier for each of the evaluated relationship types. Note that negative Shapley values indicate that the feature had a detrimental contribution.
5. Discussion
The use of dependency parse-based methods for finding the focus concept and untyped entity relations of problem descriptions was an effective approach, as shown by Tables 2 & 3. Furthermore, these tables show that performance was significantly increased by fine-tuning the pre-trained ScispaCy Biomedical parsing model. This reinforces our first methodological focus of emphasizing transfer learning and fine-tuning, as a significant increase in performance was achieved with a relatively small cost of manual training data annotation. Conversely, our data augmentation algorithm did not yield a noticeable change in performance. Kobayashi also reported minimal improvement with a similar non-synonym word replacement augmentation technique,[44] leading us to conclude that more exploration is needed to determine if data augmentation can be successfully applied to this task.
Given the extracted untyped entity relationships, Table 4 shows that our neural network model was able to classify the correct relationship type with an overall 0.89 weighted average F1 score. Although performance on several relationship types surpassed a 0.9 F1 score, some variation in performance across different types is noted. Specifically, the classifier struggled with the more semantically open-ended relationship types such as associatedSignAndSymptom.
Figure 3 shows that data programming can be an effective technique for augmenting a rule-based approach, as the neural network classifier was able to outperform the data programming rule-based classifier. This pairing of a rule-based system with a neural network model eliminated the need for creating a human-annotated training data set, a significant savings of time and effort. It also adds evidence that our second methodological focus of incorporating rule-based methods and knowledge bases is both an effective and pragmatic technique for this task.
Figure 4 and Table 5 show that BERT features dominate the Shapley value analysis of the system. It is of interest to note, however, that for two attributes dueTo and occurredFollowing, the “Dependency Parse Shortest Path” feature dominates, as shown in Figure 4. Qualitative analysis of the results shows that these two relationship types generally have indicative words between the two entities, for example “right-sided [CHF]source caused by chronic [pulmonary embolism]target” and “chronic low thoracic [pain]source after [fall]target”. This also reinforces the shortest path hypothesis[50] in that the words between source and target entities in the dependency parse tree primarily contribute to their relationship type. Also of note is the relatively small importance of the cui2vec vectors when compared to the BERT representations. This observation is in line with similar findings of Kearns et al.[81] Given our last methodological focus of explainability, we can use these insights in the future to improve our model. For example, we may use the findings in Table 5 to justify the removal of low-impact features, simplifying our architecture. More importantly, we know that feature importance in our model is not evenly distributed between relationship types – some features such as “Dependency Parse Shortest Path” may have low average overall impact but are critical to certain relationships. This will be an important consideration as we expand our model to different relationship types.
From an implementation perspective, the methods described in this framework are intended to be generalizable to any data set of clinical problem descriptions. Given the data programming approach, large-scale annotation of training of data is not necessary, but adaptation to a particular context or data set does include the following steps:
Implementation of labeling functions. Our data programming approach is heavily dependent on accurate labeling functions to produce the training data set. Implementers of this framework should expect to create a set of labeling functions using rules or heuristics specific to their data.
Fine-tuning the dependency parse model. An accurate dependency parse model is an important facet of our approach. While Tables 2 & 3 show that reasonable performance can be obtained using freely available pre-trained models, at least a small amount of fine-tuning is recommended to account for data set specific variations in problem description phrasing or structure.
6. Conclusion
In this study we have described a framework for standardizing free-text clinical problem descriptions using HL7 FHIR. We have demonstrated that by leveraging domain-specific knowledge bases and rules, we were able to combine data programming and neural networks to achieve higher performance than via a rule-based approach alone, all while minimizing the need for human-annotated training data. We also examined the feature set of our model and found that BERT language representations contribute significantly more to model performance compared to cui2vec’s concept-based vectors. These methods ultimately allow for the alignment of free-text clinical problems into the HL7 FHIR Condition resource. All source code for this framework is available via https://github.com/OHNLP/clinical-problem-standardization.
Highlights.
A multi-faceted framework for standardizing free-text clinical problem descriptions
Data programming helps to eliminate the need for manually annotated training data
Fine-tuning of existing pre-trained models improves performance
BERT embeddings are critical for classifying concept-to-concept relations
Shapley value analysis shows different relations rely on different model features
Acknowledgments
This study was funded by grant NCATS U01TR02062. We thank Sunyang Fu, Donna Ihrke, and Luke Carlson for assisting in the test corpus annotation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weed LL. Special article: Medical records that guide and teach. N Engl J Med. 1968;278(12):593–600. [DOI] [PubMed] [Google Scholar]
- 2.Salmon P, Rappaport A, Bainbridge M, Hayes G, Williams J. Taking the problem oriented medical record forward. In: Proceedings of the AMIA Annual Fall Symposium American Medical Informatics Association; 1996. p. 463. [PMC free article] [PubMed] [Google Scholar]
- 3.Acker B, Bronnert J, Brown T, Clark J, Dunagan B, Elmer T, et al. Problem list guidance in the EHR. Journal of AHIMA. 2011;82(9):52. [PubMed] [Google Scholar]
- 4.Feinstein AR. The problems of the problem-oriented medical record. Ann Intern Med. American College of Physicians; 1973;78(5):751–62. [DOI] [PubMed] [Google Scholar]
- 5.Simons SM, Cillessen FH, Hazelzet JA. Determinants of a successful problem list to support the implementation of the problem-oriented medical record according to recent literature. BMC Med Inform Decis Mak. BioMed Central; 2016;16(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: A perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. Nature Publishing Group; 2019;25(1):24. [DOI] [PubMed] [Google Scholar]
- 8.Arndt BG, Beasley JW, Watkinson MD, Temte JL, Tuan W-J, Sinsky CA, et al. Tethered to the EHR: Primary care physician workload assessment using EHR event log data and time-motion observations. Ann Fam Med. Annals of Family Medicine; 2017;15(5):419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroth PJ, Morioka-Douglas N, Veres S, Babbott S, Poplau S, Qeadan F, et al. Association of electronic health record design and use factors with clinician stress and burnout. JAMA Netw Open American Medical Association; 2019;2(8):e199609–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkin PL, Bailey KR, Ogren PV, Bauer BA, Chute CG. A randomized double-blind controlled trial of automated term dissection In: Proceedings of the AMIA Symposium. American Medical Informatics Association; 1999. pp. 62–6. [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers J, Rector AL. Terminological systems: Bridging the generation gap In: Proceedings of the AMIA Annual Fall Symposium. American Medical Informatics Association; 1997. pp. 610–4. [PMC free article] [PubMed] [Google Scholar]
- 12.Rector AL. Clinical terminology: Why is it so hard? Methods Inf Med. 1999;38(04/05):239–52. [PubMed] [Google Scholar]
- 13.Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, Savova G. SemEval-2015 task 14: Analysis of clinical text. In: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) Denver, Colorado: Association for Computational Linguistics; 2015. pp. 303–10. [Google Scholar]
- 14.Liu H, Wagholikar K, Wu ST-I. Using SNOMED-CT to encode summary level data–a corpus analysis. AMIA Summits on Translational Science Proceedings. American Medical Informatics Association; 2012;2012:30–7. [PMC free article] [PubMed] [Google Scholar]
- 15.Elkin PL, Brown SH, Husser CS, Bauer BA, Wahner-Roedler D, Rosenbloom ST, et al. Evaluation of the content coverage of SNOMED CT: Ability of SNOMED Clinical Terms to represent clinical problem lists. Mayo Clinic Proc. Elsevier; 2006;81(6):741–8. [DOI] [PubMed] [Google Scholar]
- 16.Coyle J, Heras Y, Oniki T, Huff S. Clinical Element Model. University of Utah; 2008; [Google Scholar]
- 17.Bender D, Sartipi K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In: Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems IEEE; 2013. pp. 326–31. [Google Scholar]
- 18.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In: AMIA Annual Symposium Proceedings American Medical Informatics Association; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 19.Kersloot MG, Lau F, Abu-Hanna A, Arts DL, Cornet R. Automated SNOMED CT concept and attribute relationship detection through a web-based implementation of cTAKES. J Biomed Semantics. 2019;10(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, et al. Clinical information extraction applications: A literature review. J Biomed Inform. 2018;77:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman C, Hripcsak G, DuMouchel W, Johnson SB, Clayton PD. Natural language processing in an operational clinical information system. Nat Lang Eng. Cambridge University Press; 1995;1(1):83–108. [Google Scholar]
- 22.Fillmore CJ. Frame semantics and the nature of language. Ann N Y Acad Sci. 1976;280(1):20–32. [Google Scholar]
- 23.International Health Terminology Standards Development Organization (IHTSDO). Compositional grammar specification and guide, v2.3.1. 2016.
- 24.Mead CN. Data interchange standards in healthcare IT-computable semantic interoperability: Now possible but still difficult. Do we really need a better mousetrap? J Healthc Inf Manag. 2006;20(1):71–8. [PubMed] [Google Scholar]
- 25.OpenCEM. OpenCEM Browser. http://www.opencem.org/, [Accessed: 2019–11-10];
- 26.CIMI. Clinical Information Modeling Initiative. https://cimi.hl7.org/, [Accessed: 2019–12-12];
- 27.Hong N, Wen A, Shen F, Sohn S, Liu S, Liu H, et al. Integrating structured and unstructured EHR data using an FHIR-based type system: A case study with medication data. AMIA Summits on Translational Science Proceedings. American Medical Informatics Association; 2018;2018:74–83. [PMC free article] [PubMed] [Google Scholar]
- 28.Despotou G, Korkontzelos Y, Matragkas N, Bilici E, Arvanitis TN. Structuring clinical decision support rules for drug safety using natural language processing. In: 16th International Conference on Informatics, Management, and Technology in Healthcare (ICIMTH 2018) 2018. pp. 89–92. [PubMed] [Google Scholar]
- 29.Hong N, Wen A, Stone DJ, Tsuji S, Kingsbury PR, Rasmussen LV, et al. Developing a FHIR-based EHR phenotyping framework: A case study for identification of patients with obesity and multiple comorbidities from discharge summaries. J Biomed Inform. 2019;99:103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shull JG. Digital health and the state of interoperable electronic health records. JMIR Med Inform. 2019;7(4):e12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrucci D, Lally A. UIMA: An architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. Cambridge University Press; 2004;10(3–4):327–48. [Google Scholar]
- 32.Hong N, Wen A, Shen F, Sohn S, Wang C, Liu H, et al. Developing a scalable FHIR-based clinical data normalization pipeline for standardizing and integrating unstructured and structured electronic health record data. JAMIA Open. 2019. October;2(4):570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourigault D Surface grammatical analysis for the extraction of terminological noun phrases. In: Proceedings of the 14th Conference on Computational Linguistics - Volume 3 USA: Association for Computational Linguistics; 1992. pp. 977–81. (COLING ‘92). [Google Scholar]
- 34.Plank B What to do about non-standard (or non-canonical) language in NLP. arXiv preprint arXiv:160807836. 2016; [Google Scholar]
- 35.Vadas D, Curran JR. Parsing noun phrases in the Penn Treebank. Comput Linguist. MIT Press; 2011;37(4):753–809. [Google Scholar]
- 36.Sawai Y, Shindo H, Matsumoto Y. Semantic structure analysis of noun phrases using abstract meaning representation. In: Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 2: Short Papers). Beijing, China: Association for Computational Linguistics; 2015. pp. 851–6. [Google Scholar]
- 37.Peterson KJ, Liu H. Automating the transformation of free-text clinical problems into SNOMED CT expressions In: AMIA Summits on Translational Science Proceedings. American Medical Informatics Association; 2020. pp. 497–506. [PMC free article] [PubMed] [Google Scholar]
- 38.Torii M, Yang EW, Doan S. A preliminary study of clinical concept detection using syntactic relations. In: AMIA Annual Symposium Proceedings American Medical Informatics Association; 2018. p. 1028. [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtz R, Roxbo D, Kuhlmann M. Improving semantic dependency parsing with syntactic features. In: Proceedings of the First NLPL Workshop on Deep Learning for Natural Language Processing Turku, Finland: Linköping University Electronic Press; 2019. pp. 12–21. [Google Scholar]
- 40.Sohn S, Wu S, Chute CG. Dependency parser-based negation detection in clinical narratives. AMIA Summits on Translational Science Proceedings. American Medical Informatics Association; 2012;2012:1. [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann M, King D, Beltagy I, Ammar W. ScispaCy: Fast and robust models for biomedical natural language processing. In: Proceedings of the 18th BioNLP Workshop and Shared Task Florence, Italy: Association for Computational Linguistics; 2019. pp. 319–27. [Google Scholar]
- 42.Zhang X, Zhao J, LeCun Y. Character-level convolutional networks for text classification. In: Proceedings of the 28th International Conference on Neural Information Processing Systems - Volume 1 Cambridge, MA, USA: MIT Press; 2015. pp. 649–57. (NIPS’15). [Google Scholar]
- 43.Kolomiyets O, Bethard S, Moens M-F. Model-portability experiments for textual temporal analysis. In: Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies: Short Papers - Volume 2 USA: Association for Computational Linguistics; 2011. pp. 271–6. (HLT ‘11). [Google Scholar]
- 44.Kobayashi S Contextual augmentation: Data augmentation by words with paradigmatic relations. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 2 (Short Papers) New Orleans, Louisiana: Association for Computational Linguistics; 2018. pp. 452–7. [Google Scholar]
- 45.Spasić I, Corcoran P, Gagarin A, Buerki A. Head to head: Semantic similarity of multi–word terms. IEEE Access. IEEE; 2018;6:20545–57. [Google Scholar]
- 46.Bodenreider O The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004;32(suppl_1):D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichartz F, Korte H, Paass G. Semantic relation extraction with kernels over typed dependency trees. In: Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining New York, NY, USA: Association for Computing Machinery; 2010. pp. 773–82. (KDD ‘10). [Google Scholar]
- 48.Fundel K, Küffner R, Zimmer R. RelEx—relation extraction using dependency parse trees. Bioinformatics. 2006;23(3):365–71. [DOI] [PubMed] [Google Scholar]
- 49.Gamallo P, Garcia M, Fernández-Lanza S. Dependency-based open information extraction. In: Proceedings of the Joint Workshop on Unsupervised and Semi-supervised Learning in NLP Association for Computational Linguistics; 2012. pp. 10–8. [Google Scholar]
- 50.Bunescu RC, Mooney RJ. A shortest path dependency kernel for relation extraction. In: Proceedings of the Conference on Human Language Technology and Empirical Methods in Natural Language Processing Association for Computational Linguistics; 2005. pp. 724–31. [Google Scholar]
- 51.Kim S, Liu H, Yeganova L, Wilbur WJ. Extracting drug–drug interactions from literature using a rich feature-based linear kernel approach. J Biomed Inform. Elsevier Science; 2015;55:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Wei F, Li S, Ji H, Zhou M, Wang H. A dependency-based neural network for relation classification. In: Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 2: Short Papers) Beijing, China: Association for Computational Linguistics; 2015. pp. 285–90. [Google Scholar]
- 53.Xu Y, Jia R, Mou L, Li G, Chen Y, Lu Y, et al. Improved relation classification by deep recurrent neural networks with data augmentation. arXiv preprint arXiv:160103651. 2016; [Google Scholar]
- 54.Luo Y, Uzuner Ö, Szolovits P. Bridging semantics and syntax with graph algorithms—state-of-the-art of extracting biomedical relations. Brief Bioinformatics. 2016;18(1):160–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei C-H, Peng Y, Leaman R, Davis AP, Mattingly CJ, Li J, et al. Assessing the state of the art in biomedical relation extraction: Overview of the BioCreative V chemical-disease relation (CDR) task. Database. Oxford University Press; 2016. March;2016:baw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Sun Y, Johnson RJ, Sciaky D, Wei C-H, Leaman R, et al. BioCreative V CDR task corpus: A resource for chemical disease relation extraction. Database. Oxford University Press; 2016. May;2016:baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia E, Sun W, Mei J, Xu E, Wang K, Qin Y. Mining disease-symptom relation from massive biomedical literature and its application in severe disease diagnosis. In: AMIA Annual Symposium Proceedings American Medical Informatics Association; 2018. pp. 1118–26. [PMC free article] [PubMed] [Google Scholar]
- 58.Quan C, Wang M, Ren F. An unsupervised text mining method for relation extraction from biomedical literature. PloS One. Public Library of Science; 2014. July;9(7):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalra D, Beale T, Heard S. The openEHR Foundation. Stud Health Technol Inform. 2005;115:153–73. [PubMed] [Google Scholar]
- 60.Chollet F, others. Keras. HTTPS://KERAS.IO, [Accessed: 01–02-2020]; 2015.
- 61.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805. 2018; [Google Scholar]
- 62.Alsentzer E, Murphy J, Boag W, Weng W-H, Jin D, Naumann T, et al. Publicly available clinical BERT embeddings. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop Minneapolis, Minnesota, USA: Association for Computational Linguistics; 2019. pp. 72–8. [Google Scholar]
- 63.Xiao H bert-as-service. HTTPS://GITHUB.COM/HANXIAO/BERT-AS-SERVICE, [Accessed: 01–02-2020]; 2018.
- 64.Beam AL, Kompa B, Fried I, Palmer NP, Shi X, Cai T, et al. Clinical concept embeddings learned from massive sources of multimodal medical data. arXiv preprint arXiv:180401486. 2018; [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Yang Z, Shen C, Xu J, Zhang Y, Xu H. Integrating shortest dependency path and sentence sequence into a deep learning framework for relation extraction in clinical text. BMC Med Inform Decis Mak. 2019;19(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCray AT. An upper-level ontology for the biomedical domain. Comp Funct Genomics. 2003;4(1):80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform. 2001;84(Pt 1):216–20. [PMC free article] [PubMed] [Google Scholar]
- 68.Ratner AJ, De Sa CM, Wu S, Selsam D, Ré C. Data programming: Creating large training sets, quickly. In: Advances in Neural Information Processing Systems; 2016. pp. 3567–75. [PMC free article] [PubMed] [Google Scholar]
- 69.Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Ré C. Snorkel: Rapid training data creation with weak supervision. In: Proceedings of the VLDB Endowment International Conference on Very Large Data Bases 2017. pp. 269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia F, Yetisgen-Yildiz M. Clinical corpus annotation: Challenges and strategies. In: Proceedings of the Third Workshop on Building and Evaluating Resources for Biomedical Text Mining (BioTxtM’2012) in conjunction with the International Conference on Language Resources and Evaluation (LREC), Istanbul, Turkey 2012. [Google Scholar]
- 71.Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, et al. An information extraction framework for cohort identification using electronic health records. AMIA Summits on Translational Science Proceedings. American Medical Informatics Association; 2013;2013:149. [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Sohn S, Liu S, Shen F, Wang L, Atkinson EJ, et al. A deep representation empowered distant supervision paradigm for clinical information extraction. arXiv preprint arXiv:180407814. 2018; [Google Scholar]
- 73.Chiticariu L, Li Y, Reiss FR. Rule-based information extraction is dead! Long live rule-based information extraction systems! In: EMNLP; 2013. pp. 827–32. [Google Scholar]
- 74.Wen A, Fu S, Moon S, El Wazir M, Rosenbaum A, Kaggal VC, et al. Desiderata for delivering NLP to accelerate healthcare AI advancement and a Mayo Clinic NLP-as-a-service implementation. NPJ Digit Med. Nature Publishing Group; 2019;2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shapley LS. A value for n-person games. Contributions to the Theory of Games. 1953;2(28):307–17. [Google Scholar]
- 76.Roth AE. The Shapley value: Essays in honor of Lloyd S. Shapley. Cambridge University Press; 1988. [Google Scholar]
- 77.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. In: Advances in Neural Information Processing Systems; 2017. pp. 4765–74. [Google Scholar]
- 78.Štrumbelj E, Kononenko I. Explaining prediction models and individual predictions with feature contributions. Knowledge and Information Systems. 2014;41(3):647–65. [Google Scholar]
- 79.Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii J. BRAT: A web-based tool for NLP-assisted text annotation. In: Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics Association for Computational Linguistics; 2012. pp. 102–7. [Google Scholar]
- 80.Hayes AF, Krippendorff K. Answering the call for a standard reliability measure for coding data. Communication Methods and Measures. 2007;1(1):77–89. [Google Scholar]
- 81.Kearns W, Lau W, Thomas J. UW-BHI at MEDIQA 2019: An analysis of representation methods for medical natural language inference. In: Proceedings of the 18th BioNLP Workshop and Shared Task Florence, Italy: Association for Computational Linguistics; 2019. pp. 500–9. [Google Scholar]