Abstract
Small molecule phosphoantigens such as (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate stimulate human Vγ9Vδ2 T cells after binding to the intracellular B30.2 domain of the immune receptor butyrophilin 3 isoform A1 (BTN3A1). To understand the ligand-target interaction in greater detail, we performed molecular docking. Based on the docking results, we synthesized the novel ligand (E)-(7-hydroxy-6-methylhept-5-en-1-yl) phosphonate and mutated proposed binding site residues. We evaluated the impact on butyrophilin binding of existing and novel ligands using a newly developed high-throughput fluorescence polarization assay. We also evaluated the ability of the compounds to stimulate proliferation and interferon-γ production of Vγ9Vδ2 T cells. Mutation of H381 fully blocked ligand binding, while mutations to charged surface residues impacted diphosphate interactions. Monophosphonate analogs bind similarly to BTN3A1, though differ in their antigenicity, demonstrating binding and efficacy are not linearly correlated. These results further define the structure-activity relationships underlying BTN3A1 ligand binding and antigenicity and support further structure-guided drug design.
Graphical Abstract
INTRODUCTION
Butyrophilin 3A1 (BTN3A1, Figure 1) is the molecular target of phosphoantigens, and this interaction is required for stimulation of Vγ9Vδ2 T cells1, 2 and the immune response they generate.3–5 Though it was initially reported that ligands bind to the extracellular domain, multiple studies identified a positively-charged ligand binding pocket within the B30.2 intracellular domain.6–8 Once bound, the intracellular region of BTN3A1 undergoes a conformational change involving the juxtamembrane (JM) region,9–11 which is likely associated with an extracellular change12 that leads to Vγ9Vδ2 T cell recognition. The ability of Vγ9Vδ2 T cells to detect ligand-bound BTN3A1 implicates a role against malignant cells13, 14 and has been explored as a chemoimmunotherapeutic target.15–21
Figure 1.
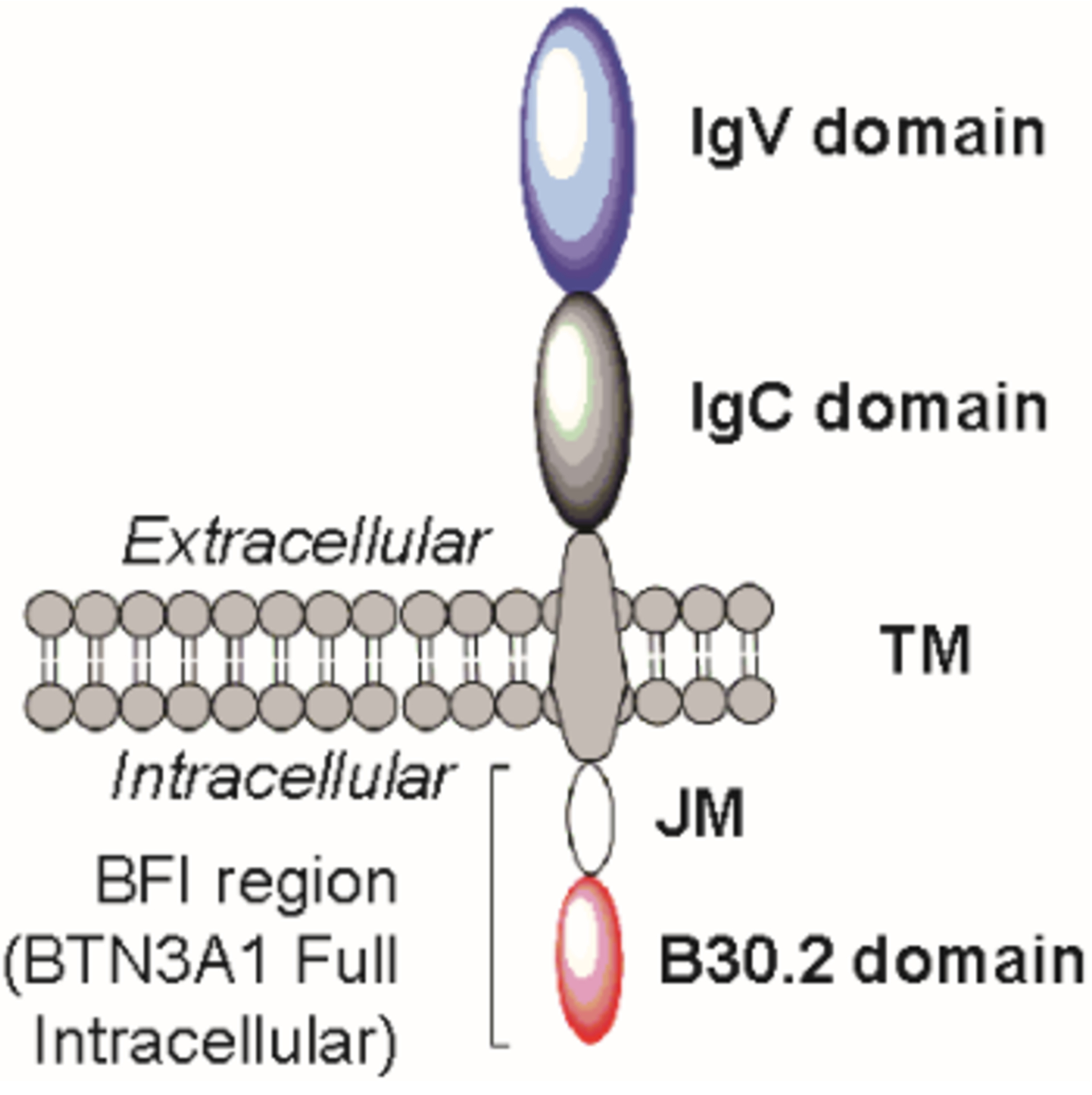
Representation of BTN3A1.
The most potent natural BTN3A1 ligand by cellular Vγ9Vδ2 T cell expansion and cytokine production is (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate (HMBPP, 1), an intermediate in the 2-C-methyl-ᴅ-erythritol 4-phosphate (MEP) pathway in plants and bacteria.22, 23 The first common products of the MEP pathway and the human mevalonate pathway are dimethylallyl diphosphate (DMAPP, 2) and isopentenyl diphosphate (IPP, 3). Diphosphates 2 and 3 (Figure 2A) also stimulate Vγ9Vδ2 T cells, albeit at lower potency compared to 1,24 while synthetic bisphosphonates such as zoledronate (4) stimulate Vγ9Vδ2 T cells by inhibiting farnesyl diphosphate synthase and elevating 2 and/or 3.25, 26 Therapeutic use of diphosphates to target BTN3A1 is limited by poor membrane permeability as they are negatively charged at physiological pH.27 This has been overcome by the use of prodrugs7, 28–30 on a phosphonate scaffold. However, prodrugs provide little additional information on the ligand binding pocket as they release the same ligand C-HMBP (5).17
Figure 2.
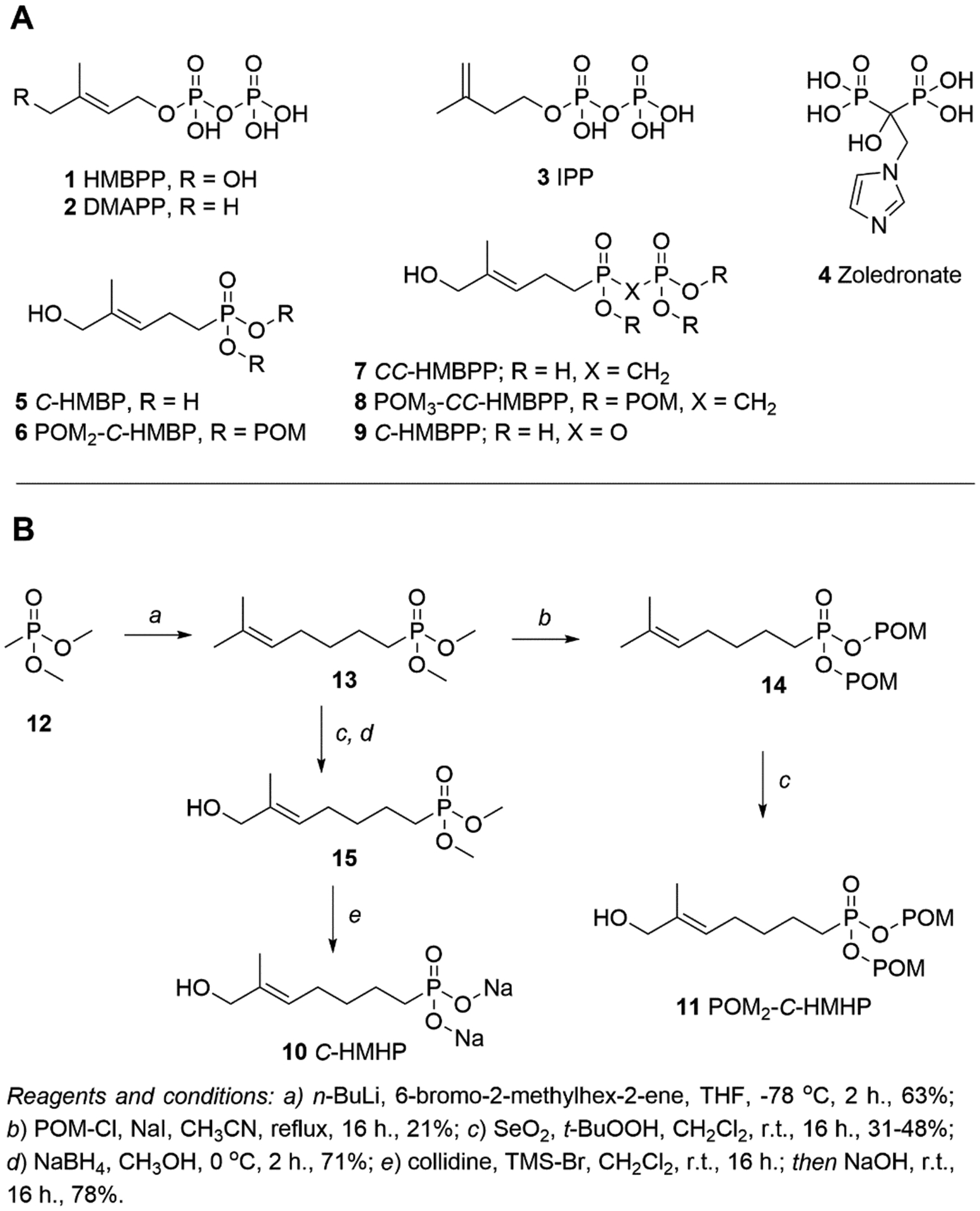
Chemical structures of compounds used in this study. (A) Natural (1 – 3) and synthetic (5 – 9) butyrophilin ligands bind to BTN3A1 and promote Vγ9Vδ2 T cell activation. Zoledronate (4) promotes indirect activation through the build-up of 2 and/or 3. POM = pivaloyloxymethyl, CH2OC(O)C(CH3)3. (B) Synthesis of C-HMHP (10) and POM2-C-HMHP (11).
Binding to BTN3A1 has been evaluated using isothermal titration calorimetry (ITC).6, 7, 31 This approach quantifies direct binding (e.g. Kd, ΔG) but has been limited to 1, 3 and C-HMBPP (9) as it requires a high amount of reagents in a low-throughput assay.32 Despite its picomolar cellular activity, 1 binds BTN3A1 only with low micromolar affinity, which complicates evaluation of less potent analogs.33 These ITC studies have focused on evaluating ligands, whereas molecular modeling4, 33 and NMR experiments9–11 have identified key residues within the ligand binding pocket. These include a histidine (H381) and a set of positively charged residues in the diphosphate binding region.6 However, prior evaluation of these residues focused on cellular activity without examining ligand binding.8
The goal of this study was to explore in more detail the relationship between ligand binding and cellular potency at the molecular level. We developed a high-throughput fluorescence polarization (FP) assay to measure binding of butyrophilin ligands. The ligand binding pocket of BTN3A1 was probed using known phosphoantigens and novel synthetic ligands, while BTN3A1 point-mutations were made to elucidate key binding interactions. These studies provide new insight on the mechanisms involving Vγ9Vδ2 T cell activation through BTN3A1 at the molecular level and will support further structure-guided drug design.
RESULTS
Design and synthesis of extended length C-HMBP analogs
Our previous studies7, 28–30 relied upon prodrug forms to activate BTN3A1 and focused on ligands in which the α-phosphate of HMBPP (1) had been replaced with a metabolically stable phosphonate and the β-phosphate group was absent. This resulted in an active ligand, C-HMBP (5), that was two atoms shorter in length than the natural ligand 1 (Figure 3A). Both the crystal structure6 and our modeling33 of 1 indicated diphosphate interactions (Figure 3B). While oxygens of both phosphates of 1 interact with R448 (salt bridges not shown), the α-phosphate interacts with W421 and M424 while the β-phosphate interacts with R442 and R499. Initially, we hypothesized that 5 would interact with W421 and M424 but not with R442 or R499. Phosphonate C-HMHP (C-hydroxymethylhexylphosphonate, 10) was proposed (Figure 2B) to reverse these interactions. These two ligands, 5 and 10, could then be used to evaluate the relative contributions of the residues. Alternatively, docking experiments predicted participation of R442, R448, and R499 for both monophosphonates (Figure 3C–D), with superior docking of 10 (score of −7.66) (Table 1) compared to 5 (-5.72), which suggests that the cell permeable prodrug POM2-C-HMHP (11) might stimulate Vγ9Vδ2 T cells more efficiently than POM2-C-HMBP (6).
Figure 3.
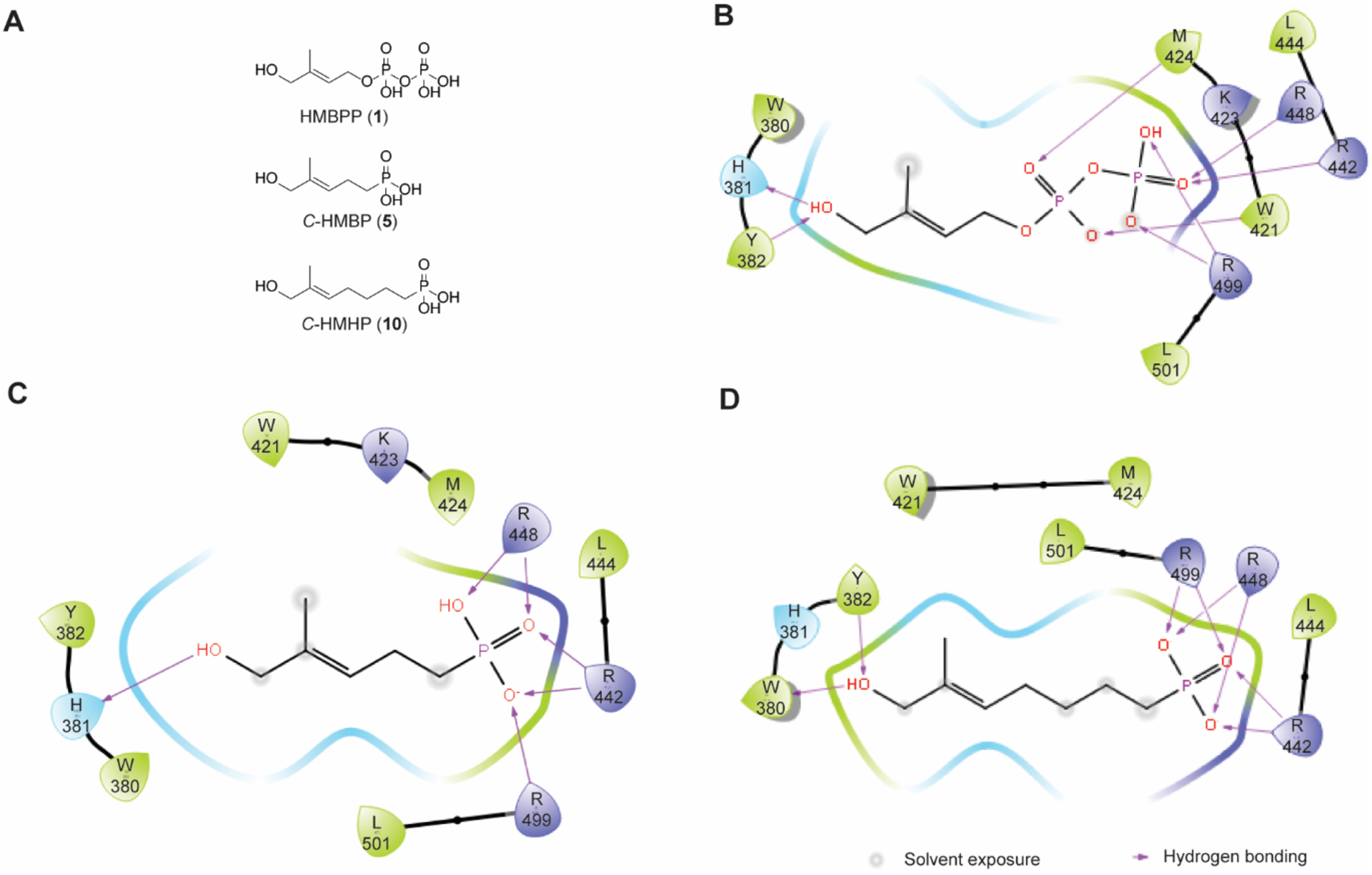
Molecular modeling of HMBPP and its monophosphonate analogs. (A) Length of butyrophilin ligands 1, 5, and 10. Molecular modeling with predicted residue interactions of (B) 1 (C) 5, and (D) 10.
Table 1.
Physical properties and activity of butyrophilin ligands (n ≥ 3).
| Cmpd. | clogP | Docking Score | Proliferation EC50 (μM) with 95% CI |
IFN-γ EC50 (μM) with 95% CI | FP IC50 (μM) with 95% CI |
|---|---|---|---|---|---|
| 1 | −2.34 | −9.73 | 0.00051a | 0.0042 (0.0033 – 0.0054) |
23.0 (16.1 – 32.8)b |
| 2 | −0.59 | −9.31 | n.d. | 1.5 (1.1 – 2.1) |
270 (220 – 340) |
| 3 | −0.59 | −8.32 | 36a | 71 (52 – 97) |
513 (390 – 660) |
| 9 | −2.38 | −9.32 | 0.086 (1.2E−4 – 63) |
0.0020 (2.8E−6 – 1.5) |
7.87 (6.17 – 10.0) |
| 7 | −2.27 | −7.01 | 26c | > 100 | 103 (48.9 – 218) |
| 5 | −1.08 | −5.72 | 4.0a | 3.4 (0.94 – 12) |
2,560 (1350 – 4870) |
| 10 | −0.02 | −7.66 | n.s. | n.s. | 1,040 (493 – 2200) |
| 8 | 3.47 | n.d. | 0.041c | > 1 | n.d. |
| 6 | 3.10 | n.d. | 0.0054a | 0.015 (0.0068 – 0.032) |
n.d. |
| 11 | 2.08 | n.d. | n.s. | n.s. | n.d. |
| NaPP | −3.03 | n.d. | n.d. | n.s. | 1,140 (421 – 3100) |
Data from Hsiao 2014.
The IC50 value for HMBPP (1) is an average from experiments presented in Figure 4.
Data from Shippy 2017.
Abbr.: n.d., not determined; n.s., no stimulation; NaPP, sodium diphosphate
The HMBPP analogs 10 and 11 were prepared (Figure 2B). Prior to alkylation of 12, 6-bromo-2-methylhex-2-ene was synthesized as reported34 and subsequently used in conjunction with n-butyllithium to reach the key intermediate 13. Prodrug 11 was obtained through a pivaloyloxymethyl (POM) protection (14) and subsequent allylic hydroxylation using selenium dioxide. Disodium salt 10 was synthesized from 13 through hydroxylation (15) and demethylation in a route similar to one previously described.7
Compounds 10 and 11 are taken up by cells but fail to stimulate Vγ9Vδ2 T cells
The new ligands 10 and prodrug 11 were assessed for stimulation of Vγ9Vδ2 T cells in a proliferation assay and an IFN-γ production assay. Ligands 1 and 6 were used as positive controls and significantly expanded T cells at 0.1 μM above a non-stimulated baseline (Figure 4A–B, Table 1). Despite the modeling having predicted a stronger interaction, neither prodrug 11 nor salt 10 (Supplemental Figure S1B) stimulated T cell proliferation at concentrations up to 100 μM. As a control, 9 was assessed (Supplemental Figure S1A) and was found to potently stimulate T cell proliferation (EC50 = 0.086 μM). Additionally, neither intermediate 14 nor 15 displayed signs of Vγ9Vδ2 T cell stimulation (Supplemental Figure S1C–D).
Figure 4.
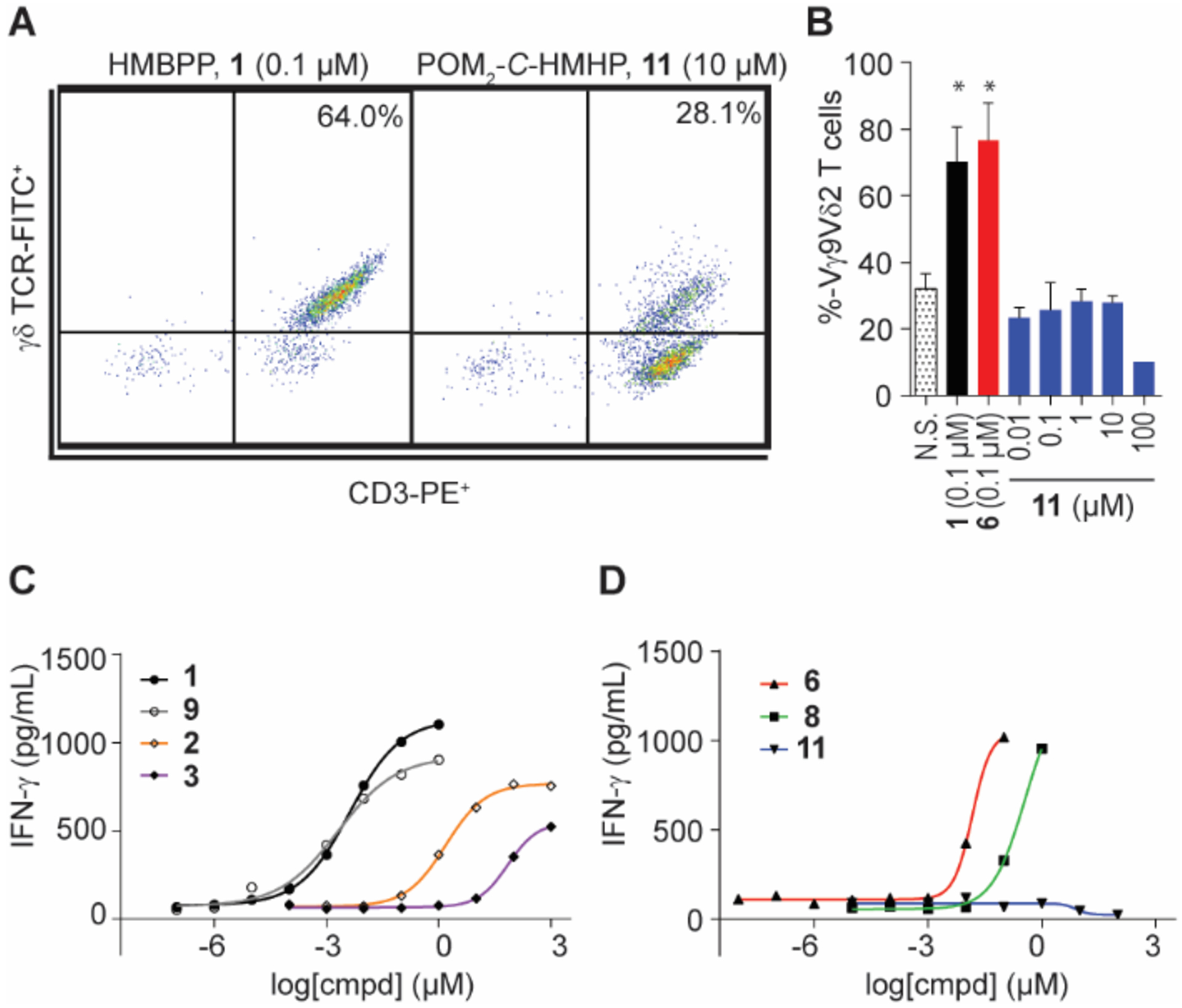
Cellular activity of butyrophilin ligands 10 and 11. (A) Representative population and (B) quantification of γδ T cell proliferation by flow cytometry analysis. Data represents mean +/− SD. (C-D) Quantification of PBMC IFN-γ production of butyrophilin ligands determined by ELISA. Results reported in Table 1; n = 5, *p < 0.05, ANOVA.
One possible explanation for the lack of stimulation could be lower cellular uptake of the prodrug 11 relative to 6. To rule out the possibility that 11 was not internalized, a cellular uptake assay was performed by incubating prodrugs 6 and 11 with peripheral blood mononuclear cells (PBMCs). Mass spectrometry was used to monitor the disappearance of each ligand from the media and compared to t = 0 (Supplemental Table S1). Following a six-hour incubation, both 6 and 11 were similarly internalized and/or degraded, with 58% and 44% remaining. By 24 hours, less than 1% of the parent ion was detected for each ligand. Compared to PBMC-free controls in which degradation is likely due to instability of the POM-group (63% and 44% remaining at 48 hours for 6 and 11, respectively), these results indicate that 11 undergoes cell internalization similar to 6.
A second cellular assay was completed to characterize the compounds. Human PBMCs containing naïve Vγ9Vδ2 T cells were incubated in a dose-response manner with ligands 1 – 11 or sodium diphosphate for 48 hours and an ELISA assay was completed to quantify IFN-γ (Figures 4C–D, S2, Table 1). Incubation with 10 or 11 did not stimulate at concentrations up to 100 μM, while 1 and prodrug 6 provided EC50 values consistent with our previous studies7, 33 in the low-nanomolar level at 4.2 and 15 nM, respectively. Interestingly, 2 was more potent than 3, which is similar to proliferation studies that found 2 more active than 324 but contrary to another report that 3 was more active.23 Neither zoledronate (4) nor sodium diphosphate stimulated IFN-γ production. The lack of response from 4 is likely due to an insufficient treatment concentration, as judged by subsequent results.35 Ligand 9 had an EC50 similar to 1. Salt 7 did stimulate IFN-γ production at 100 μM, while its prodrug 8 was 100-times more potent.
Optimization of BTN3A1 GDP-BODIPY Fluorescence Polarization Assay Conditions
Though inactive in functional assays, we suspected salt 10 might be a useful tool to explore the ligand binding pocket of BTN3A1. A fluorescence polarization (FP) assay was developed to assess the binding characteristics of butyrophilin ligands. This was chosen as an alternative to ITC, as FP methodology would allow for higher throughput using fewer reagents. Mattarei, et al.36 have synthesized a cross-linkable probe that bound to BTN3A1, but our studies required a competitively-binding probe. We have previously shown that BTN3A1 weakly binds to guanosine diphosphate (GDP),9 and ADP was also reported to interact with BTN3A1.10 As such, a commercially available GDP-BODIPY probe (Figure 5A) was incubated with the BTN3A1 full intercellular domain (BFI) and monitored using a plate reader set to an emission/excitation spectrum of 535/485 nm for FP analysis. A protein-probe interaction was observed at probe concentrations of 100 nM and greater (Figure 5B–C). Displacement studies with 1 indicated that the probe interacted with the positively-charged binding pocket (likely through the diphosphate of GDP) and identified an ideal probe concentration of 750 nM to avoid the use of excess probe (Figure 5D) while providing consistent results with low variation (Z-factor = 0.71). BFI concentrations at less than 10 μM failed to produce observable probe binding (Figure 5E).
Figure 5.
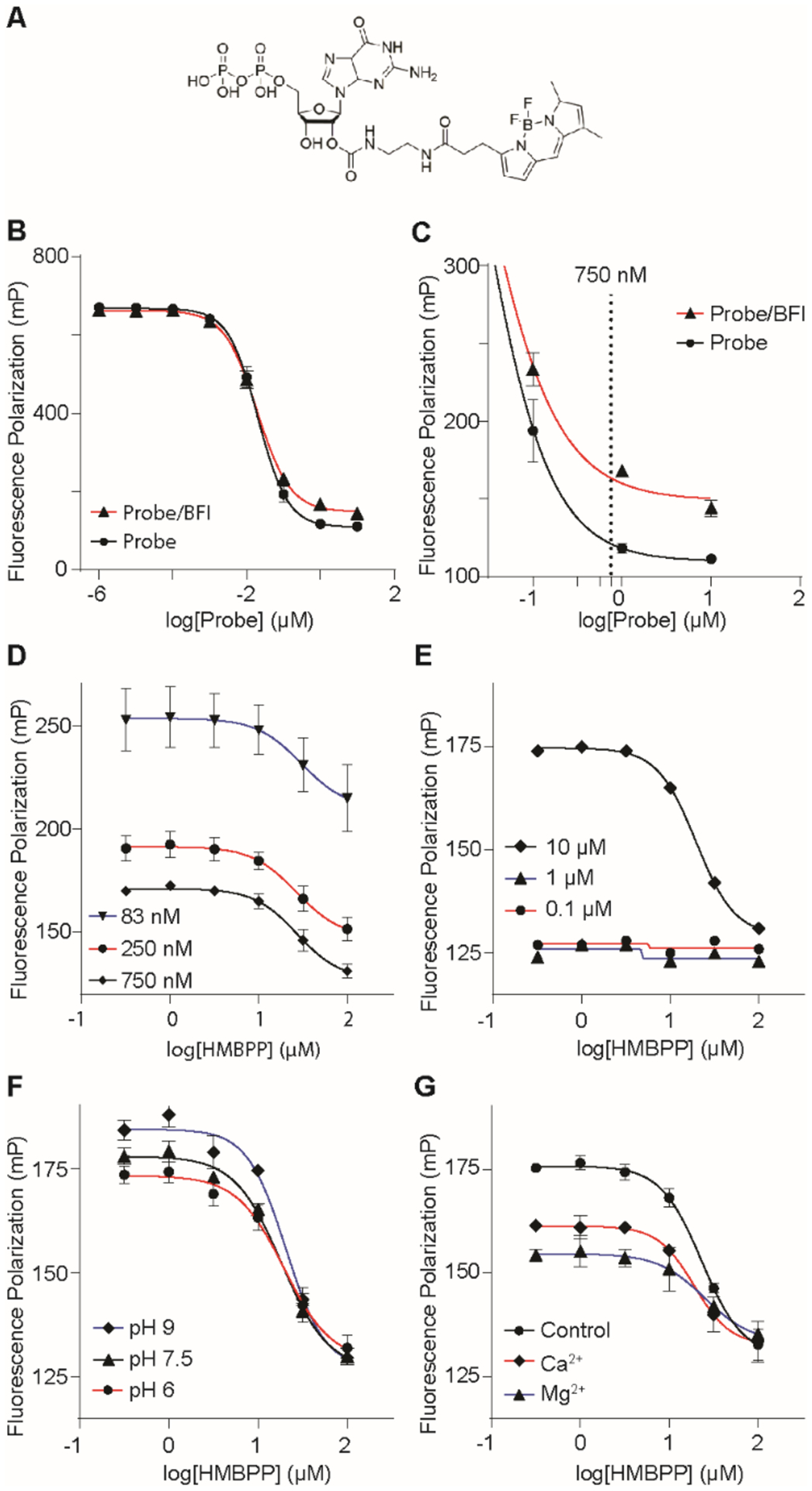
Optimization of a fluorescence polarization assay to assess BTN3A1 ligand binding. (A) Structure of the GDP-BODIPY probe. Optimization of the fluorescence polarization assay involved varying probe (B-D) and BFI (E) concentrations. pH (F) and divalent cation (G) effects were also investigated. Data represents mean +/− SD.
Further optimization studies included exploring the effects of pH and divalent cations (Figure 5F–G). These conditions altered the positive control (incubation of probe with protein in the absence of a ligand) without having an effect on the IC50 of 1. Alkaline conditions (pH 9) resulted in a higher control which indicates a greater amount of probe was able to bind to the protein that may be attributed to a more-deprotonated form of the diphosphates. Acidic conditions (pH 6) and the addition of either magnesium or calcium resulted in a lower control, potentially due to a more-protonated diphosphate or interactions with the diphosphate to limit protein binding, respectively. The IC50 value of 1 varied within a range of 18–27 μM; because of this, 1 was repeated as a control in all experiments analyzing additional ligands. Taken together, GDP-BODIPY is a useful probe that can be used in fluorescence polarization assays to monitor binding of 1 and other experimental ligands to the BTN3A1 diphosphate binding pocket.
BTN3A1 H381 is essential for HMBPP binding in the FP assay
HMBPP (1) was analyzed for its ability to displace probe binding in three separate BTN3A1 constructs; wt-BFI, BFI(H381R), and B30.2 (Figure 6A). Ligand 1 displaced the probe in wt-BFI similar to B30.2. This result reflects our previous ITC experiments9 where 1 had a slightly stronger affinity for BFI (Kd = 2.36 μM) than to B30.2 (Kd = 4.27 μM). Both constructs showed similar ranges (difference between positive control and fully displaced probe).
Figure 6.
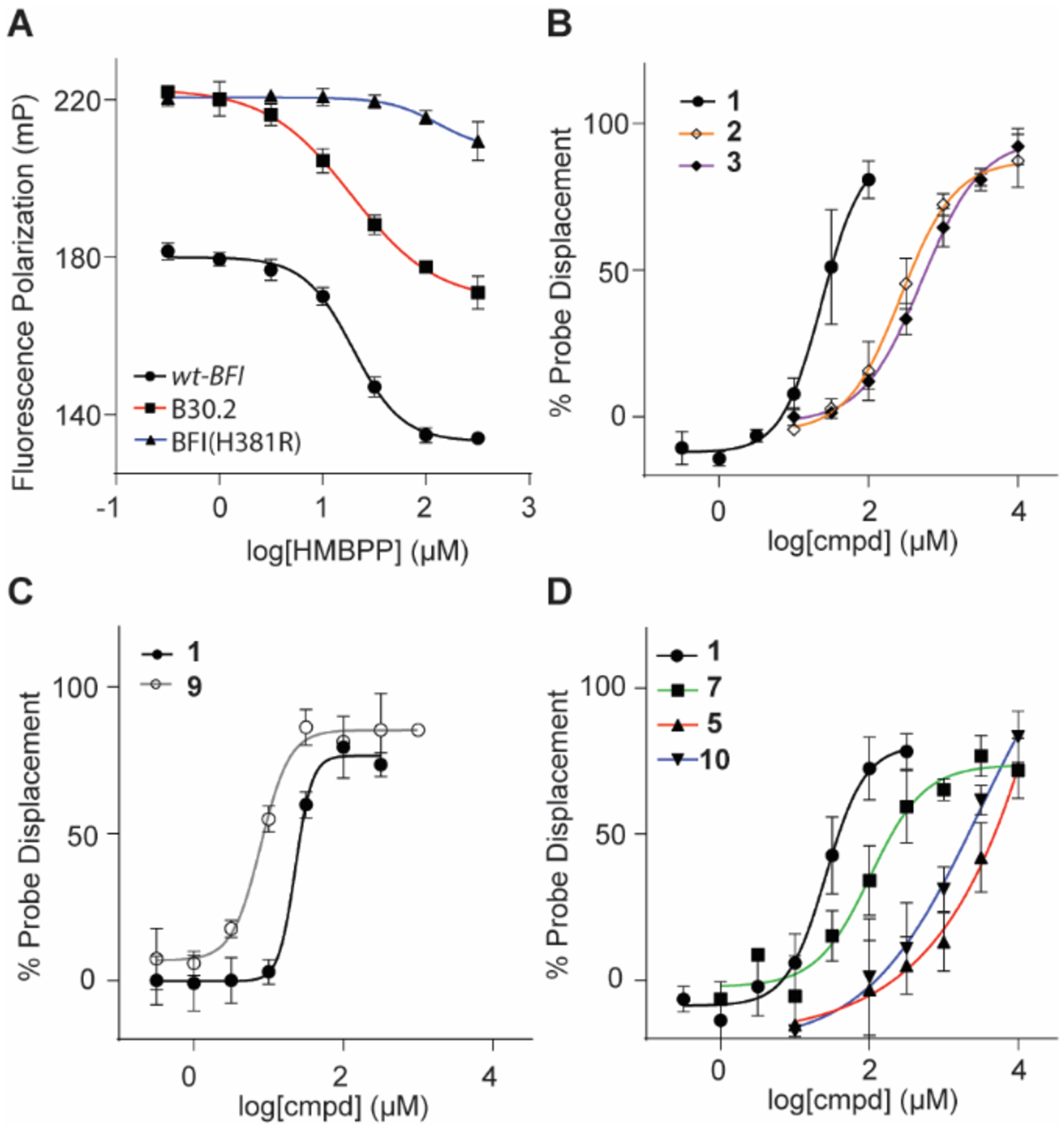
Binding of test compounds to BTN3A1-BFI as determined by fluorescence polarization assay. (A) Competitive inhibition curves HMBPP (1) in BTN3A1 domains: wt-BFI (IC50 = 19.9 μM), B30.2 (23.3 μM) and BFI(H381R) ( > 333 μM). Natural (B) and synthetic (C-D) butyrophilin ligands assayed in a competitive inhibition study. IC50 values reported in Table 1; n = 3 for all experiments. Data represents mean +/− SD.
BTN3A1 H381 has been reported to participate in a hydrogen-bond interaction with the allylic alcohol of 1 (Figure 3B).6, 33 Additionally, Yang et al. has recently reported that H381 is vital for the β → α conformation and induction of Vγ9Vδ2 T cell proliferation through molecular dynamic modeling studies.37 To confirm this interaction and further validate our approach, we generated the H381R mutation on the BFI construct. The arginine replacement was chosen due to its presence in butyrophilin isoform BTN3A3,1 which does not stimulate Vγ9Vδ2 T cell proliferation in the presence of 1. The replacement of the histidine with arginine results in a lack of binding between protein and ligand 1 (IC50 > 333 μM).
Structure activity relationships for natural and synthetic diphosphates to displace probe binding to the BTN3A1 BFI domain
We next assessed a small panel of diphosphates for their ability to displace probe from the wt-BFI domain of BTN3A1. Natural phosphoantigens 1, 2 and 3 were found to have IC50 values of 23.5-, 270-, and 513-μM, respectively (Figure 6B and Table 1). These results provide insight into two separate hypotheses. First, ligand 2 is twice as active as 3 in the FP assay but 200-fold more active for cellular IFN-γ production. This finding is not inconsistent with our prior results where we speculated that exponential correlation exists between affinity and potency of other phosphoantigens,33 though we cannot rule out the possibility that other factors affect this cellular assay. Second, 2 exhibits a moderate affinity for wt-BFI whereas 1 is does not bind to BFI(H381R). In theory, the interaction between 2 and wt-BFI should be similar to 1 and BFI(H381R) if the primary role of the histidine were to provide hydrogen bonding with the ligand; however, because of the inability of 1 to displace the probe in BFI(H381R), this indicates that the H381R mutation hinders binding in a way that is more disruptive than simple loss of hydrogen bonding with the ligand,6 which may include the loss of additional hydrogen bonds (W421 and M424).37
C-HMBPP (9) was assessed against wt-BFI and found to have an IC50 value of 7.87 μM, compared to 1 at 22.8 μM (Figure 6C). The relative potency of 1 and 9 is mildly inconsistent with our ITC results7 in wt-BFI but supports6 independent ITC results of 9 having a stronger affinity for B30.2. The crystal structure of B30.2 and 9 does not clearly define the alkyl chain. This mobility combined with mixed results may indicate that the oxygen linking the α-phosphate to the prenyl-group may hinder or enhance binding depending on the preferred conformation which could be affected by the experimental conditions. Sodium diphosphate was also assessed to determine the ability for any diphosphate to displace the probe and was found to have an IC50 value of 1.14 mM (Supplemental Figure S3A).
BTN3A1 probe displacement by monophosphonates
The synthetic monophosphonates C-HMBP (5), C-HMHP (10) and phosphinophosphonate (7) were assessed for their ability to bind to wt-BFI (Figure 6D). Previously, phosphinophosphonate 7 produced a weak dissociation constant (Kd) of 111 μM (compared to 1 at 1.54 μM) in ITC experiments while a Kd was not calculable for phosphonate 5, despite their respective prodrugs, 8 and 6, stimulating Vγ9Vδ2 T cells in proliferation assays. Phosphinophosphonate 7 was the most potent of the three ligands with an IC50 value of 103 μM. Phosphonates 5 and 10 displaced the probe in the low millimolar range; however, additional experiments (Supplemental Table S2) with various phosphates revealed that high concentrations (> 33 mM) of phosphates resulted in probe interference. Therefore, the raw data (Supplemental Figure S3B) was analyzed with constraints set to the negative control. As predicted by the modeling, the FP data indicated that phosphonate 10 has a stronger affinity for BTN3A1 (IC50 = 1.04 mM) compared to 5 (2.58 mM). This enhanced affinity may be attributed to hydrogen bond interactions with W380 and Y382 as predicted by molecular modeling. It was unexpected that the HMBPP-length 10 would bind more strongly to BFI compared to 5, while prodrug 11 was inactive in cellular assays. One possible explanation for the inactivity of 11 in cellular assays, despite a strong inaction of 10 in the competitive inhibition FP assay, may be intracellular phosphorylation of the monophosphonates following cleavage of the protecting groups. This would result in the intracellular conversion of C-HMBP (5) to C-HMBPP (9); whereas C-HMHP (10) would be phosphorylated to a ligand with a suboptimal length between the diphosphate and hydroxyl moiety. Alternatively, the longer chain length of compounds 10 and 11 may block a resulting protein: protein interaction at the ligand binding interface as has been observed with other more bulky substituants.37
Residues R442 and R448 contribute to ligand diphosphate binding while W421, M424 and R499 hinder probe interactions
Site-directed mutagenesis was employed to mutate each of the key binding BFI residues W421, M424, R442, R448, and R499 to alanine to further assess the diphosphate binding pocket. Circular dichroism was performed on all proteins which indicated no significant changes in any mutant secondary structure compared to the wild-type (Figure S4). All proteins were revealed to consist of primarily β-sheets (molar ellipticity minima at 218 nm) which is consistent with the crystal structure of BFI.6, 37 HMBPP (1) was assessed for its ability to displace the probe from wt-BFI and the five mutated proteins (Figure 7A, Table 2). The two major aspects of the assay that were analyzed were the ranges (spans) and IC50 values. Additionally, non-specific binding, which was observed due to incomplete displacement of the probe, was corrected and competitive inhibition curves were translated (bottom limit = 166 mP) for visual representation (Figure 7B). The ranges for mutants W421A, M424A, and R499A all increased compared to the wild-type. This indicates that an increased amount of probe is able to bind to BTN3A1 within the HMBPP diphosphate binding region, and may be attributed to steric hindrance of the wild-type residues. This may be especially true for M424A, as the binding interaction is expected to be through the protein backbone and the mutation would still support this binding interaction. However, major conformation changes that expose the binding pocket are also possible for each of these mutations.
Figure 7.
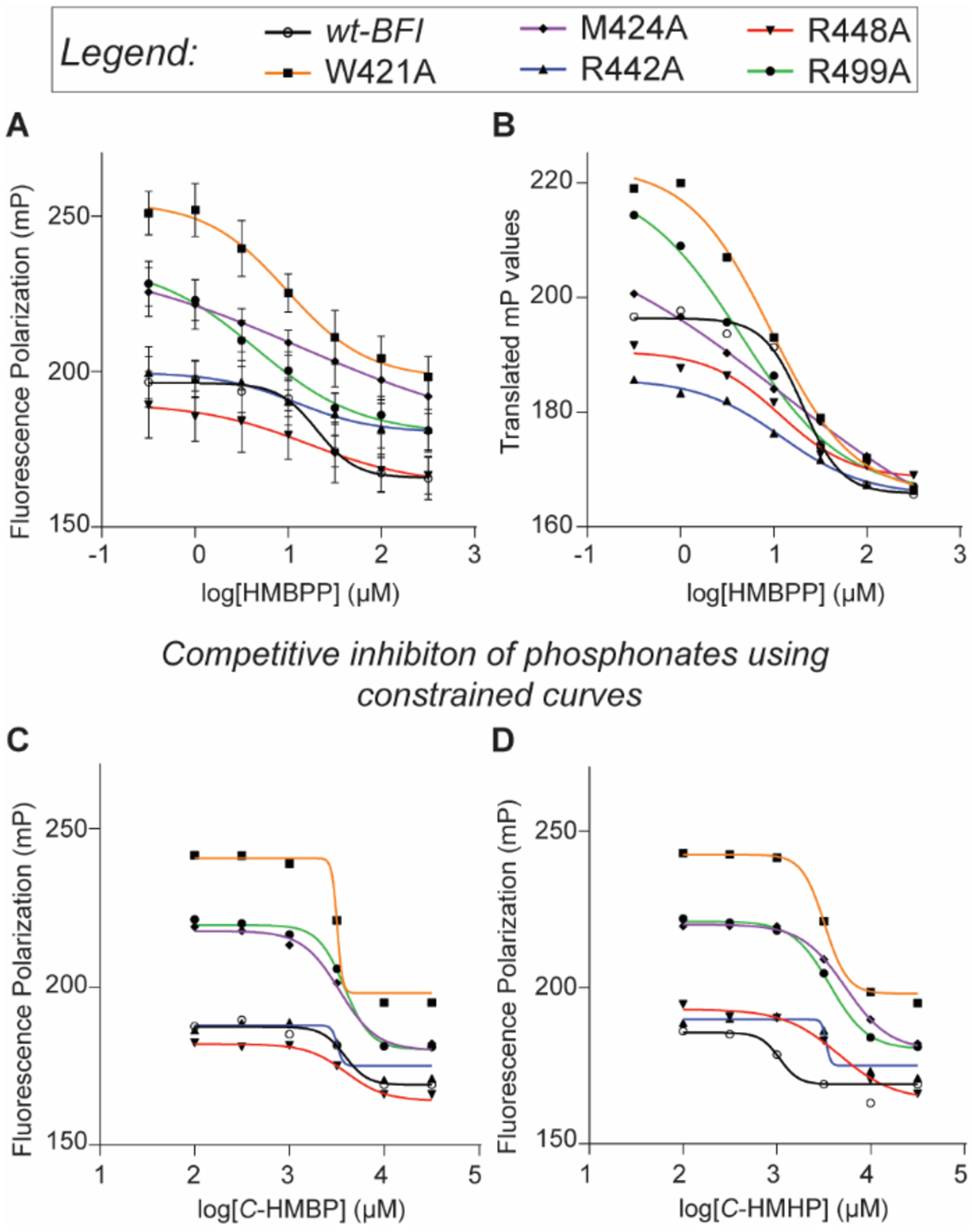
Analysis of butyrophilin ligands using mutants of the BTN3A1 diphosphate binding region. (A) HMBPP (1) displaces the GDP-BODIPY probe at different rates in mutants than in wt-BFI. (B) Curves of 1 displacement translated for range comparison. (C-D) Due to phosphonate probe interference, lower limit constraints for C-HMBP (5) and C-HMHP (10) were utilized. Lower limit seen using 1 were used as constraints for each individual mutant; raw files in Supplemental Information. Ranges and IC50 values reported in Table 2; n = 3 for all experiments.
Table 2.
Probe displacement range and comparison of competitive inhibition IC50 values (μM) of butyrophilin ligands in BTN3A1 BFI mutations (n ≥ 3).
| wt-BFI | W421A | M424A | R442A | R448A | R499A | |
|---|---|---|---|---|---|---|
| Probe displacement (mP) range with 95% CI | ||||||
| 1 | 30.6 (25 – 36) |
56.7 (40 – 73)* |
59.1a
(38 – 81)* |
19.4 (13 – 26) |
26.2 (12 – 40) |
55.0 (21 – 78)* |
| IC50 values (μM) with 95% CI | ||||||
| 1 | 20.5 (14 – 30) |
9.64 (5.3 – 18) |
n.d. | 12.1 (5.8 – 25) |
16.5 (5.8 – 47) |
4.56 (1.9 – 11)* |
| 5 | 3,850 | 3,190 | 3,390 | 3,220 | 3,930 | 3,830 |
| 10 | 1,070 | 3,240* | 5,350* | 3,430 | 4,620* | 3,720* |
Ambiguous; approximate slope estimated by GraphPad.
significant difference in mutant relative to wt-BFI for indicated ligand.
The R442A and R448A mutations appear to decrease the range compared to 1, though this result did not reach statistical significance. Regardless, because the binding pocket would theoretically have less steric hindrance with the removal of the arginine, this decreased range may be attributed to the loss of binding interaction from the arginine residues. Between these two residues, R442, which is expected to interact with the β-phosphate of 1 based on modeling, is trending towards significance and may be more important than R448 in binding to a diphosphate despite R448 having interactions with both phosphate groups.
The IC50 value of 1 in the R499A mutant was significantly lower than in the wt-BFI, while W421A, R442A, and R448A trended lower but did not reach statistical significance compared to the wild-type (Table 2). This could be explained as the probe and 1 both contain diphosphates. Decreased binding resulting from a mutation should affect each compound equally in this region. However, because 1 also interacts with the protein via its prenyl-chain and allylic alcohol, the overall effect on binding of 1 is less relative to the probe, lowering the IC50 values for 1 in the mutants compared to the wild-type. The greater the IC50 improvement in an individual mutation can be attributed to a strong binding interaction between the wild-type residue and the diphosphate. This supports the case that R442 (IC50 = 12.1 μM in the R442A mutation) is more important than R448 (16.5 μM). The IC50 values resulting from W421A (9.64 μM) and R499A (4.56 μM) could argue that the tryptophan and arginine residues in the wild-type play major roles in binding, with R499 having stronger interactions with the β-phosphate.
Effect of ligand chain modifications on binding to BTN3A1 mutants
Finally, we examined whether alterations to the ligand chain would affect binding to the different BTN3A1 mutants. Ligands C-HMBP (5) and C-HMHP (10) were utilized to determine the effects the chain-length had on the ability to competitively inhibit the fluorescent probe binding to the mutant proteins. The results from initial testing (Table 1) indicated the additional ethylene unit in 10 resulted in an improved affinity compared to 5. High concentrations of the phosphonates interfered with the probe (Figure S5), and constraints were used that were set to the displacement of the probe while using 1 in each individual protein. The constrained competitive inhibition curves (Figure 7C–D) provided IC50 values in the low millimolar range (Table 2). The values for wt-BFI were similar to those previously found for both 5 and 10; however, due to a small range in IC50 values, we were not confident that the results provided data that could be used to study the effects of the mutations in regards to the diphosphate binding pocket.
DISCUSSION AND CONCLUSIONS
The factors contributing to the antigenic activity of phosphoantigens towards Vγ9Vδ2 T cells has remained mysterious for some time, though recent studies have made it clear that ligand binding to BTN3A1 in target cells leading to conformational change in the protein structure is required for this process. Understanding the binding interaction at the molecular level would further enable design and development of novel synthetic butryophilin ligands that could overcome limited pharmacokinetic parameters of the naturally-occurring ligands for future in vivo therapeutic applications.15 In the present study, we examined factors that contribute to the binding of BTN3A1 to its phosphoantigen ligands.
Ligand binding occurs at a shallow binding pocket on the surface of the intracellular B30.2 domain of the protein. Earlier6 and recent37 X-ray studies clearly identified the basic amino acid residues lining the pocket, while cell-based studies8 had shown some of these residues were required for function. Through mutation of these residues, we directly assessed their importance to interactions with a panel of ligands. Mutation of H381 blocked HMBPP binding, and of the other mutants R499 had the strongest impact on HMBPP binding relative to the probe. Mutations to R442 and R448 decreased diphosphate probe affinity, and mutations to W421, M424, and R499 increased diphosphate probe affinity.
We evaluated the impact on butyrophilin binding of existing and novel ligands, as well as their ability to stimulate proliferation and interferon-γ production of Vγ9Vδ2 T cells. Although novel compound 10 interacted more strongly with BTN3A1 than compound 5, both 10 and prodrug 11 were inactive in functional assays. Thus monophosphonate analogs of different chain lengths bind similarly to BTN3A1, though differ in their antigenicity, providing further evidence that binding and efficacy are not linearly correlated.33,37 Because compound 10 lacks the α but not β phosphorus group, the interactions of the α-phosphate are critical to the antigenic activity of phosphoantigens. This could be due to loss of interactions with W421 and M424 in the binding pocket, decreasing binding affinity. On the other hand, because the diphosphate sits on the surface of the protein, we cannot rule out the possibility of the α-phosphate also contributing to asymmetrical interactions with a second BTN3A1 molecule in the cellular context.37
We found that our newly developed fluorescence polarization approach is a useful assay for evaluating BTN3A1 interactions. The FP approach takes advantage of the binding pocket being accessible on the surface of the protein, which enables interactions even with non-antigenic diphosphates such as GDP. The FP assay gives results that are consistent with prior ITC, while being faster and using less reagents. This approach will facilitate both characterization of novel synthetic ligands and high throughput library screening. The FP assay worked well to distinguish binding of different ligands to the wt-BFI construct defining a clear pattern of binding (C-HMBPP > HMBPP > CC-HMBPP > DMAPP > IPP) within the micromolar to millimolar range necessitated by the underlying biology of the protein. It also worked well to understand the impact of point mutations on both probe and HMBPP binding. However, it was less useful in matching specific protein amino acid residues to specific functional groups of the ligands, because both factors impact both probe and ligand binding in sometimes unpredictable ways.
Together, these findings provide rationale for the positively charged binding region (e.g., R442, R448, R499) of BTN3A1 to serve as an anchor for the HMBPP (1) diphosphate, while H381 provides antigenic specificity through binding the HMBPP hydroxyl group and W421 and M424 contribute to both binding and antigenic specificity through interactions with the α-phosphate. This data can be used to further guide BTN3A1 drug design as potent phosphoantigens would be expected to retain these properties.
EXPERIMENTAL SECTION
General Experimental Procedures.
Commercially available reagents and solvents were used without further purification. Flash column chromatography was performed on silica gel (60 Å, 230 × 400 mesh) and TLC (200 μm thickness) analyses were visualized by heating post exposure to phosphomolybdic acid. NMR (Bruker AVANCE 500) spectra were obtained at 500 MHz (1H), 125 MHz (13C), or 202 MHz (31P). HRMS and purity were obtained used UHPLC-MS (Waters Synapt G2-Si) equipped with a q-TOF and ESI. Purity of all compounds were > 95%.
Sodium (E)-(7-hydroxy-6-methylhept-5-en-1-yl) phosphonate (10).
In an oven-dried flask under argon, 15 (60 mg, 0.254 mmol) was dissolved in CH2Cl2 (anhydrous, 2 mL). The solution was cooled to 0 °C with an ice-bath, then collidine (123 mg, 1.016 mmol) and trimethysilyl bromide (155 mg, 0.1016 mmol) were added slowly by syringe. The reaction was stirred overnight at room temperature. The reaction was concentrated under reduced pressure and toluene was added and then removed under reduced pressure three times. The white solid that resulted was dissolved in NaOH (1 M aqueous solution, 0.495 mmol) and stirred at room temperature overnight. Acetone (1 mL) was added and the flask was placed in a −20 °C freezer overnight to promote precipitation of unreacted sodium hydroxide. The solution was filtered and the acetone was removed under reduced pressure. The resulting aqueous solution was washed with EtOAc (3 × 3 mL) and the water was evaporated using a lyophilizer to afford the sodium salt 10 as a white solid (51.3 mg, 82%): 1H NMR (500 MHz, D2O) δ 5.43 (t, J = 7.2 Hz, 1H), 3.92 (s, 2H), 2.02 (q, J = 7.2 Hz, 2H), 1.60 (s, 3H), 1.57 – 1.43 (m, 4H), 1.42 – 1.36 (m, 2H); 13C NMR (126 MHz, D2O) δ 134.4 (s), 126.2 (d, J = 264.7 Hz), 67.8 (s), 30.1 (d, J = 16.8 Hz), 27.6 (d, J = 133.2 Hz), 22.7 (d, J = 4.5 Hz), 18.4 (s), 13.0 (s); 31P NMR (202 MHz, D2O) δ 26.8; HRMS (ES+) calc. for C8H17O4P [M− - H] 207.0786, found 207.0793.
(E)-(((7-hydroxy-6-methylhept-5-en-1-yl)phosphoryl)bis(oxy))bis(methylene) bis(2,2-dimethylpropanoate) (11).
Compound 14 (72 mg, 0.1712 mmol) was dissolved in CH2Cl2 (2 mL) and selenium dioxide (11 mg, 0.10 mmol) followed by tert-butyl hydroperoxide (70% aq. soln., 0.13 mL, 1.010 mmol) were added. The solution was stirred vigorously at room temperature and monitored by TLC (silica gel, 1:1 EtOAc:hexanes) until starting material was consumed, about 24 hours. The reaction was quenched with NaHCO3 (sat’d aq. solution), diluted with water and organics were extracted with CH2Cl2. Organics were dried (MgSO4), combined and concentrated under reduced pressure. The residue which resulted was purified by flash column chromatography (silica gel; 15–40% EtOAc in hexanes gradient) to afford prodrug 11 as a colorless oil (23.2 mg, 31% yield): 1H NMR (500 MHz, CDCl3) δ 5.71 (d, J = 13.0 Hz, 4H), 5.42 (t, J = 7.2 Hz, 1H), 4.04 (d, J = 5.2 Hz, 1H), 2.09 (q, J = 7.3 Hz, 2H), 1.91 – 1.82 (m, 2H), 1.70 (s, 3H), 1.68 – 1.63 (m, 2H), 1.49 (dt, J = 15.2, 7.5 Hz, 2H), 1.28 (s, 18H); 13C NMR (126 MHz, CDCl3) δ 177.0 (s, 2C), 135.5 (s), 125.2 (s), 81.3 (d, J = 6.2 Hz, 2C), 68.9 (s), 38.8 (s, 2C), 30.2 (d, J = 17.6 Hz), 27.0 (d, J = 6.8 Hz), 26.9 (s, 6C), 25.9 (s), 21.6 (d, J = 5.6 Hz), 13.7 (s); 31P NMR (202 MHz, CDCl3) δ 33.2; HRMS (ES+) calc. for C20H38O8P [M+ + H] 437.2304, found 437.2318.
Dimethyl (6-methylhept-5-en-1-yl) phosphonate (13).
Compound 12 (0.68 mL, 4.348 mmol) was dissolved in THF (anhydrous, 20 mL) and cooled to −78 °C in an oven-dried flask under argon. Then n-butyllithium (2.5 M in hexanes, 1.505 mL, 3.756 mmol) was added dropwise and stirred for 10 minutes. After 6-bromo-2-methylhex-2-ene34 (700 mg, 3.953 mmol) was added dropwise, the reaction was stirred for 2 hours. The flask was removed from the dry-ice bath and allowed to warm to −20 °C; quenched with NaHCO3 (sat’d aq. solution), diluted with water and extracted with EtOAc. Combined organics were washed with brine, dried (Na2SO4), and concentrated under reduced pressure. The residue which resulted was purified by flash chromatography (silica gel; 70–100% EtOAc in hexanes) to afford 13 as a colorless oil (523.6 mg, 63% yield): 1H NMR (500 MHz, CDCl3) δ 5.13 (t, J = 7.2 Hz, 1H), 3.78 (d, J = 10.7 Hz, 6H), 2.03 (q, J = 7.3 Hz, 2H), 1.82 – 1.74 (m, 2H), 1.73 (s, 3H), 1.69 – 1.59 (m, 6H), 1.64 (s, 3H), 1.49 – 1.42 (m, 2H); 13C NMR (126 MHz, CDCl3) δ 131.9 (s), 124.0 (s), 52.2 (d, J = 6.5 Hz), 30.8 (d, J = 16.8 Hz), 27.5 (s), 25.7 (s), 24.7 (d, J = 140.3 Hz), 22.0 (d, J = 5.2 Hz), 17.7 (s); 31P NMR (202 MHz, CDCl3) δ 35.1; HRMS (ES+) calc. for C10H22O3P [M+ + H] 221.1307, found 221.1311.
(((6-Methylhept-5-en-1-yl) phosphoryl) bis(oxy))bis(methylene) bis(2,2-dimethylpropanoate) (14).
In an oven-dried flask, phosphonate 13 (200 mg, 0.9081 mmol) was dissolved in CH3CN (anhydrous, 8 mL) under argon and sodium iodide (476 mg, 3.178 mmol) and chloromethyl pivalate (0.46 mL, 3.178 mmol) were added. Reaction mixture was heated to reflux and stirred overnight. Once the starting material was consumed, reaction mixture was cooled to room temperature, diluted with water and organics extracted with EtOAc. Combined organics were washed with brine, dried (Na2SO4), and concentrated under reduced pressure. The oil which resulted was purified by flash column chromatography (silica gel; 5–15% EtOAc gradient in hexanes) to afford the POM-protected 14 as a colorless oil (80.6 mg, 21% yield): 1H NMR (500 MHz, CDCl3) δ 5.71 (d, J = 12.9 Hz, 4H), 5.12 (t, J = 6.5 Hz, 1H), 2.01 (dd, J = 14.8, 7.4 Hz, 2H), 1.91 – 1.82 (m, 2H), 1.72 (s, 3H), 1.67 – 1.60 (m, 2H), 1.63 (s, 3H), 1.48 – 1.41 (m, 2H), 1.28 (s, 18H).; 13C NMR (126 MHz, CDCl3) δ 176.9 (s, 2C), 132.0 (s), 123.8 (s), 81.3 (d, J = 6.2 Hz, 2C), 38.8 (s, 2C), 30.7 (d, J = 17.5 Hz), 27.5 (s), 27.1 (s), 26.9 (s, 6C), 25.9 (d, J = 38.6 Hz), 21.7 (d, J = 5.2 Hz), 17.7 (s); 31P NMR (202 MHz, CDCl3) δ 33.5; HRMS (ES+) calc. for C10H22O4P [M+ + H] 237.1256, found 237.1268.
Dimethyl (E)-(7-hydroxy-6-methylhept-5-en-1-yl) phosphonate (15).
Phosphonate 13 (200 mg, 0.9081 mmol) was dissolved in CH2Cl2 (2 mL) and selenium dioxide (76 mg, 0.6810 mmol) followed by tert-butyl hydroperoxide (70% aq. soln., 0.47 mL, 3.632 mmol) were added. The reaction was stirred vigorously at room temperature and monitored by TLC (silica gel, 100% EtOAc) until starting material was consumed, about 24 hours. Reaction was quenched with NaHCO3 (sat’d aq. solution), diluted with water and extracted with CH2Cl2. The extracts were dried (MgSO4), combined, and concentrated under reduced pressure. The oil which resulted was dissolved in CH3OH (8 mL) and cooled to 0 °C. Sodium borohydride (51.5 mg, 1.362 mmol) was added in portions and the reaction mixture was allowed to stir for 1.5 hours. The reaction was quenched with NaHCO3 (sat’d aq. solution), diluted with water and the organics were extracted with EtOAc. The combined organics were washed with brine, dried (Na2SO4), and concentrated under reduced pressure. The residue which resulted was purified by flash column chromatography (silica gel; 100% EtOAc) to afford the alcohol 15 as a colorless oil (72.9 mg, 34% yield): 1H NMR (500 MHz, CDCl3) δ 5.44 (t, J = 7.1 Hz, 1H), 4.05 (s, 3H), 3.78 (d, J = 10.7 Hz, 6H), 2.10 (dd, J = 14.4, 7.1 Hz, 2H), 1.86 – 1.74 (m, 2H), 1.71 (s, 3H), 1.72 – 1.61 (m, 2H), 1.54 – 1.46 (m, 2H); 13C NMR (126 MHz, CDCl3) δ 135.3 (s), 125.5 (s), 68.9 (s), 52.3 (d, J = 6.5 Hz), 30.4 (d, J = 16.7 Hz), 27.1 (s), 24.6 (d, J = 140.7 Hz), 22.0 (d, J = 5.1 Hz), 13.7 (s); 31P NMR (202 MHz, CDCl3) δ 35.0; HRMS (ES+) calc. for C20H38O7P [M+ + H] 421.2355, found 421.2340.
Reagents and Supplies.
HMBPP (Cat: I-M055), IPP (Cat: I-0050), and DMAPP (Cat: I-0051) were purchased from Echelon (Salt Lake City, UT, USA). C-HMBPP (Cat. 13151) was purchased from Cayman Chemical (Ann Arbor, MI, USA). Probe (BODIPY FL GDP, bis(triethylammonium) salt) was purchased from ThermoFisher Scientific (ref G22360). POM2-CC-HMBPP, CC-HMBPP, and POM2-C-HMBP were gifts from Prof. David Wiemer at the University of Iowa (Iowa City, IA, USA). C-HMBP was synthesized as previously described.7
Modeling of BTN3A1 ligand binding.
Molecular docking was carried out by Glide in the Schrodinger suite as previously described.33
T cell proliferation.
T cell proliferation assays were carried out as previously described.7, 29
Ligand Uptake.
Compounds POM2-C-HMBP (6), and POM2-C-HMHP (11) or control (T cell media) were added separately to PBMCs (1,000,000 cells/mL) and at a final concentration of 10 μM in 100 μL cells suspended in media in a 96-well plate. Aliquots were incubated at 37 °C with time points of 0, 6, 24, and 48 hours. At each time point, 100 μL of sample was collected, spun (600 rcf, 3 minutes), and 80 μL of the supernatant was collected. Acetonitrile (320 μL) was added to the supernatant and the samples were vortexed and spun (10,000 rcf, 2 minutes); 300 μL of the supernatant was collected and stored at −20 °C until analysis. Analysis was completed using UPLC-MS (Waters Xevo G2-XS QTOF-MS) using a C18 column. The gradient ranged from 25% acetonitrile to 80% acetonitrile over 8 minutes and was held for 1 minute before re-equilibration. Integrated value of the molecular ion is reported relative to control (t = 0).
ELISA.
Unstimulated human PBMCs (200 μL at 1,000,000 cells/mL) were incubated with test compounds at varying concentrations or control (T cell media) with IL-2 (5 ng/mL) for 48 hours in duplicate. Cell supernatants were analyzed for IFN-γ by ELISA according to the manufacturer’s protocol (BioLegend) with a standard curve.35
Protein Expression, Purification, and Mutagenesis.
The wt-BFI and B30.2 domains were expressed and purified as previously described.7 Specific point mutations to the wt-BFI construct (H381R, W421A, M424A, R442A, R448A, and R499A) were generated using the QuikChange (Stratagene) protocol with mutagenesis primers that were designed using PrimerX. The following primers were used: BTN3A1 BFI(H381R) Forward 5ʹ- GAG-AGA-TTT-AAT-TGG-CGT-TAT-TGT-GTT-CTC-GGC-3ʹ; BTN3A1 BFI(H381R) Reverse 5ʹ- GCC-GAG-AAC-ACA-ATA-ACG-CCA-ATT-AAA-TCT-CTC-3ʹ; BTN3A1 BFI(W421A) Forward 5ʹ-GTG-CAG-AGA-AAA-GGC-GCG-GTC-AAA-ATG-ACA-C-3ʹ; BTN3A1 BFI(W421A) Reverse 5ʹ-GTG-TCA-TTT-TGA-CCG-CGC-CTT-TTC-TCT-GCA-C-3ʹ; BTN3A1 BFI(M424A) Forward 5ʹ-GAG-AAA-AGG-CTG-GGT-CAA-AGC-CAG-ACC-TGA-GAA-TGG-ATT-C-3ʹ; BTN3A1 BFI(M424A) Reverse 5ʹ-GAA-TCC-ATT-CTC-AGG-TGT-CGC-TTT-GAC-CCA-GCC-TTT-TCT-C- 3ʹ; BTN3A1 BFI(R442A) Forward 5ʹ-CTG-ATG-GGA-ATA-AGT-ATG-CGA-CTC-TAA-CTG-AGC-CC-3ʹ; BTN3A1 BFI(R442A) Reverse 5ʹ-GGG-CTC-AGT-TAG-AGT-CGC-ATA-CTT-ATT-CCC-ATC-AG-3ʹ; BTN3A1 BFI(R448A) Forward 5ʹ-GAC-TCT-AAC-TGA-GCC-CGC-GAC-CAA-CCT-GAA-ACT-TC-3ʹ; BTN3A1 BFI(R448A) Reverse 5ʹ-GAA-GTT-TCA-GGT-TGG-TCG-CGG-GCT-CAG-TTA-GAG-TC-3ʹ; BTN3A1 BFI(R499A) Forward 5ʹ-CTC-TAT-ATC-CTG-TTT-TCG-CGA-TTT-TGA-CCT-TGG-AGC-CC-3ʹ; BTN3A1 BFI(R499A) Reverse 5ʹ-GGG-CTC-CAA-GGT-CAA-AAT-CGC-GAA-AAC-AGG-ATA-TAG-AG-3ʹ. All novel mutations were confirmed by Sanger Sequencing (Eurofins) of the entire open reading frame using a standard T7 terminator primer.
Butyrophilin Binding Determination by Fluorescence Polarization (FP).
Experiments were carried out in 384-well black microplates (Falcon, ref 353962). Conditions were optimized by varying reaction volume, protein concentration and probe concentration as described in the text. The final assay solution (25 μL) for testing compounds and point mutations consisted of buffer (100 mM NaCl, 50 mM Tris, and 5 mM beta-mercaptoethanol at pH 7.5), 10 μM protein (wt-BFI, B30.2, BFI(H381R), BFI(W421A), BFI(M424A), BFI(R442A), BFI(R448A), or BFI(R499A)), and 750 nM GDP-probe. Competitive inhibition studies were performed with the addition of a test compound at varying concentrations. Following set up, plates were incubated in the dark for 60 minutes and fluorescence polarization was detected at an emission / excitation wavelength of 535 / 485 nm (Perkin Elmer VICTOR X5) in three experimental replicates and repeated in at least 3 independent experiments. Probe in buffer was used as a negative control, while probe incubated with protein was used as a positive control.
Statistical Analysis.
Graphs represent mean of indicated number of independent experiments (n). Experiments were replicated at least 3 times (n = 3). Nonlinear regression was used to determine IC50 values, using a 4-parameter model in GraphPad Prism.
Circular Dichroism (CD).
CD measurements were performed on a Jasco J-815 Spectrometer using a Hellma 1.0 mm path-length QS cuvette at 20 °C from wavelengths 260 – 200 nm. Spectra of the proteins were conducted at 3 μM in a buffer consisting of 10 mM K3PO4 and 50 mM Na2SO4 at pH 7.5.
Supplementary Material
Acknowledgments
We thank Dr. Olga Vinogradova at the University of Connecticut for assistance with protein purification. We appreciate the assistance of Dr. Jeremy Balsbaugh and the University of Connecticut Proteomics & Metabolomics Facility with the LCMS analysis. M.M.P. was supported by an American Cancer Society - Kirby Foundation Postdoctoral Fellowship, PF-18-119-01-LIB. A.J.W. was supported by the National Institutes of Health under Award Number R01CA186935 and the Herman Frasch Foundation for Chemical Research, Bank of America, N.A., Trustee (HF17).
Abbreviations Used
- BFI
BTN3A1 full intracellular domain
- FP
fluorescence polarization
- HMBPP
(E)-4-hydroxy-3-methyl-but-2-enyl diphosphate
- IPP
isopentenyl diphosphate
- ITC
isothermal titration calorimetry
- JM
juxtamembrane
- MEP
2-C-methyl-ᴅ-erythritol 4-phosphate
- PBMC
peripheral blood mononuclear cell
- POM
pivaloyloxymethyl
Footnotes
Supporting Information
The supporting information is available free of charge via the Internet at http://pubs.acs.org.
T cell proliferation data for compounds 9, 10, 14, 15; cellular uptake of compounds 6 and 11; ELISA assay data for compounds 4, 5, 7, 10, and sodium diphosphate; FP assay data for sodium diphosphate and compounds 1, 5, and 10; probe MP values; CD spectra for all seven BFI constructs; Raw FP data of compounds 5 and 10 against six BFI constructs (PDF)
Molecular Formula Strings (CSV)
A.J.W. owns shares in Terpenoid Therapeutics, Inc. The current work did not involve the company. The other authors have no financial conflicts of interest.
REFERENCES
- 1.Harly C; Guillaume Y; Nedellec S; Peigne C-M; Monkkonen H; Monkkonen J; Li J; Kuball J; Adams EJ; Netzer S; Dechanet-Merville J; Leger A; Hermann T; Breathnach R; Olvie D; Bonneville M; Scotet E Key Implications of Cd277/Butyrophilin-3 (Btn3a) in Cellular Stress Sensing by a Major Human Гδ T-Cell Subset. Blood. 2012, 120, 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H; Henry O; Distefano MD; Wang Y-C; Raikkonen J; Monkkonen J; Tanaka Y; Morita CT Butyrophilin 3a1 Plays an Essential Role in Prenyl Pyrophosphate Stimulation of Vγ2vδ2 T Cells. J. Immunol 2013, 191, 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonneville M; O’Brien RL; Born WK Гδ T Cell Effector Functions: A Blend of Innate Programming and Acquired Plasticity. Nat. Rev. Immunol 2010, 10, 467–478. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes DA; Reith W; Trowsdale J Regulation of Immunity by Butyrophilins. Annu. Rev. Immunol 2016, 34, 151–172. [DOI] [PubMed] [Google Scholar]
- 5.Arnett HA; Viney JL Immune Modulation by Butyrophilins. Nat. Rev. Immunol 2014, 14, 559–569. [DOI] [PubMed] [Google Scholar]
- 6.Sandstrom A; Peigne C-M; Leger A; Crooks JE; Konczak F; Gesnel M-C; Breathnach R; Bonneville M; Scotet E; Adams EJ The Intracellular B30.2 Domain of Butyrophilin 3a1 Binds Phosphoantigens to Mediate Activation of Human Vγ9vδ2 T Cells. Immunity. 2014, 40, 409–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsiao CHC; Lin X; Barney RJ; Shippy RR; Li J; Vinogradova O; Wiemer DF; Wiemer AJ Synthesis of a Phosphoantigen Prodrug That Potently Activates Vγ9vδ2 T-Lymphocytes. Chem. Biol 2014, 21, 945–954. [DOI] [PubMed] [Google Scholar]
- 8.Wang H; Morita CT Sensor Function for Butyrophilin 3a1 in Prenyl Pyrophosphate Stimulation of Human Vgamma2vdelta2 T Cells. J. Immunol 2015, 195, 4583–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen K; Li J; Puthenveetil R; Lin X; Poe MM; Hsiao CC; Vinogradova O; Wiemer AJ The Butyrophilin 3a1 Intracellular Domain Undergoes a Conformational Change Involving the Juxtamembrane Region. FASEB J. 2017, 31, 4697–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salim M; Knowles TJ; Baker AT; Davey MS; Jeeves M; Sridhar P; Wilkie J; Willcox CR; Kadri H; Taher TE; Vantourout P; Hayday A; Mehellou Y; Mohammed F; Willcox BE Btn3a1 Discriminates Гδ T Cell Phosphoantigens from Nonantigenic Small Molecules Via a Conformational Sensor in Its B30.2 Domain. ACS Chem. Biol 2017, 12, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu S; Sachleben JR; Boughter CT; Nawrocka WI; Borowska MT; Tarrasch JT; Skiniotis G; Roux B; Adams EJ Phosphoantigen-Induced Conformational Change of Butyrophilin 3a1 (Btn3a1) and Its Implication on Vγ9vδ2 T Cell Activation. Proc. Natl. Acad. Sci 2017, 114, E7311–E7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilcollins AM; Li J; Hsiao CHC; Wiemer AJ Hmbpp Analog Prodrugs Bypass Energy-Dependent Uptake to Promote Efficient Btn3a1-Mediated Malignant Cell Lysis by Vγ9vδ2 T Lymphocyte Effectors. J. Immunol 2016, 197, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gober H-J; Kistowska M; Angman L; Jeno P; Mori L; De Libero G Human T Cell Receptor Гδ Cells Recognize Endogenous Mevalonate Metabolites in Tumor Cells. J. Exp. Med 2003, 197, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santolaria T; Robard M; Leger A; Catros V; Bonneville M; Scotet E Repeated Systemic Administrations of Both Aminobisphosphonates and Human Vγ9vδ2 T Cells Efficiently Control Tumor Development in Vivo. J. Immunol 2013, 191, 1993–2000. [DOI] [PubMed] [Google Scholar]
- 15.Wiemer DF; Wiemer AJ Opportunities and Challenges in Development of Phosphoantigens as Vγ9vδ2 T Cell Agonists. Biochem. Pharmacol 2014, 89, 301–312. [DOI] [PubMed] [Google Scholar]
- 16.Belmant C; Espinosa E; Halary F; Tang Y; Peyrat M-A; Sicard H; Kozikowski A; Buelow R; Poupot R; Bonneville M; Fournie J-J A Chemical Basis for Recognition of Nonpeptide Antigens by Human Гδ T Cells. FASEB J. 2000, 14, 1669. [DOI] [PubMed] [Google Scholar]
- 17.Boedec A; Sicard H; Dessolin J; Herbette G; Ingoure S; Raymond C; Belmant C; Kraus J-L Synthesis and Biological Activity of Phosphonate Analogues and Geometric Isomers of the Highly Potent Phosphoantigen (E)-1-Hydroxy-2-Methylbut-2-Enyl 4-Diphosphate. J. Med. Chem 2008, 51, 1747–1754. [DOI] [PubMed] [Google Scholar]
- 18.Breccia P; Angeli F; Cerbara I; Topai A; Auricchio G; Nartino A; Colizzi V; Poccia F Thiopyrophosphoantigens: Solid-Phase Synthesis and in Vitro Characterization of a New Class of Vγ9 Vδ2 T Cells Activators. J. Med. Chem 2009, 52, 3716–3722. [DOI] [PubMed] [Google Scholar]
- 19.Song Y; Zhang Y; Wang H; Raker AM; Sanders JM; Broderick E; Clark A; Morita CT; Oldfield E Synthesis of Chiral Phosphoantigens and Their Activity in Гδ T Cell Stimulation. Bioorg. Med. Chem. Lett 2004, 14, 4471–4477. [DOI] [PubMed] [Google Scholar]
- 20.Reichenberg A; Hintz M; Kletschek Y; Kuhl T; Haug C; Engel R; Moll J; Ostrovsky DN; Jomaa H; Eberl M Replacing the Pyrophosphate Group of Hmb-Pp by a Diphosphonate Function Abrogates Its Potential to Activate Human Гδ T Cells but Does Not Lead to Competitive Antagonism. Bioorg. Med. Chem. Lett 2003, 13, 1257–1260. [DOI] [PubMed] [Google Scholar]
- 21.Davey MS; Malde R; Mykura RC; Baker AT; Taher TE; Le Duff CS; Willcox BE; Mehellou Y Synthesis and Biological Evaluation of (E)-4-Hydroxy-3-Methylbut-2-Enyl Phosphate (Hmbp) Aryloxy Triester Phosphoramidate Prodrugs as Activators of Vγ9/Vδ2 T-Cell Immune Responses. J. Med. Chem 2018, 61, 2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberl M; Hintz M; Reichenberg A; Kollas A-K; Wiesner J; Jomaa H Microbial Isoprenoid Biosynthesis and Human Гδ T Cell Activation. FEBS Letters. 2003, 544, 4–10. [DOI] [PubMed] [Google Scholar]
- 23.Morita CT; Jin C; Sarikonda G; Wang H Nonpeptide Antigens, Presentation Mechanisms, and Immunological Memory of Human Vγ9vδ2 T Cells: Discriminating Friend from Foe through the Recognition of Prenyl Pyrophosphate Antigens. Immunol. Rev 2007, 215, 59–76. [DOI] [PubMed] [Google Scholar]
- 24.Amslinger S; Hecht S; Rohdich F; Eisenreich W; Adam P; Bacher A; Bauer S Stimulation of Vγ9/Vδ2 T-Lymphocyte Proliferation by the Isoprenoid Precursor, (E)-1-Hydroxy-2-Methyl-but-2-Enyl 4-Diphosphate. Immunobiology. 2007, 212, 47–55. [DOI] [PubMed] [Google Scholar]
- 25.Sicard H; Ingoure S; Luciani B; Serraz C; Fournie J-J; Bonneville M; Toillier J; Romange F In Vivo Immunomanipulation of Vγ9vδ2 T Cells with a Synthetic Phosphoanitgen in a Preclinical Nonhuman Primate Model. J. Immunol 2005, 175, 5471–5480. [DOI] [PubMed] [Google Scholar]
- 26.Thompson K; Rojas-Navea J; Rogers MJ Alkylamines Cause Vγ9vδ2 T-Cell Activation and Proliferation by Inhibiting the Mevalonate Pathway. Blood. 2006, 107, 651–654. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh S; Chan JMW; Lea CR; Meints GA; Lewis JC; Tovian ZS; Flessner RM; Loftus TC; Bruchhaus I; Kendrick H; Croft SL; Kemp RG; Kobayashi S; Nozaki T; Oldfield E Effects of Bisphosphonates on the Growth of Entamoeba Histolytica and Plamodium Species in Vitro and in Vivo. J. Med. Chem 2004, 47, 175–187. [DOI] [PubMed] [Google Scholar]
- 28.Wiemer AJ; Shippy RR; Kilcollins AM; Li J; Hsiao CHC; Barney RJ; Geng ML; Wiemer DF Evaluation of a 7-Methoxycoumarin-3-Carboxylic Acid Ester Derivative as a Fluorescent, Cell-Cleavable, Phosphonate Protecting Group. ChemBioChem. 2016, 17, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foust BJ; Poe MM; Lentini NA; Hsiao C-HC; Wiemer AJ; Wiemer DF Mixed Aryl Phosphonate Prodrugs of a Butyrophilin Ligand. ACS Med. Chem. Lett 2017, 8, 914–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lentini NA; Foust BJ; Hsiao C-HC; Wiemer AJ; Wiemer DF Phosphonamidate Prodrugs of a Butyrophilin Ligand Display Plasma Stability and Potent Vγ9vδ2 T Cell Stimulation. J. Med. Chem 2018, 61, 8658–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes DA; Chen H-C; Price AJ; Keeble AH; Davey MS; James LC; Eberl M; Trowsdale J Activation of Human Гδ T Cells by Cytosolic Interactions of Btn3a1 with Soluble Phosphoantigens and the Cytoskeletal Adaptor Periplakin. J. Immunol 2015, 194, 2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi AM; Taylor CW Analysis of Protein-Ligand Interactions by Fluorescence Polarization. Nat. Protoc 2011, 6, 365–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shippy RR; Lin X; Agabiti SS; Li J; Zangari BM; Foust BJ; Poe MM; Hsiao CHC; Vinogradova O; Wiemer DF; Wiemer AJ Phosphinophosphonates and Their Tris-Pivaloyloxymethyl Prodrugs Reveal a Negatively Cooperative Butyrophilin Activation Mechanism. J. Med. Chem 2017, 60, 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wills VS; Metzger JI; Allen C; Varney ML; Wiemer DF; Holstein SA Bishomoisoprenoid Triazole Bisphosphonates as Inhibitors of Geranylgeranyl Diphosphate Synthase. Bioorg. Med. Chem 2017, 25, 2437–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiao CC; Wiemer AJ A Power Law Function Describes the Time- and Dose-Dependency of Vgamma9vdelta2 T Cell Activation by Phosphoantigens. Biochem. Pharmacol 2018, 158, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattarei A; Enzinger M; Gu S; Karunakaran MM; Kimmel B; Berner N; Adams EJ; Herrmann T; Amslinger S A Photo-Crosslinkable Biotin Derivative of the Phosphoantigen (E)-4-Hydroxy-3-Methylbut-2-Enyl Diphosphate (Hmbpp) Activates Vγ9vδ2 T Cells and Binds to the Hmbpp Site of Btn3a1. Chem. Eur. J 2017, 23, 11945–11954. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y; Li L; Yuan L; Zhou X; Duan J; Xiao H; Cai N; Han S; Ma X; Liu W; Cehn C-C; Wang L; Li X; Chen J; Kang N; Chen J; Shen Z; Malwal SR; Liu W; Shi Y; Oldfield E; Guo R-T; Zhang Y A Structural Change in Butyrophilin Upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated Vγ9vδ2 T Cell Activation. Immunity. 2019, 50, 1043–1053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


