Cryptococcus yeast species typically display characteristics of opportunistic pathogens, with the exception of C. gattii, which can cause life-threatening respiratory and disseminated brain infections in otherwise healthy people. The pathogenesis of C. gattii is not well understood, but an important characteristic is that C. gattii is capable of evading host cell-mediated immune defenses initiated by DCs. Here, we report that when virulent C. gattii becomes ingested by a DC, the intracellular compartment containing the fungi is covered by a persistent protein cage structure consisting of F-actin. This F-actin cage acts as a barrier to prevent interaction with other intracellular compartments, and as a result, the DC fails to kill the fungi and activate important cell-mediated immune responses. We propose that this unique immune evasion mechanism permits C. gattii to remain unchallenged within host cells, leading to persistent infection.
KEYWORDS: Cryptococcus gattii, dendritic cells, immune evasion, immunoparalysis, phagosomal F-actin
ABSTRACT
Cryptococcus gattii is a major cause of life-threatening mycosis in immunocompetent individuals and responsible for the ongoing epidemic outbreak of cryptococcosis in the Pacific Northwest of North America. This deadly fungus is known to evade important host immune responses, including dendritic cell (DC) maturation and concomitant T cell immunity, via immune evasion mechanisms that remain unclear. Here, we demonstrate that primary human DCs phagocytose C. gattii but the maturation of phagosomes to phagolysosomes was blocked as a result of sustained filamentous actin (F-actin) that entrapped and concealed the phagosomes from recognition. Superresolution structured illumination microscopy (SR-SIM) revealed that the persistent phagosomal F-actin formed a cage-like structure that sterically hindered and functionally blocked the fusion of lysosomes. Blocking lysosome fusion was sufficient to inhibit phagosomal acidification and subsequent intracellular fungal killing by DCs. Retention of phagosomal F-actin by C. gattii also caused DC immunoparalysis. Disrupting the retained F-actin cage with cytochalasin D not only restored DC phagosomal maturation but also promoted DC costimulatory maturation and robust T cell activation and proliferation. Collectively, these results reveal a unique mechanism of DC immune evasion that enhances intracellular fungal pathogenicity and may explain suppressed cell-mediated immunity.
INTRODUCTION
The emergence of Cryptococcus gattii, a basidiomycete yeast, as a novel respiratory pathogen began on the west coast of Canada during the Vancouver Island outbreak nearly 2 decades ago. The outbreak, which resulted from a hypervirulent lineage of C. gattii (1), has spread to the surrounding regions encompassing the Pacific Northwest (PNW) of the United States, where there is now established endemicity among healthy residents and travelers (2, 3). In contrast, individuals with a compromised immune system are at an increased risk of becoming infected by the related opportunistic pathogen Cryptococcus neoformans (4, 5). Although C. gattii can also infect this susceptible population (6, 7), it is notorious for infecting individuals with intact immunity. The mortality rate associated with C. gattii infection remains intolerably high (up to 33%) despite the best available care (8). Consequently, endemic C. gattii continues to be a serious public health threat with a propensity for infecting the healthy population.
Virulence and evasion of host immunity are key determinants of C. gattii pathogenesis in the PNW outbreak. Infection by hypervirulent C. gattii (represented by strain R265), the major clinical isolate of the PNW outbreak, causes a failure in the early activation of inflammatory immune responses (9, 10) and the induction of protective Th1 and Th17 cells by subverting dendritic cell (DC) immune activation (11). This is consistent with protective host defense against other cryptococcal species, which are dependent on T cell-mediated immunity (5, 12, 13). Additionally, DCs fail to undergo maturation and activate T cells in response to hypervirulent C. gattii, further supporting the importance of DC-mediated immunity (14, 15). However, the mechanism of DC immune evasion is unclear.
DCs are sentinels of the immune system and have a crucial role in orchestrating T cell responses (16). During an infection, resting or immature DCs (iDCs) phagocytose invading pathogens and undergo a maturation program (17), leading to DC activation, which results in a number of cellular changes: (i) increased antigen (Ag) presentation, (ii) increased expression of costimulatory ligands such as CD80 and CD86 (18), and (iii) the production of inflammatory cytokines, such as tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6), which are all important triggers of T cell activation. Intracellularly, this occurs via a cascade of fusion events between the phagosome and the endosomal and lysosomal vesicles, a process called phagosomal maturation (19). Phagosomal maturation is essential for the killing and breakdown of the engulfed microbes to facilitate antigen processing and presentation in DCs (20, 21).
The actin cytoskeleton plays a pivotal role in phagocytosis and subsequently in phagosomal processing. Nascent phagosomes are covered with F-actin as a result of actin accumulation during phagocytosis. This F-actin coat depolymerizes almost immediately after phagocytosis, which is a crucial step preceding phagosomal maturation (22, 23). It has been reported that in macrophages, intracellular pathogens like Salmonella and Leishmania can build F-actin on their phagosomes to escape intracellular killing (24, 25). Moreover, phagocytosis of C. neoformans by macrophages has been shown to generate waves of transient phagosomal F-actin, which is referred to as the “actin flash” (26). Interestingly, the C. neoformans actin flash seems to act as a mechanism of macrophage cell defense against nonlytic exocytosis (or vomocytosis) of the pathogenic yeast, a mechanism that may contribute to C. neoformans in vivo dissemination (26). However, it is not known if the actin flash can occur in DCs containing C. neoformans or C. gattii. Furthermore, it is unknown to what degree phagosomal actin rearrangements affect DC function as antigen-presenting cells (APCs).
Here, we examined the impacts of phagosomal F-actin on the DC immune response to hypervirulent C. gattii. We show that PNW C. gattii isolates, especially hypervirulent C. gattii, retain a persistent phagosomal F-actin structure, which we call an F-actin cage. The F-actin cage blocked DC phagosomal maturation, resulting in higher intracellular fungal loads. Furthermore, suppression of the DC immune response was facilitated by the F-actin cage. Therefore, our data suggest that retention of phagosomal F-actin is important in the mechanism of DC immunoparalysis caused by hypervirulent C. gattii infection.
RESULTS
Hypervirulent Cryptococcus gattii suppresses DC immunity.
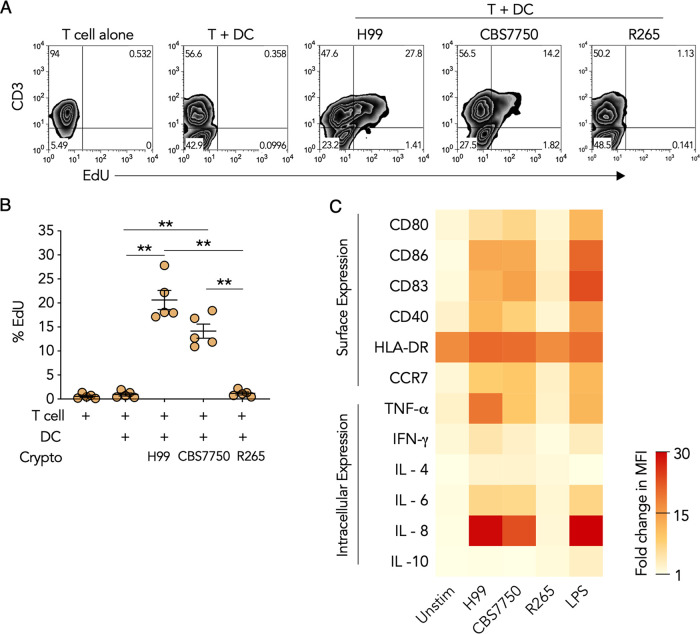
The main function of DCs is to activate T cells (27). DC-mediated T cell activation is dependent on both antigen processing and presentation as well as costimulation by APCs (28, 29). Hypervirulent C. gattii has been demonstrated to suppress DC maturation, but the mechanism and extent to which it affects different aspects of DC functions relative to other cryptococcal strains have not been comprehensively studied. To study DC suppression, we first assessed T cell activation and costimulation in DCs infected with hypervirulent C. gattii in comparison to virulent C. neoformans and avirulent C. gattii. Human monocyte-derived DCs were infected with hypervirulent C. gattii R265; C. neoformans H99, a virulent clinical isolate; and C. gattii CBS7750, an avirulent environmental isolate (30). Infected DCs were subsequently cultured with autologous T cells, and proliferation was measured by 5-ethynyl-2-deoxyuridine (EdU) uptake, which is a marker of DNA replication (31). We observed that DCs infected with CBS7750 or H99 induced robust T cell proliferation as indicated by increased EdU incorporation in CD3+ T cells (Fig. 1A and B). However, DCs infected with R265 failed to induce T cell proliferation (Fig. 1A and B).
FIG 1.
Hypervirulent Cryptococcus gattii suppresses DC immunity. (A) Representative flow cytometry plot of EdU incorporation (measure of cell proliferation) in peripheral blood T cells cultured for 6 days either alone, with resting DCs, or with DCs infected with R265, H99, or CBS7750 (6 h). (B) Quantification of data in panel A as percent EdU staining in CD3+ T cells. Data are presented as means ± standard errors of the means (SEM) from 5 donors, shown individually as dots. **, P < 0.001 by one-way ANOVA (adjusted P values). (C) Heat map of the fold changes in the mean fluorescence intensity (MFI) of surface and intracellular markers in resting (Unstim) DCs, Cryptococcus-infected DCs (6 h), and LPS-stimulated DCs (6 h). Data are presented as the average fold changes in MFI, standardized to isotype-matched controls, from 5 different donors. The color spectrum represents the minimum and maximum fold changes in the MFI.
To determine whether the lack of T cell proliferation by autologous DCs infected with hypervirulent C. gattii was associated with changes in DC activation, intracellular and surface markers in DCs were quantified by flow cytometry. In accordance with T cell activation, both CBS7750 and H99 induced proinflammatory activation and costimulatory maturation in DCs (Fig. 1C). In response to strains H99 and CBS7750, DCs increased the expression of the surface costimulatory ligands CD80, CD83, and CD86 as well as the activation receptor CD40 and the DC lymph node homing chemokine receptor CCR7 (Fig. 1C). Furthermore, DC intracellular expression levels of the proinflammatory cytokines TNF-α, interferon gamma (IFN-γ), and IL-6 as well as the neutrophil chemotactic factor IL-8 were increased, but the anti-inflammatory cytokines IL-4 and IL-10 were not (Fig. 1C). In contrast, DCs failed to increase the expression of surface costimulatory markers and intracellular cytokines in response to R265 (Fig. 1C). Together, these results pointed to a defect in the activation and maturation of DCs in response to hypervirulent C. gattii leading to nonfunctioning T cell stimulation.
Hypervirulent Cryptococcus gattii attenuates DC phagosome maturation.
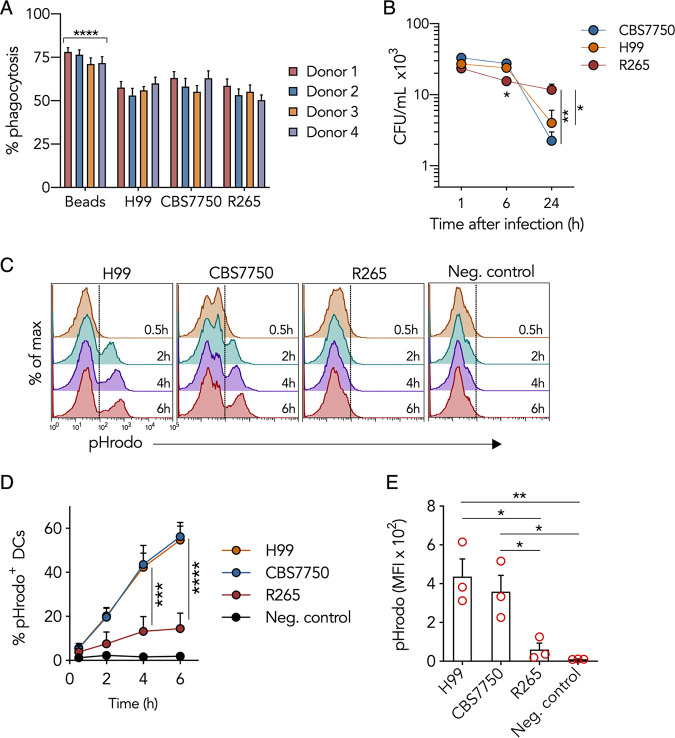
The initial interaction between DCs and Cryptococcus leads to phagocytosis. We considered the possibility that the dramatic differences in DC immune responses to the different cryptococcal strains could be a result of differences in phagocytosis. However, this was not the case since all three cryptococcal species were phagocytosed to similar degrees (Fig. 2A), revealing that hypervirulent C. gattii subverts DC immunity through an alternate mechanism. Phagocytosis rates can sometimes change with adjustments in experimental parameters. Thus, we measured phagocytic rates under different experimental conditions and observed similar results (see Fig. S1A in the supplemental material). The result of phagocytosis is microbial killing through an elaborate process of phagosomal maturation (21). Since the degrees of uptake were similar among the three species of Cryptococcus, we questioned whether intracellular killing by DCs was similar. DCs infected with the different cryptococcal strains were lysed, and the number of viable intracellular cryptococci was quantified as CFU. We observed that the CFU of CBS7750 and H99 decreased by 83% ± 9% and 92% ± 3%, respectively, at 24 h postinfection, while the CFU of R265 decreased by only 48% ± 2% (Fig. 2B), indicating that R265 was not as efficiently killed by DCs as CBS7750 and H99. The magnitudes of initial infection by the different strains were comparable since the CFU immediately (1 h) after infection were similar (Fig. 2B), and the numbers of cryptococcal cells per infected DC were also similar (Fig. S1B). Thus, the reduced killing of R265, despite normal phagocytosis, indicated that the defect in the DC response was likely somewhere along the phagosomal maturation pathway.
FIG 2.
Hypervirulent Cryptococcus gattii attenuates DC phagosomal maturation. (A) Phagocytic frequency 1 h after infection (MOI of 10 for 2 h), measured by confocal microscopy in DCs infected with Cryptococcus or fed polystyrene beads. The frequency was calculated as the number of DCs with internalized cryptococci or beads (confirmed by phalloidin labeling of cortical actin) relative to the total number of DCs in a field of view (FOV). Data are presented as means ± SEM from 25 to 30 FOVs per donor. ****, P < 0.0001 (by one-way ANOVA [adjusted P values] of beads compared to Cryptococcus). (B) Intracellular Cryptococcus survival within DCs at 1 h, 6 h, or 24 h postinfection (MOI of 10 for 2 h). Data are presented as means ± SEM on a log10 scale and are averages from 4 donors. *, P < 0.03; **, P = 0.0062 (by one-way ANOVA [adjusted P values] at each time point). (C) Representative flow cytometric histograms (gated on total DCs) of pHrodo fluorescence at different times after the 2-h infection period. DCs were cultured with pHrodo-labeled cryptococci or given unlabeled beads (negative [Neg.] control) for 2 h. (D) Percent change in pHrodo+ DCs, similar to panel C. Data are presented as means ± SEM from 3 donors. ***, P < 0.001; ****, P < 0.0001 (by two-way ANOVA [adjusted P values]). (E) MFI of pHrodo 6 h after infection in DCs cultured with cryptococci or fed unlabeled beads (negative control). *, P < 0.05; **, P < 0.01 (by one-way ANOVA [adjusted P values]). Data are presented as means ± SEM from 3 donors (represented as dots).
Ratio of Cryptococcus cells per infected DC and effect of culture plates and well size on DC phagocytosis frequency. (A) Phagocytic frequency 1 h after infection (MOI of 10 for 2 h), measured by confocal microscopy, in DCs infected with Cryptococcus or fed polystyrene beads. DCs were infected in either 24-well plastic plates, 24-well plates containing glass coverslips, 96-well plastic plates, or 96-well plates with a glass-like coverslip bottom. The frequency was calculated as the number of DCs with internalized cryptococci or beads (confirmed by phalloidin labeling of cortical actin) relative to the total number of DCs in a field of view (FOV). Data are presented as means ± SEM for 54 to 67 FOVs from 3 donors. ***, P < 0.001 (by one-way ANOVA [adjusted P values] of beads compared to Cryptococcus in each column). (B) Percentage of infected DCs containing 1, 2, or 3 or more intracellular Cryptococcus cells. DCs were infected with Cryptococcus (MOI = 10) for 2 h and washed to rinse excess yeast. Infected DCs were fixed with 4% PFA and permeabilized with saponin 1 h after infection. Fixed cells were labeled with phalloidin and imaged by confocal microscopy. Data represent means ± standard deviations (SD) for 77 to 86 FOVs pooled from 3 donors. Download FIG S1, TIF file, 0.3 MB (299.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We studied phagosomal maturation by measuring changes in DC acidity using the pH-sensitive molecular marker pHrodo, which becomes fluorescent under acidic conditions (32). DCs were infected with pHrodo-labeled Cryptococcus at physiological pH, and fluorescence was measured by flow cytometry at different times after infection, with gating parameters set on DCs (Fig. S2A). The percentage of DCs containing fluorescent pHrodo-labeled CBS7750 or H99 increased in a time-dependent manner until 6 h after infection (Fig. 2C and D). In contrast, the percentage of DCs containing fluorescent pHrodo-labeled R265 was 6- to 7-fold lower than that of DCs containing CBS7750 or H99 6 h after infection (Fig. 2D), indicating that phagosomal acidification of R265 was abnormally low. This difference was not due to potential variation in pHrodo labeling since culturing pHrodo-labeled cryptococci in cell-free acidic (pH 5.5) phosphate-buffered saline (PBS) resulted in equal fluorescence emissions (Fig. S2B and C).
Fluorescence emission from R265, CBS7750, and H99 cryptococcal strains labeled with pHrodo under acidic conditions. (A) Gating strategy for Fig. 2C. The gate (black oval) is set on total DCs as determined by forward scatter (FSC) and side scatter (SSC). (B) Representative flow cytometry histograms (gated on fungi) of cryptococcal strains labeled with pHrodo and cultured in PBS at either pH 7.4 or pH 5.5 for 15 min (room temperature [RT]). The MFI under each condition is shown below the histograms. (C) Quantification of data in panel B. Data are presented as means ± SD of the average MFIs of pHrodo-labeled cryptococci at pH 7.4 and pH 5.5. Averages of data from 3 separate experiments are shown. Download FIG S2, TIF file, 0.6 MB (608.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DC phagosomes containing PNW outbreak strains of Cryptococcus gattii are enriched with an F-actin coat.
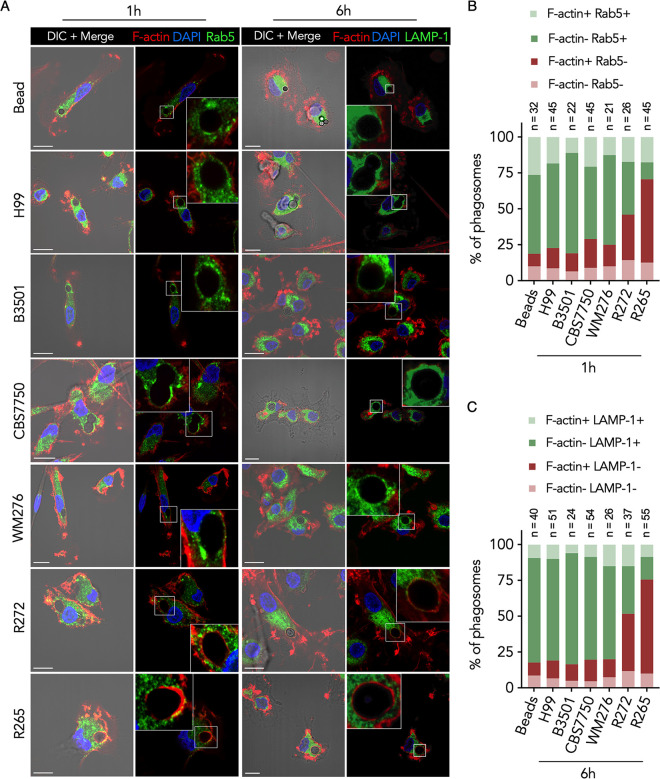
We further studied DC phagosomal maturation of Cryptococcus by immunofluorescence microscopy and measured the acquisition of the small GTPase Rab5 and lysosome-associated membrane protein 1 (LAMP-1) on phagosomes, which are markers of early phagosomal maturation (33) and phagolysosome formation (34), respectively. Consistent with the changes in phagosomal pH, phagosomes containing CBS7750 or H99 were enriched with Rab5 at the early (1 h postinfection) time point (Fig. 3A and B) and LAMP-1 at the late (6 h postinfection) time point (Fig. 3A and C), indicating sequential phagosomal maturation. Surprisingly, phagosomes containing R265 were predominantly absent in Rab5 and LAMP-1 staining and were instead enriched with F-actin at both the early and late time points (Fig. 3A to C), indicating the failure of early endosome fusion and phagolysosome formation combined with F-actin accumulation.
FIG 3.
DC phagosomes containing PNW outbreak strains of Cryptococcus gattii are enriched with an F-actin coat. (A) Confocal micrographs of DCs infected with Cryptococcus or fed polystyrene beads and immunolabeled for Rab5 (green) 1 h or LAMP-1 (green) 6 h after the 2-h infection period. In both cases, coverslips were stained with phalloidin (DyLight 554) to label F-actin (red) and DAPI to stain the nucleus (blue). Images are shown as maximum-intensity projections (MIPs) in the merge panels and as a single confocal stack in the digitally magnified region represented by a white box. DIC, differential interference contrast. Bars = 10 μm. (B) Quantification of the data in panel A at 1 h postinfection. Data are presented as stacked bar graphs of the average proportions of phagosomes per FOV that are enriched for F-actin, Rab5, both, or neither. (C) Quantification of the data in panel A at 6 h postinfection. Data are presented as stacked bar graphs of the average proportions of phagosomes per FOV that are enriched for F-actin, LAMP-1, both, or neither. The number of FOVs (n) is shown under each condition. Data are representative of results from 3 donors.
To determine whether enrichment of phagosomal F-actin might occur with other less virulent strains of C. gattii, we tested additional cryptococcal strains. The clinical strain R272 of C. gattii represents a second less virulent genotype (VGIIb) from the PNW outbreak (35). WM276 is an environmental C. gattii isolate from Australia (VGI). In addition, we also included a laboratory strain (B3501) of C. neoformans (VNIV). We observed that phagosomes containing strain B3501 or WM276 sequentially acquired Rab5 and LAMP-1 at the respective time points (Fig. 3A to C), indicating successful phagosomal maturation. Interestingly, phagosomes containing R272 demonstrated a reciprocal change, with increased F-actin and reduced Rab5 and LAMP-1 (Fig. 3A), but showed an intermediate phenotype since the frequency of F-actin-enriched phagosomes was roughly 30 to 40% lower than with R265 (Fig. 3B and C). These results demonstrate that subversion of DC phagosomal maturation is likely obscured by the presence of an F-actin structure, which is unique to hypervirulent C. gattii isolates of the PNW outbreak.
Phagosomal F-actin is retained and does not fluctuate over time in response to hypervirulent Cryptococcus gattii.
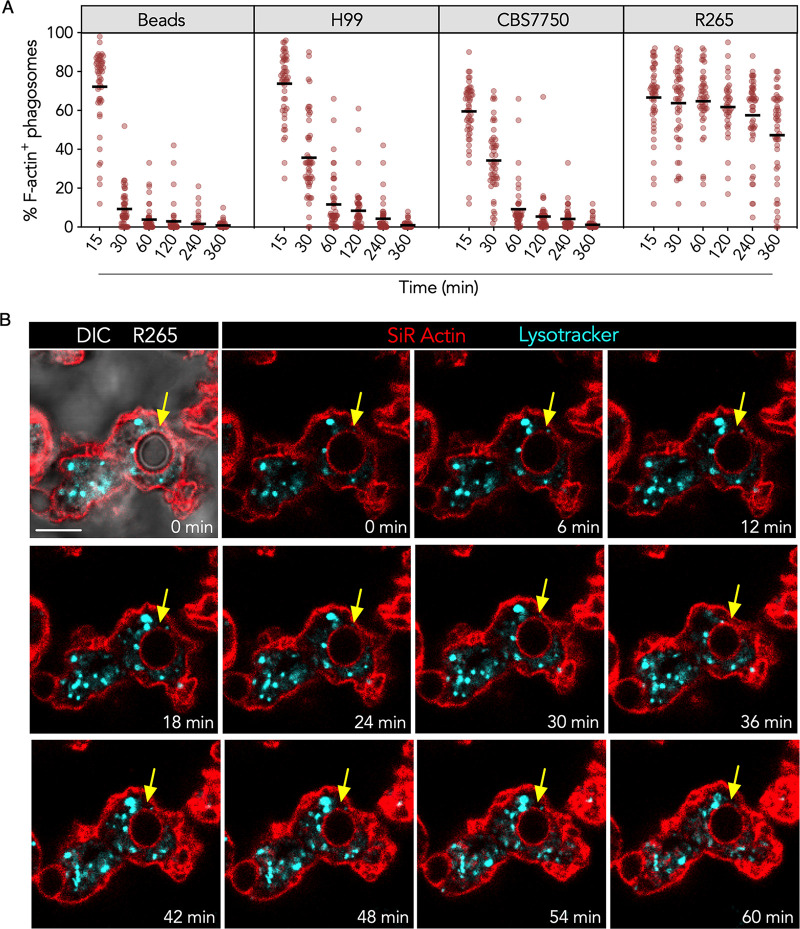
Actin dynamics is an important part of phagocytosis and, possibly, phagosomal processing. Nascent phagosomes are covered by a layer of F-actin that is rapidly dissolved, followed by subsequent membrane fusion events (22, 36). Since R265 phagosomal F-actin was present at early and late times (Fig. 3), we sought to measure the frequency of DCs expressing F-actin-positive phagosomes containing R265 at various times. We focused on strain R265 since the frequency of phagosomal F-actin was significantly higher than for R272 (Fig. 3). We observed that immediately after phagocytosis, 60% to 80% of the very early phagosomes were coated with F-actin regardless of the phagocytic cargo (Fig. 4A). However, as the time after internalization increased, the frequency of phagosomal F-actin quickly decreased in DCs containing beads, CBS7750, or H99 (Fig. 4A, first three panels), indicating nascent phagosomal F-actin resorption. In contrast, the frequency of R265-infected DCs expressing F-actin-positive phagosomes remained relatively unchanged over the course of 6 h (Fig. 4A, far-right panel).
FIG 4.
DC phagosomal F-actin is retained and does not fluctuate over time in response to hypervirulent Cryptococcus gattii. (A) Frequency of phagosomal F-actin enrichment at various times. DCs were infected with Cryptococcus or given polystyrene beads for 2 h and fixed at increasing times after infection (minutes). Fixed samples were imaged by confocal microscopy, and F-actin was labeled with DyLight 554 phalloidin. Each data point is the proportion of phagosomes per FOV (50 to 65 FOVs pooled from 3 donors) that are enriched for F-actin. (B) Time-lapse confocal micrographs of DCs infected with C. gattii R265 corresponding to Movie S1 in the supplemental material. DCs were labeled with SiR-actin (red) to stain F-actin and with LysoTracker (cyan) to stain the lysosomes prior to coculture with the cryptococci for 30 min. Images are presented as single confocal stacks at 6-min intervals over a course of 1 h. The yellow arrows point to the position of the phagosome containing R265, as determined by DIC. Data are representative of results from 3 separate experiments.
Live-cell microscopy of C. gattii R265 yeast cells engulfed by a DC, which was stained with SiR-actin (red) to label F-actin and LysoTracker (cyan) to label lysosomes prior to culture with R265. The cell was imaged every 1 min for just over an hour across 3 channels and multiple confocal stacks. The acquisition time is on the top left, and the scale bar is on the bottom right. Download Movie S1, MOV file, 5.6 MB (5.6MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Although the F-actin coat persisted on R265 phagosomes, we considered the possibility that the F-actin intensity might have changed between the measured time points (Fig. 4A), similar to the macrophage actin flash described previously (22, 26). The frequency of the actin flash is reported to be around 3 to 4 min from the moment of F-actin assembly to F-actin disassembly and is more prevalent within the first hour after phagocytosis (22, 23). To determine whether the persistent R265 phagosomal F-actin “flashed,” we performed live-cell microscopy. We observed that DCs infected with R265 displayed a strong and persistent signal of phagosomal F-actin over the course of 1 h (Fig. 4B; Movie S1), and the F-actin coat had minimal density fluctuation (Fig. S3A and Movie S1). This corroborated our above-described observation (Fig. 4A) that R265 phagosomal F-actin is persistent and not an actin flash. Furthermore, the persistent F-actin was not a projection of a newly forming phagocytic cup but a coat that completely encircled the phagosome, as determined by the confocal stacks. This F-actin coat was also not on the fungal yeast surface since the uninternalized yeasts did not emit a silicon rhodamine (SiR)-actin fluorescence signal, indicating that the persistent F-actin was part of the DC cytoskeleton (Movie S1).
C. gattii R265 phagosomal F-actin fluorescence intensity over time and size of DC phagosomes containing Cryptococcus. (A) Quantification of data in Fig. 4B. The F-actin fluorescence intensity over the course of 1 h is shown. Data are presented as means ± SD at 6-min intervals and are averages from 3 separate experiments. DCs were labeled with 200 nM SiR-actin dissolved in RPMI culture medium with the addition of verapamil for 3 to 4 h in a 37°C cell incubator. DCs were then cocultured with C. gattii R265 for 30 min at 37°C. Cells were imaged every 1 to 2 min for 1 h. (B) Phagosome size was measured as an average from two perpendicular planes at the confocal stack correlating to the widest diameter using confocal microscopy. Phagosome borders were determined by the boundary of the yeast, visualized by DIC, and LAMP-1 or F-actin staining. Data are presented as means ± SD and are averages for 65 phagosomes from 3 different donors. The mean (μ) under each condition is shown. Polystyrene beads (3.3 μm) were used as a control. Download FIG S3, TIF file, 0.2 MB (177.6KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Phagosomal F-actin dynamics can also be influenced by the size of the phagosome. In macrophages, a large phagosomal cargo is associated with a transient delay in phagosomal F-actin resorption (22). We measured the sizes of phagosomes containing different cryptococcal strains. DCs infected with R265, CBS7750, or H99 were internalized in phagosomes of similar sizes (Fig. S3B). Thus, the different rates of F-actin resorption were not due to differences in cargo size. Altogether, these results demonstrated that hypervirulent C. gattii forms a prominent F-actin coat that is continuously present on DC phagosomes.
Hypervirulent Cryptococcus gattii forms a porous cage-like F-actin structure on DC phagosomes that limits lysosome fusion.
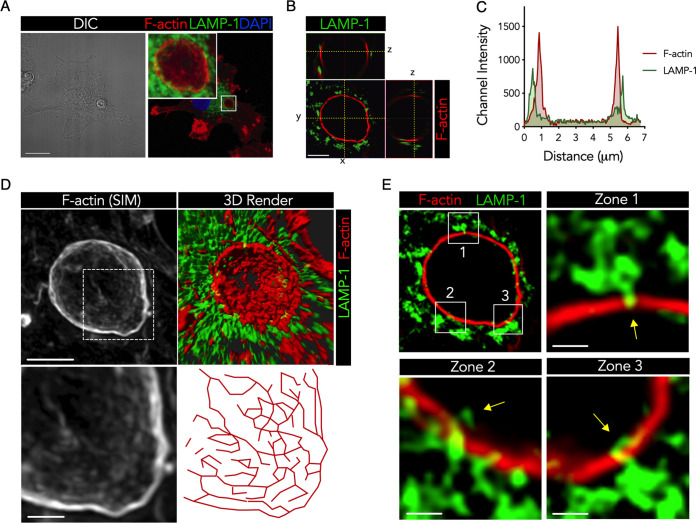
To further characterize the R265 phagosomal F-actin coat, we examined the F-actin ultrastructure by superresolution microscopy. Superresolution structured illumination microscopy (SR-SIM) micrographs revealed that the F-actin coat was in close spatial proximity to the phagosomal cargo compared to the lysosomes (Fig. 5A to C). The F-actin coat had a highly branched ultrastructure that completely wrapped around the phagosome in a cage-like fashion, with gaps between the branches, resembling pores (Fig. 5D; Movies S2 and S3). From the live-cell microscopy experiments, we observed that lysosomes (labeled with LysoTracker) were present and adjacent to the R265 phagosomal F-actin cage (Fig. 4B; Movie S1). Although it appeared that lysosome fusion was completely blocked by the phagosomal F-actin barrier, a concept that has been suggested previously (22), we observed that a small number of lysosomes had sporadically breached through the F-actin barrier (Fig. 5E, zones 1 to 3). This indicated that phagosomal F-actin is not an absolute barrier to lysosome fusion but significantly limits membrane fusion events through physical interference.
FIG 5.
Hypervirulent Cryptococcus gattii forms a porous cage-like F-actin structure on DC phagosomes that limits lysosome fusion. (A) Confocal micrograph (MIP) of R265-infected DCs and immunolabeled LAMP-1 (green), phalloidin-labeled F-actin (red), and the DAPI-stained nucleus (blue) 4 h after the 2-h infection period. Bar = 15 μm. (B) SR-SIM image (single z-stack) of the phagosomal region (white box in panel A) shown as a cross section and orthogonal sections. Bar = 2 μm. (C) Positional pixel intensity graph of LAMP-1 (green) and F-actin (red) on a single line depicted by “y” in panel B from left (0 μm) to right (7 μm). (D) SR-SIM image (MIP) of F-actin (top left) and 3D reconstruction of F-actin and LAMP-1 (top right) of the phagosomal region (white box in panel A). A digital magnification (bottom left) and a pixel map (bottom right) of the F-actin SR-SIM image (dashed square, top left) are shown. Bars = 0.5 μm. (E) SR-SIM image (single stack) of the phagosome (white box in panel A), with magnified regions indicated by the corresponding numbers. Bars = 0.2 μm. Data are representative of results from 3 donors.
Animation of the 3D reconstruction of an SR-SIM micrograph (shown in Fig. 5) showing a DC that contains C. gattii R265 within the F-actin cage (red ball). F-actin was stained with phalloidin (red), and lysosomes were stained with LAMP-1 antibodies (green). Download Movie S2, MOV file, 6.6 MB (6.6MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cropped section of the phagosomal region shown in Movie S2 in the supplemental material. F-actin was stained with phalloidin (red), and lysosomes were stained with LAMP-1 antibodies (green). Download Movie S3, MOV file, 3.5 MB (3.5MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The hypervirulent Cryptococcus gattii phagosomal F-actin cage subverts DC phagosomal processing and maturation.
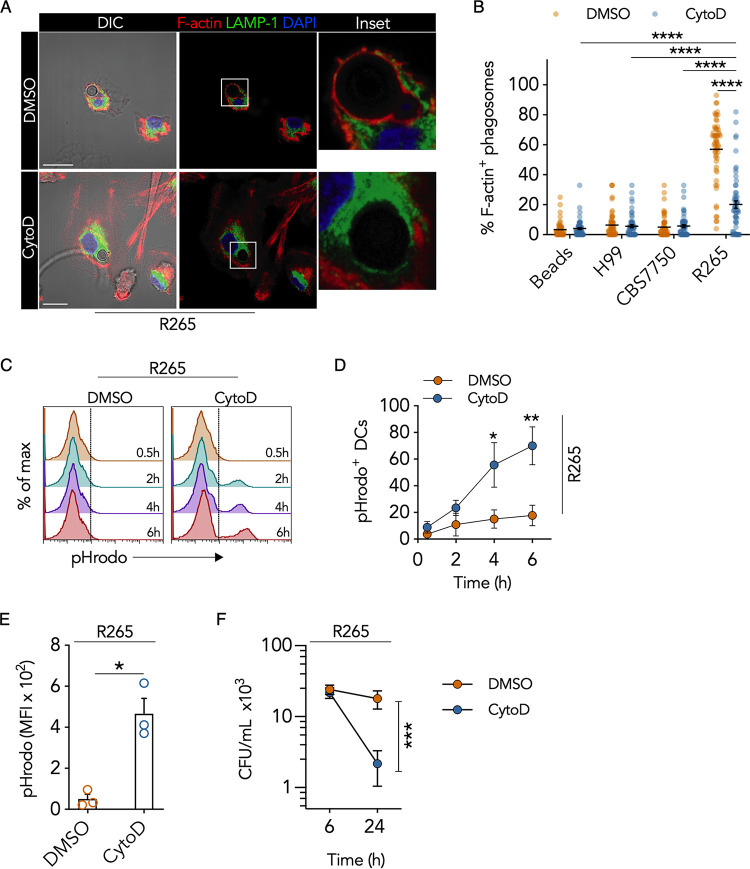
To determine the role of the R265 phagosomal F-actin cage, we took a loss-of-function approach using the F-actin inhibitor cytochalasin D (CytoD) (37). By pulse-treating DCs after the uptake of Cryptococcus, we limited the effect of the inhibitor on phagocytosis, which is an actin-dependent process. We determined that treatment with the inhibitor for 2 h was more effective than treatment for 1 h after infection with Cryptococcus (Fig. S4). Cytochalasin D treatment resulted in a 65% ± 4% reduction in the frequency of DCs containing F-actin-positive phagosomes compared to the vehicle control at 4 h posttreatment (Fig. 6A and B). This reduction corresponded to reciprocal changes in phagosomal LAMP-1 enrichment (Fig. S4B), indicating that disruption of R265 F-actin augmented phagolysosome formation. Although phagosomal F-actin was significantly decreased, the intensity of total cellular F-actin was unaffected by cytochalasin D pulse-treatment (Fig. S5A).
FIG 6.
The hypervirulent Cryptococcus gattii phagosomal F-actin cage subverts DC phagosomal processing and maturation. (A) Confocal micrographs (MIPs) and digital magnifications (white boxes) of R265-infected DCs that were pulsed with cytochalasin D (CytoD) or DMSO (vehicle control) for 2 h and cultured for an additional 4 h in medium alone. Cells were immunostained for LAMP-1 (green), phalloidin to label F-actin (red), and DAPI to stain the nucleus (blue). Bars = 10 μm. (B) Quantification of the data in panel A as the proportion of F-actin-enriched phagosomes containing cryptococci or beads per FOV in response to cytochalasin D treatment. Data are presented as a scatterplot of 50 to 55 FOVs from 3 donors (the mean is represented by a black line). ****, P < 0.0001 (by one-way ANOVA [adjusted P values]). (C) Representative flow cytometric histograms of DCs cultured with pHrodo-labeled R265 and treated with cytochalasin D or DMSO at 0.5 to 6 h posttreatment. (D) Quantification of the data in panel C. Data are presented as means ± SEM of the percentage of pHrodo+ DCs (averages from 3 donors). *, P = 0.033; **, P = 0.00053 (compared to the control [DMSO] by two-way ANOVA [adjusted P values]). (E) MFI of DCs cultured with pHrodo-labeled R265 6 h after cytochalasin D or DMSO treatment. Data are presented as means ± SEM of results from 3 donors (shown as dots). *, P = 0.023 by unpaired Welch’s t test (two-tailed P value). (F) Intracellular survival of R265 within DCs 1 h or 24 h following cytochalasin D or DMSO treatment. Data are presented as means ± SEM from 3 donors in a before-after graph. ***, P < 0.001 (by one-way ANOVA [adjusted P values]).
Effects of transient cytochalasin D treatment and metabolic activity on C. gattii R265 phagosomal F-actin frequency. (A) Representative confocal micrographs of R265-infected DCs treated with cytochalasin D for either 1 h or 2 h or with DMSO (vehicle control). After cells were washed and incubated for 4 h in the absence of the inhibitor, cells were fixed with PFA and immunostained for LAMP-1 (green), phalloidin to label F-actin (red), and DAPI to stain the nucleus (blue). The phagosome region (represented as white squares in the DIC panels) was cropped and digitally magnified to view phagosomal F-actin and LAMP-1 in single confocal stacks. Bar = 10 μm. (B) Quantification of data in panel A. Data are presented as stacked bar graphs of the averages for phagosomes per FOV that are enriched for F-actin, LAMP-1, both, or neither. The number of FOVs (n) under each condition is shown. Data are representative of results from 3 donors. (C) Comparison of live and dead Cryptococcus cells for the frequency of phagosomal F-actin. Cryptococcus cells were either heat killed (HK) for 60 min in a 60°C water bath or fixed with 4% PFA for 30 min. Data are presented as means ± SEM of the percentages of phagosomes in the FOV. Shown are averages for 10 FOVs from separate experiments. *, P < 0.05; **, P < 0.01 (by two-way ANOVA [adjusted P values]). Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effects of transient cytochalasin D treatment on total DC F-actin and Cryptococcus growth. (A) Representative histogram of DCs unlabeled or labeled with phalloidin conjugated to Alexa Fluor 555. The table on the right shows the MFI under each condition and DC viability as measured by trypan blue exclusion at the end of the experiment. DCs were treated with either 2 μg/ml cytochalasin D or DMSO for 2 h in a 37°C CO2 incubator, followed by a 4-h period of incubation in inhibitor-free culture medium after being washed. After the 4-h period, cells were fixed with 4% PFA and permeabilized with saponin. Fixed samples were labeled with phalloidin at a 1:200 (vol/vol) dilution for 30 min at room temperature. Data are representative of results from 3 donors. (B) CFU of different Cryptococcus strains at 6 and 24 h. Cryptococcus cells were either untreated or treated with 2μg/ml of CytoD or DMSO for 2 h in a 37°C 5% CO2 incubator, washed, and incubated in inhibitor-free medium for an additional 4 h. Cryptococcal yeast cells were counted, and approximately 1,000 yeast cells were plated in a 96-well plate in culture medium. Experiments were done in triplicates. CFU were measured either immediately or 24 h later. Data are representative of results from 3 donors. Download FIG S5, TIF file, 0.4 MB (407.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Although the frequency of R265 phagosomal F-actin dramatically decreased after cytochalasin D treatment, it remained higher than those of the other cryptococcal strains or control beads (Fig. 6B). Consequently, we questioned whether the F-actin reduction was sufficient to restore DC phagosomal maturation. We first investigated intracellular acidification in R265-infected DCs after cytochalasin D treatment. The percentage of DCs containing pHrodo-positive (pHrodo+) phagosomes increased in a time-dependent manner, with the most significant increase (52% ± 14%) occurring 6 h after cytochalasin D treatment (Fig. 6C and D). At 6 h, the mean fluorescence intensity (MFI) of pHrodo increased 6.6-fold after cytochalasin D treatment compared to the vehicle control (Fig. 6E), suggesting acidification of the phagosome. Cytochalasin D treatment also resulted in an 88% ± 6% reduction in R265 CFU 24 h after DC infection compared to the vehicle control (Fig. 6F). The reduction in fungal CFU was not due to cytochalasin D or dimethyl sulfoxide (DMSO) alone since treatment of Cryptococcus cells directly with the inhibitor had no significant effect on their growth (Fig. S5B). Thus, disruption of hypervirulent C. gattii phagosomal F-actin restored DC phagosomal maturation, which resulted in enhanced fungal killing.
Immunoparalysis of DCs by hypervirulent Cryptococcus gattii is caused by the retention of the phagosomal F-actin cage.
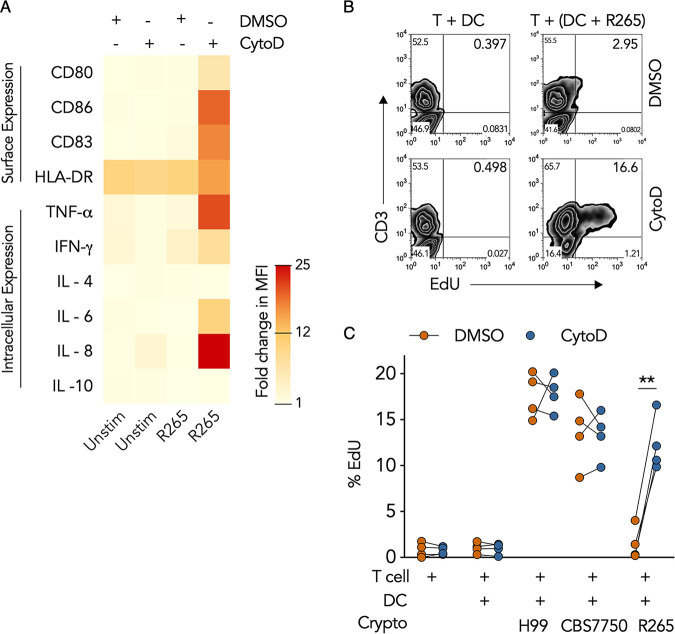
Aside from functioning as degradative, antimicrobial, and antigen-processing machinery, the phagosome is also a rich source of pathogen-associated molecular patterns (PAMPs) that can activate DC maturation and concomitant T cell immunity (38, 39). Since disruption of the R265 F-actin cage with cytochalasin D augmented phagosomal maturation, we questioned whether F-actin disruption could also restore DC immunity. We observed that DCs cultured with R265 expressed significantly higher levels of the intracellular proinflammatory markers TNF-α, IL-6, IL-8, and IFN-γ and showed increased surface expression of the costimulatory ligands CD86, CD83, and CD80 after cytochalasin D treatment (Fig. 7A), indicating that disruption of R265 phagosomal F-actin induced DC immune activation. Furthermore, T cell proliferation was 8.2-fold higher when DCs cultured with R265 were pulse-treated with cytochalasin D than with the vehicle control (Fig. 7B and C). This response was not donor dependent since all four donors exhibited similar responses to treatment (Fig. 7C), indicating that disruption of R265 phagosomal F-actin induced a strong biological response in the DCs that resulted in robust T cell proliferation. This response was not due to the inhibitor alone since treatment of uninfected DCs with cytochalasin D had no significant effect on T cell proliferation (Fig. 7C). Likewise, cytochalasin D treatment also had no significant effect on T cell proliferation in response to DCs cultured with CBS7750 or H99 (Fig. 7C). Taken together, these results revealed that the F-actin cage on hypervirulent C. gattii phagosomes plays an important role in causing DC immune evasion and is the mechanism of DC immunoparalysis.
FIG 7.
Immunoparalysis of DCs by hypervirulent Cryptococcus gattii is caused by the retention of the phagosomal F-actin cage. (A) Heat map of the fold changes in the MFIs of surface and intracellular markers in resting DCs (Unstim) or DCs infected with R265 that were pulsed with cytochalasin D or DMSO for 2 h and cultured for an additional 4 h in medium alone. Data are presented as the average fold changes in MFIs, standardized to isotype-matched controls, from 4 different donors. The color spectrum represents the minimum and maximum fold changes in MFIs. (B) Representative flow cytometry plot of EdU incorporation in peripheral blood T cells cultured for 6 days with uninfected DCs or DCs infected with R265 and pulse-treated with cytochalasin D or DMSO (similar to panel A) prior to coculture with T cells. (C) Quantification of data in panel B as percent EdU staining in CD3+ T cells in response to DCs treated with cytochalasin D or DMSO. Data are presented in a before-after graph where each connected pair of dots represents data for each donor (total of 4 donors). **, P = 0.0014 (by one-way ANOVA [adjusted P values]).
DISCUSSION
Cryptococcus is known to escape immune detection through a number of mechanisms. We previously demonstrated that the hypervirulent strain of C. gattii remains largely undetected by T cells due to a failure in T cell costimulation by DCs compared to other cryptococcal strains (14). A diminished T cell response in the context of hypervirulent C. gattii infection has been shown in other experimental systems to depend on the state of DC activation (11). These findings suggest that hypervirulent C. gattii infection causes DC immunoparalysis. Our present results extend the findings from these studies by revealing a unique mechanism by which hypervirulent C. gattii subverted DCs through the retention of a phagosomal F-actin structure, which attenuated DC proinflammatory activation and concomitant T cell activation.
The role of actin polymerization in phagosomal maturation is underexplored despite controversy in the field. Some studies have reported that actin assembly on phagosomes assists in vesicular docking and fusion (40, 41), while others have suggested that F-actin dissolution on nascent phagosomes is an important process of phagosomal maturation (22, 42). While phagosomal F-actin has been correlated with inhibition of lysosome fusion (22), the mechanism of this blockade was unclear. Others have speculated that F-actin accumulation on phagosomes acts as a barrier to block downstream phagosomal maturation by inhibiting membrane fusion events (22, 24, 42). We investigated the structural relationship between the hypervirulent C. gattii phagosomal F-actin cage and the lysosome network using superresolution microscopy. The prominent F-actin cage did not create a complete barrier for lysosomal fusion. A small number of lysosomes appeared to have penetrated the F-actin cage, presumably as a result of the cage structure, where the actin fibers are organized in a cross-linking manner such that certain areas of the phagosome are not covered by actin. This is consistent with the dimensions of smaller lysosomes, ranging between 0.1 and 1.5 μm in diameter (43, 44), which could pass through the smaller pores in the F-actin cage. Nevertheless, this level of lysosomal fusion was not sufficient to cause functional changes in the phagosome since phagosome acidification or killing of hypervirulent C. gattii remained at baseline levels in the presence of the F-actin cage.
One unique role of phagosomal F-actin has been described for a host cell evasion mechanism of C. neoformans known as vomocytosis (26, 45, 46). During this nonlytic expulsion, the macrophage phagosomes containing C. neoformans are transiently wrapped with F-actin in a manner consistent with the actin flash. The actin flash is believed to be a mechanism by which macrophages avoid vomocytosis. We performed time course and live-cell microscopy experiments to determine if hypervirulent C. gattii phagosomal F-actin had features of the actin flash and demonstrated that the actin flash and F-actin cage formation are different processes. We observed that the hypervirulent C. gattii phagosomal F-actin cage is sustained over time and does not show evidence of complete actin resolution and accumulation cycles, a key feature of the actin flash. Therefore, our data strongly suggest that the C. gattii phagosomal F-actin cage is a phenomenon that is distinct from the actin flash and likely has a different pathophysiological function. Furthermore, phagosomal actin was recruited in response to both live and killed C. gattii albeit at a lower level than with live organisms (see Fig. S4C in the supplemental material). Consequently, C. gattii growth was not required for phagosomal actin retention as it was for actin recruitment in response to C. albicans (47).
Despite the persistence of the C. gattii phagosomal F-actin cage, the actin structure was not static. Live-cell microscopy revealed focal actin intensity variation within the persistent phagosomal F-actin cage structure (Movie S4). These results have two important implications. First, the F-actin cage was actively remodeled such that the focal actin turnover did not destabilize the entire F-actin cage. Second, this active actin remodeling indicates that there must be active signaling pathways that maintain the persistent F-actin cage. These unidentified signaling pathways triggered by hypervirulent C. gattii are likely different from those triggered by C. neoformans to execute the actin flash. Future studies comparing these signaling mechanisms will help address this point.
Cropped section of the phagosomal region shown in Movie S1 in the supplemental material. The time-lapse of F-actin (labeled with SiR-actin) is shown as a heat map; red represents hot pixels, and white represents cooler pixels. The acquisition timer is on the top right. Download Movie S4, MOV file, 9.4 MB (9.4MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In DCs infected with hypervirulent C. gattii, we noticed that disruption of the persistent F-actin cage with cytochalasin D resulted in the permanent resorption of the F-actin cage. This is likely due to the rapid phagolysosome formation that followed F-actin cage disruption. Through a process that is not well understood, phagolysosome formation appears to have an inverse relationship with phagosomal F-actin (22, 23). We speculate that phagolysosome formation overrides the signaling pathways that are responsible for maintaining the persistent F-actin cage of hypervirulent C. gattii. In addition to increased phagosomal maturation, disruption of the hypervirulent C. gattii F-actin cage reversed DC immunoparalysis and induced proinflammatory activation. Cytochalasin D treatment could restore DC activation because either disruption of the F-actin allows cryptococcal ligands to engage the phagosomal pattern recognition receptor (PRR) or it leads to the liberation of proinflammatory signaling molecules. The latter is suggested when disruption of the microtubule and actin cytoskeleton spontaneously activates NF-κB signaling (48–50). Hence, future studies of signaling pathways that drive DC activation upon C. gattii phagosomal F-actin cage disruption are warranted. Cytochalasin D would affect all F-actin. Therefore, we also considered the possibility that our observations could be a result of cytochalasin D interference with other parts of the cytoskeleton or pathways dependent on the actin cytoskeleton.
In an effort to determine the specificity of the phagosomal F-actin cage, we studied various strains of C. gattii and C. neoformans. We found that the hypervirulent (R265) and less virulent (R272) PNW outbreak strains of C. gattii retained phagosomal F-actin considerably more than did non-PNW-outbreak strains. This was an important observation as it indicated that phagosomal F-actin cage retention could be an important factor driving the PNW strains’ virulence. This unique feature of PNW isolates, especially hypervirulent C. gattii, to form and retain phagosomal F-actin could also play a role in making PNW C. gattii a primary pathogen. We have demonstrated that phagosomal F-actin retention is coupled with DC immunoparalysis, and C. neoformans isolates with different degrees of virulence (51) do not retain F-actin and hence do not block DC activation. When considering new therapeutic approaches against PNW C. gattii, it will be important to study the virulence factors that trigger phagosomal F-actin retention and thereby block DC immune function.
A limitation of our study is that we analyzed a mixed population of DCs, some of which had internalized Cryptococcus and some of which had not, when measuring phagosomal acidification and DC costimulatory and cytokine expression via flow cytometry. We considered the possibility that the differences that we observed in these experiments between C. gattii R265 and other Cryptococcus strains could be a result of differences in phagocytic rates. However, the 2 to 4% difference in phagocytic rates between C. gattii R265 and other Cryptococcus strains (Fig. 2A) is very unlikely to account for the 40 to 44% difference in pHrodo+ DCs (Fig. 2D) or the 16 to 28% difference in costimulatory marker expression (Fig. 1C) between C. gattii R265 and other strains of Cryptococcus.
In summary, we have shown that hypervirulent C. gattii infects human DCs and forms a permanent F-actin cage structure that inhibits phagosomal maturation and DC immune activation, resulting in immunoparalysis. This unique mechanism of host cell evasion is likely a crucial aspect of PNW C. gattii virulence, contributing to its propensity to infect healthy hosts.
MATERIALS AND METHODS
Reagent suppliers and catalog numbers can be found in Table S1 in the supplemental material.
List of reagents. Download Table S1, DOCX file, 0.02 MB (20.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primary cells and Cryptococcus.
Human blood from healthy volunteers was obtained following informed consent via venipuncture by trained phlebotomists using heparin-containing tubes. Peripheral blood mononuclear cells (PBMCs) were separated on Ficoll Paque Plus by centrifugation for 30 min at 650 × g and then transferred, washed, and quantified in Hanks’ balanced salt solution (HBSS) medium. CD14+ monocytes and pan-T cells were isolated from PBMCs using the AutoMacs cell sorter and human CD14 microbead (positive-selection) or pan-T cell microbead (negative-selection) kits (Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer’s protocol. Immature DCs (iDCs) were generated by culturing purified CD14+ monocytes with 500 IU/ml IL-4 and 1,000 IU/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) for 6 to 7 days as previously described (14). The purity of monocytes and iDCs was routinely checked and reported as ≥95%. All cells were cultured and maintained in complete RPMI culture medium supplemented with 10% heat-inactivated fetal bovine serum, 1% sodium pyruvate, 1% nonessential amino acids, and 1% penicillin-streptomycin at 1 × 106 cells/ml.
C. gattii strains R265, R272, and WM276 were kindly supplied by Jim Kronstad (University of British Columbia, Vancouver, BC, Canada). C. neoformans strain H99 was generously donated by John Perfect (Duke University, Durham, NC). C. gattii strain CBS7750 was generously donated by Karen Bartlett (University of British Columbia, Vancouver, BC, Canada). C. neoformans strain B3501 was purchased from the ATCC. All strains were stored at −80°C in 25% glycerol. For experimental use, frozen samples were cultured on Sabouraud dextrose (SD) agar for 48 h in a 32°C incubator. Single CFU were transferred to SD broth and grown to stationary phase over 72 h at 32°C. Aliquots from the stationary phase were transferred to fresh SD broth and grown to exponential phase over 24 h at 32°C. Aliquots were then washed in RPMI culture medium and quantified using a hemocytometer.
A new nomenclature for Cryptococcus has been proposed (52). Under the new nomenclature, the VGII genotype of the C. gattii species complex, which includes the R265, R272, and CBS7750 strains, is called C. deuterogattii. The VGI genotype of the C. gattii species complex, which includes the WM276 strain, is called C. gattii. The VNI genotype of the C. neoformans species complex, which includes the H99 strain, is called C. neoformans. The VNIV genotype of the C. neoformans species complex, which includes the B3501 strain, is called C. deuterogattii. In this report, we have adhered to the traditional nomenclature for consistency with previous work.
DC infection assay.
iDCs were suspended in complete RPMI culture medium overnight in either 24-well plastic plates, 24-well plates containing glass coverslips, 96-well plastic plates, or 96-well plates with a glass-like coverslip bottom. iDCs were seeded at 0.75 × 105 cells in a volume of 100 μl per well of a 96-well plate and at 2 × 105 cells in a volume of 500 μl per well of a 24-well plate. Cryptococcus cells were prepared as described above and grown to mid-log phase. Organisms were washed twice in sterile phosphate-buffered saline (PBS) by centrifugation using 1.5-ml tubes and counted via a hemocytometer at room temperature. The desired number of yeast cells was added to a final volume of 1 ml of complete RPMI culture medium in 1.5-ml tubes and washed again by centrifugation. The final washed samples were heated to 37°C in a humidified incubator. For 96-well experiments, 50-μl aliquots of Cryptococcus (prewarmed) were added to the wells containing iDCs at a multiplicity of infection (MOI) of 10. The wells were mixed 20 times (8 wells at a time) using an electronic multichannel pipette (Rainin). For 24-well experiments, 200-μl aliquots of Cryptococcus (prewarmed) were added to the wells containing iDCs (MOI = 10). The wells were mixed individually with a single pipette. The coculture plates were incubated in a 37°C humidified incubator for 2 h. After the incubation period, 80% of the volume in each well was gently removed via vacuum suction. The wells were then washed twice with preheated (37°C) sterile PBS, the plates were gently mixed by pipetting, and 80% of the volume was again removed via vacuum suction. Finally, the volume in each well was restored with preheated (37°C) complete RPMI culture medium, and the plates were incubated for an additional 4 h in a 37°C humidified incubator. Cells were then fixed with 4% paraformaldehyde (PFA).
Fixed cells were labeled with DyLight 554 phalloidin and transferred to glass slides. Slides were analyzed by confocal microscopy at a ×40 magnification in a grid format such that the entire slide was read. Phagocytosis was determined when the target (Cryptococcus or beads) was within the DC cortical actin boundaries, confirmed by confocal z-plane imaging. The frequency was calculated as the number of DCs with internalized yeast divided by the total number of DCs for each field of view (FOV). DCs with incomplete phagocytosis, which accounted for <1% of the total DCs, were excluded from the pool of DCs with the internalized target.
DC maturation and activation assays.
iDCs were infected with Cryptococcus in 96-well plates as mentioned above. As a positive control, ultrapure lipopolysaccharide (LPS) of Escherichia coli was used at 700 ng/ml to stimulate iDCs for a total of 6 h. In some experiments, iDCs that had been infected with Cryptococcus or given beads for 2 h were treated with 2 μg/ml cytochalasin D or DMSO (vehicle control) for 1 h or 2 h to disrupt F-actin (37). Cultures were then washed to remove the inhibitor and resuspended in fresh culture medium for an additional 4 h. To detect intracellular cytokines, brefeldin A (BFA) was added (3 μg/ml) to the DC culture, and cells were permeabilized with saponin. Surface antigens on DCs were labeled with antibodies in PFA-fixed cells. DC surface and intracellular antigens were detected using Guava easyCyte flow cytometry (Millipore, Danvers, MA), and data were analyzed using Guava Incyte software and FlowJo.
T cell proliferation assay.
Autologous pan-T cells were cultured either alone, with uninfected DCs, or with Cryptococcus-infected DCs (described above) at a ratio of 1:10 (DCs to T cells) in 24-well plates for 6 days in RPMI culture medium in a 37°C cell incubator. In some experiments, T cells were cultured with DCs (infected or uninfected) that were treated with 2 μg/ml cytochalasin D or the vehicle control for 2 h, followed by a 4-h period without the inhibitor prior to culturing with T cells. On day 5, 1 μM 5-ethynyl-2-deoxyuridine (EdU) was added to the cell culture. On day 6, cells were permeabilized with saponin and labeled with anti-CD3 monoclonal antibody. EdU was labeled using the click chemistry technique, according to the manufacturer’s protocol, and cells were detected using Guava easyCyte flow cytometry.
DC intracellular killing and phagosomal pH assays.
To measure the intracellular survival of Cryptococcus, plastic-adhered iDCs in 24-well plates were infected with Cryptococcus (MOI = 10) for 2 h and washed several times with sterile PBS to remove excess yeast. Infected DCs were lysed either immediately (1 h) or after 6 or 24 h of incubation with sterile ice-cold double-distilled water (ddH2O) for 20 min with repeated pipetting. Lysates were serially diluted in sterile ice-cold ddH2O, and 10-μl aliquots were plated on SD agar and incubated at 32°C for 48 h. CFU were counted after 48 h. In some experiments, Cryptococcus-infected DCs were treated with 2 μg/ml cytochalasin D or the vehicle control for 2 h, washed, and incubated for 4 h in inhibitor-free medium. Cells were lysed at 6 or 24 h with sterile ice-cold ddH2O for 20 min. CFU were determined as described above. To determine the effect of transient cytochalasin D or DMSO treatment on fungal growth, Cryptococcus cells alone were treated with 2 μg/ml cytochalasin D or the vehicle control for 2 h in RPMI culture medium, washed, and incubated for an additional 4 h (corresponding to the 6-h CFU count) or 24 h in inhibitor-free RPMI culture medium. CFU were determined. All experiments were performed in triplicates under each condition, and the average CFU count was used for analysis.
To assess changes in phagosomal pH, 107 cryptococcal yeast cells were labeled with pHrodo green AM, according to the manufacturer’s protocol, on ice for 1 h in sterile PBS (pH 7.4) and rinsed several times. DCs were infected with pHrodo-labeled cryptococci for 2 h and washed to remove excess yeast. DCs were also cultured with unlabeled beads as a negative control. Samples were fixed with 4% PFA either 0.5, 2, 4, or 6 h after the infection period. In some experiments, DCs infected with pHrodo-labeled Cryptococcus were pulse-treated with 2 μg/ml cytochalasin D or the vehicle control for 2 h. Samples were fixed with 4% PFA either 0.5, 2, 4, or 6 h after cytochalasin D treatment. The fluorescence of pHrodo was measured using Guava easyCyte flow cytometry.
Immunofluorescence microscopy.
iDCs were grown on glass coverslips overnight and infected with Cryptococcus (MOI = 10) for 2 h in a 37°C cell incubator. In some experiments, Cryptococcus-infected or uninfected DCs were transiently treated with 2 μg/ml cytochalasin D or the vehicle control for 1 h or 2 h at 37°C. At different times after infection or treatment, coverslips were fixed with 4% PFA and permeabilized with 1× saponin. Coverslips were then blocked with 5% bovine serum albumin (BSA) in PBS following fixation and immunostained with the respective unconjugated primary antibodies (diluted 100-fold in 5% BSA in PBS) for 1 h in a 37°C cell incubator. Coverslips were incubated in PBS for 2 h (wash cycle) at room temperature and labeled with secondary antibodies conjugated to Alexa Fluor 488 and with DyLight 554-conjugated phalloidin to label F-actin (both diluted 200-fold in 5% BSA in PBS) for 30 min in a 37°C cell incubator. Coverslips were washed again for 2 h in PBS and mounted on glass slides with ProLong gold antifade containing 4′,6-diamidino-2-phenylindole (DAPI). Slides were imaged with a Zeiss microscope fitted with a 63× plan apochromat, 1.4-numerical-aperture (NA) oil immersion objective. For confocal laser scanning microscopy, images were acquired at 12 bits with a 1.26-μs/pixel dwell time on 12 to 16 optical z-sections using a Zeiss LSM780 microscope. For SR-SIM, images were acquired at 16 bits with five rotations per channel every 0.1 μm on 50 to 55 optical z-sections using a Zeiss Elyra PS.1 microscopy.
Live-cell microscopy.
DCs were grown on a 35-mm cell culture ibidi dish (ibidi, Verona, WI, USA) with a coverslip bottom at 50% confluence or lower. Cells were labeled with 200 nM SiR-actin dissolved in RPMI culture medium with the addition of verapamil for 3 to 4 h in a 37°C cell incubator. To label the intracellular lysosomes, DCs were also labeled with LysoTracker green (DND-26; Invitrogen). DCs were washed and cocultured with C. gattii R265 for 30 min at 37°C. Cells were imaged using a Zeiss LSM780 microscope with a 63× plan apochromat (1.4-NA) oil immersion objective in a 37°C chamber. Cells were imaged every 1 to 2 min for >1 h over multiple z-stacks.
Image analysis.
Confocal and SR-SIM micrographs and live-cell imaging time-lapse videos were analyzed using Zen black and blue edition software (Carl Zeiss, Oberkochen, Germany). Three-dimensional (3D) reconstruction and animation of the SR-SIM micrographs were created using Imaris (Bitplane). Pixel tracing in Fig. 3D was done manually. Although the localizations of F-actin on phagosomes are quite distinct, we used an analytic approach, adapted from the one described previously by Dingjan et al. (53), to quantify phagosomal LAMP-1, Rab5, and F-actin enrichment. First, the MFI of each channel at optical sections above and below the largest diameter of the phagosomes was calculated on the whole cell area (MFIcell), with boundaries defined by cortical F-actin. Next, the MFI of the channel intensity on individual phagosomes was calculated (MFIphag). The phagosomal signal intensity was normalized with the following expression: MFInorm = MFIphag/MFIcell. Conditions were set for each channel such that if MFInorm was >1, then a score of 1 was assigned, but if MFInorm was <1, a score of 0 was assigned for each DC per FOV. The percentage of phagosomes enriched with or “positive” for LAMP-1, Rab5, or F-actin was calculated as the total number of 1’s divided by the total number of infected DCs in an FOV.
Statistical analysis.
GraphPad Prism (v8) was used to perform statistical analysis. Statistical significance was calculated using repeated-measures (RM) or ordinary one-way analysis of variance (ANOVA) with a Sidak post hoc test, two-way ANOVA with a Tukey post hoc test, and unpaired Welch’s t test. Adjusted P values of <0.05 were considered significant. In this study, each biological replicate represents data from an individual donor.
Ethical statements.
Experimental protocols for the use of human blood were approved and conducted under the guidelines of the Conjoint Health Research Ethics Board of the University of Calgary (certificate number REB15-0600).
ACKNOWLEDGMENTS
This work was supported by research grants from the Alberta Lung Association (ALA) (principal investigator [PI], C.H.M.; co-PI, A.G.) (grant number 10017730) and the Canadian Institute for Health Research (CIHR) (PI, C.H.M.; co-PI, A.G.) (grant number 365812), the Jessie Bowden Lloyd Professorship in Immunology (C.H.M.), an equipment and infrastructure grant from the Canadian Foundation for Innovation (CFI), and a graduate studentship from the Alberta Lung Association (K.J.). M.O. was funded by U.S. Department of Veterans Affairs Merit Review and Research Career Scientist awards (1I01BX000656 and 1IK6BX003615).
Work with confocal and superresolution immunofluorescence microscopy and the Zeiss Elyra LSM780 research microscope was supported by the Microscopy and Imaging Facility, University of Calgary, with help from Priyanka Mukherjee. We thank Radhika Singh for reviewing the manuscript and Karen Bartlett, Jim Kronstad, and John Perfect for sharing resources.
The funders had no role in the decision to publish or in the preparation of the manuscript.
Footnotes
Citation Jamil K, Polyak MJ, Feehan DD, Surmanowicz P, Stack D, Li SS, Ogbomo H, Olszewski M, Ganguly A, Mody CH. 2020. Phagosomal F-actin retention by Cryptococcus gattii induces dendritic cell immunoparalysis. mBio 11:e01821-20. https://doi.org/10.1128/mBio.01821-20.
REFERENCES
- 1.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, MacDougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris JR, Lockhart SR, Sondermeyer G, Vugia DJ, Crist MB, D’Angelo MT, Sellers B, Franco-Paredes C, Makvandi M, Smelser C, Greene J, Stanek D, Signs K, Nett RJ, Chiller T, Park BJ. 2013. Cryptococcus gattii infections in multiple states outside the US Pacific Northwest. Emerg Infect Dis 19:1620–1626. doi: 10.3201/eid1910.130441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris JR, Lockhart SR, Debess E, Marsden-Haug N, Goldoft M, Wohrle R, Lee S, Smelser C, Park B, Chiller T. 2011. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin Infect Dis 53:1188–1195. doi: 10.1093/cid/cir723. [DOI] [PubMed] [Google Scholar]
- 4.Leongson K, Cousineau-Cote V, Goupil M, Aumont F, Senechal S, Gaboury L, Jolicoeur P, Kronstad JW, de Repentigny L. 2013. Altered immune response differentially enhances susceptibility to Cryptococcus neoformans and Cryptococcus gattii infection in mice expressing the HIV-1 transgene. Infect Immun 81:1100–1113. doi: 10.1128/IAI.01339-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mody CH, Lipscomb MF, Street NE, Toews GB. 1990. Depletion of CD4+ (L3T4+) lymphocytes in vivo impairs murine host defense to Cryptococcus neoformans. J Immunol 144:1472–1477. [PubMed] [Google Scholar]
- 6.Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V. 2005. Cryptococcus gattii in AIDS patients, southern California. Emerg Infect Dis 11:1686–1692. doi: 10.3201/eid1111.040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J Infect Dis 192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 8.Anonymous. . 2015. Crytococcus gattii infection statistics. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/fungal/diseases/cryptococcosis-gattii/statistics.html. [Google Scholar]
- 9.Cheng PY, Sham A, Kronstad JW. 2009. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect Immun 77:4284–4294. doi: 10.1128/IAI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ. 2012. The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3:e00103-12. doi: 10.1128/mBio.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angkasekwinai P, Sringkarin N, Supasorn O, Fungkrajai M, Wang Y-H, Chayakulkeeree M, Ngamskulrungroj P, Angkasekwinai N, Pattanapanyasat K. 2014. Cryptococcus gattii infection dampens Th1 and Th17 responses by attenuating dendritic cell function and pulmonary chemokine expression in the immunocompetent hosts. Infect Immun 82:3880–3890. doi: 10.1128/IAI.01773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huffnagle GB, Yates JL, Lipscomb MF. 1991. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J Exp Med 173:793–800. doi: 10.1084/jem.173.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mody CH, Chen GH, Jackson C, Curtis JL, Toews GB. 1993. Depletion of murine CD8+ T cells in vivo decreases pulmonary clearance of a moderately virulent strain of Cryptococcus neoformans. J Lab Clin Med 121:765–773. [PubMed] [Google Scholar]
- 14.Huston SM, Li SS, Stack D, Timm-McCann M, Jones GJ, Islam A, Berenger BM, Xiang RF, Colarusso P, Mody CH. 2013. Cryptococcus gattii is killed by dendritic cells, but evades adaptive immunity by failing to induce dendritic cell maturation. J Immunol 191:249–261. doi: 10.4049/jimmunol.1202707. [DOI] [PubMed] [Google Scholar]
- 15.Ueno K, Kinjo Y, Okubo Y, Aki K, Urai M, Kaneko Y, Shimizu K, Wang DN, Okawara A, Nara T, Ohkouchi K, Mizuguchi Y, Kawamoto S, Kamei K, Ohno H, Niki Y, Shibuya K, Miyazaki Y. 2015. Dendritic cell-based immunization ameliorates pulmonary infection with highly virulent Cryptococcus gattii. Infect Immun 83:1577–1586. doi: 10.1128/IAI.02827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, Jeffrey KL, Anthony RM, Kluger C, Nchinda G, Koh H, Rodriguez A, Idoyaga J, Pack M, Velinzon K, Park CG, Steinman RM. 2010. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell 143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalod M, Chelbi R, Malissen B, Lawrence T. 2014. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J 33:1104–1116. doi: 10.1002/embj.201488027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellman I, Steinman RM. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell 106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 19.Vieira OV, Botelho RJ, Grinstein S. 2002. Phagosome maturation: aging gracefully. Biochem J 366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savina A, Amigorena S. 2007. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev 219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 21.Yates RM, Hermetter A, Russell DG. 2005. The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic 6:413–420. doi: 10.1111/j.1600-0854.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 22.Liebl D, Griffiths G. 2009. Transient assembly of F-actin by phagosomes delays phagosome fusion with lysosomes in cargo-overloaded macrophages. J Cell Sci 122:2935–2945. doi: 10.1242/jcs.048355. [DOI] [PubMed] [Google Scholar]
- 23.Poirier MB, Fiorino C, Rajasekar TK, Harrison RE. 2020. F-actin flashes on phagosomes mechanically deform contents for efficient digestion in macrophages. J Cell Sci 133:jcs239384. doi: 10.1242/jcs.239384. [DOI] [PubMed] [Google Scholar]
- 24.Holm A, Tejle K, Magnusson KE, Descoteaux A, Rasmusson B. 2001. Leishmania donovani lipophosphoglycan causes periphagosomal actin accumulation: correlation with impaired translocation of PKCalpha and defective phagosome maturation. Cell Microbiol 3:439–447. doi: 10.1046/j.1462-5822.2001.00127.x. [DOI] [PubMed] [Google Scholar]
- 25.Meresse S, Unsworth KE, Habermann A, Griffiths G, Fang F, Martinez-Lorenzo MJ, Waterman SR, Gorvel JP, Holden DW. 2001. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell Microbiol 3:567–577. doi: 10.1046/j.1462-5822.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- 26.Johnston SA, May RC. 2010. The human fungal pathogen Cryptococcus neoformans escapes macrophages by a phagosome emptying mechanism that is inhibited by Arp2/3 complex-mediated actin polymerisation. PLoS Pathog 6:e1001041. doi: 10.1371/journal.ppat.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinman RM, Witmer MD. 1978. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc Natl Acad Sci U S A 75:5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenschow DJ, Walunas TL, Bluestone JA. 1996. CD28/B7 system of T cell costimulation. Annu Rev Immunol 14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 29.Shahinian A, Pfeffer K, Lee KP, Kundig TM, Kishihara K, Wakeham A, Kawai K, Ohashi PS, Thompson CB, Mak TW. 1993. Differential T cell costimulatory requirements in CD28-deficient mice. Science 261:609–612. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- 30.Hagen F, Ceresini PC, Polacheck I, Ma H, van Nieuwerburgh F, Gabaldon T, Kagan S, Pursall ER, Hoogveld HL, van Iersel LJ, Klau GW, Kelk SM, Stougie L, Bartlett KH, Voelz K, Pryszcz LP, Castaneda E, Lazera M, Meyer W, Deforce D, Meis JF, May RC, Klaassen CH, Boekhout T. 2013. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One 8:e71148. doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salic A, Mitchison TJ. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miksa M, Komura H, Wu R, Shah KG, Wang P. 2009. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J Immunol Methods 342:71–77. doi: 10.1016/j.jim.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol 155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. 2007. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 36.Baranov MV, Revelo NH, Dingjan I, Maraspini R, Ter Beest M, Honigmann A, van den Bogaart G. 2016. SWAP70 organizes the actin cytoskeleton and is essential for phagocytosis. Cell Rep 17:1518–1531. doi: 10.1016/j.celrep.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schliwa M. 1982. Action of cytochalasin D on cytoskeletal networks. J Cell Biol 92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasare C, Medzhitov R. 2004. Toll-dependent control mechanisms of CD4 T cell activation. Immunity 21:733–741. doi: 10.1016/j.immuni.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. 2001. Toll-like receptors control activation of adaptive immune responses. Nat Immunol 2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 40.Anes E, Kuhnel MP, Bos E, Moniz-Pereira J, Habermann A, Griffiths G. 2003. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat Cell Biol 5:793–802. doi: 10.1038/ncb1036. [DOI] [PubMed] [Google Scholar]
- 41.Defacque H, Egeberg M, Habermann A, Diakonova M, Roy C, Mangeat P, Voelter W, Marriott G, Pfannstiel J, Faulstich H, Griffiths G. 2000. Involvement of ezrin/moesin in de novo actin assembly on phagosomal membranes. EMBO J 19:199–212. doi: 10.1093/emboj/19.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lerm M, Brodin VP, Ruishalme I, Stendahl O, Sarndahl E. 2007. Inactivation of Cdc42 is necessary for depolymerization of phagosomal F-actin and subsequent phagosomal maturation. J Immunol 178:7357–7365. doi: 10.4049/jimmunol.178.11.7357. [DOI] [PubMed] [Google Scholar]
- 43.Mellman I. 1989. Organelles observed: lysosomes. Science 244:853–854. doi: 10.1126/science.244.4906.853. [DOI] [PubMed] [Google Scholar]
- 44.Bandyopadhyay D, Cyphersmith A, Zapata JA, Kim YJ, Payne CK. 2014. Lysosome transport as a function of lysosome diameter. PLoS One 9:e86847. doi: 10.1371/journal.pone.0086847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 46.Ma H, Croudace JE, Lammas DA, May RC. 2006. Expulsion of live pathogenic yeast by macrophages. Curr Biol 16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 47.Heinsbroek SE, Kamen LA, Taylor PR, Brown GD, Swanson J, Gordon S. 2009. Actin and phosphoinositide recruitment to fully formed Candida albicans phagosomes in mouse macrophages. J Innate Immun 1:244–253. doi: 10.1159/000173694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemeth ZH, Deitch EA, Davidson MT, Szabo C, Vizi ES, Hasko G. 2004. Disruption of the actin cytoskeleton results in nuclear factor-kappaB activation and inflammatory mediator production in cultured human intestinal epithelial cells. J Cell Physiol 200:71–81. doi: 10.1002/jcp.10477. [DOI] [PubMed] [Google Scholar]
- 49.Rosette C, Karin M. 1995. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-kappa B. J Cell Biol 128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C, Haynes EM, Asokan SB, Simon JM, Sharpless NE, Baldwin AS, Davis IJ, Johnson GL, Bear JE. 2013. Loss of Arp2/3 induces an NF-kappaB-dependent, nonautonomous effect on chemotactic signaling. J Cell Biol 203:907–916. doi: 10.1083/jcb.201306032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A, Wang J, Brunham R, Fyfe M, Ouellette BFF, Siddiqui A, Marra M, Jones S, Holt R, Birren BW, Galagan JE, Cuomo CA. 2011. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2:e00342-10. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, Bicanic TA, Castaneda E, Chang YC, Chen J, Cogliati M, Dromer F, Ellis D, Filler SG, Fisher MC, Harrison TS, Holland SM, Kohno S, Kronstad JW, Lazera M, Levitz SM, Lionakis MS, May RC, Ngamskulrongroj P, Pappas PG, Perfect JR, Rickerts V, Sorrell TC, Walsh TJ, Williamson PR, Xu J, Zelazny AM, Casadevall A. 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2:e00357-16. doi: 10.1128/mSphere.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dingjan I, Linders PT, van den Bekerom L, Baranov MV, Halder P, Ter Beest M, van den Bogaart G. 2017. Oxidized phagosomal NOX2 complex is replenished from lysosomes. J Cell Sci 130:1285–1298. doi: 10.1242/jcs.196931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ratio of Cryptococcus cells per infected DC and effect of culture plates and well size on DC phagocytosis frequency. (A) Phagocytic frequency 1 h after infection (MOI of 10 for 2 h), measured by confocal microscopy, in DCs infected with Cryptococcus or fed polystyrene beads. DCs were infected in either 24-well plastic plates, 24-well plates containing glass coverslips, 96-well plastic plates, or 96-well plates with a glass-like coverslip bottom. The frequency was calculated as the number of DCs with internalized cryptococci or beads (confirmed by phalloidin labeling of cortical actin) relative to the total number of DCs in a field of view (FOV). Data are presented as means ± SEM for 54 to 67 FOVs from 3 donors. ***, P < 0.001 (by one-way ANOVA [adjusted P values] of beads compared to Cryptococcus in each column). (B) Percentage of infected DCs containing 1, 2, or 3 or more intracellular Cryptococcus cells. DCs were infected with Cryptococcus (MOI = 10) for 2 h and washed to rinse excess yeast. Infected DCs were fixed with 4% PFA and permeabilized with saponin 1 h after infection. Fixed cells were labeled with phalloidin and imaged by confocal microscopy. Data represent means ± standard deviations (SD) for 77 to 86 FOVs pooled from 3 donors. Download FIG S1, TIF file, 0.3 MB (299.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Fluorescence emission from R265, CBS7750, and H99 cryptococcal strains labeled with pHrodo under acidic conditions. (A) Gating strategy for Fig. 2C. The gate (black oval) is set on total DCs as determined by forward scatter (FSC) and side scatter (SSC). (B) Representative flow cytometry histograms (gated on fungi) of cryptococcal strains labeled with pHrodo and cultured in PBS at either pH 7.4 or pH 5.5 for 15 min (room temperature [RT]). The MFI under each condition is shown below the histograms. (C) Quantification of data in panel B. Data are presented as means ± SD of the average MFIs of pHrodo-labeled cryptococci at pH 7.4 and pH 5.5. Averages of data from 3 separate experiments are shown. Download FIG S2, TIF file, 0.6 MB (608.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Live-cell microscopy of C. gattii R265 yeast cells engulfed by a DC, which was stained with SiR-actin (red) to label F-actin and LysoTracker (cyan) to label lysosomes prior to culture with R265. The cell was imaged every 1 min for just over an hour across 3 channels and multiple confocal stacks. The acquisition time is on the top left, and the scale bar is on the bottom right. Download Movie S1, MOV file, 5.6 MB (5.6MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
C. gattii R265 phagosomal F-actin fluorescence intensity over time and size of DC phagosomes containing Cryptococcus. (A) Quantification of data in Fig. 4B. The F-actin fluorescence intensity over the course of 1 h is shown. Data are presented as means ± SD at 6-min intervals and are averages from 3 separate experiments. DCs were labeled with 200 nM SiR-actin dissolved in RPMI culture medium with the addition of verapamil for 3 to 4 h in a 37°C cell incubator. DCs were then cocultured with C. gattii R265 for 30 min at 37°C. Cells were imaged every 1 to 2 min for 1 h. (B) Phagosome size was measured as an average from two perpendicular planes at the confocal stack correlating to the widest diameter using confocal microscopy. Phagosome borders were determined by the boundary of the yeast, visualized by DIC, and LAMP-1 or F-actin staining. Data are presented as means ± SD and are averages for 65 phagosomes from 3 different donors. The mean (μ) under each condition is shown. Polystyrene beads (3.3 μm) were used as a control. Download FIG S3, TIF file, 0.2 MB (177.6KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Animation of the 3D reconstruction of an SR-SIM micrograph (shown in Fig. 5) showing a DC that contains C. gattii R265 within the F-actin cage (red ball). F-actin was stained with phalloidin (red), and lysosomes were stained with LAMP-1 antibodies (green). Download Movie S2, MOV file, 6.6 MB (6.6MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cropped section of the phagosomal region shown in Movie S2 in the supplemental material. F-actin was stained with phalloidin (red), and lysosomes were stained with LAMP-1 antibodies (green). Download Movie S3, MOV file, 3.5 MB (3.5MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effects of transient cytochalasin D treatment and metabolic activity on C. gattii R265 phagosomal F-actin frequency. (A) Representative confocal micrographs of R265-infected DCs treated with cytochalasin D for either 1 h or 2 h or with DMSO (vehicle control). After cells were washed and incubated for 4 h in the absence of the inhibitor, cells were fixed with PFA and immunostained for LAMP-1 (green), phalloidin to label F-actin (red), and DAPI to stain the nucleus (blue). The phagosome region (represented as white squares in the DIC panels) was cropped and digitally magnified to view phagosomal F-actin and LAMP-1 in single confocal stacks. Bar = 10 μm. (B) Quantification of data in panel A. Data are presented as stacked bar graphs of the averages for phagosomes per FOV that are enriched for F-actin, LAMP-1, both, or neither. The number of FOVs (n) under each condition is shown. Data are representative of results from 3 donors. (C) Comparison of live and dead Cryptococcus cells for the frequency of phagosomal F-actin. Cryptococcus cells were either heat killed (HK) for 60 min in a 60°C water bath or fixed with 4% PFA for 30 min. Data are presented as means ± SEM of the percentages of phagosomes in the FOV. Shown are averages for 10 FOVs from separate experiments. *, P < 0.05; **, P < 0.01 (by two-way ANOVA [adjusted P values]). Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effects of transient cytochalasin D treatment on total DC F-actin and Cryptococcus growth. (A) Representative histogram of DCs unlabeled or labeled with phalloidin conjugated to Alexa Fluor 555. The table on the right shows the MFI under each condition and DC viability as measured by trypan blue exclusion at the end of the experiment. DCs were treated with either 2 μg/ml cytochalasin D or DMSO for 2 h in a 37°C CO2 incubator, followed by a 4-h period of incubation in inhibitor-free culture medium after being washed. After the 4-h period, cells were fixed with 4% PFA and permeabilized with saponin. Fixed samples were labeled with phalloidin at a 1:200 (vol/vol) dilution for 30 min at room temperature. Data are representative of results from 3 donors. (B) CFU of different Cryptococcus strains at 6 and 24 h. Cryptococcus cells were either untreated or treated with 2μg/ml of CytoD or DMSO for 2 h in a 37°C 5% CO2 incubator, washed, and incubated in inhibitor-free medium for an additional 4 h. Cryptococcal yeast cells were counted, and approximately 1,000 yeast cells were plated in a 96-well plate in culture medium. Experiments were done in triplicates. CFU were measured either immediately or 24 h later. Data are representative of results from 3 donors. Download FIG S5, TIF file, 0.4 MB (407.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cropped section of the phagosomal region shown in Movie S1 in the supplemental material. The time-lapse of F-actin (labeled with SiR-actin) is shown as a heat map; red represents hot pixels, and white represents cooler pixels. The acquisition timer is on the top right. Download Movie S4, MOV file, 9.4 MB (9.4MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of reagents. Download Table S1, DOCX file, 0.02 MB (20.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.