Abstract
The neuropancreatic polypeptide hormone amylin forms pancreatic islet amyloid in type-2 diabetes. Islet amyloid formation contributes to β-cell death in the disease and to the failure of islet transplants, but the features which influence amylin amyloidogenicity are not understood. We constructed an amino acid sequence alignment of 202 sequences of amylin and used the alignment to design consensus sequences of vertebrate amylins, mammalian amylins, and primate amylins. Amylin is highly conserved, but there are differences between human amylin and each consensus sequence, ranging from one to six substitutions. Biophysical analysis shows that all of the consensus sequences form amyloid, but do so more slowly than human amylin in vitro. The rate of amyloid formation by the primate consensus sequence is 3 to 4-fold slower than human amylin, the mammalian consensus sequence is approximately 20 to 25-fold slower, and the vertebrate consensus sequence approximately 6-fold slower. All of the consensus sequences are moderately less toxic than human amylin towards a cultured β-cell line, with the vertebrate consensus sequence displaying the largest reduction in toxicity of 3 to 4-fold. All of the consensus sequences activate a human amylin receptor and exhibit only modest reductions in activity, ranging from 3 to 4-fold as judged by a cAMP production assay. The analysis argues that there is no strong selective evolutionary pressure to avoid the formation of islet amyloid and provides information relevant to the design of less amyloidogenic amylin variants.
Keywords: Amylin, Amyloid, Consensus Sequence, Islet Amyloid Polypeptide, Type-2 Diabetes
INTRODUCTION
The neuropancreatic polypeptide hormone amylin, also known as islet amyloid polypeptide (IAPP), is produced by the pancreatic β-cells and is secreted in response to the same stimuli that lead to insulin secretion (1–11). Amylin has an adaptive role in metabolism, but forms pancreatic islet amyloid in type-2 diabetes. Islet amyloid formation, while not believed to be the cause of type-2 diabetes, contributes to β-cell dysfunction and death in the disease (2, 12, 13). Rapid amyloid formation has also been linked to the failure of islet transplants and could be a key factor limiting this potential treatment of type-1 diabetes (14–16). Some organisms do not develop type-2 diabetes and there is a strong correlation between those which do develop type-2 diabetes and the observation of islet amyloid in vivo and the in vitro amyloidogenicity of amylin (1, 17–20). For example, rat and mouse amylin, which have identical sequences, are not amyloidogenic under normal conditions (13) and neither species develops islet amyloid or type-2 diabetes, nor do cultured wild type mouse islets. Rat IAPP has been reported to form amyloid in vitro, but only at very high non-physiological conditions. In contrast, human amylin is aggressively amyloidogenic in vitro and mice transgenic for the human amylin gene develop islet amyloid (17,18). Pre-amyloid oligomers appear to be the most toxic species produced during amyloid formation by human amylin and a range of independent studies have demonstrated that amylin exerts its toxic effects via multiple mechanisms (11, 13, 21).
Type-2 diabetes, until relatively recently, was a disease that developed later in life, typically after childbearing years. Thus, there may be little to no selective pressure to avoid islet amyloid formation. In contrast, some animals have adapted to thrive in metabolically stressful environments and here one may expect evolutionary advantages to be provided by resistance to islet amyloid formation (14, 22, 23). For example, polar bears eat an extremely high fat diet and have a form of amylin which is less amyloidogenic than humans (23). In contrast, mice transgenic for human amylin develop islet amyloid and diabetes if fed a high fat diet, even one with a notably lower percentage of fat than the natural polar bear diet (18). In addition, porcine and bovine amylin are much less amyloidogenic than human amylin and domesticated pigs and cattle are believed not to develop type-2 diabetes (14, 23). Sequence differences between species have been exploited to develop first generation non-amyloidogenic analogs of human amylin for clinical applications. In particular, comparative analysis of human vs rat/mouse amylin has led to a variant of amylin, pramlintide, also known as Symlin which is used clinically (24–27). There is also iinterest in potential combined leptin pramlintide or GLP-1 agonist pramlintide therapy for obesity.
Human amylin lacks proline (Pro) residues while rat/mouse amylin contains prolines at position 25, 28, and 29 and a H18R replacement relative to human amylin. The multiple proline residues found in rat amylin have generally been thought to be responsible for its reduced amyloidogencity. Pramlintide is a triple mutant of human amylin with A25, S28, and S29 replaced by Pro. Pramlintide is not amyloidogenic in vitro, except at high concentrations. Nevertheless, next generation bioactive analogs of human amylin with increased solubility are desired, as the low solubility of pramlintide at neutral pH precludes co-formulation with insulin formulations. However, despite considerable work, the features which control amylin amyloidogenicity are still not fully understood and no alternative peptide analogs of amylin have been approved for clinical use.
We constructed a multiple sequence alignment using 202 sequences of amylin and examined the in vitro amyloidogenicity, cytotoxicity, and receptor activity of amylin consensus sequences, in order to gain insight into factors which modulate amyloidogenicity and to examine if there may be selective pressure to avoid islet amyloid formation. Consensus sequences are derived by selecting the most common residue found at each position in the alignment. The approach has been widely applied to soluble globular proteins and can lead to proteins with enhanced stability or other desirable traits (28–36). The analysis of consensus sequences has also been used to infer evolutionary relationships (28, 29, 35, 36). While commonly applied to globular proteins, the methodology has not been broadly used to study amyloidogenic proteins and polypeptides. We designed consensus sequences of all vertebrate amylins, all mammalian amylins, and all primate amylins in the alignment. All of the consensus sequences form amyloid, but do so more slowly than human amylin in vitro. All of the consensus sequences are moderately less toxic than human amylin towards a cultured β-cell line. All of the consensus sequences activate a human amylin receptor and exhibit only modest reductions in activity. The analysis argues that there is no strong selective evolutionary pressure to avoid the formation of islet amyloid and provides information relevant to the design of less amyloidogenic amylin variants.
MATERIALS AND METHODS
Peptide Synthesis and Purification.
Human amylin and the three consensus sequence peptides were synthesized on a 0.10 mmol scale using 9-Fluorenylmethyloxycarbonyl (Fmoc) chemistry using a microwave peptide synthesizer (CEM Liberty Blue). Fmoc-PAL-PEG-PS resin (0.18mmol/eq) was used to provide a C-terminus amide. Pseudoproline derivatives were used at residue 9 and 10 (Ala and Thr), residue 19 and 20 (Ser and Ser), and residue 27 and 28 (Leu and Ser) to minimize aggregation during synthesis (37–39). The first residue attached to the resin, β-branched amino acids, Arg, and all pseudoproline dipeptide derivatives were double coupled. TFA based cleavage cocktails (92.5% trifluoroacetic acid, 2.5% triisopropylsilane, 2.5% 3,6-Dioxa-1,8-Octanedithiol, and 2.5% water) were used to release the peptides from the resin and remove side chain protecting groups. Crude peptides were dissolved in 20% acetic acid (4 mg/ml) frozen in liquid nitrogen and lyophilized to increase their solubility. The disulfide bond between residues 2 and 7 was formed by dimethyl sulfoxide (DMSO) oxidation (10mg/ml of peptide in 100% DMSO) on a shaker at room temperature for 3 to 4 days. Peptides were purified using reverse-phase HPLC (Higgins analytical C18 preparative column, 25mm × 250mm) with buffer A (100% H2O and 0.045% HCl) and buffer B (80% Acetonitrile, 20% H2O, and 0.045% HCl). HCl was used as a counterion instead of TFA since TFA can affect thioflavin- T kinetic assays and cell toxicity experiments. A second HPLC purification was used to remove residual scavengers. Peptide was dissolved in 100% Hexafluoroisopropanol (HFIP) (10 mg/ml), allowed to sit for 4 hours and then purified via HPLC. Analytical HPLC and either Maldi-TOF or direct injection electrospray mass spectrometry were performed to confirm the mass and purity of the peptides: human amylin (WT), expected 3903.32, observed 3903.03; primate consensus sequence, expected 3922.36, observed 3921.89; mammalian consensus sequence, expected 3898.38, observed 3897.91; vertebrate consensus sequence, expected 3977.44, observed 3977.34.
Preparation of Peptide Stock Solutions for Biophysical Measurements.
Dry purified peptides were dissolved in 100% HFIP at room temperature for 4 hours. Peptide stock solutions were filtered using 0.22 μm Millex low protein binding durapore membrane filter. Concentrations were calculated using a molar extinction coefficient of 1615 M−1cm−1 at 280 nm. Aliquots of the desired amount of peptide were lyophilized for 24 hours in order to remove HFIP. Samples were reconstituted in PBS to initiate thioflavin- T amyloid assays.
Thioflavin T Kinetic Assays.
Thioflavin T kinetic assays were performed to monitor the kinetics of amyloid formation in 10 mM phosphate with 140 mM KCl, pH 7.4 using a Molecular Devices SpectraMax Gemini EM microplate reader with dual monochromators (450 nm excitation wavelength, 485 nm emission wavelength). Assays were carried out at 25 °C without shaking. The final concentration of the peptide and thioflavin-T were 16 and 32 μM respectively. This concentration of thioflavin-T has previously been shown not to perturb he kinetics of amylin amyloid formation under the conditions of these assays (40). The data were analyzed using Microsoft Excel.
Transmission Electron Microscopy (TEM).
Transmission electron microscopy (TEM) was used to confirm presence of amyloid fibrils. Images were recorded at the Life Science Microscopy Center at the State University of New York at Stony Brook. 15 μl of samples taken from the thioflavin-T kinetic assay at the end of the experiment and were blotted on a Carbon-coated Formvar 300 mesh copper grid for 1 minute and then stained negatively using 2 % saturated uranyl acetate for 1 minute. TEM images were recorded using a FEI Bio TwinG2 transmission electron microscope.
Cytotoxicity Assays.
INS-1 cells were purchased from AddexBio and cultivated in optimized RPMI-1640 (AddexBio, #C0004–02) with 10% ultra-low IgG FBS (Gibco, #16250078). CellTiter-Glo 2.0 (Promega, #G9242), assays were used to monitor the cytotoxic effects of the peptides on INS-1 cells. Peptide concentrations ranging from 4 to 200 μM were used to construct concentration response curves and estimate the value of EC50, the concentration required to achieve 1/2 of the maximal effect. Cells were seeded at ~50 % confluence on 96-well half-area clear bottom white plates (Greiner, #675083) and incubated for 36 hours at 37°C in a 5 % CO2 humidified incubator. Cultured cells were exposed to the peptide in fresh complete medium for a further 24 hours. Serial dilutions of the peptides were freshly prepared before use from lyophilized peptide aliquots. Culture plates were cooled to room temperature and an equal volume of the assay reagent was added to the treated cells for the CellTiter-Glo 2.0 assays. The plates were shaken for 1 min at 700 rpm and the luminescence intensity was measured with a Clariostar plate reader. Alamar Blue assays were independently performed by adding an equal volume of 20 % of Alamar Blue reagent in complete medium to the cells and the plates were further incubated at 37°C for ~ 1 hour. Fluorescence emission of the Alamar Blue reduction product was monitored at 590 nm with excitation at 550 nm. The CellTox Green assays were conducted by exposing the cells to the different peptides in the presence of the assay dye (1:5000 dilution) for 24 hours, after which the fluorescence intensity was measured at 525 nm with excitation at 480 nm. Calculation of EC50 values and statistical analysis were performed using Graph Pad Prism 5.
Cell Culture and Transient Transfections.
Cos-7 cells were cultured and transfected for receptor activity assays as described previously (41). Transient transfections were carried out using polyethyleneimine (PEI) and maintained at 37 °C in a humidified 95% air/5% CO2 incubator for 36–48 hours. All of the DNA constructs used in these experiments were in pcDNA3.1 vectors. The insert-negative human CTR with leucine at the polymorphic amino acid position 447 with an N-terminal hemeagglutinin (HA) tag (HA-hCTR) and myc-tagged hRAMP1 was utilized in these experiments.
Cell Signaling Assays.
Cos7 cells were serum starved in cAMP assay media for 30 minutes at 37 °C prior to peptide stimulation. Peptides were serially diluted in cAMP assay media and cells were incubated with assay media alone or with each concentration of peptide at 37 °C for 15 minutes. Media was then aspirated and the reaction was stopped with the addition of 50 μl ethanol for at least 10 minutes at −20 °C. Ethanol was evaporated in a fume hood, and the cells were lysed with cAMP detection buffer (0.35% Triton X-100, 50mM HEPES and 10mM calcium chloride in ddH2O, pH 7.4), shaken at room temperature for 15 minutes. Intracellular cAMP was measured using a time-resolved fluorescent resonance energy transfer assay (LANCE cAMP assay, PerkinElmer Life and Analystical Sciences, Waltham, MA, USA), similar to the previously described Alphascreen assay (42). Plates were read after a 4-hour incubation on an Envision plate reader (PerkinElmer Life and Analytical Sciences, Waltham, MA, USA). Data were from at least four independent experiments and all experiments were performed with two or three technical replicates. Quantification of cAMP was achieved using a standard curve included in each experiment and data were plotted using the software GraphPad Prism 8.2 (GraphPad Software Inc, San Diego, CA, USA). pEC50 values were determined using non-linear regression with a 3-parameter logistic equation with a Hill coefficient of 1. Data were combined and normalized to wild-type human amylin using the minimum (Emin) and maximum (Emax) response of human amylin and tested for statistical significance by comparing to the wild type human peptide using an unpaired Student’s t-test, with statistical significance defined as * p < 0.05.
RESULTS
Amylin sequence alignment
Amylin sequences from various species were retrieved from prior work (21) and from reported sequences found in the National Center for Biotechnology Information (NCBI) protein database. Mature amylin sequences were distinguished from pre pro and pro amylin sequences based on a number of conserved features. These include the disulfide bond between residues 2 and 7, and the presence of a C-terminal aromatic residue or His. There are also conserved residues in the pro regions which are required for the processing of pro-amylin by prohormone convertases (1, 43, 44). These include residues involved in the processing of the C-terminal pro extension and amidation of residue 37 in mature amylin. mature amylin. Residue C+1, the residue immediately C-terminal to residue 37 of the mature polypeptide, is always a Gly. This Gly acts as the nitrogen donor for amidation. Residues C+2 and C+3 are always basic (43, 44). All of the retrieved sequences shared all of these common features. A total 202 amylin sequences were aligned and categorized into three groups; primate amylins, mammalian amylins, and vertebrate amylins (Table-S1, Figure-1, Figure-2). The level of conservation is very high. At 21 of the 37 positions the most common residue is found in at least 93% of the sequences. At even the most variable positions, residues 8, 14, 22, 23, 25, 26, 27, 28, 29 and 35, the most common residue is found between 54 and 88% of the time, and the replacements are often conservative substitutions. Overall the greatest region of variability is found between residues 22 to 29, with the exception of position 24 which is almost always Gly (Table-S1, Figure-1A).
Figure-1.
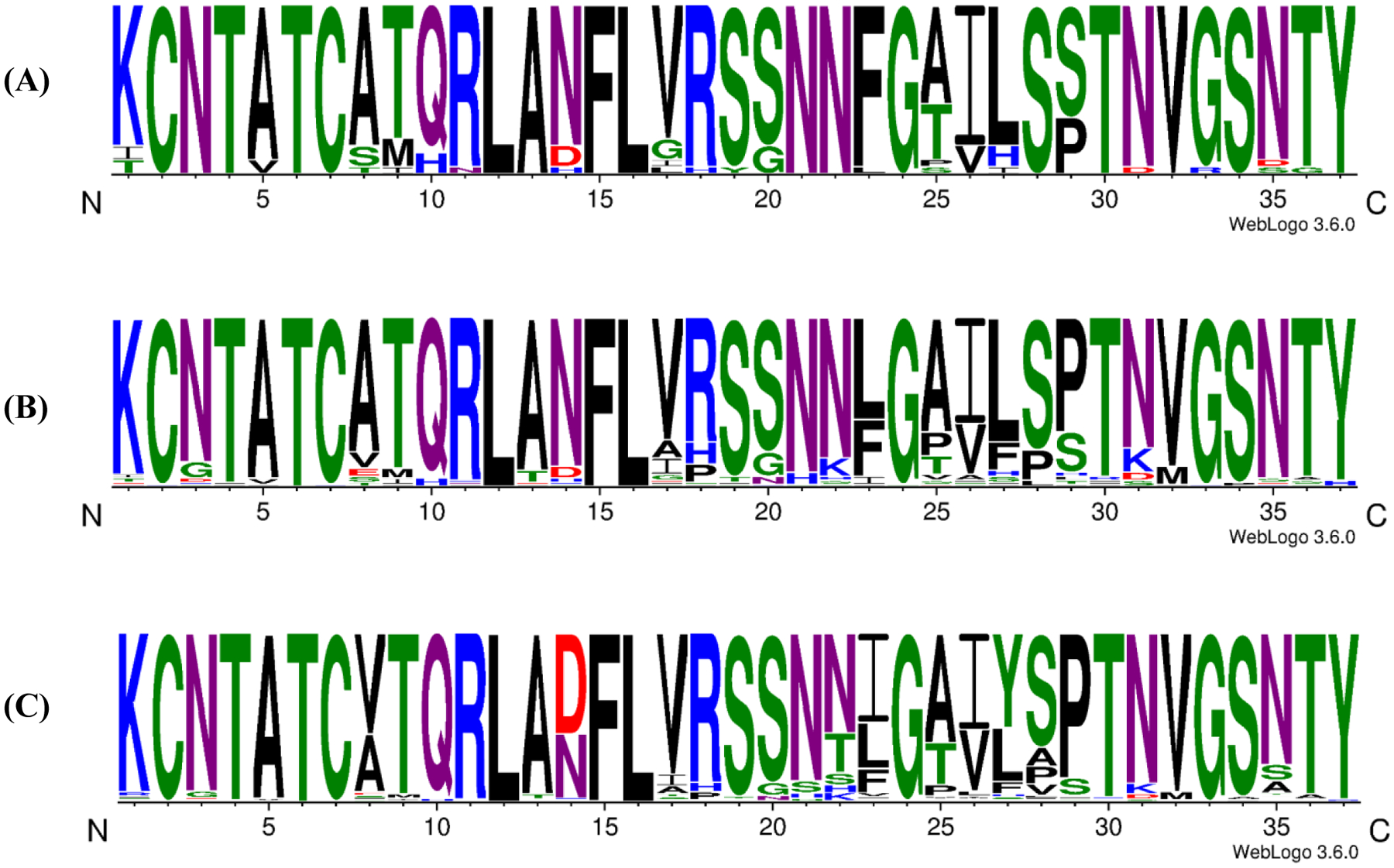
Schematic representation of the amino acid sequences of (A) Primate amylins. (B) Mammalian amylins. (C) Vertebrate amylins. The height of the letter represents the abundance of the different residues at each position. Color coding corresponds to: Blue, basic (K, R, H); Green, polar (G, S, T, Y, C); Purple amide sidechains (N, Q); Red, acidic (D, E); Black, hydrophobic (A, V, L, I, P, W, F, M).
Figure-2.
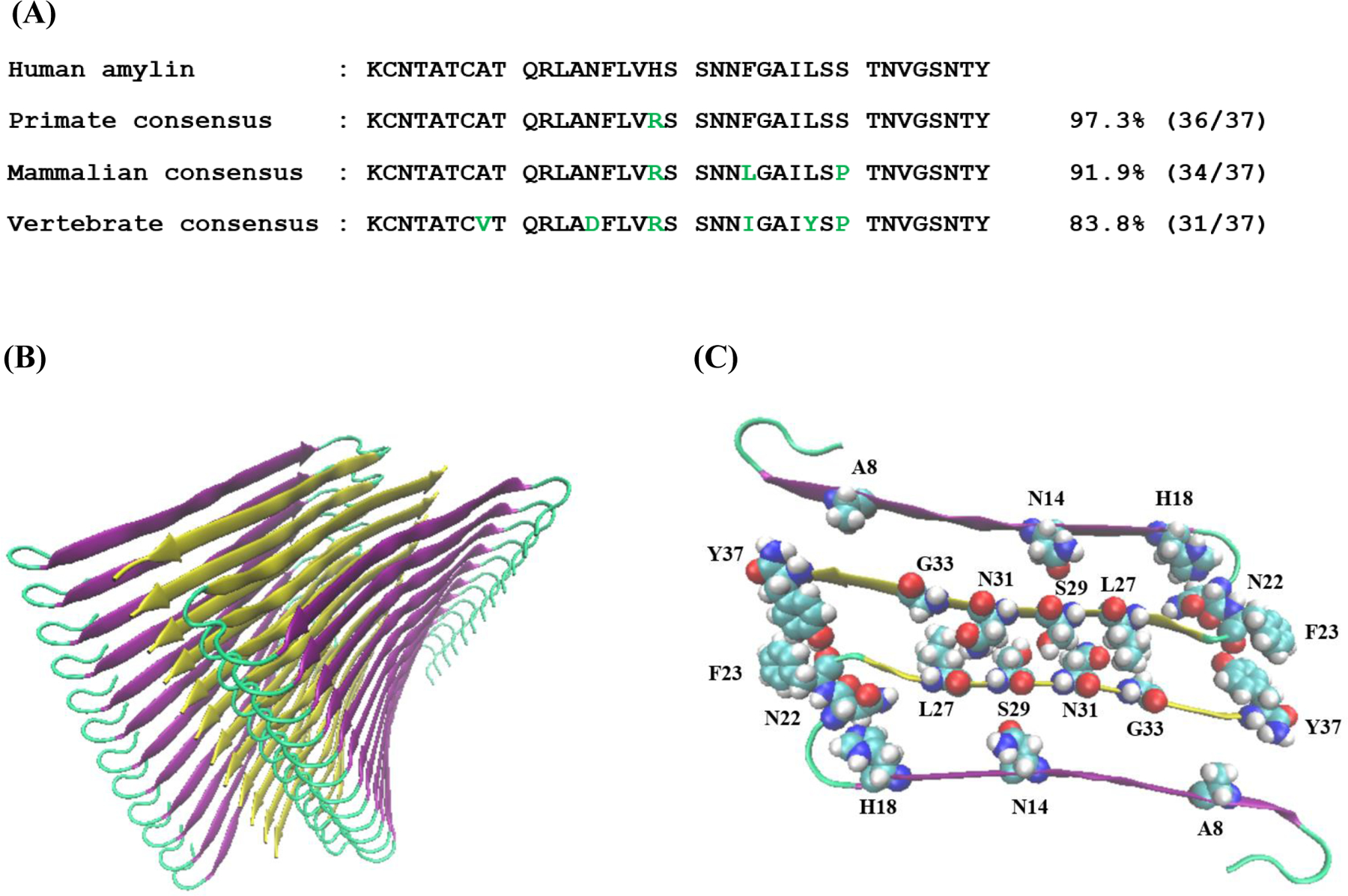
(A) Amino acid sequence of human amylin and consensus sequences derived from the sequence alignment. The numbers refer to % identity. Residues which differ from human amylin are in green. (B) A ribbon diagram of the high-resolution model of the human amylin amyloid fiber (C) A top down view of one layer of the model shown in panel (B). Residues in the human peptide which differ in the vertebrate consensus sequence are shown in space filling format as are selected residues, which pack against them.
Using this alignment, we construct three consensus sequences: one for the primate sequences, a second for the mammalian sequences, and the third for all sequences in the alignment, i.e. for vertebrate amylins (Figure-1, Figure-2). Figure 2 also shows the location of the residues which differ from those in human amylin in the context of a high-resolution model of the human amylin amyloid fibers. There are two such models available of the amyloid fibers formed at pH 7.4, one based on crystal structures of small peptide fragments of human amylin and the other based on solid state NMR studies of human amylin (45, 46). Although there are some differences in these two structures, they share many common features. Both structures consist of two columns of amylin monomers. The monomers each contribute two β-strands to the fiber with the strands linked by a partially ordered loop. There are no backbone hydrogen bonds within a monomer rather the β-sheet hydrogens occur between different monomers in the same column. The N-terminal β-strand in the x-ray derived model consists of residues 8 to 17 and the C-terminal β-strand is made up of residues 23 to 37. The two columns pack against each other via sidechain-sidechain interactions from residues in the C-terminal β-strand. Residues 23, 25, 27, 29, 31, 33, 35, and 37 project into the interface of the two columns while even numbered residues (24 through 36) project into the internal of a column. The interface in this model involves a tightly packed steric zipper (45). A recent cryo-em structure of human amylin fibers formed at pH 6.0 reveals a different structure (accessible as pdb code 6Y1A). In this structure density could be defined for residues 13 through 37. One layer of the fiber is made up of 2 monomers, similar to the structural models of the pH7.4 fibers, but the monomers adopt a compact S-shape conformation instead of the U-shaped conformation. The β-strands in this structure are comprised of residues 14 to 20 and residues 26 to 37, excluding a turn centered at residue 33. Residues 22 through 27 are in the central fibril core. The sidechain of His-18 projects into the core of the fiber in the pH 7.4 structure, but projects outward in the pH 6.0 structure, perhaps reflecting a difference in protonation state of the His-18 sidechain and it is tempting to postulate that this could contribute to the formation of the different structures. The interface between the two monomers is different between the pH 6.0 and pH 7.4 structure.
Despite the highly conserved nature of the amylin sequence, human amylin exhibits some interesting variations compared to other amylins. Consider first the primate sequences, human amylin is the only one with a His at position 18; this site is Arg in all other known primate sequences and is found in over 89% of all the sequences analyzed. Prior reports have suggested that a H18R substitution reduces the rate of amyloid formation by human amylin, in vitro (47). The consensus sequence for all mammalian amylins is identical to the human sequence at 34 of the 37 positions, differing only at position 18, 23, and 29. These residues are His, Phe, and Ser in human amylin respectively and are Arg, Leu, and Pro in the mammalian consensus sequence. The effect of individually replacing the aromatic residues in human amylin on the rate of in vitro amyloid formation has previously been examined and a Phe 23 to Leu substitution increases the time course of amylin formation by approximately a factor of 2 at pH 7.4 in vitro under the conditions studied (40). The consensus sequence derived from the complete alignment differs from human amylin at 6 out of 37 positions and contains A8V, N14D, H18R, F23I, L27Y, and S29P substitutions relative to human amylin.
Consensus sequences form amyloid, but do so more slowly than human amylin.
We compared the ability of the three consensus sequences and human amylin to form amyloid using fluorescence detected thioflavin-T binding assays and TEM. Thioflavin-T is a dye which undergoes a significant increase in fluorescence upon binding to amyloid fibers and is widely used to monitor amyloid formation kinetics (48). We used TEM to directly verify the presence of amyloid fibers as thioflavin-T assays can give false negatives (49). The four peptides all displayed typical sigmodal thioflavin-T intensity vs. time curves consisting of a lag phase, a growth phase, and a final plateau (Figure-3 Supporting Figure S2). The concentration of thioflavin-T used in these assays has been shown to not perturb the kinetics of amylin amyloid formation under the conditions of these assays. Human amylin formed amyloid most rapidly as judged by the T50 time, where T50 is the time required to achieve 50% of the fluorescence change in a thioflavin-T assay (Table-1). The T50 for human amylin was measured to be 3.3 ± 1.0 hours in phosphate buffered saline (PBS, 10 mM phosphate, 140 mM KCl, pH 7.4), at 25°C. The primate consensus sequence formed amyloid with a T50 of 10.1 ± 2.6 hours and the next slowest was the vertebrate consensus sequence with a T50 of 20.8 ± 6.8 hours. The mammalian consensus sequence was the slowest to form amyloid with a T50 of 80.1 ± 28.7 hours. The quoted uncertainties reflect the apparent standard deviation of 12 measurements. TEM images collected at the end of the kinetic experiments revealed typical amyloid fibers for all four of the peptides (Figure-3).
Figure-3.
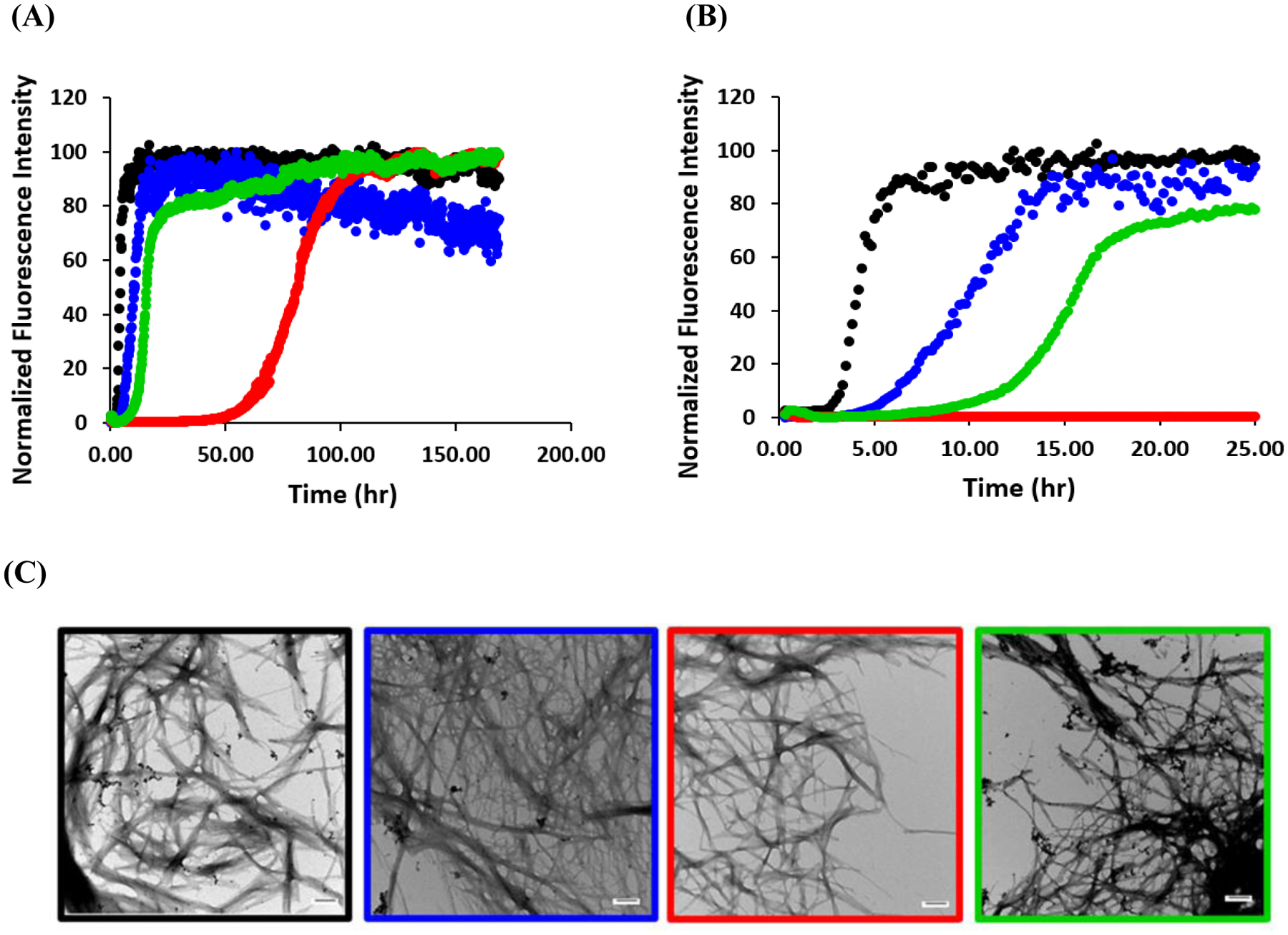
Human amylin forms amyloid faster than the consensus sequences. Representative curves from one experiment out of 12 are shown. (A) Thioflavin-T assays. Black, human amylin: Blue, primate consensus sequence: Red, mammalian consensus sequence: Green, vertebrate consensus sequence. (B) An expanded plot of the data showing the first 25 house of the time course. (C) TEM images, human (black box), primate consensus sequence (blue box), mammalian consensus sequence (red box) and vertebrate consensus sequences (green box). Scale bars represent 100 nm. Kinetic experiments were conducted at 25°C, pH 7.4 in phosphate buffered saline (10 mM phosphate, 140 mM KCl).
Table-1.
T50 values determined via thioflavin-T fluorescence assays for wild type human amylin and the three consensus sequences. Experiments were conducted at 25°C in PBS pH 7.4 at 16 μM peptide concentration. Reported uncertainties are the apparent standard deviation from 12 measurements.
| Peptide | T50 (hrs) |
|---|---|
| Human amylin | 3.3 ± 1.0 |
| Primate consensus | 10.1 ± 2.6 |
| Mammalian consensus | 80.1 ± 28.7 |
| Vertebrate consensus | 20.8 ± 6.8 |
Consensus sequences are toxic towards cultured β-cells, but have less effect than human amylin.
We next examined the effects of the three consensus sequences and human amylin on INS-1 cells, a well validated β-cell line. Concentration dependent response curves were generated using peptide concentrations ranging from 4 to 200 μM in order to determine EC50 values, where EC50 is the peptide concentration required to achieve 50 % of the maximum effect. We used three independent cell viability assays: one which measured net cellular ATP levels via luciferase (CellTiter-Glo® 2.0), a second that monitored the overall redox status of the cells via NAD(P)H-dependent resazurin reduction (Alamar Blue), and a third that monitored the permeability/integrity of the plasma membrane to a membrane impermeable DNA intercalating fluorescent probe (CellTox™ Green). The human polypeptide was consistently more cytotoxic than the consensus sequences (Figure-4) with an EC50 value approximately 2-fold less than the values measured for the primate and mammalian consensus sequences and 3.5 to 4-fold less than the value measured for the vertebrate consensus sequence (Figure-4, Table-2). The values of EC50 for the primate and mammalian consensus sequences are very similar, indicating that the two residue changes between these sequences (F23L and S29P) do not make significant contributions to toxicity, at least in a H18R background.
Figure-4.
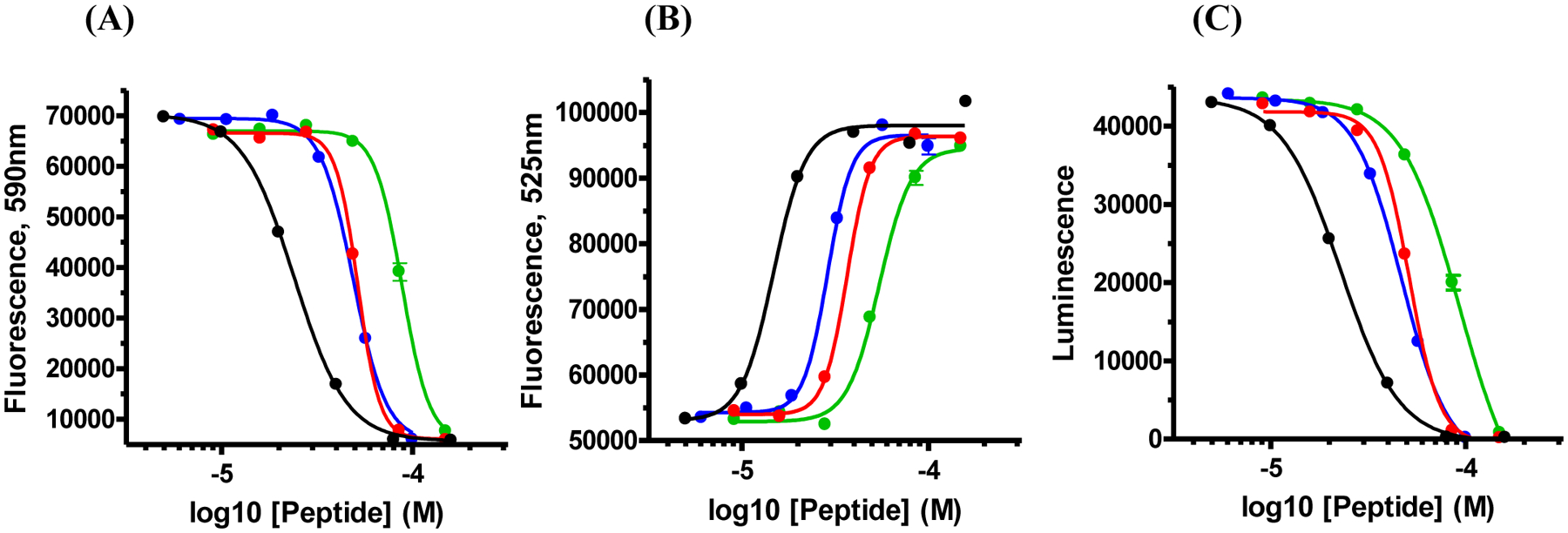
Human amylin exhibits greater toxicity towards INS-1 cells than do the consensus sequences. Black, human amylin; Blue, primate consensus sequence; Red, mammalian consensus sequence; Green, vertebrate consensus sequence. (A) The results of Alamar Blue assays. (B) The results of assays which measure global ATP levels (CellTiter Glo). (C) The results of assays which measure the permeability of the cell membrane (CellTox Green). The x-axis is plotted on a log scale. Data points represent the average of 3 measurements. The curve represents the best fit to a sigmoidal function.
Table-2.
EC50 values measured for cytotoxicity towards cultured INS-1 cells measured for wild type human amylin and the three consensus sequences. All data are presented as mean ± standard error of mean (SEM).
| EC50 (μM) ± SEM | |||
|---|---|---|---|
| Assay | Alamar Blue | CellTiter-Glo 2.0 | CellTox Green |
| Human amylin | 24.0 ± 0.6 | 22.8 ± 0.7 | 14.7 ± 1.5 |
| Primate consensus | 48.9 ± 1.4 | 46.8 ± 1.0 | 28.8 ± 1.6 |
| Mammalian consensus | 51.8 ± 1.4 | 51.3 ± 1.2 | 36.7 ± 1.2 |
| Vertebrate consensus | 88.3 ± 3.7 | 93.0 ± 7.5 | 54.5 ± 2.8 |
Lower EC50 values were consistently observed for the membrane permeability assays compare to the assays which measured ATP levels and the redox state for all peptides (Figure-4, Table-2). This effect has been observed previously and indicates that INS-1 cells display detectable permeability to the small fluorescent probe at a lower peptide concentration than leads to a detectable reduction in ATP levels or change in redox state. We note that this result does not address whether or not membrane permeability is upstream of the other effects as the assays were not conducted in a time resolved fashion.
Consensus sequences activate a human amylin receptor.
Amylin receptors are composed of the calcitonin receptor complexed with a receptor activity modifying protein (RAMP). We compared the ability of the four polypeptides to activate a typical amylin receptor, the human amylin hAMY1(a) receptor by analyzing cAMP production, a robust assay for amylin receptor activity (50, 51). Stimulation of cAMP production was observed with all of the consensus sequences and the difference from the effects induced by human amylin were modest ranging from 3 to 4-fold (Figure-5, Table-3).
Figure-5.
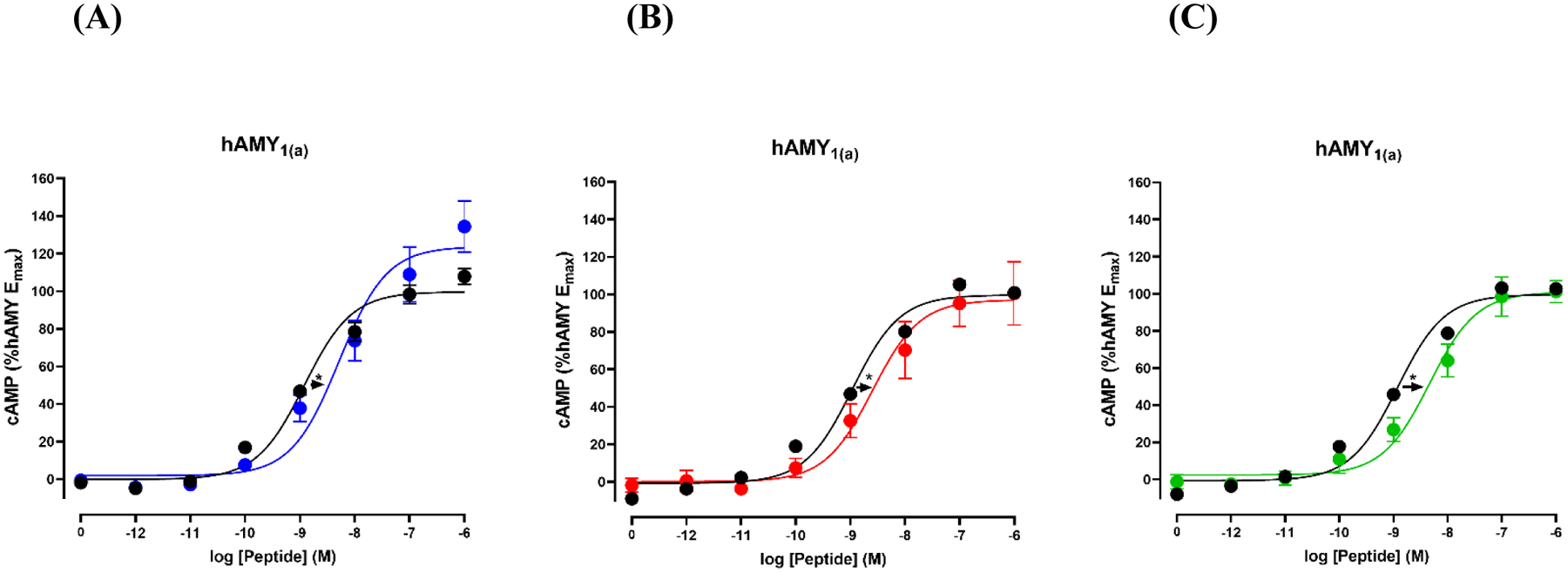
Potency of human amylin and consensus sequences at the hAMY1(a) receptor. Shown are concentration-response curves of cAMP production. Curves are plotted as percentages of maximal human amylin stimulated cAMP production and each data point is the mean ± SEM of at least three independent measurements. (A) Comparison of human amylin (black) and the primate consensus sequence. (blue) (B) Comparison of human amylin (black) and the mammalian consensus sequence (red). (C) Comparison of human amylin and the vertebrate consensus sequence (green).
Table-3.
Summary of activity data for human amylin and consensus sequences, showing pEC50 (pEC50 = −log10 (EC50) and Emax values at hAMY1(a) receptors. Separate controls with different samples of human amylin were run for each consensus sequence.
| pEC50 | Fold Change | Emax | n | |
|---|---|---|---|---|
| Human amylin | 8.91 ± 0.07 | −4 | 100 | 4 |
| Primate consensus | 8.33 ± 0.13* | 123 ± 14.2 | ||
| Human amylin | 8.94 ± 0.06 | −3 | 100 | 5 |
| Mammalian consensus | 8.50 ± 0.12* | 99.1 ± 13.3 | ||
| Human amylin | 8.90 ± 0.06 | −4 | 100 | 5 |
| Vertebrate consensus | 8.33 ± 0.14* | 102 ± 6.58 |
p < 0.05 by unpaired Student t-test compared to human amylin.
CONCLUSIONS
The analysis presented here reveals that human amylin forms amyloid more rapidly in vitro than any of the consensus sequences and is more toxic towards cultured β-cells. The receptor activity data show that the consensus sequences are only moderately less active than human amylin. This work shows that amylin can tolerate multiple non-conservative substitutions without significantly comprising activity. All of the consensus sequences contain an Arg at position 18 compared to the His residue found in human amylin. This substitution is not included in pramlintide. The slower rate of aggregation and reduced toxicity and minimal effects on receptor activity of the consensus sequences suggestions that a H18R substitution may be profitably used in next generation human amylin variants.
The mammalian consensus sequence differs from human amylin at two other positions in addition to the H18R replacement; a F23L substitution and a S29P replacement, while the mammalian and primate consensus sequences differ only by the F23L and S29P replacements. The F23L substitution is a relatively conservative substitution while the S29P replacement is not. The very minor modest change in EC50 values between the primate and mammalian consensus sequence argues that the F23L and S29P substitutions do not have a significant effect on toxicity. This in turn implies that of the three prolines in rodent amylin and pramlintide, Pro-29 does not make a critical contribution to their reduced amyloidogencity. It is interesting to note that the two substitutions between the mammalian and primate consensus sequences have a larger impact on the time scale of amyloid formation (a 5 to 8-fold change in T50) versus their effect on the EC50 values. Thus mutations in the core region of the structure can have a different impact on amyloid formation kinetics and toxicity. It is also noteworthy that the polypeptide with the N14D replacement (the vertebrate consensus sequence), is almost as active as wild type human amylin. This suggests that N14 could be another site to explore in efforts to develop new analogs of human amylin. Interestingly, an Ala substitution of this position increased human amylin activity in a prior study (52).
It is interesting to examine the position of the F23L and S29P replacements in the context of the existing high-resolution structural model of the human amylin amyloid fiber. Leu is a relatively conservative replacement for Phe although it clearly differs in shape. Pro is not a conservative replacement for Ser in shape, hydrophobicity or functionality. Both residue 23 and 29 are located at the interface between the two columns of amylin monomers in the pH 7.4 structures, forming part of the steric zipper like interface. Thus, these substitutions may be expected to perturb the steric zipper interface in the pH 7.4 structure. Residue 23 is part of the core of the pH 6.0 structure and is near residues 21, 25 and 37 in the other monomer within the same layer. In contrast, in the pH 6.0 structure residues 29 makes contacts with residues within the same monomer and is not part of the monomer monomer interface within the layer.
We used the 3D ZipperDB program developed by Eisenberg and coworkers to examine the possible effect of these replacements on steric zipper formation (53). The method makes use of the ROSETTA program to calculate an apparent energy of different steric zipper fragments using a sliding window of 6 residues. The calculations predict that the F23L and S29P replacements significantly reduce the steric zipper propensity in the intercolumn interface (Figure-S1). The fact that the all consensus sequences form amyloid at pH 7.4 indicates that significant perturbations to this interface can be tolerated, without abolishing the ability to form amyloid. Of course the substitutions may result in formation of different polymorphs which form different steric zipper interfaces. Along these lines, the pH 6.0 structure provides an interesting example of how small changes in conditions can lead to very different structures.
Overall, the fact that all three consensus sequences are slower to form amyloid compared to human amylin, but activate the hAMY1(a) receptor and are less toxic is consistent with the notion that there is no strong selective pressure to avoid amylin amyloid formation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (GM078114 and the Marsden Fund, Royal Society of New Zealand (to D.L.H.). We thank Dr Robert Tycko for providing the coordinates of the pH 7.4 solid state NMR based structural model of the amylin fiber. We thank Professors Wolfgang Hoyer and Gunnar F. Schröder for providing the coordinates of their cryo-EM structure and for helpful discussions. We also thank Professor Andisheh Abedini as well as members of the Raleigh lab for helpful discussions.
Footnotes
Supporting Information
The Supporting Information is available free of charge athttps://pubs.acs.org/doi/10.1021/acschembio.9b01050
A table of the complete sequence alignment for 202 amylin sequences. Figures showing the analysis of potential steric zipper regions in human amylin and the various consensus sequences. Figures showing 12 independent experimental thioflavin-T curves for human amylin and the various consensus sequences.
The authors declare no competing financial interest. D.P.R is a co-inventor on a patent application for soluble analogs of amylin.
REFERENCES
- 1.Westermark P, Andersson A, and Westermark GT (2011) Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol Rev 91, 795–826. [DOI] [PubMed] [Google Scholar]
- 2.Raleigh D, Zhang X, Hastoy B, and Clark A (2017) The beta-cell assassin: IAPP cytotoxicity. J Mol Endocrinol 59, R121–R140. [DOI] [PubMed] [Google Scholar]
- 3.Hay DL, Chen S, Lutz TA, Parkes DG, and Roth JD (2015) Amylin: pharmacology, physiology, and clinical potential. Pharmacol Rev 67, 564–600. [DOI] [PubMed] [Google Scholar]
- 4.Lutz TA (2010) The role of amylin in the control of energy homeostasis. Am J Physiol Regul Integr Comp Physiol 298, R1475–1484. [DOI] [PubMed] [Google Scholar]
- 5.Kahn SE, D’Alessio DA, Schwartz MW, Fujimoto WY, Ensinck JW, Taborsky GJ Jr., and Porte D Jr. (1990) Evidence of cosecretion of islet amyloid polypeptide and insulin by beta-cells. Diabetes 39, 634–638. [DOI] [PubMed] [Google Scholar]
- 6.Westermark P, Wernstedt C, Wilander E, Hayden DW, Obrien TD, and Johnson KH (1987) Amyloid fibrils in human insulinoma and islets of langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. P Natl Acad Sci USA 84, 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper GJS, Willis AC, Clark A, Turner RC, Sim RB, and Reid KBM (1987) Purification and characterization of a peptide from amyloid-rich pancreases of type-2 diabetic-patients. P Natl Acad Sci USA 84, 8628–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper GJS, Leighton B, Dimitriadis GD, Parrybillings M, Kowalchuk JM, Howland K, Rothbard JB, Willis AC, and Reid KBM (1988) Amylin found in amyloid deposits in human type-2 diabetes-mellitus may be a hormone that regulates glycogen-metabolism in skeletal-muscle. P Natl Acad Sci USA 85, 7763–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opie EL (1901) On the relation of chronic interstitial pancreatitis to the islands of langerhans and to diabetes mellitus. J Exp Med 5, 397–U119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao P, Abedini A, and Raleigh DP (2013) Aggregation of islet amyloid polypeptide: from physical chemistry to cell biology. Curr Opin Struc Biol 23, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abedini A, and Schmidt AM (2013) Mechanisms of islet amyloidosis toxicity in type 2 diabetes. FEBS Lett 587, 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zraika S, Hull RL, Verchere CB, Clark A, Potter KJ, Fraser PE, Raleigh DP, and Kahn SE (2010) Toxic oligomers and islet beta cell death: guilty by association or convicted by circumstantial evidence? Diabetologia 53, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abedini A, Plesner A, Cao P, Ridgway Z, Zhang J, Tu LH, Middleton CT, Chao B, Sartori DJ, Meng F, Wang H, Wong AG, Zanni MT, Verchere CB, Raleigh DP, and Schmidt AM (2016) Time-resolved studies define the nature of toxic IAPP intermediates, providing insight for anti-amyloidosis therapeutics. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potter KJ, Abedini A, Marek P, Klimek AM, Butterworth S, Driscoll M, Baker R, Nilsson MR, Warnock GL, Oberholzer J, Bertera S, Trucco M, Korbutt GS, Fraser PE, Raleigh DP, and Verchere CB (2010) Islet amyloid deposition limits the viability of human islet grafts but not porcine islet grafts. P Natl Acad Sci USA 107, 4305–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udayasankar J, Kodama K, Hull RL, Zraika S, Aston-Mourney K, Subramanian SL, Tong J, Faulenbach MV, Vidal J, and Kahn SE (2009) Amyloid formation results in recurrence of hyperglycaemia following transplantation of human IAPP transgenic mouse islets. Diabetologia 52, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson A, Bohman S, Borg LA, Paulsson JF, Schultz SW, Westermark GT, and Westermark P (2008) Amyloid deposition in transplanted human pancreatic islets: a conceivable cause of their long-term failure. Exp Diabetes Res 2008, 562985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betsholtz C, Christmansson L, Engstrom U, Rorsman F, Svensson V, Johnson KH, and Westermark P (1989) Sequence divergence in a specific region of islet amyloid polypeptide (IAPP) explains differences in islet amyloid formation between species. Febs Letters 251, 261–264. [DOI] [PubMed] [Google Scholar]
- 18.Verchere CB, DAlessio DA, Palmiter RD, Weir GC, BonnerWeir S, Baskin DG, and Kahn SE (1996) Islet amyloid formation associated with hyperglycemia in transgenic mice with pancreatic beta cell expression of human islet amyloid polypeptide. P Natl Acad Sci USA 93, 3492–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westermark P, Engstrom U, Johnson KH, Westermark GT, and Betsholtz C (1990) Islet amyloid polypeptide - pinpointing amino-acid-residues linked to amyloid fibril formation. P Natl Acad Sci USA 87, 5036–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburn TT, and Lansbury PT (1993) Interspecies sequence variations affect the kinetics and thermodynamics of amyloid formation - peptide models of pancreatic amyloid. Journal of the American Chemical Society 115, 11012–11013. [Google Scholar]
- 21.Akter R, Cao P, Noor H, Ridgway Z, Tu LH, Wang H, Wong AG, Zhang X, Abedini A, Schmidt AM, and Raleigh DP (2016) Islet amyloid polypeptide: structure, function, and pathophysiology. J Diabetes Res 2016, 2798269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akter R, Bower RL, Abedini A, Schmidt AM, Hay DL, and Raleigh DP (2018) Amyloidogenicity, cytotoxicity, and receptor activity of bovine amylin: implications for xenobiotic transplantation and the design of nontoxic amylin variants. ACS Chem Biol 13, 2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akter R, Abedini A, Ridgway Z, Zhang XX, Kleinberg J, Schmidt AM, and Raleigh DP (2017) Evolutionary adaptation and amyloid formation: does the reduced amyloidogenicity and cytotoxicity of ursine amylin contribute to the metabolic adaption of bears and polar bears? Isr J Chem 57, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan G, Briscoe TA, and Jobe L (2009) Review of pramlintide as adjunctive therapy in treatment of type 1 and type 2 diabetes. Drug Des Devel Ther 2, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaacs D, Yager S, Parker M, Wolfe L, Luxenburg J, and Lekic S (2019) Adjunct antihyperglycemic agents in overweight and obese adults with type 1 diabetes. Ann Pharmacother 53, 371–384. [DOI] [PubMed] [Google Scholar]
- 26.Chapman I, Parker B, Doran S, Feinle-Bisset C, Wishart J, Strobel S, Wang Y, Burns C, Lush C, Weyer C, and Horowitz M (2005) Effect of pramlintide on satiety and food intake in obese subjects and subjects with type 2 diabetes. Diabetologia 48, 838–848. [DOI] [PubMed] [Google Scholar]
- 27.Weyer C, Maggs DG, Young AA, and Kolterman OG (2001) Amylin replacement with pramlintide as an adjunct to insulin therapy in type 1 and type 2 diabetes mellitus: a physiological approach toward improved metabolic control. Curr Pharm Design 7, 1353–1373. [DOI] [PubMed] [Google Scholar]
- 28.Sternke M, Tripp KW, and Barrick D (2019) Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc Natl Acad Sci U S A 116, 11275–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okafor CD, Pathak MC, Fagan CE, Bauer NC, Cole MF, Gaucher EA, and Ortlund EA (2018) Structural and dynamics comparison of thermostability in ancient, modern, and consensus elongation factor tus. Structure 26, 118–129 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porebski BT, and Buckle AM (2016) Consensus protein design. Protein Eng Des Sel 29, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steipe B, Schiller B, Pluckthun A, and Steinbacher S (1994) Sequence statistics reliably predict stabilizing mutations in a protein domain. Journal of Molecular Biology 240, 188–192. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan BJ, Nguyen T, Durani V, Mathur D, Rojas S, Thomas M, Syu T, and Magliery TJ (2012) Stabilizing proteins from sequence statistics: the interplay of conservation and correlation in triosephosphate isomerase stability. Journal of Molecular Biology 420, 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polizzi KM, Chaparro-Riggers JF, Vazquez-Figueroa E, and Bommarius AS (2006) Structure-guided consensus approach to create a more thermostable penicillin G acylase. Biotechnol J 1, 531–536. [DOI] [PubMed] [Google Scholar]
- 34.Rath A, and Davidson AR (2000) The design of a hyperstable mutant of the Abp1p SH3 domain by sequence alignment analysis. Protein Science 9, 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, and Ranganathan R (2005) Evolutionary information for specifying a protein fold. Nature 437, 512–518. [DOI] [PubMed] [Google Scholar]
- 36.Capra JA, and Singh M (2007) Predicting functionally important residues from sequence conservation. Bioinformatics 23, 1875–1882. [DOI] [PubMed] [Google Scholar]
- 37.Marek P, Woys AM, Sutton K, Zanni MT, and Raleigh DP (2010) Efficient microwave-assisted synthesis of human islet amyloid polypeptide designed to facilitate the specific incorporation of labeled amino acids. Org Lett 12, 4848–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abedini A, and Raleigh DP (2005) Incorporation of pseudoproline derivatives allows the facile synthesis of human IAPP, a highly amyloidogenic and aggregation-prone polypeptide. Org Lett 7, 693–696. [DOI] [PubMed] [Google Scholar]
- 39.Abedini A, Singh G, and Raleigh DP (2006) Recovery and purification of highly aggregation-prone disulfide-containing peptides: application to islet amyloid polypeptide. Analytical Biochemistry 351, 181–186. [DOI] [PubMed] [Google Scholar]
- 40.Tu L-H, and Raleigh DP (2013) Role of aromatic interactions in amyloid formation by islet amyloid polypeptide. Biochemistry 52, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey RJ, and Hay DL (2006) Pharmacology of the human CGRP(1) receptor in Cos 7 cells. Peptides 27, 1367–1375. [DOI] [PubMed] [Google Scholar]
- 42.Gingell JJ, Qi T, Bailey RJ, and Hay DL (2010) A key role for tryptophan 84 in receptor activity-modifying protein 1 in the amylin 1 receptor. Peptides 31, 1400–1404. [DOI] [PubMed] [Google Scholar]
- 43.Sanke T, Bell GI, Sample C, Rubenstein AH, and Steiner DF (1988) An islet amyloid peptide is derived from an 89-amino acid precursor by proteolytic processing. Journal of Biological Chemistry 263, 17243–17246. [PubMed] [Google Scholar]
- 44.Marzban L, Trigo-Gonzalez G, and Verchere CB (2005) Processing of pro-islet amyloid polypeptide in the constitutive and regulated secretory pathways of beta cells. Mol Endocrinol 19, 2154–2163. [DOI] [PubMed] [Google Scholar]
- 45.Wiltzius JJ, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, and Eisenberg D (2008) Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin). Protein Sci 17, 1467–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luca S, Yau WM, Leapman R and Tycko R (2007) Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry 46, 13505–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khemtemourian L, Guillemain G, Foufelle F, and Killian JA (2017) Residue specific effects of human islet polypeptide amyloid on self-assembly and on cell toxicity. Biochimie 142, 22–30. [DOI] [PubMed] [Google Scholar]
- 48.LeVine H (1999) Quantification of beta-sheet amyloid fibril structures with thioflavin T. Method Enzymol 309, 274–284. [DOI] [PubMed] [Google Scholar]
- 49.Wong AG, Wu C, Hannaberry E, Watson MD, Shea JE, and Raleigh DP (2016) Analysis of the amyloidogenic potential of pufferfish (takifugu rubripes) islet amyloid polypeptide highlights the limitations of thioflavin-T assays and the difficulties in defining amyloidogenicity. Biochemistry 55, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hay DL, Christopoulos G, Christopoulos A, Poyner DR, and Sexton PM (2005) Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol Pharmacol 67, 1655–1665. [DOI] [PubMed] [Google Scholar]
- 51.Hay DL, Garelja ML, Poyner DR, and Walker CS (2018) Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Brit J Pharmacol 175, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bower RL, Yule L, Rees TA, Deganutti G, Hendrikse ER, Harris PWR, Kowalczyk R, Ridgway Z, Wong AG, Swierkula K, Raleigh DP, Pioszak AA, Brimble MA, Reynolds CA, Walker CS, and Hay DL (2018) Molecular signature for receptor engagement in the metabolic peptide hormone amylin. ACS Pharmacology & Translational Science 1, 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldschmidt L, Teng PK, Riek R, and Eisenberg D (2010) Identifying the amylome, proteins capable of forming amyloid-like fibrils. P Natl Acad Sci USA 107, 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


