Abstract
Ecological systems can often be characterised by changes among a finite set of underlying states pertaining to individuals, populations, communities or entire ecosystems through time. Owing to the inherent difficulty of empirical field studies, ecological state dynamics operating at any level of this hierarchy can often be unobservable or ‘hidden’. Ecologists must therefore often contend with incomplete or indirect observations that are somehow related to these underlying processes. By formally disentangling state and observation processes based on simple yet powerful mathematical properties that can be used to describe many ecological phenomena, hidden Markov models (HMMs) can facilitate inferences about complex system state dynamics that might otherwise be intractable. However, HMMs have only recently begun to gain traction within the broader ecological community. We provide a gentle introduction to HMMs, establish some common terminology, review the immense scope of HMMs for applied ecological research and provide a tutorial on implementation and interpretation. By illustrating how practitioners can use a simple conceptual template to customise HMMs for their specific systems of interest, revealing methodological links between existing applications, and highlighting some practical considerations and limitations of these approaches, our goal is to help establish HMMs as a fundamental inferential tool for ecologists.
Keywords: Behavioural ecology, community ecology, ecosystem ecology, hierarchical model, movement ecology, observation error, population ecology, state‐space model, time series
By formally disentangling state and observation processes based on simple yet powerful mathematical properties that can be used to describe many ecological phenomena, hidden Markov models (HMMs) can facilitate inferences about complex system state dynamics that might otherwise be intractable. We provide a gentle introduction to HMMs, establish some common terminology, review the immense scope of HMMs for applied ecological research, and provide a tutorial on some of the more technical aspects of HMM implementation and interpretation. By illustrating how practitioners can use a simple conceptual template to customise HMMs for their specific systems of interest, revealing methodological links between existing applications, and highlighting some practical considerations and limitations of these approaches, our goal is to help establish HMMs as a fundamental inferential tool for ecologists.
INTRODUCTION
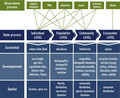
Ecological systems can often be characterised by changes among underlying system states through time. These state dynamics can pertain to individuals (e.g. birth, death), populations (e.g. increases, decreases), metapopulations (e.g. colonisation, extinction), communities (e.g. succession) or entire ecosystems (e.g. regime shifts). Gaining an understanding of state dynamics at each level of this hierarchy is a central goal of ecology and fundamental to studies of climate change, biodiversity, species distribution and density, habitat and patch selection, population dynamics, behaviour, evolution and many other phenomena (Begon et al., 2006). However, inferring ecological state dynamics is challenging for several reasons, including: (1) these complex systems often display nonlinear, non‐monotonic, non‐stationary and non‐Gaussian behaviour (Scheffer et al., 2001; Tucker and Anand, 2005; Wood, 2010; Pedersen et al., 2011a; Fasiolo et al., 2016); (2) changes in underlying states and dynamics can be rapid and drastic, but also gradual and more subtle (Beisner et al., 2003; Scheffer and Carpenter, 2003; Folke et al., 2004); and (3) the actual state of an ecological entity, be it an individual plant or animal, or a population or community, can often be difficult or impossible to observe directly (Martin et al., 2005; Kéry and Schmidt, 2008; Royle and Dorazio, 2008; Chen et al., 2013; Kellner and Swihart, 2014). Ecologists must therefore often contend with pieces of evidence believed to be informative of the state of an unobservable system at a particular point in time (see Fig. 1).
Figure 1.
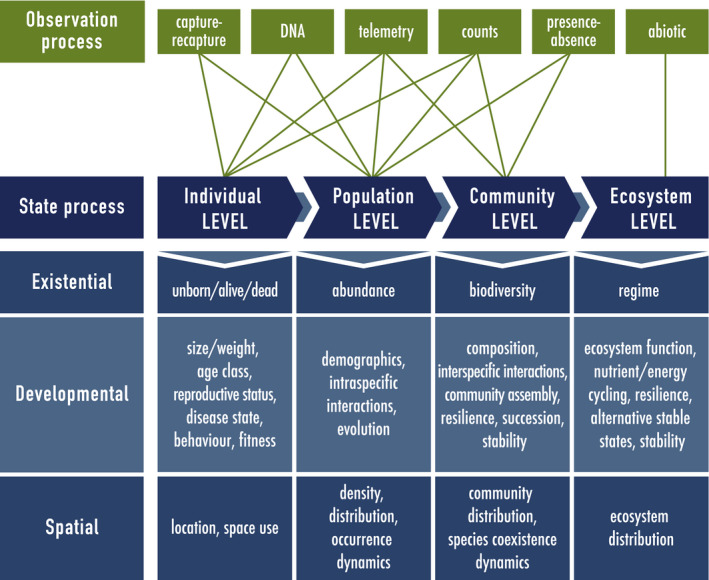
System state processes that can be difficult to observe directly, but can be uncovered from common ecological observation processes using hidden Markov models. The state process (blue) can pertain to any level within the ecological hierarchy (‘Individual’, ‘Population’, ‘Community’ or ‘Ecosystem’) and for convenience is categorised as primarily ‘Existential’, ‘Developmental’ or ‘Spatial’ in nature. The observation process (green) can provide information about state processes at different levels of the hierarchy (green lines) and includes capture–recapture, DNA sampling, animal‐borne telemetry, count surveys, presence–absence surveys and/or abiotic measurements. Observation and state processes from lower levels can be integrated for inferences at higher levels. For example, community‐level biodiversity data could be combined with environmental data to describe ecosystem‐level processes.
Whether for management, conservation or empirical testing of ecological theory, there is a need for inferential methods that seek to uncover the relationships between factors driving such systems, and thereby predict them in quantitative terms. Hidden Markov models (HMMs) constitute a class of statistical models that has rapidly gained prominence in ecology because they are able to accommodate complex structures that account for changes between unobservable system states (Ephraim and Merhav, 2002; Cappé et al., 2005; Zucchini et al., 2016). By simultaneously modelling two time series – one consisting of the underlying state dynamics and a second consisting of observations arising from the true state of the system – HMMs are able to detect state changes in noisy time‐dependent phenomena by formally disentangling the state and observation processes. For example, using HMMs and their variants:
Historical regime shifts can be identified from reconstructed chronologies;
Long‐term dynamics of populations, species, communities and ecosystems in changing environments can be inferred from dynamic biodiversity data;
Species identity and biodiversity can be determined from environmental DNA (eDNA);
Hidden evolutionary traits can be accounted for when assessing the drivers of diversification;
Species occurrence can be linked to variation in habitat, population density, land use, host–pathogen dynamics or predator–prey interactions;
Survival, dispersal, reproduction, disease status and habitat use can be inferred from capture–recapture time series;
Animal movements can be classified into foraging, migrating or other modes for inferences about behaviour, activity budgets, resource selection and physiology; and
Trade‐offs between dormancy and colonisation can be inferred from standing flora or fungal fruiting bodies.
The increasing popularity of HMMs has been fuelled by new and detailed data streams, such as those arising from modern remote sensing and geographic information systems (Viovy and Saint, 1994; Gao, 2002), eDNA (Bálint et al., 2018) and genetic sequencing (Hudson, 2008), as well as advances in computing power and user‐friendly software (Visser and Speenkenbrink, 2010). However, despite their utility and ubiquity in other fields such as finance (Bhar and Hamori, 2004), speech recognition (Rabiner, 1989) and bioinformatics (Durbin et al., 1998), the vast potential of HMMs for uncovering latent system dynamics from readily available data remains largely unrecognised by the broader ecological community. This is likely attributable to a tendency for the existing ecological literature to characterise HMMs as a subject‐specific tool reserved for a particular type of data rather than a general conceptual framework for probabilistic modelling of sequential data. This is also likely exacerbated by a tendency for HMMs to be applied and described quite differently across disciplines. Indeed, many ecologists may not recognise that some of the most well‐established inferential frameworks in population, community and movement ecology are in fact special cases of HMMs.
Catering to ecologists and non‐statisticians, we describe the structure and properties of HMMs (HIDDEN MARKOV MODELS), establish some common terminology (Table 1) and review case studies from the biological, ecological, genetics and statistical literature (ECOLOGICAL APPLICATIONS OF HIDDEN MARKOV MODELS). Central to our review and synthesis is a simple but flexible conceptual template that ecologists can use to customise HMMs for their specific systems of interest. In addition to highlighting new areas where HMMs may be particularly promising in ecology, we also demonstrate cases where these models have (perhaps unknowingly) already been used by ecologists for decades. We then identify some practical considerations, including implementation, software and potential challenges that practitioners may encounter when using HMMs (IMPLEMENTATION, CHALLENGES AND PITFALLS). Using an illustrative example, we provide a step‐by‐step tutorial on some of the more technical aspects of HMM implementation in the Supplementary Tutorial. The overall aim of our review is thus to provide a synthesis of the various ways in which HMMs can be used, reveal methodological links between existing applications and thereby establish HMMs as a fundamental inferential tool for ecologists working with sequential data.
Table 1.
Glossary
| Term | Definition | Synonyms |
|---|---|---|
| Conditional independence property | Assumption made for the state‐dependent process: conditional on the state at time t, the observation at time t is independent of all other observations and states | |
| Forward algorithm | Recursive scheme for updating the likelihood and state probabilities of an HMM through time | Filtering |
| Forward–backward algorithm | Recursive scheme for calculating state probabilities for any point in time: | Local state decoding; smoothing |
| Hidden Markov model (HMM) | A special class of state‐space model with a finite number of hidden states that typically assumes some form of the Markov property and the conditional independence property | Dependent mixture model; latent Markov model; Markov‐switching model; regime‐switching model; state‐switching model; multi‐state model |
| Initial distribution | The probability of being in any of the states at the start of the sequence: | Initial probabilities; prior probabilities |
| Markov property | Assumption made for the state process: (‘conditional on the present, the future is independent of the past’) | Memoryless property |
| Sojourn time | The amount of time spent in a state before switching to another state | Dwell time; occupancy time |
| State process | Unobserved, serially correlated sequence of states describing how the system evolves over time: for | Hidden/latent process; system process |
| State transition probability | The probability of switching from state at time to state at time , , usually represented as an transition probability matrix | |
| State‐dependent distribution | Probability distribution of an observation conditional on a particular state being active at time , usually from some parametric class (e.g. categorical, Poisson, normal) and represented as an diagonal matrix | Emission distribution; measurement model; observation distribution; output distribution; response distribution |
| State‐dependent process | The observed process within an HMM, which is assumed to be driven by the underlying unobserved state process | Observation process |
| State‐space model | A conditionally specified hierarchical model consisting of two linked stochastic processes, a latent system process model and an observation process model | |
| Viterbi algorithm | Recursive scheme for finding the sequence of states which is most likely to have given rise to the observed sequence | Global state decoding |
HIDDEN MARKOV MODELS
We begin by providing a gentle introduction to HMMs, including model formulation, inference and extensions. Although we have endeavoured to minimise technical material and provide illustrative examples wherever possible, we assume the reader has at least some basic understanding of linear algebra concepts such as matrix multiplication and diagonal matrices (e.g. see Appendix A in Caswell, 2001) and probability theory concepts such as uncertainty, random variables and probability distributions (Gotelli and Ellison, 2013, Chapters 1–2).
Basic model formulation
Hidden Markov models (HMMs) are a class of statistical models for sequential data, in most instances related to systems evolving over time. The system of interest is modelled using a state process (or system process; Table 1), which evolves dynamically such that future states depend on the current state. Many ecological phenomena can naturally be described by such a process (Fig. 1). In an HMM, the state process is not directly observed – it is a ‘hidden’ (or ‘latent’) variable. Instead, observations are made of a state‐dependent process (or observation process) that is driven by the underlying state process. As a result, the observations can be regarded as noisy measurements of the system states of interest, but they are typically insufficient to precisely determine the state. Mathematically, an HMM is composed of two sequences:
An observed state‐dependent process ; and
An unobserved (hidden) state process .
In most applications, the indices refer to observations made over time at a regular sampling interval (e.g. daily or annual rainfall measurements), but they can also refer to position (e.g. in a sequence of DNA; Henderson et al., 1997; Eddy, 2004) or order (e.g. in a sequence of marine mammal dives; DeRuiter et al., 2017). HMMs can also be formulated in continuous time (Jackson et al., 2003; Amoros et al., 2019), but these models have tended to be less frequently applied in ecology (but see Langrock et al., 2013; Choquet et al., 2017; Olajos et al., 2018). Among the many HMM formulations of relevance to ecology that we highlight in ECOLOGICAL APPLICATIONS OF HIDDEN MARKOV MODELS, some example observation sequences and underlying states include:
Observation of feeding/not feeding, with underlying state Hungry or sated;
Count of individuals, with underlying state True population abundance; and
Daily rainfall measurement, with underlying state Wet or dry season.
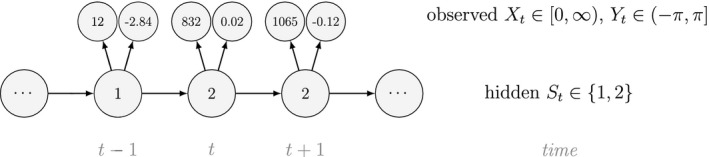
Unlike the larger class of state‐space models (see Box 1), the state process within an HMM can take on only finitely many possible values: for . The basic HMM formulation further involves two key dependence assumptions: (1) the probability of a particular state being active at any time is completely determined by the state active at time (the so‐called Markov property); and (2) the probability distribution of an observation at any time is completely determined by the state active at time (Fig. 2). The latter assumption is a conditional independence property, as this implies that is conditionally independent of past and future observations, given . Whether these simplifying assumptions can faithfully characterise the underlying dynamics for the system of interest must be carefully considered (see Challenges and pitfalls).
Box 1 Where do HMMs reside in the taxonomic zoo of latent variable models? 1.
Latent state (or latent variable) models come in many different forms, with a particular variant often evolving its own nomenclature, notation and jargon that can be confusing for non‐specialists. Here we use broad and non‐technical strokes to differentiate the HMM from its close relatives in the taxonomy of latent state models, with the aim to more clearly position HMMs relative to alternative modelling frameworks. Above all, these models are united by assuming latent states – a fundamental property of the system being modelled that is either partially, or completely, unobservable. They also tend to make a clear distinction between an observation process model – describing noise in the data – and the hidden state process model – describing the underlying patterns and dynamics of interest.
The umbrella terms mixed effects, multilevel or hierarchical models (e.g. Skrondal and Rabe‐Hesketh, 2004; Gelman and Hill, 2006; Royle and Dorazio, 2008; Lee and Song, 2012) typically include the most widely known types of latent variable models (e.g. Clogg, 1995). These often treat latent variables as random effects assumed to arise from a distribution as structural elements of a hierarchical statistical model. There is therefore not only random variation in the observations, but also in the parameters of the model itself. While there are special cases and generalisations that are not so easily classified, a simplified taxonomy for a subset of hierarchical latent variable models can be based on the structural dependence in the hidden state process and whether the state space of this hidden process is discrete (i.e. taking on finitely many values) or continuous:
|
State space |
||
|---|---|---|
|
Continuous |
Discrete |
|
|
Temporal dependence |
State‐space model |
Hidden Markov model |
|
Temporal independence |
Continuous mixture model |
Finite mixture model |
Latent variable models with a continuous state space and no temporal dependence in the hidden state process fall under the broad class of continuous mixture models (e.g. Lindsay, 1995), with ecological applications including the modelling of closed population abundance (Royle, 2004), disease prevalence (Calabrese et al., 2011) and species distribution (Ovaskainen et al., 2017). State‐space models (SSMs) are a special class of latent variable model where the observation process is conditionally specified by a (typically continuous) hidden state process with temporal dependence (e.g. Durbin and Koopman, 2012; Auger‐Méthé et al., 2020), with applications including population dynamics (Schnute, 1994; Wang, 2007; Tavecchia et al., 2009; Newman et al., 2014), disease dynamics (Rohani and King, 2010; Cooch et al., 2012) and animal movement (Patterson et al., 2008; Hooten et al., 2017; Patterson et al., 2017). An HMM is a special class of SSM where the state space is finite (see ECOLOGICAL APPLICATIONS OF HIDDEN MARKOV MODELS for many ecological examples). Finite mixture models (e.g. Frühwirth‐Schnatter, 2006) assume the state space is finite with no temporal dependence in the hidden state process (e.g. the latent states are non‐Markov or do not change over time), with examples including static species occurrence (MacKenzie et al., 2002), closed population capture–recapture (Pledger, 2000) and species distribution (Pledger and Arnold, 2014) models. HMMs and SSMs can therefore be regarded as specific variations of a hierarchical model with serial dependence, where the random effects vary over time. Furthermore, an HMM can be viewed as a discrete version of a SSM or a time‐dependent version of a finite mixture model.
It is important to note that things are not quite as simple as depicted above. For example, while an SSM with discrete latent variables can encompass features of an HMM (Jonsen et al., 2005), an SSM with a finite state space is not necessarily an HMM. An HMM might include continuous random effects on its parameters or a state‐dependent observation distribution specified as a finite mixture (Altman, 2007). If the number of states becomes very large in an HMM, then it can become a discrete approximation of an SSM with a continuous state space (Besbeas and Morgan, 2019). In the Extensions section and the Challenges and Pitfalls section, we consider circumstances where application of a standard HMM is not supported and other approaches or extensions might be required. [Correction added on 10 November 2020, after first online publication: Box 1 has been relocated to page 5.]
Figure 2.
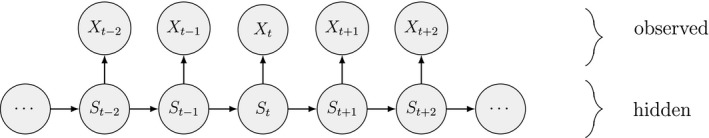
Dependence structure of a basic hidden Markov model, with an observed sequence arising from an unobserved sequence of underlying states .
As a consequence of these assumptions, HMMs generally facilitate model building and computation that might otherwise be intractable. A basic ‐state HMM that formally distinguishes the state and observation processes can be fully specified by the following three components: (1) the initial distribution, , specifying the probabilities of being in each state at the start of the sequence; (2) the state transition probabilities, , specifying the probability of switching from state at time to state at time and usually represented as an state transition probability matrix:
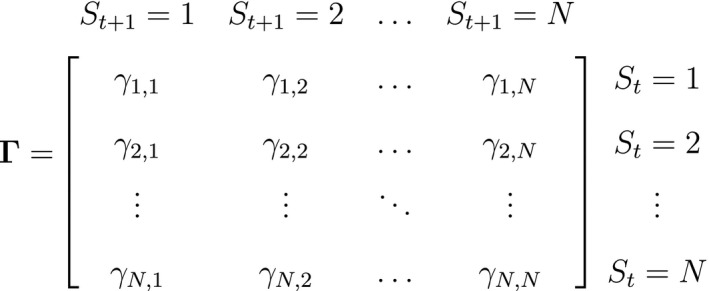
where ; and 3) the state‐dependent distributions, , specifying the probability distribution of an observation conditional on the state at time and usually represented as an diagonal matrix:
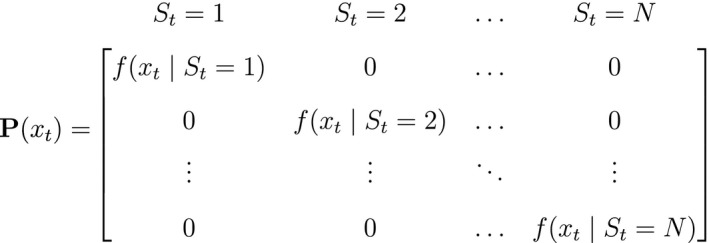
or, equivalently, for computational purposes (see Inference). These distributions can pertain to discrete or continuous observations and are generally chosen from an appropriate distributional family. For example, behavioural observation could be modelled using a categorical distribution (MacDonald and Raubenheimer, 1995), count using a non‐negative discrete distribution (e.g. Poisson; Besbeas and Morgan, 2019, and measurement using a non‐negative continuous distribution (e.g. zero‐inflated exponential; Woolhiser and Roldan, 1982). After specifying , and in terms of the particular system of interest, one can proceed to drawing inferences about unobservable state dynamics from the observation process.
We note that Markov models (Grewal et al., 2019) are commonly used for inferring community‐ or ecosystem‐level dynamics (Waggoner and Stephens, 1970; Wootton, 2001; Tucker and Anand, 2005; Breininger et al., 2010) and providing measures of stability, resilience or persistence (Li, 1995; Pawlowski and McCord, 2009; Zweig et al., 2020), especially in systems composed of sessile organisms such as plant (Horn, 1975; van Hulst, 1979; Usher, 1981; Talluto et al., 2017, but see Chen et al., 2013) or benthic communities (Tanner et al., 1994; Hill et al., 2004; Lowe et al., 2011). A Markov model can simply be viewed as an HMM where it is assumed that the state process is perfectly observed, that is, with a matrix with entry one in row , column , and otherwise zeros. For example, patch dynamics HMMs (MacKenzie et al., 2003) are simply generalisations of well‐known Markov models for patch dynamics (Hanski, 1994; Moilanen, 1999) for cases when presence–absence data are subject to imperfect detection. Likewise, any Markov model can naturally be embedded as the state process within an HMM for less observable phenomena.
Inference
In addition to the ease with which a wide variety of ecological state and observation process models can be specified (see ECOLOGICAL APPLICATIONS OF HIDDEN MARKOV MODELS), a key strength of the HMM framework is that efficient recursive algorithms are available for conducting statistical inference. Here we will briefly outline some of the most common inferential techniques for HMMs, but motivated readers can find additional technical material and a worked example on model fitting, assessment and interpretation in the Supplementary Tutorial. Using the forward algorithm (also known as filtering), the likelihood as a function of the unknown parameters given the observation sequence can be calculated at a computational cost that is (only) linear in . The parameter vector , which is to be estimated, contains any unknown parameters embedded in the three model‐defining components , and . Made possible by the relatively simple dependence structure of an HMM, the forward algorithm traverses along the time series, updating the likelihood step‐by‐step while retaining information on the probabilities of being in the different states (Zucchini et al., 2016, pp. 37–39). Application of the forward algorithm is equivalent to evaluating the likelihood using a simple matrix product expression,
| (1) |
where 1 is a column vector of ones (see Supplementary Tutorial for technical derivation).
In practice, the main challenge when working with HMMs tends to be the estimation of the model parameters. The two main strategies for fitting an HMM are numerical maximisation of the likelihood (Myung, 2003; Zucchini et al., 2016) or Bayesian inference (Ellison, 2004; Gelman et al., 2004) using Markov chain Monte Carlo (MCMC) sampling (Brooks et al., 2011). The former seeks to identify the parameter values that maximise the likelihood function (i.e. the maximum likelihood estimates ), whereas the latter yields a sample from the posterior distribution of the parameters (Ellison, 2004). Specifically for the maximum likelihood (ML) approach, the forward algorithm makes it possible to use standard optimisation methods (Fletcher, 2013) to directly numerically maximise the likelihood (eqn 1). An alternative ML approach is to employ an expectation–maximisation (EM) algorithm that uses similar recursive techniques to iterate between state decoding and updating the parameter vector until convergence (Rabiner, 1989). For MCMC, many different strategies can be used, but these tend to differ in appropriateness and efficiency in a manner that can strongly depend on the specific model and data at hand (Gilks et al., 1996; Gelman et al., 2004; Brooks et al., 2011; Robert and Casella, 2004).
The forward algorithm and similar recursive techniques can further be used for forecasting and state decoding, as well as to conduct formal model checking using pseudo‐residuals (Zucchini et al., 2016, Chapters 5 & 6). State decoding is usually accomplished using the Viterbi algorithm or the forward–backward algorithm (also known as smoothing), which respectively identify the most likely sequence of states or the probability of each state at any time , conditional on the observations. Fortunately, practitioners can often use existing software for most aspects of HMM‐based data analyses and need not dwell on many of the more technical details of implementation (see IMPLEMENTATION, CHALLENGES AND PITFALLS and Supplementary Tutorial).
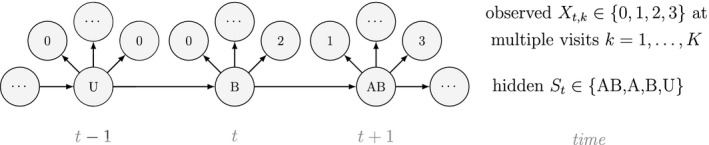
To illustrate some of the basic mechanics, we use a simple example based on observations of the feeding behaviour of a blue whale (Balaenoptera musculus; cf. DeRuiter et al., 2017). Suppose we assume that observations of the number of feeding lunges performed in each of consecutive dives ( for ) arise from states of feeding activity. Building on Fig. 2, we could for example have:
Fig. 3 displays the results for this simple two‐state HMM assuming Poisson state‐dependent (observation) distributions, for , when fitted to the full observation sequence via direct numerical maximisation of eqn 1. The rates of the state‐dependent distributions were estimated as and , suggesting states 1 and 2 correspond to ‘low’ and ‘high’ feeding activity respectively. The estimated state transition probability matrix,
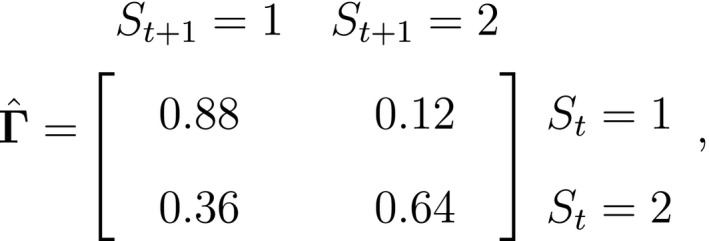
Figure 3.
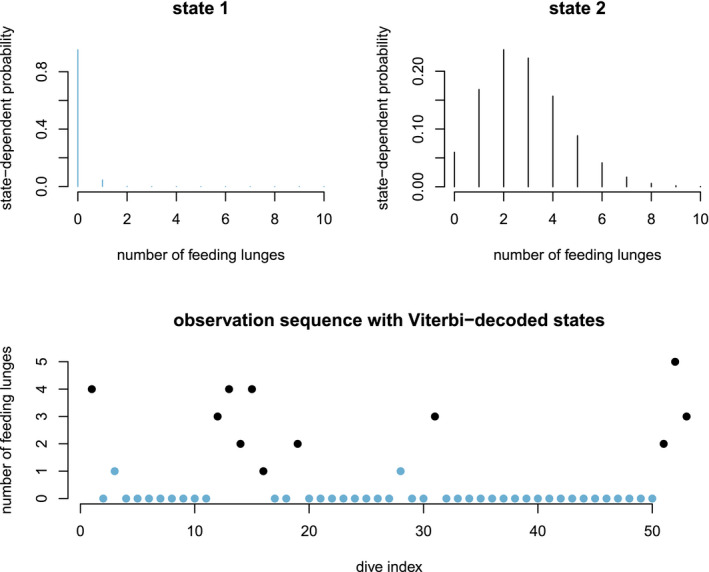
Estimated state‐dependent distributions (top row) and Viterbi‐decoded states from a two‐state HMM fitted to counts of feeding lunges performed by a blue whale during a sequence of consecutive dives. Here the most likely state sequence identifies periods of ‘low’ (state 1; blue) and ‘high’ (state 2; black) feeding activity.
suggests interspersed bouts of ‘low’ and ‘high’ feeding activity, but with bouts of ‘high’ activity tending to span fewer dives. The estimated initial distribution suggests this individual was more likely to have been in the ‘low’ activity state at the start of the sequence. Most ecological applications of HMMs involve more complex inferences related to specific hypotheses about system state dynamics, and a great strength of the HMM framework is the relative ease with which the basic model formulation can be modified to describe a wide variety of processes (Zucchini et al., 2016, Chapters 9–13). Next we highlight some extensions that we consider to be highly relevant in ecological research.
Extensions
The dependence assumptions made within the basic HMM are mathematically convenient, but not always appropriate (see Box 2). The Markov property implies that the amount of time spent in a state before switching to another state – the so‐called sojourn time – follows a geometric distribution. The most likely length of any given sojourn time hence is one unit, which may not be realistic for certain state processes. The obvious extension is to allow for th‐order dependencies in the state process (Fig. 4a), such that the state at time depends on the states at times . An alternative assumes the state process is ‘semi‐Markov’ with the sojourn time flexibly modelled using any distribution on the positive integers (Choquet et al., 2011; van de Kerk et al., 2015; King and Langrock, 2016).
Box 2 To HMM, or not to HMM, that is the question 1.
The structure of a statistical model should be congruent with the data‐generating process in question. HMMs are neither a panacea nor a black box – the appropriateness and feasibility of a particular model will be case‐dependent and requires careful consideration. In determining if HMMs are appropriate for describing a particular system, one must consider two questions:
Do the hidden state dynamics display time dependence which can be represented using Markov chains? If the current system state is not related to the previous state(s), then a latent variable model without time dependence should be considered (see Box 1). Diagnostics examining temporal patterns in residuals (Li, 2003) can help to empirically determine if the assumptions of conditional independence and Markovity are sufficient (see Supplementary Tutorial). When the first‐order Markov assumption may not be appropriate for the state process, one can further ask the question: can system memory be adequately approximated while preserving Markovity? Faithful representation of system memory may require the inclusion of informative covariates or more complex time dependence structures, and it is possible to expand HMMs to higher order Markovian or semi‐Markovian dependence (Zucchini et al., 2016, Chapter 12). While modelling this higher order temporal dependence is sometimes preferable (Hestbeck et al., 1991), it is more complex and thus less widely used. General time‐series modelling often captures complex dependence structures using autoregressive processes (Durbin and Koopman, 2012, Chapter 3), and more complicated variations of HMMs can capture some of these features (Lawler et al., 2019). However, other latent variable approaches will often be better suited for more complex temporal dependence structures. There is no foolproof or automatic way to make this determination, and we must typically rely on residual diagnostics (Li, 2003; Zucchini et al., 2016, Chapter 6) and expert knowledge of the system dynamics.
Can the system be well described by a feasibly finite set of latent states? Our review highlights a wide range of ecological scenarios where the possible states of the system of interest form (or can be approximated by) a finite set. The number of parameters and the computational burden of an HMM can become large with increases in state dimension, and this can be of particular concern when the finite set of states is a coarser approximation of a finer discrete space (e.g. population abundance) or a continuous space (e.g. spatial location). Such approximations have strengths and weaknesses. When used as discrete approximations to state‐space models with continuous support (see Box 1), HMMs can be useful when arbitrary constraints on the state space are required (e.g. restricting aquatic organisms to location states off land) or when combining both discrete and continuous state processes. However, an HMM for a large number of states with a fully parameterised transition probability matrix – where transitions between any of the states are possible – will be computationally expensive, perhaps prohibitively so. Systems with large state spaces can often be approximated by an HMM when transitions between states are local – where transitions can only occur between neighbouring states – and the transition probabilities therefore include a relatively small number of parameters that describe this local behaviour. For example, Thygesen et al. (2009), Pedersen et al. (2011b), and Glennie et al. (2019) use these properties of sparsity to make an HMM approach computationally efficient for very large state spaces. In short, large numbers of states do not necessarily prohibit application of an HMM; this is dependent on the computer resources available and the properties of the state process. Alternatively, it is possible to reduce the size of an infeasible state space by making a coarser approximation (e.g. binning abundance states together into larger states; Zucchini et al., 2016, pp. 162–163; Besbeas and Morgan, 2019). Appropriateness will depend on the sensitivity of the inference to the precise value of the state process and is best investigated by varying the coarseness of the approximation. If the set of states is too coarse‐grained, approximation might lead to spurious inference about the latent states. For example, coarse‐graining could result in masking or misclassification of meaningfully distinct states. The decision of the appropriate number of states can be challenging; there is again no foolproof or automatic way to determine this, and we must usually rely on expert knowledge of the specific system of interest. When the finite state space of an HMM is infeasible or inappropriate, it will often be better to consider other approaches (e.g. Patterson et al., 2008; Cooch et al., 2012; Patterson et al., 2017; Auger‐Méthé et al., 2020).
Figure 4.
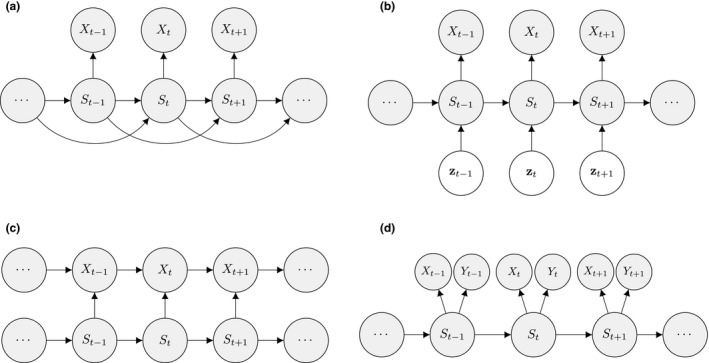
Graphical models associated with different extensions of the basic HMM formulation: (a) state sequence with memory order 2; (b) influence of covariate vectors on state dynamics; (c) observations depending on both states and previous observations; (d) bivariate observation sequence, conditionally independent given the states.
HMMs are often used to infer the drivers of ecological state processes by relating the state transition probabilities to explanatory covariates (Fig. 4b). Indeed, any of the parameters of a basic HMM can be modelled as a function of covariates (e.g. sex, age, habitat type, chlorophyll‐a) using an appropriate link function (McCullagh and Nelder, 1989). Link functions can relate the natural scale parameters to a design matrix of covariates and ‐vector of working scale parameters such that (see White and Burnham, 1999; MacKenzie et al., 2002; Patterson et al., 2009, for common examples of link functions in HMMs). When simultaneously analysing multiple observation sequences, heterogeneity across the different sequences can be modelled through explanatory covariates or mixed HMMs that include random effects (Altman, 2007; Schliehe‐Diecks et al., 2012; Towner et al., 2016).
At the level of the observation process, it is relatively straightforward to relax the conditional independence assumption. For example, it can be assumed that the observation at time depends not only on the state at time but also the observation at time (Fig. 4c; Langrock et al., 2014b; Lawler et al., 2019). It is also straightforward to model multivariate observation sequences using multivariate state‐dependent distributions (Choquet et al., 2013; Phillips et al., 2015; van Beest et al., 2019), where it is often assumed that the different variables observed are conditionally independent and a univariate distribution is specified for each of the variables (Fig. 4d). Owing to the Markov property, this does not imply that the individual components are serially independent or mutually independent (Zucchini et al., 2016, Chapter 9). However, this assumption is not required and will not always be appropriate, in which case a multivariate distribution should be considered.
ECOLOGICAL APPLICATIONS OF HIDDEN MARKOV MODELS
In their classic textbook, Begon et al. (2006) present the evolutionary foundation of ecology and its superstructure built from individual organisms to populations, communities and ecosystems. At each level of this hierarchy, we will illustrate how HMMs can be used for identifying patterns and dynamics of many different types of ecological state variables that would otherwise be difficult or impossible to observe directly. For each application, we emphasise the two principal components of any HMM – the observation process and the state process – as a conceptual template for ecologists to formulate HMMs in terms of their particular systems of interest.
The observation process in ecological studies is often driven by many factors, including the system state variable(s) of interest, the biotic and/or abiotic components of the system, and study design (Fig. 1). Among the most common types of observation processes in ecology are capture–recapture (Williams et al., 2002), DNA sampling (Bohmann et al., 2014; Rowe et al., 2017; Bálint et al., 2018), animal‐borne telemetry (Cooke et al., 2004; White and Garrott, 1990; Hooten et al., 2017), count surveys (Buckland et al., 2004; Charmantier et al., 2006; Nichols et al., 2009), presence–absence surveys (Koleff et al., 2003; MacKenzie et al., 2018) and abiotic measurement (e.g. temperature, precipitation, sediment type). These observation processes are not mutually exclusive, can contribute information at different levels of the hierarchy and can be pooled for inference (Schaub and Abadi, 2011; Gimenez et al., 2012; Evans et al., 2016).
Using Fig. 1 as our expositional roadmap, we begin with applications for individual‐level state dynamics. We then work our way up to the population, community and ecosystem levels. Within each level of the ecological hierarchy, we find it convenient to distinguish ‘existential’, ‘developmental’ and ‘spatial’ states. Although there is inevitably some degree of overlap, particularly at the higher levels of the hierarchy that are inherently spatial, we use this distinction in an attempt to separate states of being that in isolation can be viewed as essentially non‐spatial from state dynamics that are more strictly spatial in nature. We further delineate the non‐spatial states as ‘existential’ based on a fundamental measure of existence at each level of the hierarchy and ‘developmental’ based on state characteristics that can drive the dynamics of this fundamental measure of existence. We employ these categories simply for ease of exposition and view them as neither exhaustive nor mutually exclusive.
Although typically not referred to as HMMs in the ecological literature, several subfields of ecology have been using HMMs for individual‐ to community‐level inference for decades. HMMs have also become standard in biological sequence analysis and molecular ecology (Durbin et al., 1998; Barbu and Limnios, 2009; Yoon, 2009), and there is much crossover potential for state‐of‐the‐art bioinformatic methods to other applications in ecology (Jones et al., 2006; Tucker and Duplisea, 2012). HMMs are also used for very specialised tasks of relevance to ecology, such as counting annual layers in ice cores (Winstrup et al., 2012) or characterising plant architectures (Durand et al., 2005). There are therefore many example HMM applications within some areas of ecology, of which only a handful can be covered in the material that follows. However, in other areas the promise of HMMs has only just begun to be recognised.
Individual level
Existential state
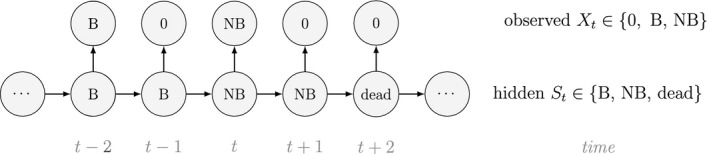
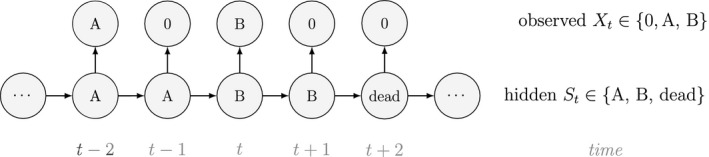
At the level of an individual organism, a fundamental measure of existence is to be alive or not (i.e. dead or unborn). We will therefore begin by demonstrating that one of the oldest and most popular inferential tools in wildlife ecology, the Cormack‐Jolly‐Seber (CJS) model of survival (Williams et al., 2002), is a special case of an HMM. The CJS model estimates survival probabilities () from capture–recapture data. Capture–recapture data consist of sequences of encounter histories for marked individuals collected through time, where for each individual the observed data are represented as a binary series of ones and zeros. For the CJS model, indicates a marked individual was alive and detected at time , while indicates non‐detection. Marked individuals can either be alive or dead at time , but the ‘alive’ state is only partially observable and the ‘dead’ state is completely unobservable. Under this observation process, if it is known that the individual survived from time to time (with probability ) and was detected with probability . However, when there are two possibilities: (1) the individual survived to time (with probability ) but was not detected (with probability ); or (2) the individual did not survive from time to time (with probability ).
Although not originally described as such, the CJS model is simply a two‐state HMM that conditions on first capture. Framing the observed and hidden processes within the dependence structure of a basic HMM (Fig. 2), we could for example have:
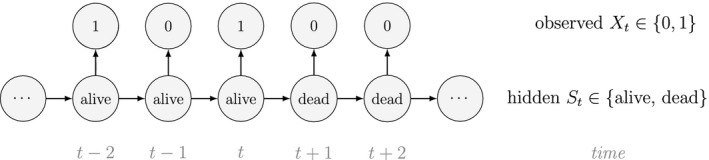
The state‐dependent observation distribution for is a simple Bernoulli (i.e. a coin flip) with success probability if alive and success probability if dead:
We thus have the initial distribution
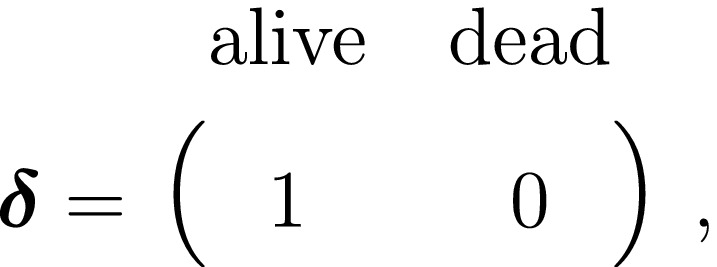
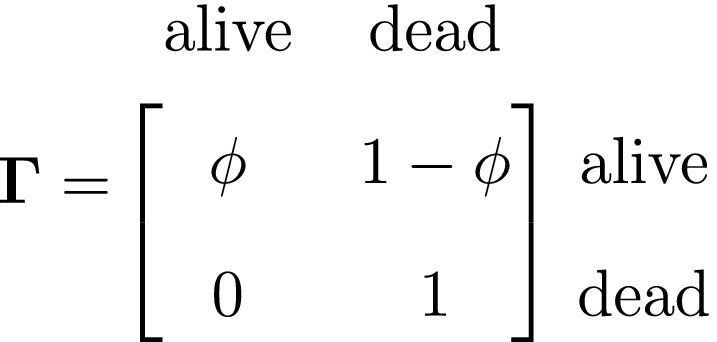
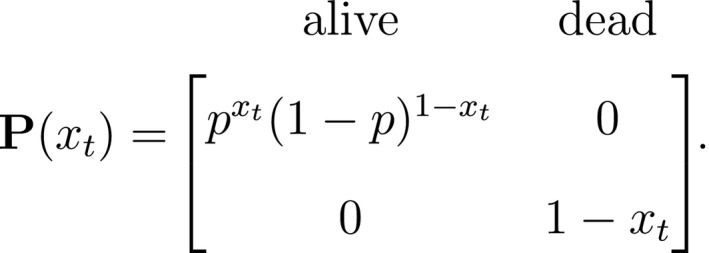
state transition probability matrix and state‐dependent observation distribution matrix
The CJS model is thus a very simple HMM with an absorbing ‘dead’ state and only two unknown parameters ( and ). As an HMM, it can not only be used to estimate survival, but also the point in time when any given individual was most likely to have died (based on local or global state decoding; see Table 1).
The classic Jolly‐Seber capture–recapture model and its various extensions (Pradel, 1996; Williams et al., 2002) go a step further by incorporating both birth and death processes. It simply involves extending the two‐state model to an additional ‘unborn’ (UB) state. We could for example now have:
To formulate a three‐state HMM with an additional ‘unborn’ state, we must extend our components for the hidden and observed processes accordingly:
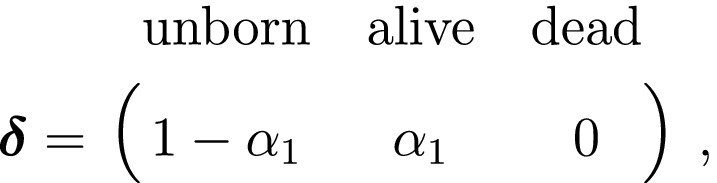
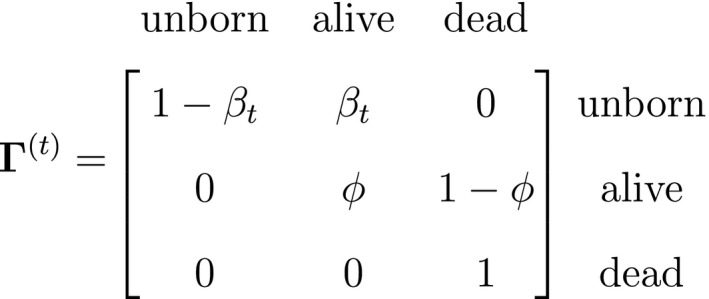
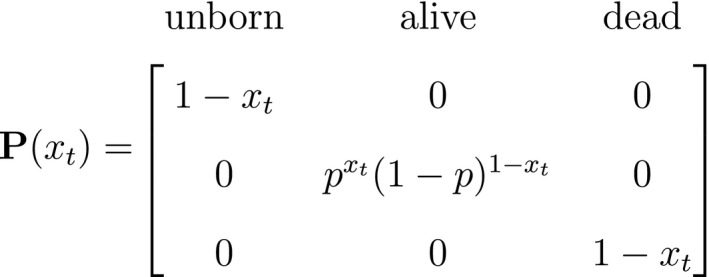
and where
is the probability that an individual was already in the population at the beginning of the study, is the probability that any given individual was born at time , and is the probability that an individual entered the population on occasion given it had not already entered up to that time. Importantly, note that the two‐state and three‐state HMMs rely on the exact same binary data , but we are able to make additional inferences in the three‐state model by re‐formulating the observed and hidden processes in terms of both birth and death. While we have employed these well‐known individual‐level capture–recapture models to initially demonstrate the key idea of linking observed state‐dependent processes to the underlying state dynamics via HMMs, these types of inferences are not limited to traditional capture–recapture observation processes. For example, telemetry and count data can also be used in HMMs describing individual‐level birth and death processes (Schmidt et al., 2015; Cowen et al., 2017).
Developmental state
Individual‐level data often contain additional information about developmental states such as those related to size (Nichols et al., 1992), reproduction (Nichols et al., 1994), social groups (Marescot et al., 2018) or disease (Benhaiem et al., 2018). However, assigning individuals to states can be difficult when traits such as breeding (Kendall et al., 2012), infection (Chambert et al., 2012), sex (Pradel et al., 2008) or even species (Runge et al., 2007) are ascertained through observations in the field. This difficulty has motivated models for individual histories that can not only account for multiple developmental states (Lebreton et al., 2009), but also uncertainty arising from partially or completely unobservable states (Pradel, 2005). Such multi‐state models can be used for testing a broad range of formal biological hypotheses, including host–pathogen dynamics in disease ecology (Lachish et al., 2011), reproductive costs in evolutionary ecology (Garnier et al., 2016) and social dominance in behavioural ecology (Dupont et al., 2015). For example, it is straightforward to extend the capture–recapture HMM to multiple ‘alive’ states parameterised in terms of state‐specific survival probabilities and transition probabilities between these ‘alive’ states . Consider a ‐state HMM for capture–recapture data that incorporates reproductive status, where indicates ‘alive and breeding’ and indicates ‘alive and non‐breeding’:
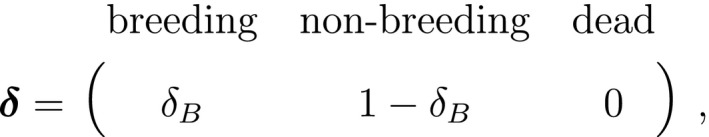

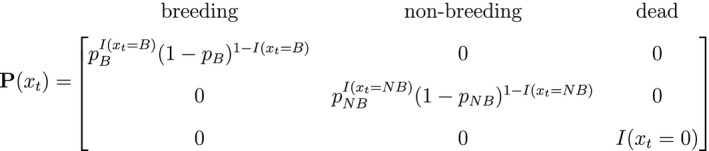
and where is an indicator function taking the value 1 when and 0 otherwise. To assess the costs of reproduction, a biologist will be interested in the probability of breeding in year , given breeding or not in year , as well as assessing any differences in survival probability between breeders and non‐breeders . By simply re‐expressing the , and components in terms of the specific state and observation processes of interest, such models can be used to infer the dynamics of conjunctivitis in house finches (Conn and Cooch, 2009), senescence in deer (Choquet et al., 2011), reproduction in Florida manatees (Kendall et al., 2012), interspecific competition between ungulates (Gamelon et al., 2020) and life‐history trade‐offs in elephant seals (Lloyd et al., 2020). Similar HMMs can also be used to investigate relationships between life‐history traits and demographic parameters that are important in determining the fitness of phenotypes or genotypes (Stoelting et al., 2015). Several measures of individual fitness have been proposed, but one commonly used for field studies is lifetime reproductive success (Rouan et al., 2009; Gimenez and Gaillard, 2018). These approaches can be readily adapted to quantify other measures of fitness (McGraw and Caswell, 1996; Link et al., 2002; Coulson et al., 2006; Marescot et al., 2018).
Inferences about developmental states are of course not limited to traditional capture–recapture data, and significant advancements in animal‐borne biotelemetry technology have brought many new and exciting opportunities (Cooke et al., 2004; Hooten et al., 2017; Patterson et al., 2017). For example, telemetry location data can be used to identify migratory phases (Weng et al., 2007), predation events (Franke et al., 2006) or the torpor‐arousal cycle of hibernation (Hope and Jones, 2012). The multi‐state (i.e. hidden Markov) movement model is often used to infer these types of movement behaviour modes from trajectories in two‐dimensional space, where the observations are typically expressed in terms of the bivariate sequence of Euclidean distances (or ‘step lengths’) and turning angles between consecutive locations (Franke et al., 2004; Morales et al., 2004). For a model involving states that assumes conditional independence between step length (; in meters) and turning angle (; in radians) as in Fig. 4d, we could for example have:
These states could correspond to ‘resident’ (state 1) and ‘transient’ (state 2) behavioural phases, such that within state 2 the movements tend to be longer and directionally persistent (i.e. with turning angles concentrated near zero). When assuming conditional independence of the observations, the bivariate state‐dependent distribution for is simply the product of two univariate state‐dependent distributions,
These univariate distributions are typically assumed to be the gamma or Weibull distribution for step length and the von Mises or wrapped Cauchy distribution for turning angle. Unlike our previous examples so far, the number of underlying states in these types of HMMs is generally not clear a priori and needs to be selected based on both biological and statistical criteria (Pohle et al., 2017). Another difference is that there is often no predetermined structure in the state transition probability matrix,

and all entries are freely estimated (but still subject to ). As a consequence, the characteristics of the model states as represented by the state‐dependent distributions are fully data driven, and hence may not correspond exactly to biologically meaningful entities (see IMPLEMENTATION, CHALLENGES AND PITFALLS).
Similar HMMs for animal movement have been used, inter alia, to identify wolf kill‐sites (Franke et al., 2006), the relationship between southern bluefin tuna behaviour and ocean temperature (Patterson et al., 2009), activity budgets for harbour seals (McClintock et al., 2013), hunting strategies of white sharks (Towner et al., 2016), the behavioural response of northern gannets to frontal activity (Grecian et al., 2018) and how common noctules adjust their space use to the lunar cycle (Roeleke et al., 2018). Driven by the influx of new biotelemetry sensor technology, HMMs have also been used to analyse the sequences of dives of marine animals (Hart et al., 2010; Quick et al., 2017; DeRuiter et al., 2017; van Beest et al., 2019). The remote collection of activity data at potentially very high temporal resolutions using accelerometers is another emerging application area (Diosdado et al., 2015; Leos‐Barajas et al., 2017b; Papastamatiou et al., 2018a,b; Adam et al., 2019b). These HMM formulations are conceptually very similar to the movement model outlined above, with the state process corresponding to behavioural modes (or at least proxies thereof), and the activity data represented by the state‐dependent process. Fig. 5 illustrates a possible workflow for inferring four behavioural modes from high‐resolution accelerometer data collected from a striated caracara (Phalcoboenus australis) over a period of 1 hour. Here the vector of dynamic body acceleration was used as a univariate summary of the three‐dimensional raw acceleration data, and a gamma distribution was used for the state‐dependent observation process. In this example, the HMM can be regarded as a clustering scheme which maps observed input data to unobserved underlying classes with biological interpretations roughly corresponding to ‘resting’, ‘minimal activity’ (e.g. preening), ‘moderate activity’ (e.g. walking, digging) and ‘flying’. Complete details of this analysis, including each step of the workflow and example R (R Core Team, 2019) code, can be found in the Supplementary Tutorial.
Figure 5.
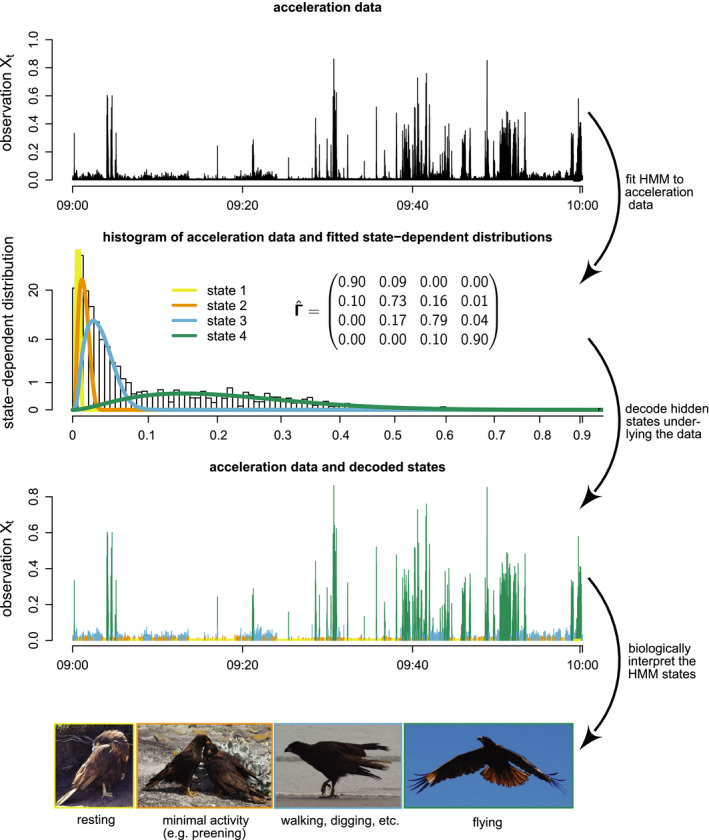
Illustration of a possible workflow when using an HMM to infer behavioural modes from the vector of dynamic body acceleration data of a striated caracara (Phalcoboenus australis) over a period of 60 min (see Fahlbusch & Harrington, 2019, for data details). Four behavioural modes were identified and biologically interpreted to be associated with resting (yellow), minimal activity (orange), moderate activity (blue) and flying (green).
Spatial state
HMMs can also be used for inferences about the unobserved spatial location of an individual. For example, capture–recapture data can consist of sequences of observations arising from a set of discrete spatial states, where these often refer to ecologically important geographic areas, such as wintering and breeding sites for migratory birds (Brownie et al., 1993) or spawning sites for fish (Schwarz et al., 1993). For a ‐state HMM with two sites (A and B), where indicates ‘alive at site A’ and indicates ‘alive at site B’, we could for example have:
Clearly, this discrete‐space HMM is structurally identical to the multi‐state capture–recapture HMMs already described in the previous section; the only difference is the state transition probability parameters are now interpreted as site‐specific survival and movement probabilities between the sites (e.g. fidelity or dispersal; Lagrange et al., 2014; Cayuela et al., 2020). Based on global state decoding, these HMMs can therefore also be used to infer the most likely spatial state for periods when an individual was alive but its location was not observed.
Another important application of HMMs is for geolocation based on indirect measurements that vary with space, such as light, pressure, temperature and tidal patterns (Thygesen et al., 2009; Rakhimberdiev et al., 2015). Although too technical to be described in detail here, geolocation HMMs can be particularly useful for inferring individual location from archival tag data (Basson et al., 2016). These HMMs have even been extended to include state‐switching behaviours such as those described in the previous section (Pedersen et al., 2008, 2011b). Animal movement behaviour HMMs have also been extended to accommodate partially observed location data common to marine mammal satellite telemetry studies (Jonsen et al., 2005; McClintock et al., 2012).
Population level
We consider two ways that inference on the population level can arise: (1) an individual‐level model, based on data from multiple individuals (e.g. capture–recapture), quantitatively connected to a population‐level concept through an explicit model; or (2) a population‐level model, based on population‐level data (e.g. counts or presence–absence), with no explicit model for processes at the individual level.
Existential state
A fundamental existential state at the population level is abundance, the number of individuals alive in a population at a particular point in time. A common way to infer this using capture–recapture HMMs is to formally link abundance to the individual‐level processes (e.g. survival, recruitment) that drive its dynamics. Intuitively, the abundance model specifies how many individuals go through the life history specified by the HMM. For the abundance component, the key pieces of information are the number of individuals in the population that were detected at least once and the probability of being detected at least once, given an individual was alive at any time during the study . The former is observed while the latter can be calculated as
using notation for the Jolly‐Seber HMM presented in Individual level. This HMM formulation is equivalent to the original Jolly‐Seber open population model (shown in Glennie et al., 2019), where population abundance at each time is derived from the individual‐level process parameters.
Instead of inducing changes in abundance through individual‐level HMMs, abundance itself can be modelled as the hidden state within an HMM (Schmidt et al., 2015; Cowen et al., 2017; Besbeas and Morgan, 2019). Here population dynamics are inferred from population‐level surveys (Buckland et al., 2004), where the observation process can include counts or other quantities that are noisy measurements of the true abundance (the hidden state), and the state transition probability matrix is naturally formulated in terms of the well‐known Leslie matrix for population growth (Caswell, 2001). For example, for imperfect count data that were collected from a population of true size (note the requirement to specify a maximum possible population size ), we could have:
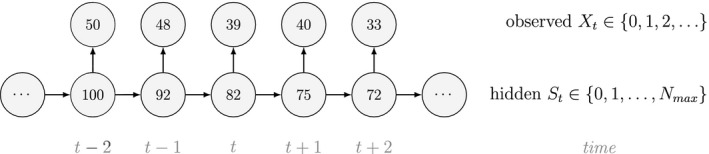
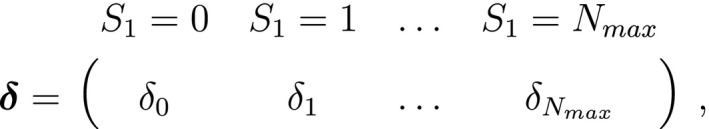
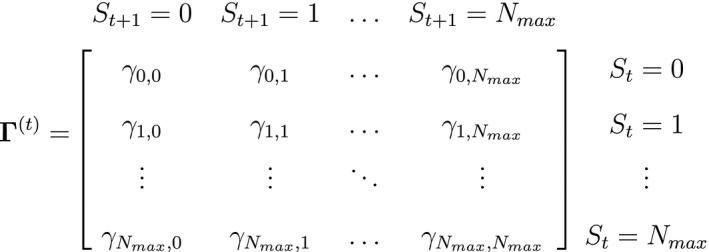
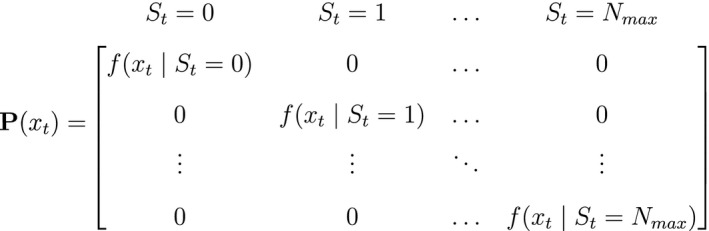
and
Each state transition probability describes the population dynamics from time to time and can be parameterised in terms of survival, reproduction, emigration, the current population size and any additional population structure (e.g. sex or age classes; see Population level ‐ Developmental state). The state‐dependent distributions can take many different forms depending on the specific observation process, but common choices for count data are binomial or Poisson models (Schmidt et al., 2015; Besbeas and Morgan, 2019). Sometimes count data alone can be insufficient for describing complex population processes, and this has led to integrated population modelling (Schaub and Abadi, 2011) that uses auxiliary data such as capture–recapture, telemetry or productivity data (Schmidt et al., 2015; Besbeas and Morgan, 2019).
Developmental state
Populations have more structure than simply their overall abundance or density. Sex, age demographics, size of breeding sub‐population, fitness of individuals, and behavioural or genetic heterogeneity all have an impact on the development of a population (Seber and Schofield, 2019). Many of these processes can be accounted for within the HMM framework presented in the previous section for individual‐level data. As before, the idea is to extend the ‘alive’ state to a more complex network of states whose state‐dependent distributions and transitions match the structure in the population. Combinations of these individual attributes provide the opportunity to build a rich state process to describe the population dynamics. This framework is built around the idea that individuals are the singular units that together drive population change, but there has also been increasing use of HMMs from a different viewpoint: that of evolutionary processes at lower levels of organisation (e.g. genes).
With recent advances in genetic sequencing, the need for interpreting and modelling biological sequences (e.g. protein or DNA) has boosted the development of HMMs in molecular ecology (Durbin et al., 1998; Boitard et al., 2009; Yoon, 2009; Ghosh et al., 2012). Many of these applications use HMMs strictly as a tool for biological sequence analysis (e.g. identifying species from DNA barcodes; Hebert et al., 2016) and are too technical to delve into detail here, but HMMs for molecular sequence data are commonly formulated in terms of evolutionary state dynamics, including for example speciation and extinction (Hobolth et al., 2007; Soria‐Carrasco et al., 2014; Crampton et al., 2018; Olajos et al., 2018), hybridisation (Schumer et al., 2018; Palkopoulou et al., 2018), mutualism (Werner et al., 2018), hidden drivers of diversification (Caetano et al., 2018) and evolutionary rates among sites (Felsenstein and Churchill, 1996).
Telemetry locations are another form of individual‐level data that, when combined across individuals, can provide population‐level inferences about movement, space use and resource selection (Hooten et al., 2017). As such, telemetry data can be well suited for addressing hypotheses related to intraspecific interactions. While such applications are still relatively rare, location data have been used in HMMs investigating intraspecific competition in marine mammals (Breed et al., 2013), herding in ungulates (Langrock et al., 2014a) and social behaviour in fish (Bode and Seitz, 2018).
Similar to approaches for inferring population‐level developmental states from individual‐level data, a rich structure can also be specified within an HMM for population‐level data. Multiple states and processes can be represented: age classes/survival, size classes/growth, sex/birth, genotypes and metapopulations are all states or networks of states with specified connections (Newman et al., 2014). Such HMMs can be informed by a wide variety of population‐level observations, for example counts of plants (Borgy et al., 2015) or animals (Schmidt et al., 2015), as well as auxiliary individual‐level observations (Besbeas and Morgan, 2019). From this general viewpoint, HMMs can be seen as the structure behind open population N‐mixture models (Schmidt et al., 2015; Cowen et al., 2017), distance sampling models (Sollmann et al., 2015) and approximate state‐space population dynamics models (Besbeas and Morgan, 2019).
Spatial state
The spatial state of a population can be conceived as a surface (or map) quantifying density at each point in space, and population models for individual‐level data can be extended to allow density to change over space (Borchers and Efford, 2008). Inferring density as a spatial population state, however, requires spatial information within the data. Spatial capture–recapture surveys (Royle et al., 2013), an extension of capture–recapture, collect precisely these data. Spatial capture–recapture HMMs can be formulated in terms of survival, recruitment, movement and population density (Royle et al., 2018; Glennie et al., 2019) and are readily extendable for relating environment and population distribution across space, including how distribution is affected by landscape connectivity, dispersal, resource selection or environmental impacts such as oil spills (McDonald et al., 2017; Royle et al., 2018).
A different viewpoint is to consider population‐level data that are commonly collected over both space and time: presence–absence data. These data provide information on a population's spatial state that is not derived from abundance and arise from the monitoring of spatial units for the (apparent) presence or absence of a species. One of the most popular tools for analysing these data are patch (or site) occupancy models, which can be used to infer patterns and dynamics of species occurrence while accounting for imperfect detection (MacKenzie et al., 2018). As with capture–recapture models, patch occupancy models are also HMMs (Royle and Kéry, 2007; Gimenez et al., 2014) where, instead of the state dynamics of individual organisms, the hidden process describes the state dynamics of sites. Let indicate ‘occupied’ and indicate ‘unoccupied’, where the species can be detected or not during multiple visits to each site, with the following representation:
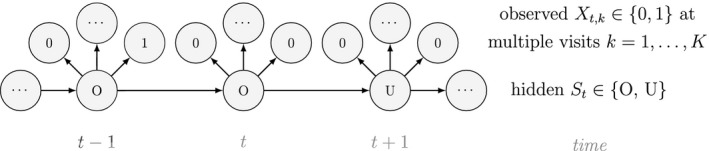
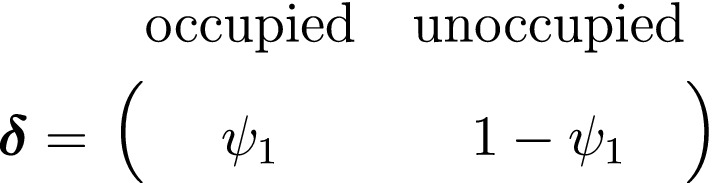
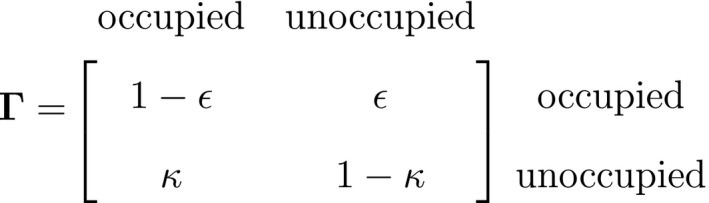
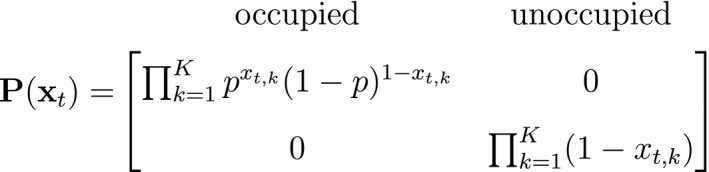
and where is the initial patch occupancy probability at time , is the species detection probability at each occupied patch and is composed of the local colonisation and extinction probabilities. Single‐season (or static) occupancy models (MacKenzie et al., 2002) are obtained as a special case with or (Gimenez et al., 2014). This HMM can not only be used to estimate patch occupancy, extinction and colonisation probabilities, but also the most likely state and times of any colonisation or extinction events within a patch. The flexibility of the HMM formulation allows patch occupancy to be conveniently extended to cope with site‐level heterogeneity in detection using finite mixtures (Louvrier et al., 2018) or a discrete measure of population density (Gimenez et al., 2014; Veran et al., 2015) and even false positives due to species misidentification (Miller et al., 2011; Louvrier et al., 2019). Just as with multi‐state capture–recapture HMMs (see Population level ‐ Developmental state), species occurrence HMMs can be readily extended to multiple ‘occupied’ states accommodating reproduction (MacKenzie et al., 2009; Martin et al., 2009), disease (McClintock et al., 2010) and other (meta‐)population dynamics (Lamy et al., 2013).
Inferences from HMMs for presence–absence data are not limited to occupancy models that account for imperfect species detection. For example, Pluntz et al., (2018) developed an HMM characterising seed dormancy, colonisation and germination in annual plant metapopulations based entirely on presence–absence observations of standing flora. In their study, the presence of a completely unobservable soil seed bank was the hidden state of interest, and they modified the dependence structure of a basic HMM such that the seed bank state dynamics at time depended not only on the seed bank state at time , but also on the presence or absence of standing flora at time . Let indicate ‘seed bank absent at time , flora absent at time ’, indicate ‘seed bank present at time , flora absent at time ’ and indicate ‘seed bank present at time , flora present at time ’, where standing flora is present or not during visit to each site and is assumed to be detected without error. We could for example have:
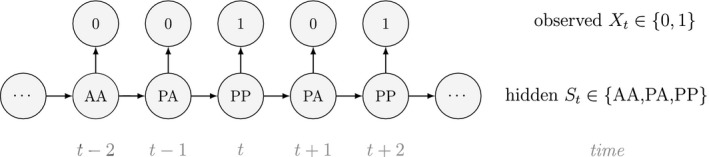
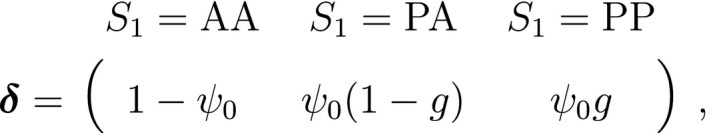
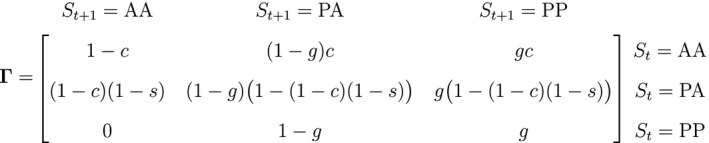
where is the probability that a seed bank was present the year before the first observation, is the probability of germination and survival to reproduction, is the probability of seed bank survival, is the probability of external colonisation and is a diagonal matrix of ones. Similar formulations could be applied to other organisms with dormant life stages (e.g. fungi, crustaceans).
Community level
Community‐level studies often focus on a subset of species based on taxonomy, trophic position or particular interactions of interest, and the diversity of topics addressed in community ecology reflects its large scope (Vellend, 2010, 2016). Here we will only scratch the surface of two study systems that can be formulated as HMMs for multi‐species presence–absence data commonly collected from field surveys or (e)DNA samples: (1) patch systems composed of (potentially) many species; and (2) patch systems composed of a few (possibly interacting) species.
Existential state
A fundamental measure of biodiversity is the number of species within a community (species richness). This community‐level state is often unobservable in studies of natural systems (Dorazio et al., 2006), even for communities composed entirely of sessile organisms (Conway‐Cranos and Doak, 2011; Chen et al., 2013). Multi‐species occupancy HMMs expand single‐species occupancy HMMs (see Population level) to the community level using presence–absence data for each species that could (potentially) occupy the sampling units within a study area (MacKenzie et al., 2018, Chapter 15). By combining single‐species HMMs, either independently or by sharing common parameters among species (Evans et al., 2016; Guillera‐Arroita, 2017), community‐level attributes (e.g. species richness) and species‐level attributes (e.g. patch occupancy) can be integrated within a single modelling framework (Royle and Dorazio, 2008, Chapter 12). By jointly modelling species‐ and community‐level processes, the approach proposed by Dorazio and Royle (2005) and its extensions (reviewed by Kery and Royle, 2015, Chapter 11) facilitate the simultaneous testing of formal hypotheses about factors influencing occupancy (Rich et al., 2016; Tenan et al., 2017), species richness (Sutherland et al., 2016) and their dynamics through time (Russell et al., 2009; Dorazio et al., 2010), with important consequences for conservation and management (Zipkin et al., 2010). Although these community dynamics models are typically fitted using hierarchical Bayesian methods and not explicitly referred to as HMMs, they share the same properties and can be similarly decomposed in terms of , and . Viewing the species richness of a community as analogous to the abundance of a population, HMM formulations similar in spirit to those described in Population level could account for species that were never detected (sensu Dorazio et al., 2006).
Developmental state
Many community‐level attributes can be constructed from ‘metacommunity’ HMMs for species richness at both the community and metacommunity level (Dorazio and Royle, 2005; Kery and Royle, 2015, Chapter 11). Species richness at each site is the diversity metric, and total richness in the whole metacommunity is the diversity (Magurran, 2004, Chapter 6). A possible metric for the diversity is the similarity Jaccard index: the proportion of species that occur at two sites among the species that occur at either site. Multi‐species occupancy models have also been used to address variation in community attributes within distinct regions using Hill numbers for species richness, Shannon diversity and Simpson diversity (Broms et al., 2015; Sutherland et al., 2016; Tenan et al., 2017; Boron et al., 2019). Dynamic multi‐species occupancy HMMs can provide inferences about changes in community composition and structure over time, entry (or ‘turnover’) probabilities of ‘new’ species into the community and species ‘extinction’ probabilities from the community (Russell et al., 2009; Dorazio et al., 2010). Although to our knowledge this has not yet been attempted, community assembly or succession dynamics could naturally be parameterised in terms of such quantities within a multi‐state, multi‐species HMM describing transitions among different community states (e.g. disturbed, climax). Community structure and composition also depend on interspecific interactions, and multi‐species occupancy HMMs can empirically test for any such evidence (Gimenez et al., 2014; Rota et al., 2016; Davis et al., 2018; MacKenzie et al., 2018; Marescot et al., 2020). To date these co‐occurrence models have mostly been used to infer predator–prey interactions (Miller et al., 2018b; Murphy et al., 2019). Other emerging frameworks for inferences about processes that structure communities could also potentially be formulated as HMMs to account for observation error in presence–absence or count data (Ovaskainen et al., 2017).
Spatial state
Understanding geographic variation in the size and structure of communities is one of the major goals in ecology. While we have so far focused on some of the more ‘non‐spatial’ aspects of community‐level inference, all multi‐species presence–absence HMMs are of course inherently spatial and also describe community distribution. Dynamic multi‐species occupancy models provide inferences about changes in community distributions (Russell et al., 2009; Dorazio et al., 2010), and, when spatio‐temporal interactions between species are of primary interest, dynamic co‐existence HMMs can incorporate local species extinction and colonisation to investigate interspecific drivers of co‐occurrence dynamics and community distribution (Fidino et al., 2019; Marescot et al., 2020). As a final illustrative example, suppose we have the states (respectively and ) for ‘site occupied by species A’ (respectively by species B and by both species) and indicates ‘unoccupied site’. Define , where indicates neither species was detected, indicates only species A was detected, indicates only species B was detected and indicates both species were detected on the th visit at time . We could for example have:
This model is more complex than previous examples, but it can still be readily expressed in terms of , and for inferring patterns and drivers of species co‐existence distribution dynamics (see Appendix A in Supplementary Material).
Ecosystem level
Despite the well‐recognised need for reliable inferences about broad‐scale ecological dynamics in the face of climate change and other challenges (Turner et al., 1995), HMMs have thus far seldom been applied at the ecosystem level. This is likely attributable to many factors, including the difficulty of obtaining and integrating observational data at the large spatio‐temporal scales required (Jones et al., 2006; Bohmann et al., 2014; Dietze et al., 2018; Estes et al., 2018; Compagnoni et al., 2019). Although there are fewer examples in the literature, HMMs have been used to make ecosystem‐level inferences about stability and regime shifts (Gal and Anderson, 2010; Gennaretti et al., 2014; Economou and Menary, 2019), climate‐driven community and disease dynamics (Moritz et al., 2008; Martinez et al., 2016; Miller et al., 2018a), the effects of management action on habitat dynamics (Breininger et al., 2010), climatic niches (Tingley et al., 2009) and ecosystem health (Xiao et al., 2019). HMMs are also frequently used by atmospheric scientists, hydrologists and landscape ecologists to describe regional‐ to global‐scale ecosystem processes such as precipitation (Zucchini and Guttorp, 1991; Srikanthan and McMahon, 2001), streamflow (Jackson, 1975; Bracken et al., 2014), wetland dynamics (Siachalou et al., 2014) and land cover dynamics (Aurdal et al., 2005; Lazrak et al., 2010; Trier and Salberg, 2011; Abercrombie and Friedl, 2015; Siachalou et al., 2015). While many of these examples tend to focus on a few specific biotic and/or abiotic components in which to frame ecosystem state dynamics, we can envision future applications adopting a more holistic approach that integrates increasingly more complex ecosystem‐level processes with observational data arising from a variety of sources and spatio‐temporal scales (see FUTURE DIRECTIONS).
IMPLEMENTATION, CHALLENGES AND PITFALLS
Software
Recent advances in computing power and user‐friendly software have made the implementation of HMMs much more feasible for practitioners. However, the features and capabilities of the software are varied, and it can be challenging to determine which software may be most appropriate for a specific objective. We briefly describe some of the HMM software currently available, limiting our treatment to freely available R (R Core Team, 2019) packages and stand‐alone programs that we believe are most accessible to ecologists and non‐statisticians. While most HMM packages in R include data simulation, parameter estimation and state decoding for an arbitrary number of system states, they differ in many key respects (Table 2). Some of the more general packages provide greater flexibility for specifying state‐dependent probability distributions (Visser and Speenkenbrink, 2010; Jackson, 2011; Harte, 2017; McClintock and Michelot, 2018). One of the earliest and most flexible HMM packages, depmixS4 (Visser and Speenkenbrink, 2010), can accommodate multivariate HMMs, multiple observation sequences, parameter covariates, parameter constraints and missing observations. Similar to depmixS4 in terms of features and flexibility, momentuHMM (McClintock and Michelot, 2018) can also be used to implement mixed HMMs (DeRuiter et al., 2017), hierarchical HMMs (Leos‐Barajas et al., 2017a; Adam et al., 2019a), zero‐inflated probability distributions (Martin et al., 2005) and partially observed state sequences. In addition to the R packages presented in Table 2, there are numerous R and stand‐alone software packages that are less general and specialise on particular HMM applications in ecology, as well as general statistical programs with which these types of models can be relatively easily implemented (see Appendix B in Supplementary Material).
Table 2.
Features of HMM packages available in the R environment for statistical computing, including capabilities for multiple observation sequences (‘Multiple sequences’), multivariate HMMs (‘Multivariate’), mixed HMMs (‘Mixed’), hierarchical HMMs (‘Hierarchical’), hidden semi‐Markov models (‘Semi‐Markov’), parameter covariate modelling (‘Covariates’), parameter constraints (‘Constraints’), missing observations (‘Missing data’) and state‐dependent probability distributions
| Package | Multiple sequences | Multivariate | Mixed | Hierarchical | Semi‐Markov | Covariates | Constraints | Missing data | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aphid | ✓ | Wilkinson (2019) | |||||||||
| depmixS4 | ✓ | ✓ |
|
|
✓ | Visser and Speenkenbrink (2010) | |||||
| HiddenMarkov |
|
Harte (2017) | |||||||||
| HMM | Himmelmann (2010) | ||||||||||
| hsmm | ✓ | Bulla and Bulla (2013) | |||||||||
| LMest | ✓ | ✓ | ✓ | or | ✓ | Bartolucci et al., (2017) | |||||
| mhsmm | ✓ | ✓ | ✓ | O'Connell and Hojsgaard (2011) | |||||||
| momentuHMM | ✓ | ✓ | ✓ | ✓ |
|
|
✓ | McClintock and Michelot (2018) | |||
| msm | ✓ | ✓ |
|
|
✓ | Jackson (2011) | |||||
| RcppHMM | Cardenas‐Ovando et al., (2017) | ||||||||||
| seqHMM | ✓ | ✓ | ✓ |
|
|
✓ | Helske and Helske (2019) |
| State‐dependent probability distributions | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bernoulli | Beta | Binomial | Categorical | Custom | Exponential | Gamma | Lognormal | Logistic | Negative binomial | Normal | Multivariate normal | Truncated normal | Poisson | Student's | Von Mises | Weibull | Wrapped Cauchy | |
| aphid | ✓ | |||||||||||||||||
| depmixS4 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||
| HiddenMarkov | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||
| HMM | ✓ | |||||||||||||||||
| hsmm | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| LMest | ✓ | ✓ | ✓ | |||||||||||||||
| mhsmm | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| momentuHMM | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| msm | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||
| RcppHMM | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| seqHMM | ✓ | |||||||||||||||||
‘Covariates’ and ‘Constraints’ can pertain to initial distribution , state‐dependent probability distribution , state transition probability and/or mixture probability parameters. Several packages facilitate extensions for user‐specified state‐dependent probability distributions that require no modifications to the package source code (‘custom’).
Covariates are only permitted on state‐dependent distribution location parameters for the binomial, gamma, normal and Poisson distributions.
Covariates are only permitted on state‐dependent categorical distribution parameters.
Covariates are only permitted on state‐dependent distribution location parameters.
[Corrections added on 10 November 2020, after first online publication: Table 2 has been updated.]
Challenges and pitfalls
HMMs are natural candidates for conducting inference related to a wide range of ecological phenomena, but they are not a panacea (see Box 2). There are many ecological processes that cannot be faithfully characterised under the simplifying assumptions of HMMs, in which case other latent variable models may be more appropriate (see Box 1). When HMMs are appropriate, it can be challenging to tailor HMMs to real data, even when using user‐friendly software packages. Here we briefly highlight those issues that, based on our experience, constitute the key challenges when using HMMs to analyse ecological data. Other important aspects of statistical practice that are not unique to HMMs, such as model checking and selection (e.g. Zucchini et al., 2016, Chapter 6), are covered in more detail in the Supplementary Tutorial.
Depending on the complexity of the state and observation processes, various modelling decisions may need to be made. Among these are the number of states to include, whether to incorporate covariates for the model parameters and whether the basic dependence structure is sufficient. These decisions tend to be case‐dependent and require expert knowledge of the system of interest, so we make no attempt to provide general guidance in this respect. However, in some cases the model structure may be a direct consequence of the ecological process. For example, in the CJS model, the two states (alive or dead) and also the state‐dependent (Bernoulli) distributions follow immediately from the capture–recapture process. In situations with more complex data, such as multivariate time series related to animal behaviour (DeRuiter et al., 2017; Ngô et al., 2019; van Beest et al., 2019), it takes experience and a good intuition both for the data and for the HMM framework to identify an adequate model formulation (Pohle et al., 2017).
Unlike other statistical models such as linear regression, there is no analytical solution for HMM parameter estimation. One must therefore resort to numerical procedures, all of which involve technical challenges: local maxima for maximum likelihood estimation (Myung, 2003), or label switching (Jasra et al., 2005) and poor mixing (Brooks et al., 2011) for MCMC sampling. Any increase in model complexity with respect to the number of states or the parameters tends to rapidly exacerbate these problems. When working with HMMs, it is thus important to develop an appreciation for these challenges and the associated risks. For maximum likelihood estimation, the risk of false convergence to a local rather than the global maximum of the likelihood must not be underestimated. In addition to the general advice to avoid overly complex models (Lavine, 2010; Cole, 2019), the main strategy to reduce this risk is to try many initial parameter vectors within the maximisation.
While it is tempting to interpret the states of an HMM fitted to ecological data as biologically meaningful entities, this is not always justifiable. Outside the standard capture–recapture or species occurrence applications, HMMs are often applied in an unsupervised learning context (see Figs 3 and 5, Supplementary Tutorial), such that the state characteristics are completely data driven rather than pre‐defined (Leos‐Barajas et al., 2017b). The model then picks up the statistically most relevant modal patterns in the data, and these may or may not correspond closely to ecologically meaningful states. It is thus important not to over‐interpret the model states, as in some cases they may only be crude proxies for the ecological system states of interest. A classic example is the simple state HMM for animal movement behaviour based on step lengths and turning angles (Morales et al., 2004), where evidence of an area‐restricted search‐type state is often labelled as ‘foraging’. Although for many animals area‐restricted search is commonly associated with foraging, one usually cannot definitively conclude when an individual was actually foraging based solely on location data. Furthermore, while it can be useful to refer to these modalities using descriptive terms such as ‘foraging’ (or ‘resident’) and ‘searching’ (or ‘transient’), this does not mean that an animal has only two modes of behaviour.
FUTURE DIRECTIONS
We have highlighted many realised and potential applications of HMMs in ecology. We anticipate increased application and development of HMMs as ecologists continue to discover how this relatively simple and flexible class of statistical models can reveal complex state dynamics that are inherently difficult to observe. Indeed, a Web of Science search for ‘hidden Markov’ suggests a rapidly increasing awareness of these models within the ecological community (Fig. 6). Given differences in terminology and a tendency for ecologists to use HMMs without explicitly referring to them as such, the use of HMMs is surely becoming even more widespread.
Figure 6.
Number of publications (left axis) and total number of times these publications were cited (right axis) per year based on a Web of Science search for ‘hidden Markov’ conducted within the categories of ‘Biology’, ‘Ecology’, ‘Marine Freshwater Biology’ and ‘Zoology’ on 7 July 2020.
In order for the power and flexibility of HMMs to be harnessed by the broader ecological community, researchers must first be able to recognise the limitations of their data and how these can be leveraged by formally linking observable phenomena to the actual ecological processes of interest. Such hierarchical modelling exercises are critical to reliable inference (Royle and Dorazio, 2008; Kery and Royle, 2015), and it is no coincidence that HMMs have independently ‘evolved’ in different ecological contexts over the years. By assuming a discrete state space with basic dependence structures, HMMs can easily capture complex system processes, such as those involving serial correlation, nonlinearity, non‐normality and non‐stationarity, in a tractable manner that goes well beyond the examples highlighted here. Instead of viewing these as a series of disparate domain‐specific applications of HMMs, we view them as a synthesis of the process by which ecologists can begin to critically think about their own sequential data, relate them to their particular system of interest and formulate an HMM for their specific domain using a simple conceptual template.
We foresee HMMs being more frequently used to integrate biotic and abiotic observations at large spatio‐temporal scales to investigate complex ecosystem‐level processes. The state process of the HMM could itself be at the ecosystem level (e.g. alternative stable states), or it could simply be used to account for unobservable state dynamics at lower levels of the hierarchy as a component of a larger (non‐Markovian) ecosystem‐level process model. Recent HMM methodological developments such as hierarchical formulations that allow data collection and/or state transitions to occur at multiple temporal resolutions (Fine et al., 1998; Leos‐Barajas et al., 2017a; Adam et al., 2019a), nonparametric approaches avoiding restrictive distributional assumptions (Yau et al., 2011; Langrock et al., 2018) and coupled HMMs for interacting state processes associated with different sequences (Sherlock et al., 2013; Touloupou et al., 2020) extend our capability to incorporate complex data structures and hierarchical relationships scaled from the individual to ecosystem level.
Despite this great potential, there remain several hurdles to the widespread implementation of HMMs describing long‐term, broad‐scale ecological dynamics (Turner et al., 1995; Lindenmayer et al., 2012; Haller, 2014). First, much like regression and analysis of variance, HMMs must become a familiar and accessible instrument within the ecologist's statistical ‘toolbox’. This has been the primary motivation for our review, and we hope our illustrative examples have provided a template by which researchers can begin to formulate HMMs according to their specific state and observation processes of interest. Second, although this challenge is by no means unique to HMMs, ecosystem‐level inferences continue to be limited by data availability, accessibility and compatibility (Jones et al., 2006; Dietze et al., 2018; Estes et al., 2018; Compagnoni et al., 2019; Halbritter et al., 2020), which can compromise our ability to empirically link observation and state processes operating at different spatio‐temporal scales. Third, as with any application of HMMs, such endeavours will require a faithful conceptualisation of ecosystem dynamics that is amenable to this discrete‐state modelling framework, as well as the identification and integration of observation processes that can provide information about the underlying system.
Data Availability Statement
No new data were used.
AUTHORSHIP
All authors conceived and wrote the manuscript.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1111/ele.13610.
Supporting information
Supplementary Material
Supplementary Material
ACKNOWLEDGEMENTS
We thank the editors and three anonymous referees for helpful comments that improved the manuscript. This research was inspired in part by the SFB TRR 212 (NC3), which is funded by the German Research Foundation (DFG). We thank K. Harrington and J. Fahlbusch for providing the striated caracara data. The scientific results and conclusions, as well as any views or opinions expressed herein, are those of the author(s) and do not necessarily reflect those of NOAA or the Department of Commerce.
References
- Abercrombie, S.P. & Friedl, M.A. (2015). Improving the consistency of multitemporal land cover maps using a hidden Markov model. IEEE Trans. Geosci. Remote Sens., 54, 703–713. [Google Scholar]
- Adam, T. , Griffiths, C.A. , Leos‐Barajas, V. , Meese, E.N. , Lowe, C.G. , Blackwell, P.G. et al (2019a). Joint modelling of multi‐scale animal movement data using hierarchical hidden Markov models. Methods Ecol. Evol., 10, 1536–1550. [Google Scholar]
- Adam, T. , Langrock, R. & Weiß, C.H. (2019b). Penalized estimation of flexible hidden Markov models for time series of counts. METRON, 77, 87–104. [Google Scholar]
- Altman, R.M. (2007). Mixed hidden Markov models: an extension of the hidden Markov model to the longitudinal data setting. Journal of the American Statistical Association, 102, 201–210. [Google Scholar]
- Amoros, R. , King, R. , Toyoda, H. , Kumada, T. , Johnson, P.J. & Bird, T.G. (2019). A continuous‐time hidden Markov model for cancer surveillance using serum biomarkers with application to hepatocellular carcinoma. METRON, 77, 67–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger‐Méthé, M. , Newman, K. , Cole, D. , Empacher, F. , Gryba, R. & King, A.A. et al (2020). An introduction to state‐space modeling of ecological time series. arXiv preprint arXiv:200202001. [Google Scholar]
- Aurdal, L. , Huseby, R.B. , Eikvil, L. , Solberg, R. , Vikhamar, D. & Solberg, A. (2005). Use of hidden Markov models and phenology for multitemporal satellite image classification: Applications to mountain vegetation classification In: Proceedings of the Third International Workshop on the Analysis of Multi‐temporal Remote Sensing Images, Biloxi, USA (eds King R.L., Younan N.H.). IEEE, pp. 220–224. [Google Scholar]
- Bálint, M. , Pfenninger, M. , Grossart, H.‐P. , Taberlet, P. , Vellend, M. & Leibold, M.A. et al, (2018). Environmental DNA time series in ecology. Trends Ecol. Evol., 33, 945–957. [DOI] [PubMed] [Google Scholar]
- Barbu, V.S. & Limnios, N. (2009). Semi‐Markov Chains and Hidden Semi‐Markov Models Toward Applications: Their Use in Reliability and DNA Analysis, Vol. 191 of Lecture Notes in Statistics. Springer.
- Bartolucci, F. , Pandolfi, S. & Pennoni, F. (2017). LMest: An R package for latent Markov models for longitudinal categorical data. J. Stat. Softw., 81, 1–38. [Google Scholar]
- Basson, M. , Bravington, M.V. , Hartog, J.R. & Patterson, T.A. (2016). Experimentally derived likelihoods for light‐based geolocation. Methods Ecol. Evol., 7, 980–989. [Google Scholar]
- van Beest, F.M. , Mews, S. , Elkenkamp, S. , Schuhmann, P. , Tsolak, D. , Wobbe, T. et al (2019). Classifying grey seal behaviour in relation to environmental variability and commercial fishing activity – a multivariate hidden Markov model. Sci. Rep., 9, 5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon, M. , Harper, J. & Townsend, C. (2006). Ecology: From Individuals to Ecosystems, 4th edn Malden, MA: Blackwell Publishing. [Google Scholar]
- Beisner, B. , Haydon, D. & Cuddington, K. (2003). Alternative stable states in ecology. Front. Ecol. Environ., 1, 376–382. [Google Scholar]
- Benhaiem, S. , Marescot, L. , Hofer, H. , East, M.L. , Lebreton, J.‐D. , Kramer‐Schadt, S. et al (2018). Robustness of eco‐epidemiological capture‐recapture parameter estimates to variation in infection state uncertainty. Frontiers in Veterinary Science, 5, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besbeas, P. & Morgan, B.J.T. (2019). Exact inference for integrated population modelling. Biometrics, 75, 475–484. [DOI] [PubMed] [Google Scholar]
- Bhar, R. & Hamori, S. (2004). Hidden Markov Models: Applications to Financial Economics, Vol. 40 of Advanced Studies in Theoretical and Applied Econometrics. Dordrecht: Springer. [Google Scholar]
- Bode, N.W. & Seitz, M.J. (2018). Using hidden Markov models to characterise intermittent social behaviour in fish shoals. The Science of Nature, 105, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann, K. , Evans, A. , Gilbert, M.T.P. , Carvalho, G.R. , Creer, S. , Knapp, M. et al (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol., 29, 358–367. [DOI] [PubMed] [Google Scholar]
- Boitard, S. & Schlötterer, C. & Futschik, A. (2009). Detecting selective sweeps: a new approach based on hidden Markov models. Genetics, 181, 1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers, D.L. & Efford, M. (2008). Spatially explicit maximum likelihood methods for capture–recapture studies. Biometrics, 64, 377–385. [DOI] [PubMed] [Google Scholar]
- Borgy, B. , Reboud, X. , Peyrard, N. , Sabbadin, R. & Gaba, S. (2015). Dynamics of weeds in the soil seed bank: a hidden Markov model to estimate life history traits from standing plant time series. PLoS One, 10, e0139278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron, V. , Deere, N.J. , Xofis, P. , Link, A. , Quiñones‐Guerrero, A. , Payan, E. & et al(2019). Richness, diversity, and factors influencing occupancy of mammal communities across human‐modified landscapes in Colombia. Biol. Cons., 232, 108–116. [Google Scholar]
- Bracken, C. , Rajagopalan, B. & Zagona, E. (2014). A hidden Markov model combined with climate indices for multidecadal streamflow simulation. Water Resour. Res., 50, 7836–7846. [Google Scholar]
- Breed, G.A. , Don Bowen, W. & Leonard, M.L. (2013). Behavioral signature of intraspecific competition and density dependence in colony‐breeding marine predators. Ecol. Evol., 3, 3838–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breininger, D.R. , Nichols, J.D. , Duncan, B.W. , Stolen, E.D. , Carter, G.M. , Hunt, D.K. et al (2010). Multistate modeling of habitat dynamics: factors affecting Florida scrub transition probabilities. Ecology, 91, 3354–3364. [DOI] [PubMed] [Google Scholar]
- Broms, K.M. , Hooten, M.B. & Fitzpatrick, R.M. (2015). Accounting for imperfect detection in Hill numbers for biodiversity studies. Methods Ecol. Evol., 6, 99–108. [Google Scholar]
- Brooks, S. , Gelman, A. , Jones, G. & Meng, X.‐L. (2011). Handbook of Markov Chain Monte Carlo. London, UK: CRC Press. [Google Scholar]
- Brownie, C. , Hines, J.E. , Nichols, J.D. , Pollock, K.H. & Hestbeck, J.B. (1993). Capture‐recapture studies for multiple strata including non‐Markovian transitions. Biometrics, 49, 1173–1187. [Google Scholar]
- Buckland, S.T. , Newman, K.B. , Thomas, L. & Koester, N. (2004). State‐space models for dynamics of wild animal populations. Ecol. Model., 171, 157–175. [Google Scholar]
- Bulla, J. & Bulla, I. (2013). hsmm: Hidden Semi Markov Models. Available at: https://CRAN.R‐project.org/package=hsmm. R package version 0.4.
- Caetano, D.S. , O’Meara, B.C. & Beaulieu, J.M. (2018). Hidden state models improve state‐dependent diversification approaches, including biogeographical models. Evolution, 72, 2308–2324. [DOI] [PubMed] [Google Scholar]
- Calabrese, J.M. , Brunner, J.L. & Ostfeld, R.S. (2011). Partitioning the aggregation of parasites on hosts into intrinsic and extrinsic components via an extended poisson‐gamma mixture model. PLoS One, 6, e29215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappé, O. , Moulines, E. & Rydén, T. (2005). Inference in Hidden Markov Models. Springer, New York. [Google Scholar]
- Cardenas‐Ovando, R.A. , Noguez, J. & Rangel‐Escareno, C. (2017). RcppHMM: Rcpp Hidden Markov Model. Available at:https://CRAN.R‐project.org/package=RcppHMM. R package version 1.2.2.
- Caswell, H. (2001). Matrix Population Models, 2nd edn. Sinauer, Sunderland, MA. [Google Scholar]
- Cayuela, H. , Besnard, A. , Cote, J. , Laporte, M. , Bonnaire, E. , Pichenot, J. et al (2020). Anthropogenic disturbance drives dispersal syndromes, demography, and gene flow in amphibian populations. Ecol. Monogr., 90, e01406. [Google Scholar]
- Chambert, T. , Staszewski, V. , Lobato, E. , Choquet, R. , Carrie, C. , McCoy, K.D. et al (2012). Exposure of black‐legged kittiwakes to Lyme disease spirochetes: dynamics of the immune status of adult hosts and effects on their survival. J. Anim. Ecol., 81, 986–995. [DOI] [PubMed] [Google Scholar]
- Charmantier, A. , Perrins, C. , McCleery, R.H. & Sheldon, B.C. (2006). Evolutionary response to selection on clutch size in a long‐term study of the mute swan. Am. Nat., 167, 453–465. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Kéry, M. , Plattner, M. , Ma, K. & Gardner, B. (2013). Imperfect detection is the rule rather than the exception in plant distribution studies. J. Ecol., 101, 183–191. [Google Scholar]
- Choquet, R. , Carrié, C. , Chambert, T. & Boulinier, T. (2013). Estimating transitions between states using measurements with imperfect detection: application to serological data. Ecology, 94, 2160–2165. [DOI] [PubMed] [Google Scholar]
- Choquet, R. , Garnier, A. , Awuve, E. & Besnard, A. (2017). Transient state estimation using continuous‐time processes applied to opportunistic capture–recapture data. Ecol. Model., 361, 157–163. [Google Scholar]
- Choquet, R. , Viallefont, A. , Rouan, L. , Gaanoun, K. & Gaillard, J.‐M. (2011). A semi‐Markov model to assess reliably survival patterns from birth to death in free‐ranging populations. Methods Ecol. Evol., 2, 383–389. [Google Scholar]
- Clogg, C.C. (1995). Latent class models In: Handbook of Statistical Modeling for the Social and Behavioral Sciences(eds Arminger G., Clogg C.C., Sobel M.E.), Boston, MA: Springer, pp. 311–359. [Google Scholar]
- Cole, D.J. (2019). Parameter redundancy and identifiability in hidden Markov models. METRON, 77, 105–118. [Google Scholar]
- Compagnoni, A. , Bibian, A.J. , Ochocki, B.M. , Levin, S. , Zhu, K. & Miller, T.E. (2019). popler: an R package for extraction and synthesis of population time series from the long‐term ecological research (LTER) network. Methods Ecol. Evol., 11, 258–264. [Google Scholar]
- Conn, P.B. & Cooch, E.G. (2009). Multistate capture–recapture analysis under imperfect state observation: an application to disease models. J. Appl. Ecol., 46, 486–492. [Google Scholar]
- Conway‐Cranos, L.L. & Doak, D.F. (2011). Sampling errors create bias in Markov models for community dynamics: the problem and a method for its solution. Oecologia, 167, 199–207. [DOI] [PubMed] [Google Scholar]
- Cooch, E.G. , Conn, P.B. , Ellner, S.P. , Dobson, A.P. & Pollock, K.H. (2012). Disease dynamics in wild populations: modeling and estimation: a review. J. Ornithol., 152, 485–509. [Google Scholar]
- Cooke, S.J. , Hinch, S.G. , Wikelski, M. , Andrews, R.D. , Kuchel, L.J. , Wolcott, T.G. et al (2004). Biotelemetry: a mechanistic approach to ecology. Trends Ecol. Evol., 19, 334–343. [DOI] [PubMed] [Google Scholar]
- R Core Team , (2019). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R‐project.org/. [Google Scholar]
- Coulson, T. , Benton, T.G. , Lundberg, P. , Dall, S.R.X. , Kendall, B.E. & Gaillard, J.M. (2006). Estimating individual contributions to population growth: evolutionary fitness in ecological time. Proceedings of the Royal Society B Biological Sciences, 273, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen, L.L.E. , Besbeas, P. , Morgan, B.J.T. & Schwarz, C.J. (2017). Hidden Markov models for extended batch data. Biometrics, 73, 1321–1331. [DOI] [PubMed] [Google Scholar]
- Crampton, J.S. , Meyers, S.R. , Cooper, R.A. , Sadler, P.M. , Foote, M. & Harte, D. (2018). Pacing of Paleozoic macroevolutionary rates by Milankovitch grand cycles. Proc. Natl Acad. Sci., 115, 5686–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C.L. , Rich, L.N. , Farris, Z.J. , Kelly, M.J. , Di Bitetti, M.S. , Blanco, Y.D. et al (2018). Ecological correlates of the spatial co‐occurrence of sympatric mammalian carnivores worldwide. Ecol. Lett., 21, 1401–1412. [DOI] [PubMed] [Google Scholar]
- DeRuiter, S.L. , Langrock, R. , Skirbutas, T. , Goldbogen, J.A. , Calambokidis, J. , Friedlaender, A.S. et al (2017). A multivariate mixed hidden Markov model to analyze blue whale diving behaviour during controlled sound exposures. The Annals of Applied Statistics, 11, 362–392. [Google Scholar]
- Dietze, M.C. , Fox, A. , Beck‐Johnson, L.M. , Betancourt, J.L. , Hooten, M.B. , Jarnevich, C.S. et al (2018). Iterative near‐term ecological forecasting: Needs, opportunities, and challenges. Proc. Natl Acad. Sci., 115, 1424–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diosdado, J.A.V. , Barker, Z.E. , Hodges, H.R. , Amory, J.R. , Croft, D.P. , Bell, N.J. et al (2015). Classification of behaviour in housed dairy cows using an accelerometer‐based activity monitoring system. Animal Biotelemetry, 3, 15. [Google Scholar]
- Dorazio, R.M. , Kery, M. , Royle, J.A. & Plattner, M. (2010). Models for inference in dynamic metacommunity systems. Ecology, 91, 2466–2475. [DOI] [PubMed] [Google Scholar]
- Dorazio, R.M. & Royle, J.A. (2005). Estimating size and composition of biological communities by modeling the occurrence of species. Journal of the American Statistical Association, 100, 389–398. [Google Scholar]
- Dorazio, R.M. , Royle, J.A. , Söderström, B. & Glimskär, A. (2006). Estimating species richness and accumulation by modeling species occurrence and detectability. Ecology, 87, 842–854. [DOI] [PubMed] [Google Scholar]
- Dupont, P. , Pradel, R. , Lardy, S. , Allainé, D. & Cohas, A. (2015). Litter sex composition influences dominance status of Alpine marmots (Marmota marmota). Oecologia, 179, 753–763. [DOI] [PubMed] [Google Scholar]
- Durand, J.‐B. , Guédon, Y. , Caraglio, Y. & Costes, E. (2005). Analysis of the plant architecture via tree‐structured statistical models: the hidden Markov tree models. New Phytol., 166, 813–825. [DOI] [PubMed] [Google Scholar]
- Durbin, R. , Eddy, S.R. , Krogh, A. & Mitchison, G. (1998). Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. New York, NY: Cambridge University Press. [Google Scholar]
- Durbin, J. & Koopman, S.J. (2012). Time Series Analysis by State Space Methods, 2nd edn Oxford, UK: Oxford University Press. [Google Scholar]
- Economou, T. & Menary, M.B. (2019). A hidden semi‐Markov model for characterizing regime shifts in ocean density variability. J. Roy. Stat. Soc.: Ser. C (Appl. Stat.), 68, 1529–1553. [Google Scholar]
- Eddy, S.R. (2004). What is a hidden Markov model? Nat. Biotechnol., 22, 1315. [DOI] [PubMed] [Google Scholar]
- Ellison, A.M. (2004). Bayesian inference in ecology. Ecol. Lett., 7, 509–520. [Google Scholar]
- Ephraim, Y. & Merhav, N. (2002). Hidden Markov processes. IEEE Trans. Inf. Theory, 48, 1518–1569. [Google Scholar]
- Estes, L. , Elsen, P.R. , Treuer, T. , Ahmed, L. , Caylor, K. , Chang, J. et al (2018). The spatial and temporal domains of modern ecology. Nature Ecology & Evolution, 2, 819. [DOI] [PubMed] [Google Scholar]
- Evans, M.E. , Merow, C. , Record, S. , McMahon, S.M. & Enquist, B.J. (2016). Towards process‐based range modeling of many species. Trends Ecol. Evol., 31, 860–871. [DOI] [PubMed] [Google Scholar]
- Fahlbusch, J.A. & Harrington, K.J. (2019). A low‐cost, open‐source inertial movement GPS logger for eco‐physiology applications. J. Exp. Biol., 222, jeb211136. [DOI] [PubMed] [Google Scholar]
- Fasiolo, M. , Pya, N. & Wood, S.N. (2016). A comparison of inferential methods for highly nonlinear state space models in ecology and epidemiology. Stat. Sci., 31, 96–118. [Google Scholar]
- Felsenstein, J. & Churchill, G.A. (1996). A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol., 13, 93–104. [DOI] [PubMed] [Google Scholar]
- Fidino, M. , Simonis, J.L. & Magle, S.B. (2019). A multistate dynamic occupancy model to estimate local colonization–extinction rates and patterns of co‐occurrence between two or more interacting species. Methods Ecol. Evol., 10, 233–244. [Google Scholar]
- Fine, S. , Singer, Y. & Tishby, N. (1998). The hierarchical hidden Markov model: Analysis and applications. Mach. Learn, 32, 41–62. [Google Scholar]
- Fletcher, R. (2013). Practical Methods of Optimization. Hoboken, NJ: Wiley. [Google Scholar]
- Folke, C. , Carpenter, S. , Walker, B. , Scheffer, M. , Elmqvist, T. , Gunderson, L. (2004). Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst., 35, 557–581. [Google Scholar]
- Franke, A. , Caelli, T. & Hudson, R.J. (2004). Analysis of movements and behavior of caribou (Rangifer tarandus) using hidden Markov models. Ecol. Model., 173, 259–270. [Google Scholar]
- Franke, A. , Caelli, T. , Kuzyk, G. & Hudson, R.J. (2006). Prediction of wolf (Canis lupus) kill‐sites using hidden Markov models. Ecol. Model., 197, 237–246. [Google Scholar]
- Frühwirth‐Schnatter, S. (2006). Finite Mixture and Markov Switching Models. New York, NY: Springer. [Google Scholar]
- Gal, G. & Anderson, W. (2010). A novel approach to detecting a regime shift in a lake ecosystem. Methods Ecol. Evol., 1, 45–52. [Google Scholar]
- Gamelon, M. , Filli, F. , Sæther, B.‐E. & Herfindal, I. (2020). Multi‐event capture‐recapture analysis in Alpine chamois reveals contrasting responses to interspecific competition, within and between populations. J. Anim. Ecol.. 10.1111/1365-2656.13299 [DOI] [PubMed] [Google Scholar]
- Gao, J. (2002). Integration of GPS with remote sensing and GIS: reality and prospect. Photogramm. Eng. Remote Sensing, 68, 447–454. [Google Scholar]
- Garnier, A. , Gaillard, J.‐M. , Gauthier, D. & Besnard, A. (2016). What shapes fitness costs of reproduction in long‐lived iteroparous species? a case study on the Alpine ibex. Ecology, 97, 205–214. [DOI] [PubMed] [Google Scholar]
- Gelman, A. , Carlin, J.B. , Stern, H.S. & Rubin, D.B. (2004). Bayesian Data Analysis, 2nd edn Boca Raton, LA: Chapman and Hall. [Google Scholar]
- Gelman, A. & Hill, J. (2006). Data Analysis Using Regression and Multilevel/Hierarchical Models. New York, NY: Cambridge University Press. [Google Scholar]
- Gennaretti, F. , Arseneault, D. , Nicault, A. , Perreault, L. & Bégin, Y. (2014). Volcano‐ induced regime shifts in millennial tree‐ring chronologies from northeastern North America. Proc. Natl Acad. Sci., 111, 10077–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, T.S. , Gajjalla, P. , Mohammed, M.H. & Mande, S.S. (2012). C16S — a hidden Markov model based algorithm for taxonomic classification of 16S rRNA gene sequences. Genomics, 99, 195–201. [DOI] [PubMed] [Google Scholar]
- Gilks, W. , Richardson, S. & Spiegelhalter, D. (1996). Markov Chain Monte Carlo in Practice. London, UK: Chapman and Hall. [Google Scholar]
- Gimenez, O. , Blanc, L. , Besnard, A. , Pradel, R. , Doherty, P.F. Jr , Marboutin, E. et al (2014). Fitting occupancy models with E‐SURGE: hidden Markov modelling of presence– absence data. Methods Ecol. Evol., 5, 592–597. [Google Scholar]
- Gimenez, O. & Gaillard, J.‐M. (2018). Estimating individual fitness in the wild using capture‐ recapture data. Popul. Ecol., 60, 101–109. [Google Scholar]
- Gimenez, O. , Lebreton, J.‐D. , Gaillard, J.‐M. , Choquet, R. & Pradel, R. (2012). Estimating demographic parameters using hidden process dynamic models. Theor. Popul. Biol., 82, 307–316. [DOI] [PubMed] [Google Scholar]
- Glennie, R. , Borchers, D.L. , Murchie, M. , Harmsen, B.J. & Foster, R.J. (2019). Open population maximum likelihood spatial capture‐recapture. Biometrics, 75, 1345–1355. [DOI] [PubMed] [Google Scholar]
- Gotelli, N.J. & Ellison, A.M. (2013). A Primer of Ecological Statistics, 2nd edn. Sunderland, MA: Sinauer Associates Publishers. [Google Scholar]
- Grecian, W.J. , Lane, J.V. , Michelot, T. , Wade, H.M. & Hamer, K.C. (2018). Understanding the ontogeny of foraging behaviour: insights from combining marine predator bio‐logging with satellite‐derived oceanography in hidden Markov models. J. R. Soc. Interface, 15, 20180084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, J.K. , Krzywinski, M. & Altman, N.S. (2019). Markov models —Markov chains. Nat. Methods, 16, 663–664. [DOI] [PubMed] [Google Scholar]
- Guillera‐Arroita, G. (2017). Modelling of species distributions, range dynamics and communities under imperfect detection: advances, challenges and opportunities. Ecography, 40, 281–295. [Google Scholar]
- Halbritter, A.H. , De Boeck, H.J. , Eycott, A.E. , Reinsch, S. , Robinson, D.A. , Vicca, S. et al (2020). The handbook for standardised field and laboratory measurements in terrestrial climate‐change experiments and observational studies (ClimEx). Methods Ecol. Evol., 11, 22–37. [Google Scholar]
- Haller, B.C. (2014). Theoretical and empirical perspectives in ecology and evolution: a survey. Bioscience, 64, 907–916. [Google Scholar]
- Hanski, I. (1994). A practical model of metapopulation dynamics. J. Anim. Ecol., 63, 151–162. [Google Scholar]
- Hart, T. , Mann, R. , Coulson, T. , Pettorelli, N. & Trathan, P. (2010). Behavioural switching in a central place forager: patterns of diving behaviour in the macaroni penguin (Eudyptes chrysolophus). Mar. Biol., 157, 1543–1553. [Google Scholar]
- Harte, D. (2017). HiddenMarkov: Hidden Markov Models. Statistics Research Associates, Wellington. R package version 1.8‐1 http://www.statsresearch.co.nz/dsh/sslib/.
- Hebert, P.D. , Ratnasingham, S. , Zakharov, E.V. , Telfer, A.C. , Levesque‐Beaudin, V. , Milton, M.A. et al (2016). Counting animal species with DNA barcodes: Canadian insects. Philosophical Transactions of the Royal Society B: Biological Sciences, 371, 20150333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helske, S. & Helske, J. (2019). Mixture hidden Markov models for sequence data: The seqHMM package in R. J. Stat. Softw., 88, 1–32. [Google Scholar]
- Henderson, J. , Salzberg, S. & Fasman, K.H. (1997). Finding genes in DNA with a hidden Markov model. J. Comput. Biol., 4, 127–141. [DOI] [PubMed] [Google Scholar]
- Hestbeck, J.B. , Nichols, J.D. & Malecki, R.A. (1991). Estimates of movement and site fidelity using mark‐resight data of wintering Canada geese. Ecology, 72, 523–533. [Google Scholar]
- Hill, M.F. , Witman, J.D. & Caswell, H. (2004). Markov chain analysis of succession in a rocky subtidal community. Am. Nat., 164, E46–E61. [DOI] [PubMed] [Google Scholar]
- Himmelmann, L. (2010). HMM: Hidden Markov Models. https://CRAN.R‐project.org/package=HMM. R package version 1.0.
- Hobolth, A. , Christensen, O.F. , Mailund, T. & Schierup, M.H. (2007). Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet., 3, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooten, M.B. , Johnson, D.S. , McClintock, B.T. & Morales, J.M. (2017). Animal Movement: Statistical Models for Telemetry Data. Baton Rouge, LA: CRC Press. [Google Scholar]
- Hope, P.R. & Jones, G. (2012). Warming up for dinner: torpor and arousal in hibernating Natterer’s bats (Myotis nattereri) studied by radio telemetry. J. Comp. Physiol. B., 182, 569–578. [DOI] [PubMed] [Google Scholar]
- Horn, H.S. (1975). Markovian properties of forest succession In: Ecology and Evolution of Communities (eds Cody M., & Diamond J.). Cambridge, MA: Harvard University Press, pp. 196–211. [Google Scholar]
- Hudson, M.E. (2008). Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Resour., 8, 3–17. [DOI] [PubMed] [Google Scholar]
- van Hulst, R. (1979). On the dynamics of vegetation: Markov chains as models of succession. Vegetatio, 40, 3–14. [Google Scholar]
- Jackson, B.B. (1975). Markov mixture models for drought lengths. Water Resour. Res., 11, 64–74. [Google Scholar]
- Jackson, C.H. (2011). Multi‐state models for panel data: the msm package for R. J. Stat. Softw., 38, 1–29. [Google Scholar]
- Jackson, C.H. , Sharples, L.D. , Thompson, S.G. , Duffy, S.W. & Couto, E. (2003). Multistate Markov models for disease progression with classification error. Journal of the Royal Statistical Society: Series D (The Statistician), 52, 193–209. [Google Scholar]
- Jasra, A. , Holmes, C.C. & Stephens, D.A. (2005). Markov chain Monte Carlo methods and the label switching problem in Bayesian mixture modeling. Stat. Sci., 20, 50–67. [Google Scholar]
- Jones, M.B. , Schildhauer, M.P. , Reichman, O. & Bowers, S. (2006). The new bioinformatics: integrating ecological data from the gene to the biosphere. Annu. Rev. Ecol. Evol. Syst., 37, 519–544. [Google Scholar]
- Jonsen, I.D. , Flemming, J.M. & Myers, R.A. (2005). Robust state–space modeling of animal movement data. Ecology, 86, 2874–2880. [Google Scholar]
- Kéry, M. & Schmidt, B. (2008). Imperfect detection and its consequences for monitoring for conservation. Community Ecology, 9, 207–216. [Google Scholar]
- Kellner, K.F. & Swihart, R.K. (2014). Accounting for imperfect detection in ecology: a quantitative review. PLoS One, 9, e111436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, W.L. , White, G.C. , Hines, J.E. , Langtimm, C.A. & Yoshizaki, J. (2012). Estimating parameters of hidden Markov models based on marked individuals: use of robust design data. Ecology, 93, 913–920. [DOI] [PubMed] [Google Scholar]
- van de Kerk, M. , Onorato, D.P. , Criffield, M.A. , Bolker, B.M. , Augustine, B.C. , McKinley, S.A. et al (2015). Hidden semi‐Markov models reveal multiphasic movement of the endangered Florida panther. J. Anim. Ecol., 84, 576–585. [DOI] [PubMed] [Google Scholar]
- Kery, M. & Royle, J.A. (2015). Applied Hierarchical Modeling in Ecology: Analysis of distribution, abundance and species richness in R and BUGS. Academic Press Inc, Amsterdam. [Google Scholar]
- King, R. & Langrock, R. (2016). Semi‐Markov Arnason‐Schwarz models. Biometrics, 72, 619–628. [DOI] [PubMed] [Google Scholar]
- Koleff, P. , Gaston, K.J. & Lennon, J.J. (2003). Measuring beta diversity for presence–absence data. J. Anim. Ecol., 72, 367–382. [Google Scholar]
- Lachish, S. , Knowles, S.C. , Alves, R. , Wood, M.J. & Sheldon, B.C. (2011). Infection dynamics of endemic malaria in a wild bird population: parasite species‐dependent drivers of spatial and temporal variation in transmission rates. J. Anim. Ecol., 80, 1207–1216. [DOI] [PubMed] [Google Scholar]
- Lagrange, P. , Pradel, R. , Bélisle, M. & Gimenez, O. (2014). Estimating dispersal among numerous sites using capture–recapture data. Ecology, 95, 2316–2323. [DOI] [PubMed] [Google Scholar]
- Lamy, T. , Gimenez, O. , Pointier, J.‐P. , Jarne, P. & David, P. (2013). Metapopulation dynamics of species with cryptic life stages. Am. Nat., 181, 479–491. [DOI] [PubMed] [Google Scholar]
- Langrock, R. , Adam, T. , Leos‐Barajas, V. , Mews, S. , Miller, D.L. & Papastamatiou, Y.P. (2018). Spline‐based nonparametric inference in general state‐switching models. Stat. Neerl., 72, 179–200. [Google Scholar]
- Langrock, R. , Borchers, D.L. & Skaug, H.J. (2013). Markov‐modulated nonhomogeneous Poisson processes for modeling detections in surveys of marine mammal abundance. Journal of the American Statistical Association, 108, 840–851. [Google Scholar]
- Langrock, R. , Hopcraft, G. , Blackwell, P. , Goodall, V. , King, R. , Niu, M. et al (2014a). Modelling group dynamic animal movement. Methods Ecol. Evol., 5, 190–199. [Google Scholar]
- Langrock, R. , Marques, T.A. , Baird, R.W. & Thomas, L. (2014b). Modeling the diving behavior of whales: a latent‐variable approach with feedback and semi‐Markovian components. Journal of Agricultural, Biological, and Environmental Statistics, 19, 82–100. [Google Scholar]
- Lavine, M. (2010). Living dangerously with big fancy models. Ecology, 91, 3487. [DOI] [PubMed] [Google Scholar]
- Lawler, E. , Whoriskey, K. , Aeberhard, W.H. , Field, C. & Flemming, J.M. (2019). The conditionally autoregressive hidden Markov model (carhmm): Inferring behavioural states from animal tracking data exhibiting conditional autocorrelation. Journal of Agricultural, Biological and Environmental Statistics, 24(4), 651–668. [Google Scholar]
- Lazrak, E. , Mari, J. , Benoît, M. (2010). Landscape regularity modelling for environmental challenges in agriculture. Landscape Ecol., 25, 169–183. [Google Scholar]
- Lebreton, J.‐D. , Nichols, J.D. , Barker, R.J. , Pradel, R. & Spendelow, J.A. (2009). Modeling individual animal histories with multistate capture–recapture models. Adv. Ecol. Res., 41, 87–173. [Google Scholar]
- Lee, S.‐Y. & Song, X.‐Y. (2012). Basic and Advanced Bayesian Structural Equation Modeling: With Applications in the Medical and Behavioral Sciences. New York, NY: Wiley. [Google Scholar]
- Leos‐Barajas, V. , Gangloff, E.J. , Adam, T. , Langrock, R. , Van Beest, F.M. , Nabe‐Nielsen, J. (2017a). Multi‐scale modeling of animal movement and general behavior data using hidden Markov models with hierarchical structures. Journal of Agricultural, Biological and Environmental Statistics, 22, 232–248. [Google Scholar]
- Leos‐Barajas, V. , Photopoulou, T. , Langrock, R. , Patterson, T.A. , Watanabe, Y.Y. , Murgatroyd, M. et al (2017b). Analysis of animal accelerometer data using hidden Markov models. Methods Ecol. Evol., 8, 161–173. [Google Scholar]
- Li, B.‐L. (1995). Stability analysis of a nonhomogeneous Markovian landscape model. Ecol. Model., 82, 247–256. [Google Scholar]
- Li, W.K. (2003). Diagnostic Checks in Time Series. London, UK: CRC Press. [Google Scholar]
- Lindenmayer, D.B. , Likens, G.E. , Andersen, A. , Bowman, D. , Bull, C.M. , Burns, E. et al (2012). Value of long‐term ecological studies. Austral Ecol., 37, 745–757. [Google Scholar]
- Lindsay, B.G. (1995). Mixture models: Theory, geometry and applications. NSF‐CBMS Regional Conference Series in Probability and Statistics, 5, i–163. [Google Scholar]
- Link, W.A. , Cooch, E. & Cam, E. (2002). Model‐based estimation of individual fitness. Journal of Applied Statistics, 29, 207–224. [Google Scholar]
- Lloyd, K.J. , Oosthuizen, W.C. , Bester, M.N. & de Bruyn, P.N. (2020). Trade‐offs between age‐related breeding improvement and survival senescence in highly polygynous elephant seals: Dominant males always do better. J. Anim. Ecol., 89, 897–909. [DOI] [PubMed] [Google Scholar]
- Louvrier, J. , Chambert, T. , Marboutin, E. & Gimenez, O. (2018). Accounting for misidentification and heterogeneity in occupancy studies using hidden Markov models. Ecol. Model., 387, 61–69. [Google Scholar]
- Louvrier, J. , Molinari‐Jobin, A. , Kéry, M. , Chambert, T. , Miller, D. & Zimmermann, F. et al (2019). Use of ambiguous detections to improve estimates from species distribution models. Conserv. Biol., 33, 185–195. [DOI] [PubMed] [Google Scholar]
- Lowe, P.K. , Bruno, J.F. , Selig, E.R. & Spencer, M. (2011). Empirical models of transitions between coral reef states: effects of region, protection, and environmental change. PLoS One, 6, e26339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, I.L. & Raubenheimer, D. (1995). Hidden Markov models and animal behaviour. Biom. J., 37, 701–712. [Google Scholar]
- MacKenzie, D.I. , Nichols, J.D. , Hines, J.E. , Knutson, M.G. & Franklin, A.B. (2003). Estimating site occupancy, colonization, and local extinction when a species is detected imperfectly. Ecology, 84, 2200–2207. [Google Scholar]
- MacKenzie, D.I. , Nichols, J.D. , Lachman, G.B. , Droege, S. , Andrew Royle, J. & Langtimm, C.A. (2002). Estimating site occupancy rates when detection probabilities are less than one. Ecology, 83, 2248–2255. [Google Scholar]
- MacKenzie, D.I. , Nichols, J.D. , Royle, J.A. , Pollock, K.H. , Bailey, L. & Hines, J.E. (2018). Occupancy Estimation and Modeling: Inferring Patterns and Dynamics of Species Occurrence, 2nd edn London, UK: Academic Press. [Google Scholar]
- MacKenzie, D.I. , Nichols, J.D. , Seamans, M.E. & Gutiérrez, R. (2009). Modeling species occurrence dynamics with multiple states and imperfect detection. Ecology, 90, 823–835. [DOI] [PubMed] [Google Scholar]
- Magurran, A.E. (2004). Measuring biological diversity. Blackwell Science, Oxford. [Google Scholar]
- Marescot, L. , Benhaiem, S. , Gimenez, O. , Hofer, H. , Lebreton, J.‐D. , Olarte‐Castillo, X.A. et al (2018). Social status mediates the fitness costs of infection with canine distemper virus in Serengeti spotted hyenas. Funct. Ecol., 32, 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marescot, L. , Lyet, A. , Singh, R. , Carter, N. & Gimenez, O. (2020). Inferring wildlife poaching in southeast Asia with multispecies dynamic occupancy models. Ecography, 43, 239–250. [Google Scholar]
- Martin, J. , McIntyre, C.L. , Hines, J.E. , Nichols, J.D. , Schmutz, J.A. & MacCluskie, M.C. (2009). Dynamic multistate site occupancy models to evaluate hypotheses relevant to conservation of golden eagles in Denali National Park, Alaska. Biol. Cons., 142, 2726–2731. [Google Scholar]
- Martin, T.G. , Wintle, B.A. , Rhodes, J.R. , Kuhnert, P.M. , Field, S.A. , Low‐Choy, S.J. et al (2005). Zero tolerance ecology: improving ecological inference by modelling the source of zero observations. Ecol. Lett., 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- Martinez, P.P. , King, A.A. , Yunus, M. , Faruque, A. & Pascual, M. (2016). Differential and enhanced response to climate forcing in diarrheal disease due to rotavirus across a megacity of the developing world. Proc. Natl Acad. Sci., 113, 4092–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B.T. , King, R. , Thomas, L. , Matthiopoulos, J. , McConnell, B.J. & Morales, J.M. (2012). A general discrete‐time modeling framework for animal movement using multistate random walks. Ecol. Monogr., 82, 335–349. [Google Scholar]
- McClintock, B.T. & Michelot, T. (2018). momentuHMM: R package for generalized hidden Markov models of animal movement. Methods Ecol. Evol., 9, 1518–1530. [Google Scholar]
- McClintock, B.T. , Nichols, J.D. , Bailey, L.L. , MacKenzie, D.I. , Kendall, W.L. & Franklin, A.B. (2010). Seeking a second opinion: uncertainty in disease ecology. Ecol. Lett., 13, 659–674. [DOI] [PubMed] [Google Scholar]
- McClintock, B.T. , Russell, D.J. , Matthiopoulos, J. & King, R. (2013). Combining individual animal movement and ancillary biotelemetry data to investigate population‐level activity budgets. Ecology, 94, 838–849. [Google Scholar]
- McCullagh, P. & Nelder, J.A. (1989). Generalized Linear Models. Chapman and Hall, New York. [Google Scholar]
- McDonald, T.L. , Hornsby, F.E. , Speakman, T.R. , Zolman, E.S. , Mullin, K.D. , Sinclair, C. et al (2017). Survival, density, and abundance of common bottlenose dolphins in Barataria Bay (USA) following the Deepwater Horizon oil spill. Endangered Species Research, 33, 193–209. [Google Scholar]
- McGraw, J.B. & Caswell, H. (1996). Estimation of individual fitness from life‐history data. Am. Nat., 147, 47–64. [Google Scholar]
- Miller, D.A. , Grant, E.H.C. , Muths, E. , Amburgey, S.M. , Adams, M.J. , Joseph, M.B. et al (2018a). Quantifying climate sensitivity and climate‐driven change in North American amphibian communities. Nat. Commun., 9, 3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.A. , Nichols, J.D. , McClintock, B.T. , Grant, E.H.C. , Bailey, L.L. & Weir, L.A. (2011). Improving occupancy estimation when two types of observational error occur: non‐detection and species misidentification. Ecology, 92, 1422–1428. [DOI] [PubMed] [Google Scholar]
- Miller, J.R.B. , Pitman, R.T. , Mann, G.K.H. , Fuller, A.K. & Balme, G.A. (2018b). Lions and leopards coexist without spatial, temporal or demographic effects of interspecific competition. J. Anim. Ecol., 87, 1709–1726. [DOI] [PubMed] [Google Scholar]
- Moilanen, A. (1999). Patch occupancy models of metapopulation dynamics: efficient parameter estimation using implicit statistical inference. Ecology, 80, 1031–1043. [Google Scholar]
- Morales, J.M. , Haydon, D.T. , Frair, J. , Holsinger, K.E. & Fryxell, J.M. (2004). Extracting more out of relocation data: building movement models as mixtures of random walks. Ecology, 85, 2436–2445. [Google Scholar]
- Moritz, C. , Patton, J.L. , Conroy, C.J. , Parra, J.L. , White, G.C. & Beissinger, S.R. (2008). Impact of a century of climate change on small‐mammal communities in Yosemite National Park, USA. Science, 322, 261–264. [DOI] [PubMed] [Google Scholar]
- Murphy, A. , Kelly, M.J. , Karpanty, S.M. , Andrianjakarivelo, V. & Farris, Z.J. (2019). Using camera traps to investigate spatial co‐occurrence between exotic predators and native prey species: a case study from northeastern Madagascar. J. Zool., 307, 264–273. [Google Scholar]
- Myung, I.J. (2003). Tutorial on maximum likelihood estimation. J. Math. Psychol., 47, 90–100. [Google Scholar]
- Newman, K.B. , Buckland, S.T. , Morgan, B.J.T. , King, R. , Borchers, D.L. , Cole, D.J. et al (2014). Modelling Population Dynamics: Model Formulation, Fitting and Assessment Using State‐Space Methods. New York, NY: Springer. [Google Scholar]
- Ngô, M.C. , Heide‐Jørgensen, M.P. & Ditlevsen, S. (2019). Understanding narwhal diving behaviour using hidden Markov models with dependent state distributions and long range dependence. PLoS Comput. Biol., 15, e1006425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, J.D. , Hines, J.E. , Pollock, K.H. , Hinz, R.L. & Link, W.A. (1994). Estimating breeding proportions and testing hypotheses about costs of reproduction with capture‐ recapture data. Ecology, 75, 2052–2065. [Google Scholar]
- Nichols, J. , Sauer, J.R. , Pollock, K.H. & Hestbeck, J.B. (1992). Estimating transition probabilities for stage‐based population projection matrices using capture–recapture data. Ecology, 73, 306–312. [Google Scholar]
- Nichols, J.D. , Thomas, L. & Conn, P.B. (2009). Inferences about landbird abundance from count data: recent advances and future directions Modeling Demographic Processes in Marked Populations. Springer, pp. 201–235. [Google Scholar]
- O’Connell, J. & Højsgaard, S. (2011). Hidden semi Markov models for multiple observation sequences: The mhsmm package for R. J. Stat. Softw., 39, 1–22. [Google Scholar]
- Olajos, F. , Bokma, F. , Bartels, P. , Myrstener, E. , Rydberg, J. , Öhlund, G. et al(2018). Estimating species colonization dates using DNA in lake sediment. Methods Ecol. Evol., 9, 535–543. [Google Scholar]
- Ovaskainen, O. , Tikhonov, G. , Norberg, A. , Guillaume Blanchet, F. , Duan, L. , Dunson, D. et al (2017). How to make more out of community data? a conceptual framework and its implementation as models and software. Ecol. Lett., 20, 561–576. [DOI] [PubMed] [Google Scholar]
- Palkopoulou, E. , Lipson, M. , Mallick, S. , Nielsen, S. , Rohland, N. , Baleka, S. et al (2018). A comprehensive genomic history of extinct and living elephants. Proc. Natl Acad. Sci., 115, E2566–E2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papastamatiou, Y.P. , Iosilevskii, G. , Leos‐Barajas, V. , Brooks, E.J. , Howey, L.A. , Chapman, D.D. et al (2018a). Optimal swimming strategies and behavioral plasticity of oceanic whitetip sharks. Sci. Rep., 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papastamatiou, Y.P. , Watanabe, Y.Y. , Demˇsar, U. , Leos‐Barajas, V. , Bradley, D. , Langrock, R. et al (2018b). Activity seascapes highlight central place foraging strategies in marine predators that never stop swimming. Movement Ecology, 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, T.A. , Basson, M. , Bravington, M.V. & Gunn, J.S. (2009). Classifying movement behaviour in relation to environmental conditions using hidden Markov models. J. Anim. Ecol., 78, 1113–1123. [DOI] [PubMed] [Google Scholar]
- Patterson, T.A. , Parton, A. , Langrock, R. , Blackwell, P.G. , Thomas, L. & King, R. (2017). Statistical modelling of individual animal movement: an overview of key methods and a discussion of practical challenges. AStA Advances in Statistical Analysis, 101(4), 399–438. 10.1007/s10182-017-0302-7. [DOI] [Google Scholar]
- Patterson, T.A. , Thomas, L. , Wilcox, C. , Ovaskainen, O. & Matthiopoulos, J. (2008). State–space models of individual animal movement. Trends Ecol. Evol., 23, 87–94. [DOI] [PubMed] [Google Scholar]
- Pawlowski, C.W. & McCord, C. (2009). A Markov model for assessing ecological stability properties. Ecol. Model., 220, 86–95. [Google Scholar]
- Pedersen, M.W. , Berg, C.W. , Thygesen, U.H. , Nielsen, A. & Madsen, H. (2011a). Estimation methods for nonlinear state‐space models in ecology. Ecol. Model., 222, 1394–1400. [Google Scholar]
- Pedersen, M.W. , Patterson, T.A. , Thygesen, U.H. & Madsen, H. (2011b). Estimating animal behavior and residency from movement data. Oikos, 120, 1281–1290. [Google Scholar]
- Pedersen, M.W. , Righton, D. , Thygesen, U.H. , Andersen, K.H. & Madsen, H. (2008). Geolocation of North Sea cod (Gadus morhua) using hidden Markov models and behavioural switching. Can. J. Fish. Aquat. Sci., 65(11), 2367–2377. 10.1139/F08-144. [DOI] [Google Scholar]
- Phillips, J.S. , Patterson, T.A. , Leroy, B. , Pilling, G.M. & Nicol, S.J. (2015). Objective classification of latent behavioral states in bio‐logging data using multivariate‐normal hidden Markov models. Ecol. Appl., 25, 1244–1258. [DOI] [PubMed] [Google Scholar]
- Pledger, S. (2000). Unified maximum likelihood estimates for closed capture–recapture models using mixtures. Biometrics, 56, 434–442. [DOI] [PubMed] [Google Scholar]
- Pledger, S. & Arnold, R. (2014). Multivariate methods using mixtures: Correspondence analysis, scaling and pattern‐detection. Comput. Stat. Data Anal., 71, 241–261. [Google Scholar]
- Pluntz, M. , Coz, S.L. , Peyrard, N. , Pradel, R. , Choquet, R. & Cheptou, P.‐O. (2018). A general method for estimating seed dormancy and colonisation in annual plants from the observation of existing flora. Ecol. Lett., 21, 1311–1318. [DOI] [PubMed] [Google Scholar]
- Pohle, J. , Langrock, R. , van Beest, F.M. & Schmidt, N.M. (2017). Selecting the number of states in hidden Markov models: pragmatic solutions illustrated using animal movement. Journal of Agricultural, Biological and Environmental Statistics, 22, 270–293. [Google Scholar]
- Pradel, R. (1996). Utilization of capture‐mark‐recapture for the study of recruitment and population growth rate. Biometrics, 52, 703–709. [Google Scholar]
- Pradel, R. (2005). Multievent: An extension of multistate capture‐recapture models to uncertain states. Biometrics, 61, 442–447. [DOI] [PubMed] [Google Scholar]
- Pradel, R. , Maurin‐Bernier, L. , Gimenez, O. , Genovart, M. , Choquet, R. & Oro, D. (2008). Estimation of sex‐specific survival with uncertainty in sex assessment. Canadian Journal of Statistics, 36, 29–42. [Google Scholar]
- Quick, N.J. , Isojunno, S. , Sadykova, D. , Bowers, M. , Nowacek, D.P. & Read, A.J. (2017). Hidden Markov models reveal complexity in the diving behaviour of short‐finned pilot whales. Sci. Rep., 7, 45765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner, L.R. (1989). A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE, 77, 257–286. [Google Scholar]
- Rakhimberdiev, E. , Winkler, D.W. , Bridge, E. , Seavy, N.E. , Sheldon, D. , Piersma, T. et al (2015). A hidden Markov model for reconstructing animal paths from solar geolocation loggers using templates for light intensity. Movement Ecology, 3, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich, L.N. , Miller, D.A.W. , Robinson, H.S. , McNutt, J.W. & Kelly, M.J. (2016). Using camera trapping and hierarchical occupancy modelling to evaluate the spatial ecology of an African mammal community. J. Appl. Ecol., 53, 1225–1235. [Google Scholar]
- Robert, C. & Casella, G. (2004). Monte Carlo Statistical Methods. 2nd edn, New York, NY: Springer. [Google Scholar]
- Roeleke, M. , Teige, T. , Hoffmeister, U. , Klingler, F. & Voigt, C.C. (2018). Aerial‐hawking bats adjust their use of space to the lunar cycle. Movement Ecology, 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani, P. & King, A.A. (2010). Never mind the length, feel the quality: the impact of long‐term epidemiological data sets on theory, application and policy. Trends Ecol. Evol., 25, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota, C.T. , Ferreira, M.A.R. , Kays, R.W. , Forrester, T.D. , Kalies, E.L. , McShea, W.J. et al (2016). A multispecies occupancy model for two or more interacting species. Methods Ecol. Evol., 7, 1164–1173. [Google Scholar]
- Rouan, L. , Gaillard, J.‐M. , Guédon, Y. & Pradel, R. (2009). Estimation of lifetime reproductive success when reproductive status cannot always be assessed In Modeling Demographic Processes In Marked Populations (eds Thomson D.L., Cooch E.G., & Conroy M.J.). Boston, MA: Springer, pp. 867–879. [Google Scholar]
- Rowe, G. , Sweet, M. & Beebee, T.J.C. (2017). An Introduction to Molecular Ecology. New York, NY: Oxford University Press. [Google Scholar]
- Royle, J.A. (2004). N‐mixture models for estimating population size from spatially replicated counts. Biometrics, 60, 108–115. [DOI] [PubMed] [Google Scholar]
- Royle, J.A. , Chandler, R.B. , Sollmann, R. & Gardner, B. (2013). Spatial Capture‐Recapture. Boston, MA: Academic Press. [Google Scholar]
- Royle, J.A. & Dorazio, R.M. (2008). Hierarchical Modeling and Inference in Ecology: the analysis of data from populations, metapopulations and communities. Burlington, MA: Academic Press. [Google Scholar]
- Royle, J.A. , Fuller, A.K. & Sutherland, C. (2018). Unifying population and landscape ecology with spatial capture–recapture. Ecography, 41, 444–456. [Google Scholar]
- Royle, J.A. & Kéry, M. (2007). A Bayesian state‐space formulation of dynamic occupancy models. Ecology, 88, 1813–1823. [DOI] [PubMed] [Google Scholar]
- Runge, J.P. , Hines, J.E. & Nichols, J.D. (2007). Estimating species‐specific survival and movement when species identification is uncertain. Ecology, 88, 282–288. [DOI] [PubMed] [Google Scholar]
- Russell, R.E. , Royle, J.A. , Saab, V.A. , Lehmkuhl, J.F. , Block, W.M. & Sauer, J.R. (2009). Modeling the effects of environmental disturbance on wildlife communities: avian responses to prescribed fire. Ecol. Appl., 19, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Schaub, M. & Abadi, F. (2011). Integrated population models: a novel analysis framework for deeper insights into population dynamics. J. Ornithol., 152, 227–237. [Google Scholar]
- Scheffer, M. & Carpenter, S.R. (2003). Catastrophic regime shifts in ecosystems: linking theory to observation. Trends Ecol. Evol., 18, 648–656. [Google Scholar]
- Scheffer, M. , Carpenter, S. , Foley, J.A. , Folke, C. & Walker, B. (2001). Catastrophic shifts in ecosystems. Nature, 413, 591. [DOI] [PubMed] [Google Scholar]
- Schliehe‐Diecks, S. , Kappeler, P. & Langrock, R. (2012). On the application of mixed hidden Markov models to multiple behavioural time series. Interface Focus, 2, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J.H. , Johnson, D.S. , Lindberg, M.S. & Adams, L.G. (2015). Estimating demographic parameters using a combination of known‐fate and open N‐mixture models. Ecology, 96, 2583–2589. [DOI] [PubMed] [Google Scholar]
- Schnute, J.T. (1994). A general framework for developing sequential fisheries models. Can. J. Fish. Aquat. Sci., 51, 1676–1688. [Google Scholar]
- Schumer, M. , Xu, C. , Powell, D.L. , Durvasula, A. , Skov, L. , Holland, C. et al (2018). Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science, 360, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, C.J. , Schweigert, J.F. & Arnason, A.N. (1993). Estimating migration rates using tag‐recovery data. Biometrics, 49, 177–193. [Google Scholar]
- Seber, G.A. & Schofield, M.R. (2019). Capture‐Recapture: Parameter Estimation for Open Animal Populations. Cham, Switzerland: Springer. [Google Scholar]
- Sherlock, C. , Xifara, T. , Telfer, S. & Begon, M. (2013). A coupled hidden Markov model for disease interactions. J. Roy. Stat. Soc.: Ser. C (Appl. Stat.), 62, 609–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siachalou, S. , Doxani, G. & Tsakiri‐Strati, M. (2014). Time‐series analysis of high temporal remote sensing data to model wetland dynamics: A hidden Markov model approach. In: Proceedings of the SENTINEL‐2 for Science Workshop–ESA–ESRINFrascati, Italy20–22 May 2014
- Siachalou, S. , Mallinis, G. & Tsakiri‐Strati, M. (2015). A hidden Markov models approach for crop classification: Linking crop phenology to time series of multi‐sensor remote sensing data. Remote Sens., 7, 3633–3650. [Google Scholar]
- Skrondal, A. & Rabe‐Hesketh, S. (2004). Generalized Latent Variable Modeling: Multilevel, Longitudinal, and Structural Equation Models. Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- Sollmann, R. , Gardner, B. , Chandler, R.B. , Royle, J.A. & Sillett, T.S. (2015). An open‐ population hierarchical distance sampling model. Ecology, 96, 325–331. [DOI] [PubMed] [Google Scholar]
- Soria‐Carrasco, V. , Gompert, Z. , Comeault, A.A. , Farkas, T.E. , Parchman, T.l. , Johnston, J.S. et al (2014). Stick insect genomes reveal natural selection's role in parallel speciation. Science, 344(6185), 738–742. 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- Srikanthan, R. & McMahon, T. (2001). Stochastic generation of annual, monthly and daily climate data: A review. Hydrology and Earth System Sciences Discussions, 5, 653–670. [Google Scholar]
- Stoelting, R.E. , Gutierrez, R.J. , Kendall, W.L. & Peery, M.Z. (2015). Life‐history tradeoffs and reproductive cycles in spotted owls. Auk, 132, 46–64. [Google Scholar]
- Sutherland, C. , Brambilla, M. , Pedrini, P. & Tenan, S. (2016). A multiregion community model for inference about geographic variation in species richness. Methods Ecol. Evol., 7, 783–791. [Google Scholar]
- Talluto, M.V. , Boulangeat, I. , Vissault, S. , Thuiller, W. & Gravel, D. (2017). Extinction debt and colonization credit delay range shifts of eastern North American trees. Nature Ecology & Evolution, 1, 0182. [Google Scholar]
- Tanner, J.E. , Hughes, T.P. & Connell, J.H. (1994). Species coexistence, keystone species, and succession: a sensitivity analysis. Ecology, 75, 2204–2219. [Google Scholar]
- Tavecchia, G. , Besbeas, P. , Coulson, T. , Morgan, B.J.T. & Clutton‐Brock, T.H. (2009). Estimating population size and hidden demographic parameters with state‐space modeling. Am. Nat., 173, 722–733. [DOI] [PubMed] [Google Scholar]
- Tenan, S. , Brambilla, M. , Pedrini, P. & Sutherland, C. (2017). Quantifying spatial variation in the size and structure of ecologically stratified communities. Methods Ecol. Evol., 8, 976–984. [Google Scholar]
- Thygesen, U.H. , Pedersen, M.W. & Madsen, H. (2009). Geolocating fish using hidden Markov models and data storage tags In: Tagging and Tracking of Marine Animals with Electronic Devices(edsNielsen J.L., Arrizabalaga H., Fragoso N., Hobday A., Lutcavage M. & Sibert J.), New York, NY: Springer, pp. 277–293. [Google Scholar]
- Tingley, M.W. , Monahan, W.B. , Beissinger, S.R. & Moritz, C. (2009). Birds track their Grinnellian niche through a century of climate change. Proc. Natl Acad. Sci., 106, 19637–19643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touloupou, P. , Finkenstädt, B. & Spencer, S.E. (2020). Scalable Bayesian inference for coupled hidden Markov and semi‐Markov models. Journal of Computational and Graphical Statistics, 29, 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner, A.V. , Leos‐Barajas, V. , Langrock, R. , Schick, R.S. , Smale, M.J. , Kaschke, T. et al (2016). Sex‐specific and individual preferences for hunting strategies in white sharks. Funct. Ecol., 30, 1397–1407. [Google Scholar]
- Trier, Ø.D. & Salberg, A.‐B. (2011). Time‐series analysis of satellite images for forest cover change monitoring in Tanzania. In: 1st EARSeL Workshop on Operational Remote Sensing in Forest Management.
- Tucker, B.C. & Anand, M. (2005). On the use of stationary versus hidden Markov models to detect simple versus complex ecological dynamics. Ecol. Model., 185, 177–193. [Google Scholar]
- Tucker, A. & Duplisea, D. (2012). Bioinformatics tools in predictive ecology: applications to fisheries. Philosophical Transactions of the Royal Society B: Biological Sciences, 367, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, M.G. , Gardner, R.H. & O’Neill, R.V. (1995). Ecological dynamics at broad scales. Bioscience, 45, S29–S35. [Google Scholar]
- Usher, M. (1981). Modelling ecological succession, with particular reference to Markovian models Vegetation Dynamics in Grasslands, Healthlands and Mediterranean Ligneous Formations. Springer, pp. 11–18. [Google Scholar]
- Vellend, M. (2010). Conceptual synthesis in community ecology. Q. Rev. Biol., 85, 183–206. [DOI] [PubMed] [Google Scholar]
- Vellend, M. (2016). The Theory of Ecological Communities. Princeton: Princeton University Press. [Google Scholar]
- Veran, S. , Simpson, S.J. , Sword, G.A. , Deveson, E. , Piry, S. , Hines, J.E. et al (2015). Modeling spatiotemporal dynamics of outbreaking species: influence of environment and migration in a locust. Ecology, 96, 737–748. [DOI] [PubMed] [Google Scholar]
- Viovy, N. & Saint, G. (1994). Hidden Markov models applied to vegetation dynamics analysis using satellite remote sensing. IEEE Trans. Geosci. Remote Sens., 32, 906–917. [Google Scholar]
- Visser, I. & Speekenbrink, M. (2010). depmixS4: an R package for hidden Markov models. J. Stat. Softw., 36, 1–21. [Google Scholar]
- Waggoner, P.E. & Stephens, G.R. (1970). Transition probabilities for a forest. Nature, 225, 1160. [DOI] [PubMed] [Google Scholar]
- Wang, G. (2007). On the latent state estimation of nonlinear population dynamics using Bayesian and non‐Bayesian state‐space models. Ecol. Model., 200, 521–528. [Google Scholar]
- Weng, K.C. , Boustany, A.M. , Pyle, P. , Anderson, S.D. , Brown, A. & Block, B.A. (2007). Migration and habitat of white sharks (Carcharodon carcharias) in the eastern Pacific Ocean. Mar. Biol., 152, 877–894. [Google Scholar]
- Werner, G.D. , Cornelissen, J.H. , Cornwell, W.K. , Soudzilovskaia, N.A. , Kattge, J. , West, S.A. et al (2018). Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proc. Natl Acad. Sci., 115, 5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, G.C. & Burnham, K.P. (1999). Program MARK: Survival estimation from populations of marked animals. Bird Study, 46, S120–S138. [Google Scholar]
- White, G.C. & Garrott, R.A. (1990). Analysis of Wildlife Radio‐Tracking Data. San Diego: Academic Press. [Google Scholar]
- Wilkinson, S. (2019). aphid: an R package for analysis with profile hidden Markov models. Bioinformatics, 35, 3829–3830. [DOI] [PubMed] [Google Scholar]
- Williams, B.K. , Nichols, J.D. & Conroy, M.J. (2002). Analysis and Management of Animal Populations. Academic Press, San Diego, CA. [Google Scholar]
- Winstrup, M. , Svensson, A. , Rasmussen, S.O. , Winther, O. , Steig, E. & Axelrod, A. (2012). An automated approach for annual layer counting in ice cores. Climate of the Past Discussions, 8, 1881–1895. [Google Scholar]
- Wood, S.N. (2010). Statistical inference for noisy nonlinear ecological dynamic systems. Nature, 466, 1102. [DOI] [PubMed] [Google Scholar]
- Woolhiser, D.A. & Roldan, J. (1982). Stochastic daily precipitation models: 2. a comparison of distributions of amounts. Water Resour. Res., 18, 1461–1468. [Google Scholar]
- Wootton, J.T. (2001). Prediction in complex communities: analysis of empirically derived Markov models. Ecology, 82, 580–598. [Google Scholar]
- Xiao, R. , Yu, X. , Shi, R. , Zhang, Z. , Yu, W. , Li, Y. et al (2019). Ecosystem health monitoring in the Shanghai‐Hangzhou Bay Metropolitan Area: A hidden Markov modeling approach. Environ. Int., 133, 105170. [DOI] [PubMed] [Google Scholar]
- Yau, C. , Papaspiliopoulos, O. , Roberts, G.O. & Holmes, C. (2011). Bayesian non‐parametric hidden Markov models with applications in genomics. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 73, 37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, B.‐J. (2009). Hidden Markov models and their applications in biological sequence analysis. Curr. Genomics, 10, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkin, E.F. , Andrew Royle, J. , Dawson, D.K. & Bates, S. (2010). Multi‐species occurrence models to evaluate the effects of conservation and management actions. Biol. Cons., 143, 479–484. [Google Scholar]
- Zucchini, W. & Guttorp, P. (1991). A hidden Markov model for space‐time precipitation. Water Resour. Res., 27, 1917–1923. [Google Scholar]
- Zucchini, W. , MacDonald, I.L. & Langrock, R. (2016). Hidden Markov Models for Time Series: An Introduction Using R, 2nd edn. Boca Raton: CRC Press. [Google Scholar]
- Zweig, C.L. , Newman, S. & Saunders, C.J. (2020). Applied use of alternate stable state modeling in restoration ecology. Ecol. Appl.. 10.1002/eap.2195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material
Data Availability Statement
No new data were used.


