Summary
It remains unknown whether and to what extent marine prokaryotic communities are capable of degrading plastic in the ocean. To address this knowledge gap, we combined enrichment experiments employing low‐density polyethylene (LDPE) as the sole carbon source with a comparison of bacterial communities on plastic debris in the Pacific, the North Atlantic and the northern Adriatic Sea. A total of 35 operational taxonomic units (OTUs) were enriched in the LDPE‐laboratory incubations after 1 year, of which 20 were present with relative abundances > 0.5% in at least one plastic sample collected from the environment. From these, OTUs classified as Cognatiyoonia, Psychrobacter, Roseovarius and Roseobacter were found in the communities of plastics collected at all oceanic sites. Additionally, OTUs classified as Roseobacter, Pseudophaeobacter, Phaeobacter, Marinovum and Cognatiyoonia, also enriched in the LDPE‐laboratory incubations, were enriched on LDPE communities compared to the ones associated to glass and polypropylene in in‐situ incubations in the northern Adriatic Sea after 1 month of incubation. Some of these enriched OTUs were also related to known alkane and hydrocarbon degraders. Collectively, these results demonstrate that there are prokaryotes capable of surviving with LDPE as the sole carbon source living on plastics in relatively high abundances in different water masses of the global ocean.
Introduction
Between 4.8 and 12.7 million metric tons of plastic are entering the ocean each year (Jambeck et al., 2015). Like all solid surfaces in the ocean, plastics are colonized by microbial communities referred to as the plastisphere (Zettler et al., 2013), which is different in its composition from the microbial communities found in the surrounding environment (Lobelle and Cunliffe, 2011). This has been shown for plastics collected in different locations (Amaral‐Zettler et al., 2015; Oberbeckmann et al., 2015; De Tender et al., 2017; Kettner et al., 2017; C Dussud et al., 2018b; Harrison et al., 2018), and has been observed in both seawater (SW) (Amaral‐Zettler et al., 2015) and sediment habitats (Harrison et al., 2014).
The composition of the plastisphere is also shaped by biogeographic and environmental factors, such as salinity and nutrient availability (Oberbeckmann et al., 2014; 2018; Amaral‐Zettler et al., 2015; Kesy et al., 2019). Surface characteristics of the plastic, such as surface free energy, hydrophobicity, presence of additives, weathering state and molecular composition might also play a role in determining the community composition of plastic‐associated biofilms (Gross et al., 2016; Cai et al., 2019; Hossain et al., 2019), especially in earlier stages of biofilm development (Pinto et al., 2019). Because some marine bacteria have the ability to degrade hydrocarbons, it has been suggested that a certain fraction of the microbial community colonizing plastics could also be capable of degrading plastics (Dussud et al., 2018a; Jacquin et al., 2019; Roager and Sonnenschein, 2019). For example, the bacterium Ideonella sakaiensis produces enzymes capable of efficiently converting poly(ethylene‐terephthalate) (PET) into benign monomers (Yoshida et al., 2016).
The two most abundant plastics in the ocean, polyethylene (PE) and polypropylene (PP), show signs of degradation when incubated with specific bacteria or fungi under controlled laboratory conditions (Sudhakar et al., 2008; Harshvardhan and Jha, 2013; Paço et al., 2017). Whether these organisms also degrade marine plastics in the oceanic environment, however, remains unclear. The majority of the tested microorganisms, such as Bacillus sp. (Harshvardhan and Jha, 2013) and the fungus Zalerion maritimum (Paço et al., 2017), while isolated from SW, were not particularly abundant in plastic‐associated biofilms, indicating that they likely play only a minor role in plastic degradation in the marine environment.
However, weight loss has been reported for PE and PP during in situ incubations in the marine environment, indicating either an abiotic loss due to fragmentation and leaching or biotically via microbial action on plastics (Sudhakar et al., 2007). The systematic association of bacteria capable of degrading hydrocarbons, such as members of the families Alcanivoraceae, Rhodobacteraceae and Sphingomonadaceae (Oberbeckmann and Labrenz, 2019; Roager and Sonnenschein, 2019) to plastic surfaces further suggests that specific members of plastic‐colonizing microbial communities associated with this plastic might be capable of degrading plastic and/or plastic‐derived compounds in the ocean. For example, the family Alcanivoraceae, well known for including alkane‐degrading taxa (Schneiker et al., 2006; Kim et al., 2018), has been found enriched on PET surfaces compared to glass surfaces (Oberbeckmann et al., 2016). Alcanivoraceae also increase in relative abundance in incubations where plastics, such as low‐density polyethylene (LDPE) and polystyrene, are the only carbon sources (Delacuvellerie et al., 2019; Syranidou et al., 2019). Weight loss of plastics and changes of the chemical properties of their surface, when incubated with specific microorganisms, are indicative of plastic biodegradation (Oberbeckmann and Labrenz, 2019). However, except for I. sakaiensis producing PET‐degrading enzymes (Yoshida et al., 2016), the metabolic pathways and associated mechanistic processes involved in the biodegradation of other plastics are yet to be characterized. Therefore, whether oceanic bacteria are capable to degrade plastics remains to be shown.
Recently, the taxonomy of plastic‐associated microbial communities has been studied in the Mediterranean Sea (Dussud et al., 2018), the Baltic Sea (Oberbeckmann et al., 2014; 2018; Kettner et al., 2017; Ogonowski et al., 2018; Kesy et al., 2019), the North Sea (Oberbeckmann et al., 2014; 2016; De Tender et al., 2017; Kirstein et al., 2019), the northern Adriatic Sea (Pinto et al., 2019), the Caribbean Sea (Dudek et al., 2020), the western Atlantic Ocean (Zettler et al., 2013; Amaral‐Zettler et al., 2015; Debroas et al., 2017) and the North Pacific Gyre (Amaral‐Zettler et al., 2015; Bryant et al., 2016). The composition of plastic‐associated bacterial communities has been compared using materials collected in the North Sea, the Baltic Sea and Yangtze Estuary (Oberbeckmann and Labrenz, 2019). The community composition of the samples from each of these locations was obtained by the taxonomic classification of the V4 region of the 16S rRNA gene (De Tender et al., 2017; Jiang et al., 2018; Ogonowski et al., 2018; Oberbeckmann et al., 2018; Kesy et al., 2019). While the biofilm samples from different locations did share operational taxonomic units (OTUs), mostly affiliated to Rhodobacteraceae and Sphingomonadaceae families, the majority of OTUs were location‐specific. This suggests that environmental variables are the main factor determining the composition of prokaryotic plastic‐associated communities within marine habitats. Most of these studies, however, used different DNA extraction methods, different primers and different sequencing technologies, which might introduce biases in the results and might explain some of the differences in community composition found between locations (Wintzingerode et al., 1997).
In this study, we compare the composition of prokaryotic communities associated with plastics collected in the Pacific, the North Atlantic and the northern Adriatic Sea (Fig. S1; Table S1). If biodegradability of plastics is a factor influencing the composition of plastic‐colonizing prokaryotic communities in the ocean, we expect the presence of plastic degraders in plastic‐associated biofilms to be independent of the oceanic region. Furthermore, it has been proposed that metabolic pathways involved in the biodegradation of plastics differ between different plastic polymers (Devi et al., 2016). Hence, we hypothesized that different plastic types (e.g. PE, PP) would harbour different putative plastic‐degrading organisms. To determine whether specific bacteria originating from the natural microbial community can utilize LDPE‐derived compounds, we incubated two PE biofilm communities with LDPE as their sole carbon source, one for 1 year and the other for 2 years. We then investigated the presence of the bacteria increasing in relative abundance in these incubations and on plastics collected in different regions of the global ocean. We also examined whether the bacteria enriched in the LDPE incubations were present in biofilms associated with PE, PP and glass during an in‐situ incubation experiment taking place in the northern Adriatic Sea (described in Pinto et al. 2019). In this previous study, we characterized the composition of bacterial communities on glass and different plastic types after 1 week, 1 month and 2 months using the same molecular and sequencing tools and protocols as used in this study.
Results
Community composition across different oceanic regions
We analysed the 16S rRNA gene sequences of biofilms associated with 13, 23 and 40 polymer (plastic) items collected in the North Atlantic, Pacific and northern Adriatic Sea respectively (Table S2). We could not identify the polymer type of 1 and 6 pieces from the North Atlantic and Pacific respectively. Thus, they were classified as not identified (NI).
Overall, OTU diversity and richness were significantly higher on plastics collected in the northern Adriatic than in the North Atlantic and Pacific, independent of the size of the plastics (Fig. S2; Tables S3 and S4).
There was no difference between the taxonomic composition of the bacterial communities colonizing PP and PE (PERMANOVA: F = 0.97, p = 0.527). The composition of bacterial communities on plastics, however, significantly differed from that of the surrounding SW at all sites (Fig. 1; Table S5). Furthermore, the bacterial community composition in plastic‐associated biofilms differed between the oceanic basins, i.e. North Atlantic, Pacific and northern Adriatic, and between sampling stations, albeit not in all of them (Fig. 1; Table S5). The bacterial communities on plastics collected in the Pacific at Stations 8 and 10 were relatively similar but were significantly different from the communities on plastics collected at any other Pacific stations (Table S5). The bacterial community composition of the plastic‐associated biofilms collected in the North Atlantic was relatively similar, with the exception of plastics collected at Stations 1 and 5, which significantly differed from each other (PERMANOVA: F = 1.50, p < 0.05).
Fig 1.

Non‐metric multidimensional scaling (NMDS) representation of the prokaryotic community of all plastic and seawater samples collected in the northern Adriatic, North Atlantic and Pacific using Bray‐Curtis as the distance measurement.
Each symbol represents one sample and the letters next to each symbol indicate the type of plastic (PE, PP, PET and nylon), unidentified polymers (NI) or seawater (SW). The environmental factor vectors were fitted to the NMDS through the envfit function. Envfit results are given in Table S6.
When we fitted environmental vectors to the non‐metric multidimensional scaling (NMDS) scores of all samples, we obtained significant correlations for temperature and salinity (p < 0.05), but only when ambient SW communities were included (Fig. 1; Table S6). When considering only the northern Adriatic and North Atlantic biofilm samples, NMDS scores were significantly correlated with temperature and NO2, NO3 and PO4 concentrations (Table S6). However, when examining the correlation between environmental factors and the overall (combined) community composition of biofilm samples from each station, Mantel tests indicated that there was no significant correlation with any of the environmental factors (Table S7). The bacterial community composition of the plastic‐associated biofilm collected in the North Atlantic was significantly correlated with the distance between stations (R 2 = 0.697, p < 0.05). For more details on the taxonomy of the bacterial community on plastics and SW from the different sampling sites, see Supporting Information S1.
We identified 462 OTUs shared between plastics across the different oceanic regions, 57 of them were present only on plastics (Fig. S6A; for taxonomy, see Table S9). When considering only PE biofilms, there were 427 shared OTUs. A total of 471 OTUs with > 1% relative abundance was present in at least one sample at each site, distributed between plastics and SW from the Atlantic, Pacific and northern Adriatic (Fig. S6B; for taxonomy; see Table S10). Out of these 481 OTUs, only 22 OTUs exhibited a relative abundance of > 1% in the plastic biofilm at all three oceanic basins (Fig. S6B; for taxonomy, see Table S11) and only 10 OTUs were also shared with ambient SW. Together, these 19 OTUs contributed a varying percentage to the total classified 16S rRNA gene sequences on plastics collected at the different locations, with a minimum of 7% in one plastic sample collected at Station 4 in the North Atlantic and a maximum of 51% ± 0.04% (n = 3) on plastics collected at Station 13 in the Pacific.
Bacterial communities associated with plastics collected in the northern Adriatic exhibited a higher number of unique OTUs with a relative abundance > 1% (260 OTUs) than corresponding samples collected in the North Atlantic and Pacific, where only 13 and 41 unique OTUs were found respectively (Fig. S6).
Taxa enriched on plastic‐associated communities across different oceanic regions
We identified taxa enriched on plastics in comparison to SW in the different water masses using DESeq2 analysis. Communities associated to plastics collected in the northern Adriatic harboured more significantly enriched families compared to SW than plastics collected in the North Atlantic and Pacific. The family Rhodobacteraceae was the only family enriched on plastics in all three oceanic regions (Fig. 2). The families Phormidesmiaceae and Hyphomonadaceae were also enriched on plastics collected in the northern Adriatic and North Atlantic compared to the corresponding SW. The families Saprospiraceae, Pirellulaceae and Oleiphilaceae were enriched only on plastics collected in the northern Adriatic compared to the ambient water, while Sphingomonadaceae and Rhizobiaceae were enriched only on plastics collected in the North Atlantic, and Comamonadaceae were enriched only on plastics collected in the Pacific compared to the corresponding ambient water (Fig. 2).
Fig 2.
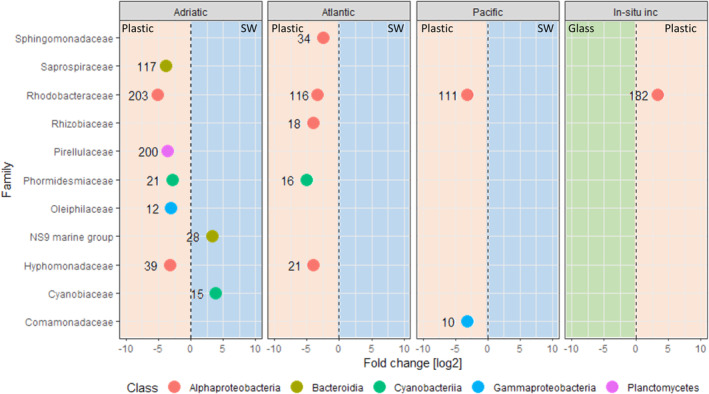
Enriched bacterial families between plastics and seawater in the northern Adriatic, the North Atlantic and the Pacific, and between plastics (HDPE + LDPE + PP) and glass in the in‐situ incubations (in situ inc.) in the northern Adriatic.
Only significantly enriched taxa were included (adjusted p < 0.05). Families enriched on plastics, glass and in seawater (SW) are represented in the red, green and blue areas respectively. The number associated with each symbol represents the number of OTUs in the family.
The bacterial community associated with PE and PP in the northern Adriatic was enriched in some OTUs (Fig. S7). Four Flavobacteriaceae OTUs (OTU 2231, OTU 243, OTU 300 and OTU 5577), one Microtrichaceae OTU (OTU 1048), one Oleiphilaceae OTU (OTU 49), one Rhodobacteraceae OTU (OTU 1457) and three Sphingomonadaceae OTUs (OTU 36, OTU 393 and OTU 6729) were enriched on PE, and one Phormidesmiaceae OTU (OTU 38), one Flavobacteriaceae OTU (OUT 137) and one Saccharospirillaceae OTU (OUT 61) were enriched on PP.
Taxa enriched on PE, PP or glass in in‐situ incubations in the northern Adriatic
To identify taxa enriched on PE versus PP versus glass, we also performed the DESeq2 analysis on 16S rRNA amplicon sequences from the 2 month incubation experiment of Pinto et al. (2019). Rhodobacteraceae was the only enriched family on plastic (PE and PP) compared to glass in the in‐situ incubations.
Comparing the bacterial communities on PE (LDPE+HDPE) and glass of the three sampling dates combined, there were no OTUs enriched on either one of the surfaces (DESeq2 analysis, p > 0.05). When performing the same analysis comparing the bacterial communities of PE and PP, OTU 6540, classified as Primorskyibacter sedentarius, was enriched in PE (log2 Fold Change = −5.53, p < 0.05). However, when analysing only the samples incubated for 1 month in the in‐situ incubations, there were 10 OTUs enriched on the PE (LDPE + HDPE) over glass, and two OTUs enriched on glass over PE. Interestingly, there were four Rhodobacteraceae OTUs (OTU 731, OTU 15, OTU 95 and OTU 23) enriched in LDPE biofilms over both PP and glass biofilms (Fig. 3).
Fig 3.

Enriched bacterial families between different surfaces incubated in the northern Adriatic after a 1 month incubation (Pinto et al., 2019).
Only significantly enriched taxa are included (adjusted p < 0.05). Each symbol represents one OTU. OTUs enriched on glass are represented in the blue area, OTUs enriched on PE (LDPE + HDPE) and LDPE are represented in the white area and OTUs enriched in PP are represented in the orange area. The red circles indicate OTUs also enriched in abundance in the LDPE laboratory incubations after 1 year.
LDPE‐laboratory incubations
We incubated two plastics (Fig. S8) collected in the northern Adriatic (incubations A and B) in artificial SW amended with ammonium and phosphate for 2 years with LDPE as the sole carbon source. A total of 35 OTUs (Table S12) increased in relative abundance in these incubations after 1 year compared to the composition of the initial inoculum. These OTUs were also still present in incubation B after 2 years. Out of these 35 OTUs, only 19 were present in the incubations after one or 2 years with relative abundances > 0.5% (Fig. S9). Operational taxonomic units (OTUs) classified as Cognatiyoonia (OTU 15), Parvibaculaceae (OTU 28), Cognatishimia (OTU 40), Methyloligellaceae (OTU 55), Ketobacter (OTU 60) and Sneathiella (OTU 273) were present in both incubations A and B in relative abundances > 1% after 1 year and also after 2 years in incubation B (Fig. S8).
Scanning electron microscopy (SEM) analysis revealed the presence of a biofilm on both incubations A and B after one and 2 years (Fig. 4B–E). After removing the biofilm from a plastic piece from incubation A after 1 year, holes on the plastics surface were visible (Fig. 4F).
Fig 4.

Scanning electron microscopy (SEM) pictures of (A) a control plastic incubated in artificial seawater without the addition of a plastic previously covered with a biofilm; (B) plastic A 1 year after the initial incubation; (C) plastic B 1 year after the initial incubation; (D) plastic A 2 years after the initial incubation; (E) plastic B 2 years after the initial incubation and (F) plastic A 1 year after the initial incubation after biofilm removal.
Presence of the OTUs enriched in the LDPE‐laboratory incubations on plastics in the ocean
We searched for the 35 OTUs enriched in our LDPE‐laboratory incubations on the plastics and SW collected in the three oceanic regions and on the in‐situ incubation in the northern Adriatic Sea. These OTUs represented 4.1 ± 3.2%, 4.8 ± 4.8% and 7.4 ± 6.4% of the total bacterial community associated with plastics collected in the northern Adriatic, the North Atlantic and the Pacific respectively. Sixteen OTUs were present on at least one plastic at all three sampling sites (Fig. S6C).
From the 35 OTUs, representatives of Cognatiyoonia, Psychrobacter, Acinetobacter, Roseovarius, Roseobacter and Marinovum were found to be relatively abundant on plastics at all three locations (Fig. 5). OTU 15, affiliated to Cognatiyoonia, was found on almost all plastic at all stations (Fig. 6), however, with varying relative abundances of up to 7%. In the ambient SW, the highest relative abundance of this representative (OTU 15) of the Cognatiyoonia was only 0.8%.
Fig 5.

Relative abundance of OTUs enriched in the LDPE laboratory long‐term incubations in seawater and different surfaces across the different sampling sites.
The numbers in red indicate the number of samples. The OTUs represented on the y‐axis are the ones enriched on LDPE biofilms after 1 year of incubation in artificial seawater with LDPE as their sole carbon source.
Fig 6.
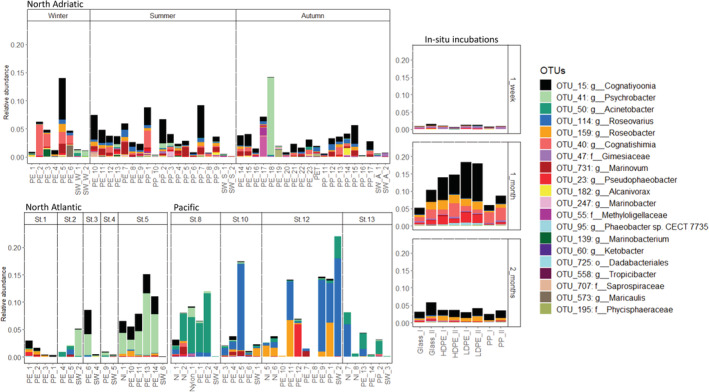
Stacked plots of the relative abundance of the OTUs enriched in the long‐term LDPE incubations after 1 year incubated in ASW with LDPE as sole carbon source.
Only OTUs that occurred with > 0.5% relative abundance in at least one of the samples are shown. The x‐axis represents each individual sample collected in the northern Adriatic, North Atlantic, Pacific and on the different surfaces incubated for 2 months in the northern Adriatic (Pinto et al., 2019). The y‐axis indicates the relative abundance of the classified 16S rRNA genes from 0% to 23%. The labels represent the number of the OTU followed by its lowest taxonomic classification.
This difference, however, was not detectable anymore after 2 months in the in‐situ incubation. From the 35 OTUs, representatives of Cognatiyoonia and Psychrobacter (OTUs 15 and 41 respectively) were the two most abundant OTUs in the plastic associated biofilm in the North Atlantic (Fig. 5). Acinetobacter and Roseovarius (OTUs 50 and 114 respectively) were the two most abundant OTUs in the biofilm from plastics collected in the Pacific (Fig. 5).
The four Rhodobacteraceae OTUs enriched on LDPE sheets in the in‐situ incubations (OTU 731, OTU 15, OTU 95 and OTU 23) when compared to PP and glass after 1 month of incubation were also enriched in the laboratory LDPE‐incubations (Fig. 6). However, this enrichment was not detectable anymore after 2 months of incubation. OTU 15 was particularly abundant on LDPE after 1 month of incubation compared to the biofilm of PP and glass. OTU 159, also enriched in the laboratory incubations, was also enriched on LDPE compared to PP. OTU 36, classified as Erythrobacter, was enriched on PE over PP in both the Adriatic and in the in‐situ incubations after 1 month of incubation (DESeq2 plot PE versus PP Adriatic).
Discussion
There has been discussion on whether there is a core plastic biofilm community and whether plastic degraders are part of it (Debroas et al., 2017). Our study reveals that while there is high variability in bacterial community composition even between plastics collected at the same site, certain taxa are consistently present on plastics even when collected in different oceanic regions (Fig. 1). Some of these taxa that are ubiquitous on plastics were enriched in our incubations with LDPE as the sole carbon source, suggesting that they might utilize LDPE‐derived compounds as a carbon source.
The bacterial community composition on PE and PP was different from that of the ambient water (Fig. 1), similar to what has been shown in previous studies (Dussud et al., 2018; Amaral‐Zettler et al., 2020) but was relatively similar between the two polymers at the different sites. For a more detailed discussion on the comparison of the bacterial community composition of plastic‐associated biofilms collected at the different sites, see Supporting Information S2. Some OTUs, however, were preferentially colonizing or growing on either PE or PP in the northern Adriatic (Fig. S6). For example, OTUs classified as Erythrobacter sp. (OTU 36, OTU 6729 and OTU 393) and Oleiphilus sp. (OTU 49) were enriched in PE over PP. Members of these genera have been shown to harbour genes encoding alkane monooxygenases and to play an important role in oil degradation in the ocean (Wang et al., 2010; Toshchakov et al., 2017). Alkane degradation pathways have been suggested as the most probable PE degradation pathway given the similar chemical structure of PE and alkanes (Devi et al., 2016). Interestingly, OTU 36 (affiliated to Erythrobacter sp.) was also enriched on PE compared to PP in the in situ incubations after 1 month (Fig. 4). Furthermore, Erythrobacter sp. has been previously found to be more abundant on PE than on any other surfaces. In the Baltic Sea, Erythrobacter sp. was also abundant especially on PE (Oberbeckmann et al., 2018), and one Erythrobacter OTU was exclusively associated with PE collected in the Baltic and the North Sea (Oberbeckmann and Labrenz, 2019). The enrichment of alkane‐degraders on PE in comparison to PP is consistent with the possibility that certain organisms discriminate for certain plastics either due to their ability to degrade plastic‐derived hydrocarbons or utilize specific plastic additives.
Members of the Sphingomonadaceae, such as Erythrobacter sp., and Rhodobacteraceae have been regularly found associated with plastics (Bryant et al., 2016; Debroas et al., 2017; Curren and Leong, 2019). Additionally, Oberbeckmann and Labrenz (2019) found that some OTUs from these families made up more than half of the only 45 OTUs present on plastics collected in four different studies. Our study provides further evidence that members of the Rhodobacteraceae family are in fact enriched on plastics compared to SW and other surfaces across different water masses. This is not surprising since members of the Rhodobacteraceae family are key members of biofilms developing on different surfaces in the marine environment (Elifantz et al., 2013). As revealed in the in‐situ incubations, however, some of them, such as Cognatiyoonia sp. (OTU 15), Pseudophaeobacter sp. (OTU 23), Phaeobacter sp. (OTU 95) and Roseobacter sp. (OTU 159) have a preference for LDPE over glass and PP. This might be due to their ability to degrade LDPE‐related hydrocarbons as indicated by their enrichment in our laboratory incubations with LDPE as the sole carbon source after 1 year. However, whether they have the ability to degrade the LDPE hydrocarbon backbone, associated plastic additives (Wright et al., 2020) or passively released compounds (Romera‐Castillo et al., 2018) remains unknown. Furthermore, it is important to note that only duplicate samples were collected in the in situ incubations in the northern Adriatic, precluding rigorous statistical analyses.
Cognatiyoonia is a new genus within the Rhodobacteraceae family (Wirth and Whitman, 2018) to reclassify Loktanella koreensis and Loktanella sediminum. The genus Loktanella has been previously found to be enriched on plastics over non‐plastic surfaces (Ogonowski et al., 2018) and particularly abundant on PET and LDPE (Delacuvellerie et al., 2019). In our study, OTU 15, classified as Cognatiyoonia, was present in the biofilm of almost all plastics (Fig. 6). Also, OTU 15 exhibited a high relative abundance in the laboratory incubations with LDPE as the sole organic carbon source after 1 year. This might indicate that OTU 15 has the ability to utilize plastic‐derived hydrocarbons, especially those related to alkanes. Certain Loktanella strains have the ability to degrade n‐alkanes between C10 and C22 (Harwati et al., 2007). Furthermore, they have been found enriched in oil degradation experiments (Lanfranconi et al., 2010; Sanni et al., 2015). The potential of Cognatiyoonia sp. to degrade hydrocarbons and plastics is uncertain, but its ubiquitous presence on plastics collected at different locations and the ability to survive on LDPE as sole carbon source suggest that it might play a role in the degradation of plastics in the ocean.
The above‐mentioned genera Pseudophaeobacter sp. (OTU 23), Phaeobacter sp. (OTU 95) and Roseobacter sp. (OTU 159), also enriched in the long‐term incubations with LDPE as the only carbon source, have also been associated with hydrocarbon degradation (Bacosa et al., 2018; Brakstad et al., 2018). Even though they were present in lower relative abundance than OTU 15, they were still present on plastics collected at all the different oceanic regions (Fig. 6).
As suggested by the in‐situ incubation, the relative abundance of the OTUs enriched in the LDPE‐laboratory incubations varies between different stages of biofilm development, especially on LDPE. The putative LDPE‐degrading OTUs were most prevalent during the early stages of biofilm development after about 1 month of incubation and significantly decreased in relative abundance after 2 months (Fig. 6). Also, after 1 month of incubation the largest differences in bacterial community composition between different plastic types were observed in the in situ incubations (Pinto et al., 2019). Microbes with a preference for a specific type of plastic are dominant in the biofilm layers which are in close contact with the plastic surface (Kirstein et al., 2019). In subsequently formed layers when the biofilm thickens, the bacterial community is most likely shaped by internal biofilm processes and micro‐environmental factors rather than by microbes selected due to surface properties. For example, Pinto et al. (2019) reported that after a 1 and 2 months incubation period the composition of the bacterial communities growing on different plastics were more similar to each other than after 1 week of incubation. This might explain the decrease in relative abundance of organisms potentially capable of degrading LDPE‐originating compounds in the later stages of biofilm development. This agrees with studies reporting similar bacterial community compositions on different polymers after prolonged incubation periods (Kirstein et al., 2018). The decrease in the number of OTUs with preference for a specific surface in later stages of the biofilm development might be the result of changes in the biofilm environment, no longer favouring microbes with the ability to metabolize plastic‐originating compounds. The development of a thicker biofilm creates a diverse habitat with a plethora of organic carbon compounds (Dang and Lovell, 2016) probably more readily accessible to bacteria than plastic‐derived hydrocarbons (Min et al., 2020). Nevertheless, in the in‐situ incubations even after 2 months, OTUs enriched in our LDPE‐laboratory incubations were still present in the biofilms obtained from all plastics (Fig. 6).
Taken together, degraders of plastic‐derived compounds are recruited early in the development of the biofilm on plastic surfaces (Kirstein et al., 2019) and attain high relative abundance at the early stage of biofilm formation, but decrease in relative abundance with further biofilm development. These putative plastic‐compound degraders remain in the plastic‐associated biofilm in low relative abundances, potentially in the layers that are tightly connected to the plastic surface. Whether these bacteria do degrade the plastics remains unknown. For example, members of the Rhodobacteraceae family are known to exhibit high metabolic versatility (Michael et al., 2016) and are capable of occupying different metabolic niches when environmental conditions change.
In conclusion, through a combination of in situ incubations, laboratory long‐term incubations with LDPE as the sole organic carbon source and analyses of collected plastics from three oceanic basins, strong evidence is provided for the ubiquitous presence of bacteria capable of utilizing LDPE‐derived compounds as a carbon source within plastic biofilms in the global ocean.
Experimental procedures
Sampling
Sampling was conducted at four stations in the Pacific, five stations in the North Atlantic and at one location in the northern Adriatic Sea (Fig. S1; Table S1). The Pacific samples were collected aboard R/V Sonne during the SO248 expedition with a bongo net and a mesh size of 300 μm in May–June 2016. The net was kept at the sea surface by attaching one float on each side of the net (Fig. S8). The North Atlantic samples were collected aboard the R/V Rámon Margalef during the RadProf 2017 expedition belonging to the time series RADIALES‐PROFUNDOS in July 2017 (Prieto et al., 2015). The northern Adriatic Sea samples were collected in February, August and November of 2016. Samples from the North Atlantic and the northern Adriatic Sea were collected with a net for microplastic sampling (Hydrobios, Germany) with a mesh size of 500 μm. At all locations, the net was towed with 2 knots for 30–45 min (detailed information of sampling at each station is given in Table S1). Water samples (2–5 l) from all locations were also collected and immediately filtered through 0.2 μm polycarbonate filters and stored at −80°C.
The plastics were collected from the cod end of the net into a bucket filled with SW from the same location and immediately taken to the laboratory for processing. To separate the collected plastic material by size, the samples were passed through a series of sieves with mesh size of 9.5 mm, 4.5 mm, 1.4 mm and 300 μm and rinsed with 0.2 μm freshly filtered SW. The content collected in each sieve was immediately transferred to petri dishes with 0.2 μm filtered SW. Each plastic piece was collected from the Petri dishes, washed three times in 0.2 μm filtered SW to remove all organisms not tightly attached to the plastic biofilm, put in a Greiner tube and immediately stored at −80°C.
Inorganic nutrient analyses and environmental data
Inorganic nutrient concentrations (NO3 −, NO2 − and PO4 3−) were determined only on SW samples collected with Niskin bottles just below the sea surface. In the northern Adriatic Sea, surface SW was filtered through combusted Whatman GF/F filters and stored in PE bottles at −20°C. Nutrient analyses were performed within 1 month following standard protocols (Strickland and Parsons, 1972). Surface SW samples collected in the North Atlantic were frozen on board until the inorganic nutrients were measured in the laboratory using a QuAAtri auto‐analyser (SEAL Analytical) following published protocols (Coverly et al., 2012). Most nutrient concentrations at the Pacific stations were below the detection limit using standard methods. Therefore, they were excluded from this study.
Temperature and salinity measurements from the northern Adriatic Sea were made available from the sampling station RV001 located off the coast of Rovinj by the Ruđer Bošković Centre for Marine Research at Rovinj, Croatia. Temperature and salinity data from the stations in the Pacific and North Atlantic were obtained from the sensors mounted on the CTD sampling rosette deployed at each station. Data from the Pacific are available at Pangaea (https://doi.pangaea.de/10.1594/PANGAEA.864673).
Colonization pattern of LDPE and other plastics incubated in situ in the northern Adriatic Sea
To investigate the presence of putative LDPE degraders in biofilms on different surfaces over a 2 month incubation period in the northern Adriatic Sea, we used 16S rRNA amplicon sequencing of the prokaryotic communities collected during incubations exposed to solar radiation, downloaded from the NCBI Sequence Read Archive (SRA) database (BioProject: PRJNA515271). Details of the experimental setup are given elsewhere (Pinto et al., 2019).
The samples comprised duplicates of 4 cm2 sized pieces of virgin PP, LDPE and HDPE, with approximately 0.5 mm thickness, glass slides of 1 cm2, with approximately 1 mm thickness and SW samples. The different substrates were mounted on a floating frame in the northern Adriatic Sea, approximately 500 m off the coast of Rovinj, Croatia, over a 3 month period (November 2016–January 2017) and the samples were taken after 1 week, 1 month and 2 months of incubation. Processing of the samples, the DNA extraction method, PCR primers and sequencing technology and specifications used were the same as described previously (Pinto et al., 2019).
Enrichment of bacterial communities living on LDPE as sole carbon source
Plastic material was collected in August 2016 off the coast of Rovinj, Croatia, and rinsed with 0.2 μm filtered SW. Two large plastic pieces (Fig. S8) were incubated each in a 1 l borosilicate bottle filled with artificial SW (ASW) amended with 10 μM NH4Cl and 2 μM Na2PO4 (incubations A and B), however, without an organic carbon source other than that contained in the plastic material. The incubations were held at room temperature in the dark. For a scheme of the experimental setup, see Fig. S9. After 3 weeks, each plastic was transferred to a new bottle with the same conditions. After repeating this procedure for 3 months, a small piece of the plastic of approximately 2.25 cm2 was cut and transferred to a new bottle with ASW amended with ammonium and phosphate added in the form of NH4Cl and Na2PO4, respectively, using the same concentrations as mentioned above, and 1 cm2 sterile LDPE sheets were added as a carbon source. The LDPE sheets were purchased from Goodfellow. The same transfer procedure was repeated with new LDPE sheets every 3 weeks for a total period of 2 years. After 1 and 2 years of the initial incubation, one square from each flask which had the biofilm developing for 1 month was taken for 16S rRNA gene amplicon sequencing of the biofilm community. Due to low quality sequencing output, the sample after 2 years of incubation A was excluded from further analysis. Two plastic pieces from the same flasks were also taken for SEM analysis after one and 2 years. One piece was used to analyse the intact biofilm, and the second was used to analyse the surface of the plastic after removal of the biofilm. To remove the biofilm, the piece was submerged in Milli‐Q water, vortexed for 30 s, then submerged in 4% SDS buffer at room temperature for 4 h. Subsequently, the plastic was submerged again two times in fresh Milli‐Q water, vortexed for 30 s and gently dried on clean paper. A control plastic that was incubated in a separate bottle with the same ASW as incubations A and B for 1 month was also stored for SEM analysis.
SEM analysis
Samples for SEM were incubated in 2% glutaraldehyde for 10 min and then stored at −80°C until processing. The samples were then dehydrated with an ethanol dilution series of 30%, 50%, 70%, 80%, 90%, 95% each for 10 min and three times in 100% absolute ethanol for 10 min. The dehydrated samples were CO2 critical‐point dried with a CPD 300 auto critical‐point dryer (Leica Microsystems). The dried pieces were gold‐coated using a JFC‐2300HR sputter coater (JEOL) for 80 s. Pictures were taken using a secondary electron detector with a JEOL JSM‐IT300 scanning electron microscope with a 15 kV acceleration voltage in ultra‐high vacuum.
DNA extraction and 16S rRNA sequencing
DNA extraction of all samples was performed using the modified bead‐beating approach in combination with the Puregene Tissue DNA extraction kit (Qiagen, Valencia, CA) (Debeljak et al., 2017). DNA extraction and sequencing were performed in four batches. The first one comprised all the northern Adriatic samples, the second the laboratory LDPE incubations after 1 year incubation and the Pacific samples, and the third the laboratory LDPE incubations after 2 years incubation and the North Atlantic samples. For each batch, we added a blank that was extracted, PCR amplified and sequenced at the same time as the samples using the same protocols (Fig. S12).
The primer pair 341F (5′‐CCT ACG GGN GGC WGC AG‐3′) and 802R (5′‐TAC HVG GGT ATC TAA TCC‐3′) was used to PCR amplify the V3‐V4 hypervariable region of the 16S rRNA gene, resulting in fragments of ~460 bp. Each PCR was performed in a total volume of 52 μl containing 25 μl Library Amplification Ready Mix 2xMM (Kapa Biosystems), 1 μl of each 25 μM primer, 23 μl of sterile water and 2 μl of extracted DNA. Amplification was done on a Mastercycler Pro (Eppendorf AG, Germany). The conditions consisted of an initial denaturation at 95°C for 3 min, followed by 20 cycles at 98°C for 20 s, at 56°C for 30 s and at 72°C for 30 s, and a final elongation at 72°C for 5 min.
PCR products were then purified in a 96‐well plate using the Agencourt AMPure XP Purification kit and protocol (Beckman Coulter Life Sciences). These purified PCR products were barcoded and an additional 10‐cycle PCR was done, followed by paired‐end next‐generation amplicon sequencing (2 × 250 bp) using Illumina MiSeq technology at Microsynth AG, Switzerland. All raw sequence files were submitted to the NCBI Sequence Read Archive (SRA) database (BioProject: PRJNA623496).
Identification of plastic types using Raman microspectroscopy
After DNA extraction, each plastic was dried on a clean paper and its chemical nature was identified using a LabRAM HR800 confocal Raman spectroscopy system (Horiba Jobin‐Yvon). Excitation for Raman scattering was provided by a 532 nm NdYAG laser. The Raman spectrum was obtained for the range of 400–3200 cm−1. Each sample was compared with reference scans from plastics of known composition. The polymers, which could not be identified were classified as NI. Most NI polymers were coloured.
Processing of sequencing data
Merging of paired‐end reads, quality filtering (max. error of 1 in 100 nucleotides), elimination of singletons, chimera filtering, mapping and clustering of OTU were performed using usearch tools, with OTU clustering being performed by UPARSE (Edgar, 2013). Taxonomic assignments of OTUs were done using blastn against the SILVA_138_SSURef_NR99 reference database with a threshold of 97% similarity between the query and reference sequences. Samples with less than 1000 classified reads were excluded from further analysis. All OTUs classified as chloroplasts were excluded from further analysis.
Statistical analysis
All statistical analyses were performed using R version 3.5.1 in RStudio version 1.1.456. All plots were done using R package ‘ggplot2’ version 3.3.1 (Wickham, 2016), unless stated otherwise. Venn diagrams were built using the R package ‘VennDiagram’ version 1.6.20 (Chen and Boutros, 2011). OTU diversity and richness indexes were calculated with the R package ‘iNext’ version 2.0.20 (Hsieh et al., 2016) using a coverage base rarefaction of 97%, corresponding to the lower coverage calculated for a sample.
To visualize the similarity of the prokaryotic communities of all samples, not including the in situ or the laboratory incubation samples, NMDS was performed with Bray–Curtis dissimilarity as the distance measurement. The environmental factors (temperature, salinity and PO4, NO3 and NO2 concentrations) were fitted to the ordination plots using the ‘envfit’ function of the vegan package in R. A Mantel test was performed to evaluate correlations between the prokaryotic community composition of the biofilms of polymers and the different environmental factors. An additional Mantel test was performed to determine correlations between the relative abundance of the 35 OTUs enriched in the laboratory incubations and environmental factors. Euclidian distance matrices were used for the environmental factors and a Bray–Curtis dissimilarity matrix was used for the community composition. A PERMANOVA was performed to determine whether the prokaryotic community composition was significantly different between sampling regions (e.g. Atlantic Ocean, Pacific Ocean and northern Adriatic Sea), sampling stations or sample type (e.g. PE, PP, PET, nylon and ambient SW). PERMANOVA analysis was also performed just with the 35 OTUs enriched in the laboratory experiments after 1 year of incubation. Samples from polymers remaining unclassified were not considered in this analysis. We also evaluated differences in community composition between different sites, stations and plastic types using homogeneity of dispersions tests on Bray–Curtis dissimilarity matrices (‘betadisper’ from R package ‘vegan’ version 2.5.6). Pair‐wise PERMANOVAs were performed to each of the above‐mentioned factors to determine differences between stations, oceanic basins or sample types. Differences were considered significant if p < 0.05.
The fold‐change in abundance of each OTU between sample types for each location was calculated using the R package ‘DESeq2’ (Love et al., 2014), which fits negative binomial generalized linear models for each OTU and tests significance using the Wald test.
Supporting information
Appendix S1. Supporting Information.
Table S1. Information on the sampling site. (.xlsx)
Table S2. Information on the samples. (.xlsx)
Table S5. Results of the PERMANOVA and pair‐wise PERMANOVAs comparing the prokaryotic community composition between different polymer types, sampling site, sampling station and season in the case of the North Adriatic samples. (.xlsx)
Table S8. Relative abundance of families present with more than 1% relative abundance in at least one samples across the three sampling sites. (.xlsx)
Table S9. OTUs found in plastics from all sampling sites, but not in SW. (.xlsx)
Table S10. OTUs with more than 1% relative abundance in at least one sample. (xlsx)
Table S11. OTUs with more than 1% relative abundance in at least one sample of each sampling site. (.xlsx)
Table S12. OTUs that were enriched in the laboratory LDPE incubations after one year incubation, and that were still present in incubation B after two years. (.xlsx)
Acknowledgements
We thank the crew and onboard scientists of R/V Rámon Margalef and the R/V Sonne assisting us during the Radprof 2017 and the SO248 campaigns respectively. We acknowledge César González Pola for organizing the RAD‐PROF 2017 expedition. We also thank the Ruđer Bošković Center for Marine Research for accommodating us during our stays in Rovinj, Croatia. We especially thank Mirjana, Dario, Ingrid, Marino and Paolo for all the help provided during sampling in the Adriatic. Financial support was provided by a Doctoral Fellowship (DOC scholarship) from the Austrian Academy of Sciences (ÖAW) awarded to Maria Pinto, the ARTEMIS project of the Austrian Science Fund (P 28781‐B21) to Gerhard J. Herndl and the research platform PLENTY financially supported by the University of Vienna, Austria. Cruise SO248 was funded by the German Federal Ministry of Education and Research (BMBF) within the BacGeoPac project (03G0248A) and by the Deutsche Forschungsgemeinschaft within the Collaborative Research Center Roseobacter (TRR51) awarded to Meinhard Simon. The RAD‐PROF campaign was funded by the RADIALES‐PROFUNDAS project (RAD‐PROF, Instituto Español de Ocenografia). Furthermore, we thank the anonymous reviewers for their constructive criticism, which helped to improve the manuscript.
References
- Amaral‐Zettler, L.A. , Zettler, E.R. , and Mincer, T.J. (2020) Ecology of the plastisphere. Nat Rev Microbiol 18: 139–151. [DOI] [PubMed] [Google Scholar]
- Amaral‐Zettler, L.A. , Zettler, E.R. , Slikas, B. , Boyd, G.D. , Melvin, D.W. , Morrall, C.E. , et al (2015) The biogeography of the Plastisphere: implications for policy. Front Ecol Environ 13: 541–546. [Google Scholar]
- Bacosa, H.P. , Erdner, D.L. , Rosenheim, B.E. , Shetty, P. , Seitz, K.W. , Baker, B.J. , and Liu, Z. (2018) Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern Gulf of Mexico to light Louisiana sweet crude oil. ISME J 12: 2532–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakstad, O.G. , Davies, E.J. , Ribicic, D. , Winkler, A. , Brönner, U. , and Netzer, R. (2018) Biodegradation of dispersed oil in natural seawaters from Western Greenland and a Norwegian fjord. Polar Biol 41: 2435–2450. [Google Scholar]
- Bryant, J.A. , Clemente, T.M. , Viviani, D.A. , Fong, A.A. , Thomas, K.A. , Kemp, P. , et al (2016) Diversity and activity of communities inhabiting plastic debris in the North Pacific gyre. mSystems 1: e00024‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, L. , Wu, D. , Xia, J. , Shi, H. , and Kim, H. (2019) Influence of physicochemical surface properties on the adhesion of bacteria onto four types of plastics. Sci Total Environ 671: 1101–1107. [Google Scholar]
- Chen, H. , and Boutros, P.C. (2011) VennDiagram: a package for the generation of highly‐customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverly, S. , Kérouel, R. , and Aminot, A. (2012) A re‐examination of matrix effects in the segmented‐flow analysis of nutrients in sea and estuarine water. Anal Chim Acta 712: 94–100. [DOI] [PubMed] [Google Scholar]
- Curren, E. , and Leong, S.C.Y. (2019) Profiles of bacterial assemblages from microplastics of tropical coastal environments. Sci Total Environ 655: 313–320. [DOI] [PubMed] [Google Scholar]
- Dang, H. , and Lovell, C.R. (2016) Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80: 91–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeljak, P. , Pinto, M. , Proietti, M. , Reisser, J. , Ferrari, F.F. , Abbas, B. , et al (2017) Extracting DNA from ocean microplastics: a method comparison study. Anal Methods 9: 1521–1526. [Google Scholar]
- Debroas, D. , Mone, A. , and Ter Halle, A. (2017) Plastics in the North Atlantic garbage patch: a boat‐microbe for hitchhikers and plastic degraders. Sci Total Environ 599: 1222–1232. [DOI] [PubMed] [Google Scholar]
- Delacuvellerie, A. , Cyriaque, V. , Gobert, S. , Benali, S. , and Wattiez, R. (2019) The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low‐density polyethylene degradation. J Hazard Mater 380: 120899. [DOI] [PubMed] [Google Scholar]
- Devi, R.S. , Kannan, V.R. , Natarajan, K. , Nivas, D. , Kannan, K. , Chandru, S. , and Antony, A.R. (2016) The role of microbes in plastic degradation. Environ Waste Manag, 341–370. [Google Scholar]
- Dudek, K.L. , Cruz, B.N. , Polidoro, B. , and Neuer, S. (2020) Microbial colonization of microplastics in the Caribbean Sea. Limnol Oceanogr Lett 5, 5–17. [Google Scholar]
- Dussud, C. , Hudec, C. , George, M. , Fabre, P. , Higgs, P. , Bruzaud, S. , et al (2018a) Colonization of non‐biodegradable and biodegradable plastics by marine microorganisms. Front Microbiol 9: 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussud, C. , Meistertzheim, A.L. , Conan, P. , Pujo‐Pay, M. , George, M. , Fabre, P. , et al (2018b) Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ Pollut 236: 807–816. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Elifantz, H. , Horn, G. , Ayon, M. , Cohen, Y. , and Minz, D. (2013) Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol Ecol 85: 348–357. [DOI] [PubMed] [Google Scholar]
- Gross, M. , Zhao, X. , Mascarenhas, V. , and Wen, Z. (2016) Effects of the surface physico‐chemical properties and the surface textures on the initial colonization and the attached growth in algal biofilm. Biotechnol Biofuels 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J.P. , Hoellein, T.J. , Sapp, M. , Tagg, A.S. , Ju‐Nam, Y. , and Ojeda, J.J. (2018) Microplastic‐associated biofilms: a comparison of freshwater and marine environments In Freshwater Microplastics. Cham, Switzerland: Springer, pp. 181–201. [Google Scholar]
- Harrison, J.P. , Schratzberger, M. , Sapp, M. , and Osborn, A.M. (2014) Rapid bacterial colonization of low‐density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol 14: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshvardhan, K. , and Jha, B. (2013) Biodegradation of low‐density polyethylene by marine bacteria from pelagic waters, Arabian Sea, India. Mar Pollut Bull 77: 100–106. [DOI] [PubMed] [Google Scholar]
- Harwati, T.U. , Kasai, Y. , Kodama, Y. , Susilaningsih, D. , and Watanabe, K. (2007) Characterization of diverse hydrocarbon‐degrading bacteria isolated from Indonesian seawater. Microbes Environ 22: 412–415. [Google Scholar]
- Hossain, M.R. , Jiang, M. , Wei, Q. , and Leff, L.G. (2019) Microplastic surface properties affect bacterial colonization in freshwater. J Basic Microbiol 59: 54–61. [DOI] [PubMed] [Google Scholar]
- Hsieh, T. , Ma, K. , and Chao, A. (2016) iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). 1451–1456.
- Jacquin, J. , Cheng, J. , Odobel, C. , Pandin, C. , Conan, P. , Pujo‐Pay, M. , et al (2019) Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the “Plastisphere”. Front Microbiol 10: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambeck, J.R. , Geyer, R. , Wilcox, C. , Siegler, T.R. , Perryman, M. , Andrady, A. , et al (2015) Plastic waste inputs from land into the ocean. Science 347: 768–771. [DOI] [PubMed] [Google Scholar]
- Jiang, P. , Zhao, S. , Zhu, L. , and Li, D. (2018) Microplastic‐associated bacterial assemblages in the intertidal zone of the Yangtze Estuary. Sci Total Environ 624: 48–54. [DOI] [PubMed] [Google Scholar]
- Kesy, K. , Oberbeckmann, S. , Kreikemeyer, B. , and Labrenz, M. (2019) Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in Baltic Sea mesocosms. Front Microbiol 10: 1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner, M.T. , Rojas‐Jimenez, K. , Oberbeckmann, S. , Labrenz, M. , and Grossart, H.‐P. (2017) Microplastics alter composition of fungal communities in aquatic ecosystems. Environ Microbiol 19: 4447–4459. [DOI] [PubMed] [Google Scholar]
- Kim, S.‐H. , Kim, J.‐G. , Jung, M.‐Y. , Kim, S.‐J. , Gwak, J.‐H. , Yu, W.‐J. , et al (2018) Ketobacter alkanivorans gen. nov., sp. nov., an n‐alkane‐degrading bacterium isolated from seawater. Int J Syst Evol Microbiol 68: 2258–2264. [DOI] [PubMed] [Google Scholar]
- Kirstein, I.V. , Wichels, A. , Krohne, G. , and Gerdts, G. (2018) Mature biofilm communities on synthetic polymers in seawater ‐ specific or general? Mar Environ Res 142: 147–154. [DOI] [PubMed] [Google Scholar]
- Kirstein, I.V. , Wichels, A. , Gullans, E. , Krohne, G. , and Gerdts, G. (2019) The Plastisphere – uncovering tightly attached plastic “specific” microorganisms. PLoS One 14: e0215859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranconi, M.P. , Bosch, R. , and Nogales, B. (2010) Short‐term changes in the composition of active marine bacterial assemblages in response to diesel oil pollution. J Microbial Biotechnol 3: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobelle, D. , and Cunliffe, M. (2011) Early microbial biofilm formation on marine plastic debris. Mar Pollut Bull 62: 197–200. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. , and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, V. , Frank, O. , Bartling, P. , Scheuner, C. , Göker, M. , Brinkmann, H. , and Petersen, J. (2016) Biofilm plasmids with a rhamnose operon are widely distributed determinants of the ‘swim‐or‐stick’ lifestyle in roseobacters. ISME J 10: 2498–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, K. , Cuiffi, J.D. , and Mathers, R.T. (2020) Ranking environmental degradation trends of plastic marine debris based on physical properties and molecular structure. Nat Commun 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogonowski, M. , Motiei, A. , Ininbergs, K. , Hell, E. , Gerdes, Z. , Udekwu, K.I. , et al (2018) Evidence for selective bacterial community structuring on microplastics. Environ Microbiol 20: 2796–2808. [DOI] [PubMed] [Google Scholar]
- Oberbeckmann, S. , Kreikemeyer, B. , and Labrenz, M. (2018) Environmental factors support the formation of specific bacterial assemblages on microplastics. Front Microbiol 8: 2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbeckmann, S. , and Labrenz, M. (2019) Marine microbial assemblages on microplastics: diversity, adaptation, and role in degradation. Ann Rev Mar Sci 12: 209–232. [DOI] [PubMed] [Google Scholar]
- Oberbeckmann, S. , Löder, M.G.J. , and Labrenz, M. (2015) Marine microplastic‐associated biofilms—a review. Environ Chem 12: 551–562. [Google Scholar]
- Oberbeckmann, S. , Loeder, M.G.J. , Gerdts, G. , and Osborn, A.M. (2014) Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol Ecol 90: 478–492. [DOI] [PubMed] [Google Scholar]
- Oberbeckmann, S. , Osborn, A.M. , and Duhaime, M.B. (2016) Microbes on a bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS One 11: e0159289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paço, A. , Duarte, K. , da Costa, J.P. , Santos, P.S.M. , Pereira, R. , Pereira, M.E. , et al (2017) Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum . Sci Total Environ 586: 10–15. [DOI] [PubMed] [Google Scholar]
- Pinto, M. , Langer, T.M. , Hüffer, T. , Hofmann, T. , and Herndl, G.J. (2019) The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS One 14: e0217165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, E. , González‐Pola, C. , Lavín, A. , and Holliday, N.P. (2015) Interannual variability of the northwestern Iberia deep ocean: response to large‐scale North Atlantic forcing. J Geophys Res Ocean 120: 832–847. [Google Scholar]
- Roager, L. , and Sonnenschein, E.C. (2019) Bacterial candidates for colonization and degradation of marine plastic debris. Environ Sci Technol 53: 11636–11643. [DOI] [PubMed] [Google Scholar]
- Romera‐Castillo, C. , Pinto, M. , Langer, T.M. , Álvarez‐Salgado, X.A. , and Herndl, G.J. (2018) Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat Commun 9: 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanni, G.O. , Coulon, F. , and McGenity, T.J. (2015) Dynamics and distribution of bacterial and archaeal communities in oil‐contaminated temperate coastal mudflat mesocosms. Environ Sci Pollut Res 22: 15230–15247. [DOI] [PubMed] [Google Scholar]
- Schneiker, S. , dos Santos, V.A.P.M. , Bartels, D. , Bekel, T. , Brecht, M. , Buhrmester, J. , et al (2006) Genome sequence of the ubiquitous hydrocarbon‐degrading marine bacterium Alcanivorax borkumensis . Nat Biotechnol 24: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland, J.D.H. , and Parsons, T.R. (1972) A Practical Handbook of Seawater Analysis. Ottawa, Canada: Fisheries Research Board of Canada. [Google Scholar]
- Sudhakar, M. , Doble, M. , Murthy, P.S. , and Venkatesan, R. (2008) Marine microbe‐mediated biodegradation of low‐ and high‐density polyethylenes. Int Biodeter Biodegr 61: 203–213. [Google Scholar]
- Sudhakar, M. , Trishul, A. , Doble, M. , Suresh Kumar, K. , Syed Jahan, S. , Inbakandan, D. , et al (2007) Biofouling and biodegradation of polyolefins in ocean waters. Polym Degrad Stab 92: 1743–1752. [Google Scholar]
- Syranidou, E. , Karkanorachaki, K. , Amorotti, F. , Avgeropoulos, A. , Kolvenbach, B. , Zhou, N.‐Y. , et al (2019) Biodegradation of mixture of plastic films by tailored marine consortia. J Hazard Mater 375: 33–42. [DOI] [PubMed] [Google Scholar]
- De Tender, C. , Devriese, L.I. , Haegeman, A. , Maes, S. , Vangeyte, J. , Cattrijsse, A. , et al (2017) Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ Sci Technol 51: 7350–7360. [DOI] [PubMed] [Google Scholar]
- Toshchakov, S.V. , Korzhenkov, A.A. , Chernikova, T.N. , Ferrer, M. , Golyshina, O.V. , Yakimov, M.M. , and Golyshin, P.N. (2017) The genome analysis of Oleiphilus messinensis ME102 (DSM 13489T) reveals backgrounds of its obligate alkane‐devouring marine lifestyle. Mar Genomics 36: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Wang, W. , Lai, Q. , and Shao, Z. (2010) Gene diversity of CYP153A and AlkB alkane hydroxylases in oil‐degrading bacteria isolated from the Atlantic Ocean. Environ Microbiol 12: 1230–1242. [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2016) ggplot2: Elegant Graphics for Data Analysis, New York: Springer International Publishing AG. [Google Scholar]
- Wintzingerode V., F., Göbel, U.B. , and Stackebrandt, E. (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR‐based rRNA analysis FEMS Microbiol Rev 21: 213–229. [DOI] [PubMed] [Google Scholar]
- Wirth, J.S. , and Whitman, W.B. (2018) Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int J Syst Evol Microbiol 68: 2393–2411. [DOI] [PubMed] [Google Scholar]
- Wright, R.J. , Bosch, R. , Gibson, M.I. , and Christie‐Oleza, J.A. (2020) Plasticizer degradation by marine bacterial isolates: a proteogenomic and metabolomic characterization. Environ Sci Technol 54: 2244–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takehana, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. , et al (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science (80‐) 351: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Zettler, E.R. , Mincer, T.J. , and Amaral‐Zettler, L. (2013) Life in the “plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol 47: 7137–7146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Table S1. Information on the sampling site. (.xlsx)
Table S2. Information on the samples. (.xlsx)
Table S5. Results of the PERMANOVA and pair‐wise PERMANOVAs comparing the prokaryotic community composition between different polymer types, sampling site, sampling station and season in the case of the North Adriatic samples. (.xlsx)
Table S8. Relative abundance of families present with more than 1% relative abundance in at least one samples across the three sampling sites. (.xlsx)
Table S9. OTUs found in plastics from all sampling sites, but not in SW. (.xlsx)
Table S10. OTUs with more than 1% relative abundance in at least one sample. (xlsx)
Table S11. OTUs with more than 1% relative abundance in at least one sample of each sampling site. (.xlsx)
Table S12. OTUs that were enriched in the laboratory LDPE incubations after one year incubation, and that were still present in incubation B after two years. (.xlsx)


