Abstract
Disturbed intrauterine development increases the risk of renal disease. Various studies have reported that Notch signalling plays a significant role in kidney development and kidney diseases. A disintegrin and metalloproteinase domain 10 (ADAM10), an upstream protease of the Notch pathway, is also reportedly involved in renal fibrosis. However, how ADAM10 interacts with the Notch pathway and causes renal fibrosis is not fully understood. In this study, using a prenatal chlorpyrifos (CPF) exposure mouse model, we investigated the role of the ADAM10/Notch axis in kidney development and fibrosis. We found that prenatal CPF‐exposure mice presented overexpression of Adam10, Notch1 and Notch2, and led to premature depletion of Six2+ nephron progenitors and ectopic formation of proximal tubules (PTs) in the embryonic kidney. These abnormal phenotypic changes persisted in mature kidneys due to the continuous activation of ADAM10/Notch and showed aggravated renal fibrosis in adults. Finally, both ADAM10 and NOTCH2 expression were positively correlated with the degree of renal interstitial fibrosis in IgA nephropathy patients, and increased ADAM10 expression was negatively correlated with decreased kidney function evaluated by serum creatinine, cystatin C, and estimated glomerular filtration rate. Regression analysis also indicated that ADAM10 expression was an independent risk factor for fibrosis in IgAN. © 2020 The Authors. The Journal of Pathology published by John Wiley & Sons, Ltd. on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: ADAM10, Notch, kidney development, renal fibrosis, chlorpyrifos, proximal tubules, IgA nephropathy
Introduction
Chronic kidney disease (CKD) is a global public health problem [1, 2], and its prevalence exceeds 10% worldwide [3, 4]. Abnormal intrauterine development can increase the risk of renal disease in adulthood [5, 6]. Kidney development can be aberrantly programmed in response to adverse conditions and result in permanent structural and functional changes [7, 8].
Environmental pollution is presently regarded as a risk factor for kidney diseases [9]. In addition to air pollution and heavy metal contamination, pesticide pollution is also a prominent problem in China [10, 11, 12, 13]. Chlorpyrifos (CPF) is a widely used organophosphorus pesticide worldwide. While a rat study verified that CPF can be distributed in various body tissues, including the kidneys [14], there are no reports on kidney development upon prenatal CPF exposure.
The Notch signalling pathway is involved in kidney development [15, 16]. This pathway is a major regulator of nephron segmentation, which is the core process of functional nephron formation [17, 18]. It is believed that NOTCH2 promotes the formation of PTs [19, 20]. Conditional knockout mouse models have shown that Notch regulates the segmentation of other nephron structures [21, 22, 23]. Notch signalling is increased in the glomeruli and tubuli of CKD patients [24], and inhibition of this pathway can mitigate kidney fibrosis in CKD mouse models [25].
Notch activation requires proteolytic cleavage by ADAM family enzymes [15]. ADAM10 mediates ectodomain shedding of numerous receptors, including Notch [26, 27, 28]. The Notch intracellular domain (NICD), produced by ADAM10 cleavage, translocates from the cytoplasm to the cell nucleus to activate the transcription of downstream genes. ADAM10 was also found to be involved in organ fibrosis; one study showed that overexpression of ADAM10 contributed to epithelial‐to‐mesenchymal transition (EMT) in rat renal tubular epithelia [29, 30]. However, the interaction of ADAM10 with the Notch signalling pathway and the mechanisms through which they affect kidney development and fibrosis are not well understood.
In a previous study [31], using a prenatal CPF exposure mouse model, we reported weak expression of NOTCH2 and JAGGED1 as assessed by immunohistochemistry and concluded that the NOTCH2–JAGGED1 pathway was broken. However, in this study, by use of more sophisticated methods, we have further investigated the role of the ADAM10/Notch axis in kidney development and kidney fibrosis.
Materials and methods
Human renal biopsy samples
The study was approved by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent for scientific research statements was obtained from all participants. Ninety‐five formalin‐fixed and paraffin‐embedded kidney tissue specimens, including 81 of IgA nephropathy (IgAN), three donor kidneys, three of mesangial proliferative glomerulonephritis, three of minimal‐change disease, three of lupus nephritis, and two of membranous nephropathies, were obtained from the Department of Pathology, Kidney Disease Center, the First Affiliated Hospital, College of Medicine, Zhejiang University from January 2018 to March 2019. All cases were confirmed by pathological diagnosis. The clinical data of the 81 IgAN patients were collected.
Mouse model
We bred Six2‐TGCtg mice [JAX009606; Tg(Six2‐EGFP/cre)1Amc/J] with ROSAmT/mG mice [JAX007676; B6.129(Cg)‐Gt(ROSA)26Sortm4(ACTB‐tdTomato,‐EGFP)Luo/J] to generate Six2‐eGFP/Rosa26mT mice for embryonic kidney culturing.
Cell culture
Inner medullary collecting duct (IMCD) cells were kindly provided by the Department of Physiology and Pathophysiology, School of Basic Medical Sciences, Dalian Medical University. Human 293T cells were purchased from the ATCC (Manassas, VA, USA). The IMCD cells were cultured in DMEM/F12 (Gibco, Thermo Fisher Scientific, Ann Arbor, MI, USA) containing 10% FBS (Sigma, St Louis, MO, USA) and 1% penicillin–streptomycin (Gibco), incubated at 37 °C in a humidified atmosphere containing 5% CO2, and routinely passaged every 2 or 3 days. Human 293T cells were cultured in DMEM (Gibco) containing 10% FBS (Sigma) and 1% penicillin–streptomycin stock solution (Gibco) and were routinely passaged every 3 or 4 days.
Small interfering (si) RNA and plasmid transfection
siRNA‐ADAM10 and siRNA‐ADAM17 were obtained from Hanbio Biotechnology (Shanghai, PR China). pcDNA3‐ADAM10‐HA was a gift from Axel Ullrich (Addgene plasmid # 65106; http://n2t.net/addgene:65106; RRID: Addgene_65 106) [32]. siRNA was transfected using RNAiMAX (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's recommendations. The plasmid was transfected using PEI (Sigma) following the manufacturer's recommendations. The sequences of target genes were as follows:
siRNA‐ADAM10‐1: 5′‐CAGCAGAGAGAUAUAUUAAdTd‐3′;
siRNA‐ADAM10‐2: 5′‐CGCAUAAGAAUCAAUACAAdTd‐3′;
siRNA‐ADAM17‐1: 5′‐CCAUGAAGAACACGUGUAAdTdT‐3′;
siRNA‐ADAM17‐2: 5′‐GGAACUCUUGGAUUAGCUUdTdT‐3′.
Animal samples
The Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University approved the experimental protocols. All experiments were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The mice were maintained at the Zhejiang University according to animal care regulations. ICR mice, aged 8 weeks, were bred using timed mating; the pregnant mice were randomly divided into two groups. CPF (96% purity) was dissolved in dimethyl sulphoxide (DMSO), and 5 mg/kg CPF was injected subcutaneously daily into the mice of the CPF group during gestation days 7.5–11.5 [33, 34]. The controls were simultaneously injected with DMSO. Isolation of the mouse embryonic kidneys was performed following the Cold Spring Harbor Laboratory protocol [35].
RNA sequencing
Total RNA was extracted from samples of different embryonic days of both the CPF‐treated and the control embryonic kidneys using a miRNeasy Mini Kit (Qiagen, Thermo Fisher Scientific). A total amount of 3 μg of RNA was used as input material for the RNA sample preparations. Sequencing libraries were generated using the rRNA‐depleted RNA by NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, New England Biolabs, Beverly, MA, USA) following the manufacturer's recommendations. The libraries were sequenced on an Illumina Hiseq 2500 platform and 125 bp paired‐end reads were generated. The cleaned reads were mapped to the mouse genome mm10 using STAR [36]. Gene expression was quantified, and differentially expressed genes (DEGs) were identified by Cuffdiff 2, with p < 0.05 between two groups [37]. Gene set enrichment analysis (GSEA) was performed as described previously [38]. The RNA‐seq data sets have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession number GSE131263.
Oligo GEArray assay
Total RNA was extracted from the samples of different embryonic days of untreated mouse embryonic kidneys using Trizol (Thermo Fisher Scientific). cRNA labelling and synthesis were performed using a TrueLabeling‐AMP™ Linear RNA Amplification Kit (SuperArray Bioscience, Frederick, MD, USA). Hybridisation was performed with Oligo GEArray (SuperArray Bioscience) and GEAhyb Hybridisation Solution (SuperArray Bioscience) in disposable hybridisation tubes. Gene expression was analysed using the GEArray Expression Analysis Suite (SuperArray Bioscience).
Histology and immunostaining
Kidney tissues fixed in formalin and embedded in paraffin wax were subjected to haematoxylin and eosin staining, Masson's trichrome staining, and immunohistochemistry (IHC). Kidney tissues fixed with 4% paraformaldehyde were subjected to immunofluorescence (IF) staining as described previously [19]. Images were acquired using a Leica DM4000 microscope (Leica, Frankfurt, Germany), Leica TCS‐SP8 confocal microscope, or a Zeiss AxioCam MRm digital camera (Zeiss, Oberkochen, Germany). Z‐scan fluorescence images were acquired using a Zeiss LSM 880 with AiryScan and analysed with Imaris 9.3.1 software (Oxford Instruments Imaris, Abingdon, UK). The antibodies used were SIX2 (11562‐1‐AP, 1:200; Proteintech, Rosemont, IL, USA); AQP1 (ab168387, 1:400), ADAM10 (ab1997, 1:200), ADAM17 (ab57484, 1:200) and presenilin‐1 (ab15458, 1:200) (all from Abcam, Cambridge, MA, USA); AQP2 (NB110‐74682, 1:200; Novus Biologicals, Centennial, CO, USA); WT1 (sc‐192, 1:200; Santa Cruz Biotechnology, Dallas, TX, USA); podocalyxin‐like (MAB1556, 1:300; R&D Systems, Minneapolis, MN, USA); E‐cadherin (#3195, 1:200), vimentin (#5741, 1:100), α‐SMA (#19245, 1:200), NOTCH2 (#5732, 1:1600), NOTCH1 (#3608, 1:400), and Hes1 (#11988, 1:200) ( all from Cell Signaling Technology, Danvers, MA, USA); cytokeratin (C2562, 1:400; Sigma); and Krt8 (TROMA‐I‐s, 1:200) and NCAM1 (5B8‐s, 1:200) (both from Developmental Studies Hybridoma Bank, Iowa City, IA, USA). Biotinylated Lotus tetragonolobus lectin was from Vector Laboratories, Burlingame, CA, USA (B1325, 1:400). Isotype‐matched antibodies were used as negative controls.
Reverse transcription‐quantitative PCR (RT‐qPCR)
RNA was extracted using Trizol (Thermo Fisher Scientific). Total RNA (1 μg) was reverse‐transcribed into cDNA using the PrimeScript™ RT Reagent Kit (Takara, Dalian, PR China). Quantitative PCR was performed using a CFX96 TouchTM System (Bio‐Rad, Hercules, CA, USA) using iQTM SYBR Green Supermix (Bio‐Rad). The oligonucleotide primers are listed in supplementary material, Table S4. Relative mRNA levels were normalised to Gapdh.
Western blotting
Kidneys were lysed in RIPA Lysis Buffer (Beyotime, Shanghai, PR China) with a proteinase inhibitor cocktail, and SDS‐PAGE and western blotting were performed. Results were quantified with ImageJ (National Institutes of Health, Bethesda, MD, USA). Antibodies were used to detect SIX2, NOTCH2, NOTCH1, AQP1, ADAM10, ADAM17, vimentin, E‐cadherin, AQP2, podocalyxin‐like, Hes1, ephrin B2 (ab131536; Abcam), ERK1/2 (#4695; Cell Signaling Technology), phospho ERK1/2 (#4370; Cell Signaling Technology), α‐tubulin (T9026; Sigma), and β‐actin (sc‐8432; Santa Cruz Biotechnology). All primary antibodies were diluted 1:1000.
Adam10 morpholino design
The Adam10 morpholino (MO) antisense oligos were designed by Gene Tools (Philomath, OR, USA). The 151 bases of exon 2 of Adam10 was skipped. The Adam10‐MO sequence is 5'‐CTGAGAGCAATGAATTTACTCACCT‐3′. The Adam10‐MO and standard control MO were added to kidney culture medium at a final concentration of 7.5 and 10 μm, respectively. Adam10 knockout efficiency was confirmed by PCR and western blotting.
Kidney explants
Isolated E12.5 embryonic kidneys were temporarily placed in pre‐cooled PBS and then transferred to 24‐mm diameter Transwell plates with 0.4‐μm pore polyester membrane inserts (Corning, Corning, NY, USA). They were then transferred to Transwell filters in 12‐well cell culture plates (Corning) with 10% FBS, 1% penicillin/streptomycin stock solution, and 1% l‐glutamine in D‐MEM/F12 medium (one filter per well; 420 μl of medium per well). The embryonic kidneys were cultured at the air–fluid interface for 72 h in a 37 °C incubator with 5% CO2. Images were acquired using an inverted Carl Zeiss LSM 710 ConfoCor 3 confocal microscope every 24 h.
Serum analysis
Mice were anaesthetised and blood samples were collected from the orbital sinus. Blood was centrifuged and serum was isolated. Serum creatinine (Scr) and blood urea nitrogen (BUN) were analysed using FUJI DRI‐CHEM CRE and BUN slides in a FUJI DRI‐CHEM 7000i biochemistry analyser (Fujifilm, Tokyo, Japan).
Cell proliferation and migration
Cell indexes of proliferation and migration were obtained using a Real‐Time Cell Analyzer (RTCA)‐DP xCELLigence system (Roche Applied Science, Basel, Switzerland) following the manufacturer's recommendations. Five thousand cells per well were added to an E‐plate 16 (Roche) for cell proliferation detection. Five thousand cells per well were added to a MIC‐plate 16 (Roche) for cell migration detection. Cells were cultured for 50 h for both experiments.
Statistical analyses
Results are displayed as means ± SD. Statistical analysis using two‐tailed t‐tests and correlation analysis using Pearson's correlation coefficient were performed with GraphPad Prism 6 (GraphPad Inc, San Diego, CA, USA). The χ2 test was performed using SPSS 23.0 (IBM, Armonk, NY, USA), and all parameters except fibrosis were categorised by their medians. p < 0.05 was considered statistically significant. Regression analysis was performed with SPSS 23.0, and the degree of renal fibrosis was categorised into two groups (< 25% and ≥ 25%) according to the Banff Classification of IgAN.
Results
Prenatal CPF‐exposed mouse embryonic kidneys exhibit the ectopic formation of PTs and Notch receptor overexpression
The prenatal CPF exposure mouse model was established as previously reported (Figure 1A) [31]. CPF exposure has no effect on kidney function in pregnant mice (supplementary material, Figure S1A–C). More proximal tubule structures were found in the CPF group at E18.5 (Figure 1B,C). However, the kidney to body weight ratio showed no difference between the groups (Figure 1D).
Figure 1.
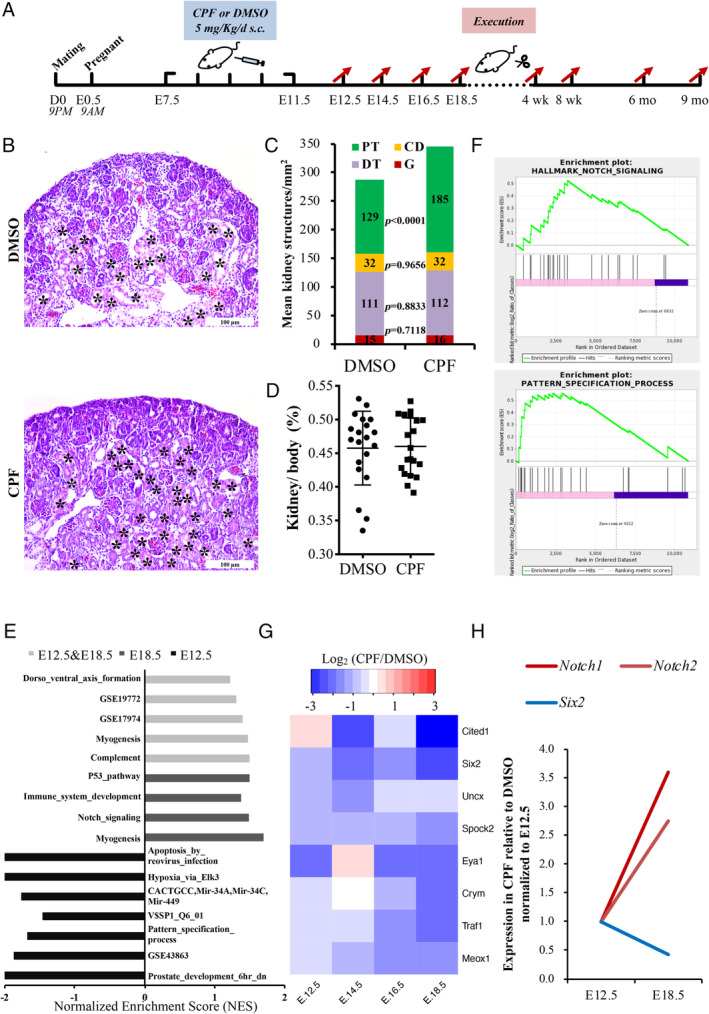
Prenatal CPF‐exposed embryonic kidneys exhibit ectopic PT formation. (A) Flow chart of the prenatal CPF exposure mouse model. (B) H&E stains of kidneys sectioned at E18.5. Asterisks indicate PTs. Scale bars: 100 μm. (C) The numbers of proximal tubules (PT), distal tubules (DT), collecting ducts (CD), and glomeruli (G) were quantified from H&E results. p < 0.0001; n = 6. (D) Comparison of kidney weight to body weight ratio at E18.5 (n = 6). (E) Gene set enrichment analysis (GSEA) of Notch‐related gene sets in RNA‐sequencing data of E12.5 and E18.5. Gene sets of p < 0.01, FDR q < 0.25 and |NES| > 1 were identified as meaningful gene sets. (F) Representative images of GSEA. (G) Heatmap of NPC genes in RNA‐sequencing data. Log2(CPF/DMSO) values are shown. (H) Trends of fold‐change (CPF/DMSO) of Notch1, Notch2, and Six2 from E12.5 to E18.5. CPF, chlorpyrifos; D, DMSO; C, CPF; TPM, transcripts per million; DT, distal tubule; mo, months; NES, normalised enrichment score.
Next, we used bulk tissue RNA‐sequencing to explore the transcriptomic changes of prenatal CPF‐exposed embryonic kidneys. CPF‐treated embryonic kidneys and DMSO‐treated embryonic kidneys at four developmental time points were sequenced and analysed (supplementary material, Figure S1D). GSEA focusing on developmental signalling pathways indicated that development‐related gene sets were mainly enriched in E12.5 and E18.5 samples (data not shown). Further analysis of E12.5 and E18.5 data revealed that gene sets containing Notch pathway genes were upregulated in the CPF group of E18.5 data and integrated data of E12.5 and E18.5, respectively (Figure 1E,F), especially the Notch receptors Notch1and Notch2. The role of differentially expressed genes (DEGs) in significantly enriched gene ontology functions and processes is shown in supplementary material, Figure S1E. Additionally, nephron progenitor cell (NPC) markers showed a reduction in the CPF group (Figure 1G) [39]; among them, Six2 and Eya1 were DEGs (supplementary material, Table S1). Six2 is a recognised marker of NPCs, and the fold‐change (CPF/DMSO) trends from E12.5 to E18.5 revealed a negative correlation between Six2, and Notch 1 and Notch 2 expression (Figure 1H).
Prenatal CPF exposure has a long‐lasting effect on mouse kidneys
To study the effects of prenatal CPF exposure on kidney development, we performed an in‐depth analysis of changes in renal phenotype. RT‐qPCR results of E18.5 kidneys were consistent with RNA sequencing. NPC markers were significantly decreased and cleaved Notch1 and Notch2 were significantly increased (Figure 2A,B). The protein levels of Notch1, Notch2, and Six2 were consistent with their mRNA levels (Figure 2C,D).
Figure 2.
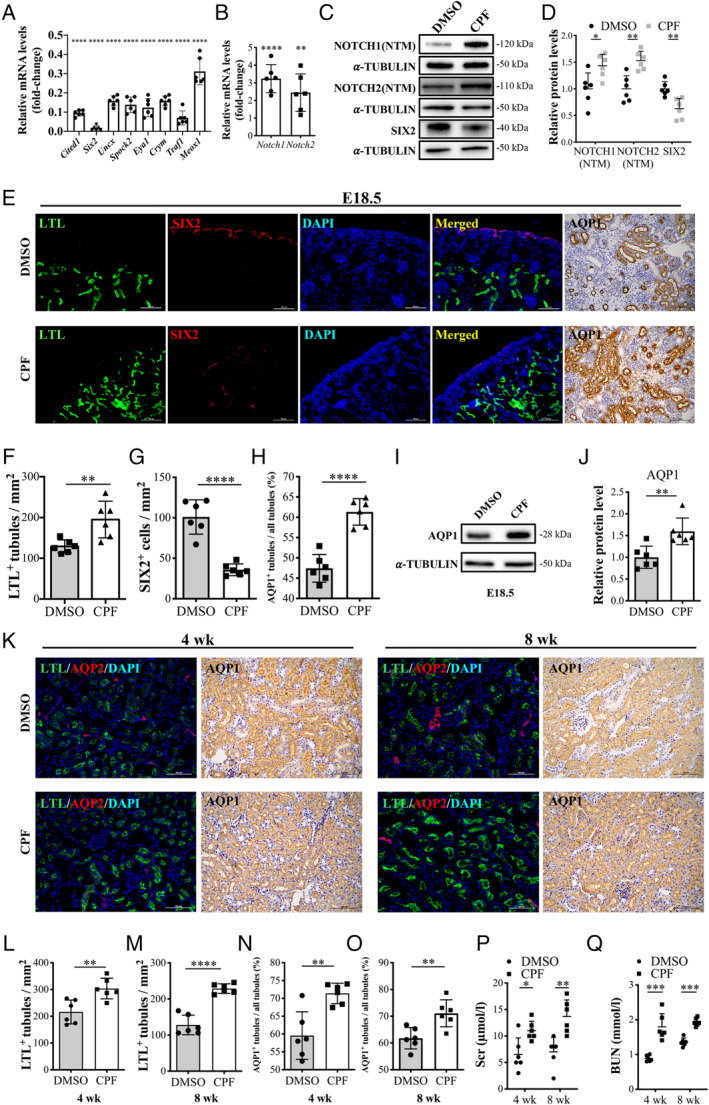
Overexpression of Notch1 and Notch2 in the CPF group results in ectopic formation of PTs. (A) mRNA levels of NPC genes and (B) Notch1 and Notch2. n = 6. (C, D) Representative immunoblot images and quantification analysis of Notch1, Notch2, and Six2 of E18.5 kidneys. n = 6. (E) E18.5 kidney fluorescence detection of Lotus tetragonolobus lectin (LTL) binding (green) and Six2 (red), and immunohistochemistry for Aqp1. Scale bars: 100 μm. (F–H) Quantification of the LTL+ tubule numbers, Six2+ cell numbers, and Aqp1+ tubule numbers (n = 6). (I, J) The protein level of Aqp1 in E18.5 kidneys. n = 6. (K) Four‐ and 8‐week immunofluorescence detection of LTL binding (green) and Aqp2 (red), and immunohistochemistry for Aqp1. Scale bars: 100 μm. (L–O) Quantification of LTL+ tubule numbers and Aqp1+ tubule numbers in 4‐ and 8‐week kidneys for the two groups (n = 6). (P, Q) Serum creatinine and blood urea nitrogen levels of 4‐ and 8‐week mice of the CPF group compared with DMSO controls. Mean ± SD from three to five experiments. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
Fluorescence staining using the proximal tubule marker Lotus tetragonolobus lectin (LTL) and immunofluorescence for SIX2 in E18.5 kidneys confirmed increased PTs and decreased SIX2+ NPCs in the CPF group (Figure 2E–G). GSEA of aquaporins revealed that aquaporin 1 (Aqp1), another proximal tubule marker, had the most obvious enrichment in the CPF group (supplementary material, Figure S1F,G). IHC staining against AQP1 showed a similar change (Figure 2E,H). Protein levels of AQP1 were also significantly increased in the CPF group (Figure 2I,J). Cell markers for other kidney cell types, E‐cadherin (CDH1) for distal tubules, aquaporin 2 (AQP2) for collecting ducts, and podocalyxin‐like (PODXL) for podocytes, showed no differences between the groups (supplementary material, Figure S2A,B).
Additionally, we found a similar phenotypic change for 4‐ and 8‐week kidneys. Expression of NOTCH1, NOTCH2, and AQP1 was increased in the CPF group (supplementary material, Figure S2C–E). Fluorescence staining with LTL revealed increased LTL+ PTs in the CPF group (Figure 2K–M). IHC staining for AQP1 also showed an increase in the CPF group (Figure 2K,N,O). Scr and BUN were significantly increased in CPF mice at 4 and 8 weeks (Figure 2P,Q). The kidney to body weight ratio and kidney size showed no differences between the groups (supplementary material, Figure S2F,G). Taken together, these data show that prenatal CPF exposure has a long‐lasting effect on mouse kidney structures.
ADAM10 is indispensable for embryonic kidney development and regulates the expression of Notch1 and Notch2
To understand the mechanism underlying Notch1 and Notch2 activation, we performed Notch signalling GeneChip analysis of normal embryonic kidneys. DEGs were screened based on fold‐change (fold‐difference; FD) analysis. The average expression values of each group were extracted and compared with E12.5; a gene would be defined as a DEG if any of its three FD values were greater than 1.5 or less than 0.67 (supplementary material, Table S2). Notch1 and Notch2 were both DEGs. Adam10, which cleaves the Notch ligands at the S2 site and produces the NICD to activate Notch signalling [15], had the same expression trend with Notch1 and Notch2 (Figure 3A). The mRNA and protein levels of Adam10 were also increased in the CPF group (Figure 3B–D).
Figure 3.
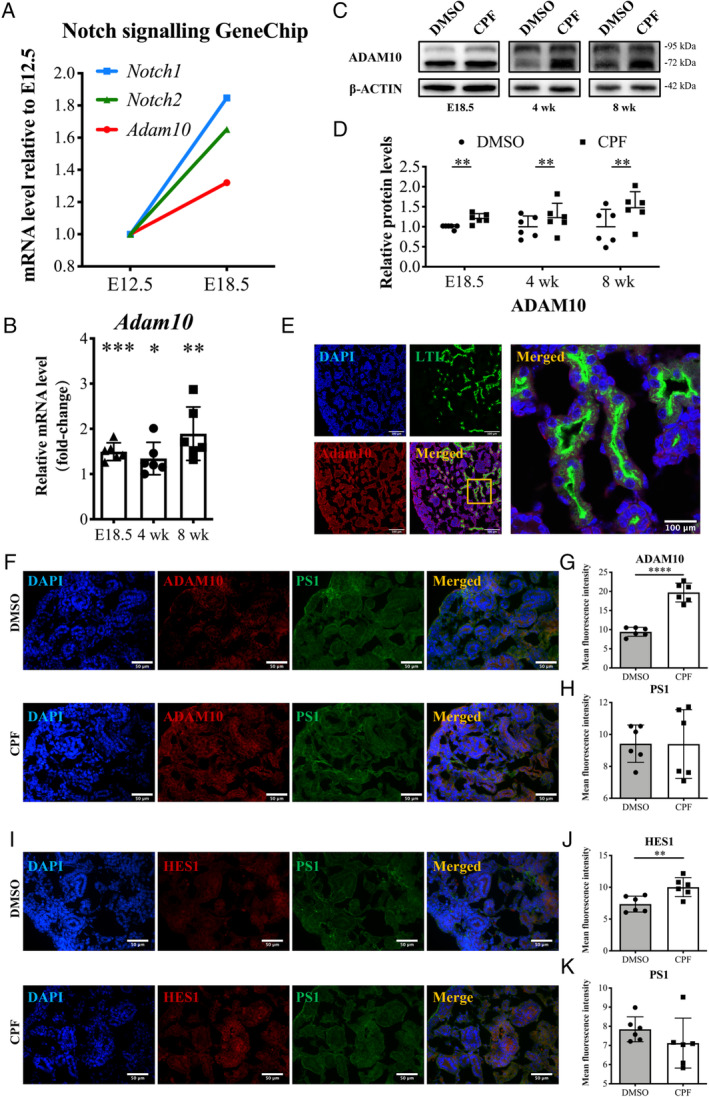
ADAM10 is upregulated in the kidneys of CPF‐exposed mice. (A) Relative expression trends of Notch1, Notch2, and Adam10 in GeneChip data from E12.5 to E18.5 normal embryonic kidneys. (B) Comparision of the mRNA levels of Adam10 in E18.5 and 4‐ and 8‐week kidneys between the two groups (n = 6). (C, D) Effect of prenatal CPF exposure on the protein levels of ADAM10 in E18.5 and 4‐ and 8‐week kidneys. n = 6. (E) E18.5 immunofluorescence staining with antibodies against LTL (green) and ADAM10 (red). Scale bars: 100 μm. (F) Co‐staining for ADAM10 (red) and presenilin‐1 (PS1) (green) at E18.5 kidneys. Quantification data are shown in panels G and H. n = 6. Scale bars: 50 μm. (I) Co‐staining for HES1 (red) and PS1 (green) at E18.5 kidneys. Quantification data are shown in panels J and K. n = 6. Scale bars: 50 μm. Mean ± SD from three to five experiments. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
IF was performed to observe the localisation of ADAM10 in normal E18.5 kidneys. Fluorescence detection of ADAM10 and kidney cell markers indicated its extensive expression in the basal lateral membranes of PTs (LTL), metanephric mesenchyme and its derivatives (NCAM1), ureteric bud and ureters (KTR8), and podocytes (WT1) (Figure 3E and supplementary material, Figure S3A). Gamma‐secretase is an alternative pathway controlling the Notch cascade [40]. Dual‐staining for ADAM10 and the gamma‐secretase component presenilin‐1 (PS1) showed that ADAM10 expression was upregulated in the CPF group, while there was no difference in PS1 expression between the groups (Figure 3F–H). Dual‐staining for the Notch effectors HES1 and PS1 showed similar results (Figure 3I–K). Protein levels of PS1 showed no difference between the groups (supplementary material, Figure S3B). Therefore, the Notch activity was specifically affected by Adam10 in our model.
To determine the role of ADAM10 in kidney development, we performed a morpholino (MO) experiment against Adam10 in Six2‐eGFP/Rosa26mT kidney explants at E12.5. IMCD cells treated with Adam10‐MO confirmed sufficient knockdown (Figure 4A–C). After in vitro culture for 72 h in the presence or absence of Adam10, branching morphogenesis was obviously hindered in Adam10‐MO‐treated embryonic kidneys compared with controls (Figure 4D and supplementary material, Figure S3C,D). These results indicate that ADAM10 is essential for kidney development. Additionally, normal E12.5 kidneys were separated and cultured in vitro for 72 h in the presence or absence of Adam10‐MO. Compared with the control group, the Adam10‐MO group showed a significant developmental constraint and decrease in Notch activity; cleaved NOTCH1 (N1ICD), NOTCH2 (N2ICD), and HES1 were all inhibited. Reduced ADAM10 also resulted in a reduction of SIX2+ NPCs and LTL+ PTs, indicating that Adam10 is indispensable for proximal tubule development (Figure 4E and supplementary material, Figure S4).
Figure 4.

Adam10 knockdown causes abnormal ex vivo cultured embryonic kidney development. (A) Schematic diagram of the Adam10 morpholino (MO) design. (B, C) Adam10‐MO knockdown efficiency was confirmed by PCR and western blotting using IMCD cells. (D) Whole‐mount fluorescence microscope images of ex vivo cultured E12.5 Six2‐eGFP/Rosa26mT mice kidneys treated with Adam10‐MO (right) or not (left). Scale bars: 200 μm. (E) Whole‐mount fluorescence microscope images of ex vivo cultured E12.5 normal mice kidneys treated with Adam10‐MO (right) or not (left). The expression of SIX2, LTL, N1ICD, N2ICD, and HES1 was detected. Scale bars of other columns: 100 μm.
Prenatal CPF‐exposed mouse kidneys exhibit aggravated renal fibrosis
To assess the long‐term effects of prenatal CPF exposure on renal function, we investigated kidneys at 6 and 9 months. NOTCH1, NOTCH2, AQP1, and ADAM10 remained upregulated in the CPF group (supplementary material, Figure S5A–D). Renal fibrosis was assessed using Masson's trichrome staining and the CPF kidneys showed significantly increased fibrosis (Figure 5A–C). The mesenchymal marker vimentin (VIM) was evaluated and showed an increase in the CPF group (Figure 5D–F). Fluorescence detection of LTL binding and α‐SMA showed that more proximal tubule cells expressed α‐SMA in the CPF group (Figure 5G,H and supplementary material, Figure S5E,F). Fluorescence detection of LTL and α‐SMA in 4‐ and 8‐week kidneys showed no significant differences between the groups; however, slight fibrosis was observed in 8‐week kidneys (supplementary material, Figure S5G–J). We also detected the protein level of the epithelial cell marker CDH1, and the results showed no difference between the groups (Figure 5I,J and supplementary material, Figure S5K,L). Thus, overexpression of the Adam10/Notch axis in CPF‐exposed mice can promote renal fibrosis.
Figure 5.

Elevation of Adam10 and Notch2 promotes renal fibrosis in the CPF group. (A) Masson trichrome staining of kidneys sectioned at 6 and 9 months. Quantification is shown in panels B and C. n = 6. Scale bars: 50 μm. (D) 6 and 9 months immunostaining for VIM. Quantification is shown in panels E and F. n = 6. Scale bars: 50 μm. (G) Six‐month kidneys: fluorescence detection of LTL binding (green) and α‐SMA (red), and immunohistochemistry for α‐SMA. Scale bars: 100 μm. (H) Nine‐month kidneys: fluorescence detection of LTL binding (green) and α‐SMA (red) and immunostaining for α‐SMA. Scale bars: 50 μm. (I, J) Protein levels of the epithelial cell marker Cdh1 in 6‐ and 9‐month kidneys (n = 6). Mean ± SD from three to five experiments. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
ADAM10 regulates Notch activity and promotes cell proliferation and migration in 293T cells
Several other ADAM10 substrates are reportedly involved in fibrosis, such as ERK and ephrin‐B2 [30, 41]. Ephrin‐B2 expression was too low to detect (data not shown) and the phosphorylation level of Erk was not affected in E18.5 kidneys (supplementary material, Figure S6A), suggesting that ADAM10 specifically regulated Notch activity in our mouse model.
Overexpression of ADAM10 in 293T cells led to high expression of cleaved NOTCH1, cleaved NOTCH2, and HES1 (Figure 6A,B). We analysed the nuclear expression ratio of NOTCH1 and NOTCH2, and the results showed that compared with the normal control, the pc‐DNA‐ADAM10‐HA group had a significant increase in nuclear expression of NOTCH1 and NOTCH2 (Figure 6C–E). Surprisingly, knocking down ADAM10 alone did not reduce the nuclear expression of NOTCH1 or NOTCH2 (Figure 6D,E and supplementary material, Figure S6B,C). Considering that ADAM17 also regulates Notch activity [42], we speculated that ADAM10 and ADAM17 have complementary effects on downstream Notch activation. Only when both ADAM10 and ADAM17 were inhibited did the nuclear expression of NOTCH1 and NOTCH2 decrease (Figure 6C–E). Expression of the Notch effector HES1 showed similar results (supplementary material, Figure S6D).
Figure 6.
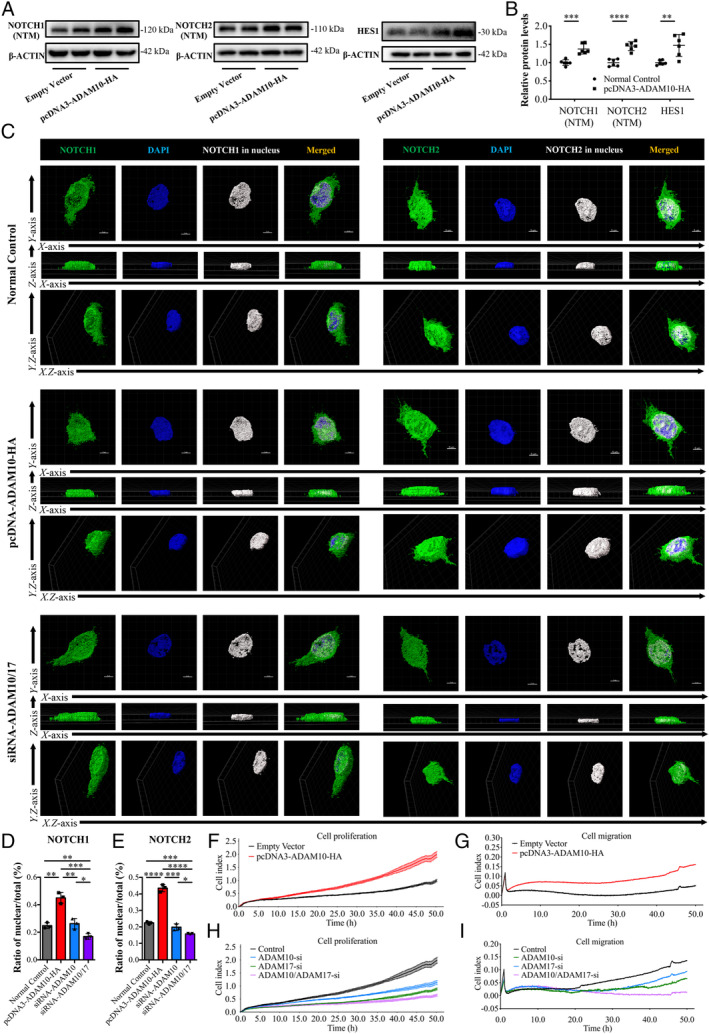
ADAM10 regulates Notch expression and promotes cell proliferation and migration in human 293T cells. (A, B) Effect of pcDNA3‐ADAM10‐HA plasmid on the protein levels of NOTCH1 (NTM), NOTCH2 (NTM), and HES1 (n = 6). (C–E) Immunofluorescence staining for NOTCH1 and NOTCH2 in 293T cells. The ratio of nuclear expression to total expression was analyzed using Imaris software (n = 3). Scale bars: 5 μm. (F, G) Effect of pcDNA3‐ADAM10‐HA plasmid on the cell proliferation index and migration index of 293T cells. (H, I) Effect of siRNA‐ADAM10 and siRNA‐ADAM17 on cell proliferation and migration of 293T cells.
Overexpression of ADAM10 promoted the proliferation and migration of 293T cells; knocking down ADAM10 or ADAM17 alone partially inhibited proliferation and migration, while simultaneous knockdown of ADAM10 and ADAM17 significantly inhibited proliferation and migration (Figure 6F–I). Overexpression of ADAM10 reduced CDH1 expression and increased α‐SMA expression compared with the control; knockdown of ADAM10 alone made no difference. Knockdown of both ADAM10 and ADAM17 increased CDH1 expression and reduced α‐SMA expression (supplementary material, Figure S6E–H). IHC staining showed that ADAM17 expression was not significantly different between the groups of E18.5, 4 and 8 weeks, and 6 and 9 months, although there was a trend for an increase of ADAM17 in the CPF group (supplementary material, Figure S7A,B). Protein levels of ADAM17 also showed no differences between the groups of 6‐ and 9‐month kidneys (supplementary material, Figure S7C,D). These results suggest that ADAM10 regulated the Notch activity and promotes cell proliferation and migration in 293T cells, and that ADAM10 and ADAM17 may have complementary effects on downstream Notch activation. The overexpression and knockdown were confirmed by PCR, western blotting, and IF (supplementary material, Figure S7E–J).
ADAM10 expression has a positive correlation with renal interstitial fibrosis and a negative correlation with renal function in kidney disease patients
To explore the role of ADAM10 in renal fibrosis, we studied renal biopsy specimens from patients with kidney disease. Eighty‐one patients with IgAN were included for correlational analysis. General information is shown in supplementary material, Table S3. Renal interstitial fibrosis was assessed using Masson's trichrome staining, and the degree of interstitial fibrosis was stratified according to the percentage of positive staining area to the total renal interstitial area (5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, > 50%). IHC staining was performed to evaluate ADAM10 and NOTCH2 expression. NOTCH1 expressed in human renal tissues showed no significant differences among different degrees of renal interstitial fibrosis (supplementary material, Figure S8A). As the degree of renal interstitial fibrosis increased, the expression of ADAM10 and NOTCH2 increased (supplementary material, Figure S8B). Fluorescence detection of LTL binding and α‐SMA revealed that a proximal tubule epithelial–myofibroblast transition appeared in IgAN patients with a renal interstitial fibrosis area greater than 50% (supplementary material, Figure S8B).
To determine the significance of ADAM10 and NOTCH2 expression on renal interstitial fibrosis, we performed a retrospective analysis. Eighty‐one renal biopsy samples of IgAN were immunostained for ADAM10, and 61 of the 81 samples were also stained for NOTCH2. Chi‐square tests were used to assess associations between ADAM10 expression or NOTCH2 expression and clinicopathological parameters. All parameters except fibrosis were categorised by their medians. ADAM10 expression had an association with NOTCH2 expression in IgAN patients (Table 1). ADAM10 expression was significantly associated with Scr (χ2 = 7.711, p = 0.005), BUN (χ2 = 7.711, p = 0.005), cystatin C (χ2 = 4.464, p = 0.035), eGFR (χ2 = 9.011, p = 0.003), and fibrosis (χ2 = 24.994, p < 0.001) (Table 2). NOTCH2 expression was significantly associated with fibrosis (χ2 = 8.658, p = 0.003) (Table 3). Correlational analysis showed the same results; ADAM10 expression had a positive correlation with NOTCH2 expression (r = 0.5267, p < 0.0001). Both ADAM10 and NOTCH2 expression had positive correlations with fibrosis (r = 0.8523, p < 0.0001; r = 0.5546, p < 0.0001). ADAM10 expression also had a positive correlation with Scr (r = 0.4561, p < 0.0001), cystatin C (r = 04992, p < 0.0001), and BUN (r = 04579, p < 0.0001), and a negative correlation with eGFR (r = −0.5327, p < 0.0001) (supplementary material, Figure S8C–I). Univariable regression analysis indicated that ADAM10 expression, Scr, BUN, eGFR, and cystatin C were associated with renal fibrosis in IgAN. Further multivariate analysis showed that ADAM10 expression and eGFR were risk factors (Table 4). Overall, our data indicate that both ADAM10 and NOTCH2 expression are important in renal interstitial fibrosis, and that ADAM10 may play a more significant role in fibrosis and kidney function. Additionally, ADAM10 and NOTCH2 expression was found to increase as the degree of fibrosis increased in various renal diseases, such as mesangial proliferative glomerulonephritis, minimal‐change disease, lupus nephritis, and membranous nephropathy, and even in donor kidneys with fibrosis (supplementary material, Figure S9A–D).
Table 1.
Correlation of ADAM10 expression with NOTCH2 expression on renal biopsy specimens from patients with the pathological diagnosis of IgA nephropathy (IgAN).
| NOTCH2 expression | n | ADAM10 expression | χ2 value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Low | 30 | 22 | 8 | 6.146 | 0.013 |
| High | 31 | 13 | 18 | ||
p < 0.05 was considered to have statistical significance. Values in bold are statistically significant.
Table 2.
Correlation of ADAM10 expression on renal biopsy specimens with major clinicopathological parameters in patients with the pathological diagnosis of IgA nephropathy (IgAN).
| Clinicopathological parameters | n | ADAM10 expression | χ2 value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | |||||
| < 41 | 39 | 19 | 20 | 0.013 | 0.908 |
| ≥ 41 | 42 | 21 | 21 | ||
| Gender | |||||
| Male | 42 | 22 | 20 | 0.314 | 0.575 |
| Female | 39 | 18 | 21 | ||
| Baseline systolic blood pressure (mmHg) | |||||
| < 133 | 40 | 21 | 19 | 0.307 | 0.579 |
| ≥ 133 | 41 | 19 | 22 | ||
| Baseline diastolic blood pressure (mmHg) | |||||
| < 82 | 37 | 19 | 18 | 0.595 | 0.441 |
| ≥ 82 | 44 | 21 | 23 | ||
| Baseline GLU (mm) | |||||
| < 4.41 | 40 | 21 | 19 | 0.307 | 0.579 |
| ≥ 4.41 | 41 | 19 | 22 | ||
| Baseline TC (mm) | |||||
| < 4.23 | 38 | 17 | 21 | 0.618 | 0.432 |
| ≥ 4.23 | 43 | 23 | 20 | ||
| Baseline TG (mm) | |||||
| < 1.47 | 40 | 23 | 17 | 2.083 | 0.149 |
| ≥ 1.47 | 41 | 17 | 24 | ||
| Baseline Scr (μm) | |||||
| < 104 | 40 | 26 | 14 | 7.711 | 0.005 |
| ≥ 104 | 41 | 14 | 27 | ||
| Baseline BUN (mm) | |||||
| < 6.65 | 40 | 26 | 14 | 7.711 | 0.005 |
| ≥ 6.65 | 41 | 14 | 27 | ||
| Baseline cystatin C (mg/l) | |||||
| < 1.39 | 41 | 25 | 16 | 4.464 | 0.035 |
| ≥ 1.39 | 40 | 15 | 25 | ||
| Baseline eGFR (EPI‐Cr) (ml/min) | |||||
| < 68.61 | 40 | 13 | 27 | 9.011 | 0.003 |
| ≥ 68.61 | 41 | 27 | 14 | ||
| Baseline urinary protein (g/l) | |||||
| < 0.87 | 40 | 23 | 17 | 2.083 | 0.149 |
| ≥ 0.87 | 41 | 17 | 24 | ||
| Fibrosis (%) | |||||
| < 25 | 40 | 31 | 9 | 24.994 | < 0.001 |
| ≥ 25 | 41 | 9 | 32 | ||
p < 0.05 was considered to have statistical significance. Values in bold are statistically significant. GLU, glucose; TC, total cholesterol; TG, triglyceride; Scr, serum creatinine; BUN, blood urea nitrogen; eGFR, estimated glomerular filtration rate.
Table 3.
Correlation of NOTCH2 expression on renal biopsy specimens with major clinicopathological parameters in patients with the pathological diagnosis of IgA nephropathy (IgAN).
| Clinicopathological parameters | n | NOTCH2 expression | χ2 value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | |||||
| < 41 | 28 | 13 | 15 | 0.157 | 0.692 |
| ≥ 41 | 33 | 17 | 16 | ||
| Gender | |||||
| Male | 33 | 14 | 14 | 0.014 | 0.906 |
| Female | 28 | 16 | 17 | ||
| Baseline systolic blood pressure (mmHg) | |||||
| < 139 | 38 | 18 | 20 | 0.132 | 0.716 |
| ≥ 139 | 23 | 12 | 11 | ||
| Baseline diastolic blood pressure (mmHg) | |||||
| < 83 | 28 | 15 | 13 | 0.399 | 0.527 |
| ≥ 83 | 33 | 15 | 18 | ||
| Baseline GLU (mm) | |||||
| < 4.37 | 30 | 16 | 14 | 0.407 | 0.523 |
| ≥ 4.37 | 31 | 14 | 17 | ||
| Baseline TC (mm) | |||||
| < 4.23 | 28 | 14 | 14 | 0.014 | 0.906 |
| ≥ 4.23 | 33 | 16 | 17 | ||
| Baseline TG (mm) | |||||
| < 1.44 | 30 | 17 | 13 | 1.324 | 0.250 |
| ≥ 1.44 | 31 | 13 | 18 | ||
| Baseline Scr (μm) | |||||
| < 101 | 29 | 16 | 13 | 0.794 | 0.373 |
| ≥ 101 | 32 | 14 | 18 | ||
| Baseline BUN (mm) | |||||
| < 6.39 | 30 | 18 | 12 | 2.765 | 0.096 |
| ≥ 6.39 | 31 | 12 | 19 | ||
| Baseline cystatin C (mg/l) | |||||
| < 1.37 | 30 | 17 | 13 | 1.324 | 0.250 |
| ≥ 1.37 | 31 | 13 | 18 | ||
| Baseline eGFR (EPI‐Cr) (ml/min) | |||||
| < 72.45 | 30 | 11 | 19 | 3.699 | 0.054 |
| ≥ 72.45 | 31 | 19 | 12 | ||
| Baseline urinary protein (g/l) | |||||
| < 0.85 | 30 | 15 | 15 | 0.016 | 0.900 |
| ≥ 0.85 | 31 | 15 | 15 | ||
| Fibrosis (%) | |||||
| < 25 | 32 | 22 | 10 | 8.658 | 0.003 |
| ≥ 25 | 29 | 9 | 20 | ||
p < 0.05 was considered to have statistical significance. Values in bold are statistically significant. GLU, glucose; TC, total cholesterol; TG, triglyceride; Scr, serum creatinine; BUN, blood urea nitrogen; eGFR, estimated glomerular filtration rate.
Table 4.
Logistic regression analysis of clinicopathological factors and expression levels of ADAM10 for renal fibrosis in IgAN patients.
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| ADAM10 expression | 12.247 | 4.94–30.36 | < 0.0001 | 10.799 | 3.782–30.838 | < 0.0001 |
| SBP (mmHg) | 1.91 | 0.888–4.107 | 0.098 | |||
| DBP (mmHg) | 2.119 | 0.983–4.567 | 0.055 | |||
| Scr (μm) | 9.3 | 3.875–22.317 | < 0.0001 | 2.625 | 0.533–12.926 | 0.236 |
| BUN (mm) | 7.19 | 3.077–16.801 | < 0.0001 | 0.953 | 0.192–4.728 | 0.953 |
| Urinary protein (g/l) | 1.91 | 0.888–4.107 | 0.098 | |||
| eGFR (EPI‐Cr) (ml/min) | 0.094 | 0.038–0.23 | < 0.0001 | 0.18 | 0.034–0.951 | 0.044 |
| Cystatin C (mg/l) | 5.639 | 2.467–12.891 | < 0.0001 | 0.903 | 0.226–3.607 | 0.885 |
| GLU (mm) | 1.28 | 0.6–2.729 | 0.523 | |||
| TC (mm) | 1.048 | 0.491–2.234 | 0.904 | |||
| TG (mm) | 1.562 | 0.73–3.341 | 0.251 | |||
p < 0.05 was considered to have statistical significance. Values in bold are statistically significant. HR, hazard ratio; CI, confidence interval.
Discussion
Here, we found that prenatal CPF exposure abnormally activates the ADAM10/Notch signalling axis in mouse embryonic kidneys, resulting in premature depletion of Six2+ NPCs and triggering the differentiation of PTs. Moreover, continuous overexpression of Adam10 and Notch2 had a long‐term effect on kidneys and promoted fibrosis in mature kidneys.
Early life adverse events such as undernutrition, ethanol consumption, drug use, and glucocorticoid exposure can cause permanent kidney changes, which is the concept of renal programming [43, 44, 45, 46]. Prenatal CPF exposure is associated with developmental neurotoxicity [47]. CPF has been shown to cause kidney injury, although little is known about its effects on kidney development prenatally. In our previous study, we reported weak staining for NOTCH2 in renal tubules when using IHC and concluded that the NOTCH2–JAGGED1 pathway was interrupted in prenatal CPF‐exposed mouse kidneys. However, the previous analysis relied on image analysis and semi‐quantitative methods only.
In this study, by use of more sophisticated and quantitative molecular approaches, we further investigated the role of the ADAM10/Notch axis in kidney development and kidney fibrosis. We found that there was an increase in the number of PTs in the CPF group, and no differences in other nephron structures, compared with the control. RNA sequencing was performed to explore changes in the transcriptome; GSEA indicated the enrichment and upregulation of gene sets containing Notch pathway genes in E18.5 kidneys of the CPF group, and NPC markers were reduced. Further experiments revealed that prenatal CPF‐exposed E18.5 kidneys exhibited ectopic formation of PTs by activating Notch1 and Notch2 to trigger the premature depletion of Six2+ NPCs. We then carried out these experiments on postnatal mouse kidneys. Our results indicated that the phenotypic change of ectopic PT formation caused by Notch1 and Notch2 overexpression was persistent and irreversible in mature kidneys. These findings verify that intrauterine adverse events can permanently affect the kidneys.
ADAM10 mediates the S2 site cleavage of Notch ligands [15, 31, 32, 33]. Notch GeneChip analysis of normal embryonic kidneys showed that Adam10 had the same expression trend from E12.5 to E18.5, compared with Notch1 and Notch2. Adam10 expression was increased in CPF kidneys. Fluorescence detection of ADAM10 and kidney cell markers indicated its extensive expression in embryonic kidneys, including the PTs. We conclude that the phenotypic changes related to overexpression of Notch1 and Notch2 were mediated by ADAM10. Self et al [48] reported that branching morphogenesis was obviously restrained in E12.5 Six2‐nullizygous mouse kidneys. However, Combes et al [49] found an increase in ureteric branching and the final nephron number in Six2 heterozygotes. The authors explained this as due to a dose response of nephron progenitors to the level of SIX2 protein. In our study, after treatment with Adam10‐MO, Six2 was hardly detectable in in vitro cultured embryonic kidneys. We believed that the expression level of Six2 in Adam10‐MO‐treated kidneys was close to that of Six2‐nullizygous mouse kidneys. Therefore, similar to Six2‐nullizygous kidneys [48], branching morphogenesis was also inhibited in Adam10‐MO‐treated kidneys.
Fibrosis is a typical histological manifestation of CKD [50], and Notch signalling is involved in kidney fibrosis. To observe the long‐term effect on kidneys of Notch activation in the prenatal CPF exposure mouse model, we investigated the renal fibrosis of 6‐ and 9‐month kidneys. Kidney fibrosis was aggravated in the CPF group. Both the mesenchymal marker vimentin and the myofibroblast marker α‐SMA were found to be increased. The protein levels of ADAM10, NOTCH1, and NOTCH2 remained increased in the CPF group at 6 and 9 months. Furthermore, ADAM10 and NOTCH2 expression increased with the degree of renal interstitial fibrosis in IgAN patients. Correlational analysis revealed that ADAM10 and NOTCH2 expression had a positive correlation to renal interstitial fibrosis, and that ADAM10 expression was negatively correlated to renal function, including Scr, cystatin C, and eGFR.
We observed increased expression of ADAM10 and NOTCH2 in other kidney diseases as renal interstitial fibrosis worsened. These data indicate that the overexpression of ADAM10 and NOTCH2 in renal interstitial fibrosis is a common phenomenon.
In summary, our study supports the notion that the ADAM10/Notch axis is activated in the prenatal CPF exposure mouse model and causes abnormal phenotypic changes in kidney development. The overexpression of Adam10 and Notch2 has a long‐term effect on the kidneys and promotes renal fibrosis. Additionally, ADAM10 may have a role in renal interstitial fibrosis in kidney disease patients.
Author contributions statement
BL and HJ designed the study. BL, CZ, LD, WX, SF, YW, SX, ChW, ClW, TZ, LT and SJ carried out experiments. LB and XW analysed the data. LB made the figures and drafted the paper. LD, HJ and JW revised the paper. JQ contributed to the analysis and interpretation of RNA‐sequencing data. CE supported the Adam10‐morpholino experiment and provided intellectual input to the project. AD provided intellectual input to the project. JC and HJ supported the whole project. All the authors approved the final version of the manuscript.
Supporting information
Figure S1. Prenatal CPF exposure mouse model has no effect on kidney function of pregnant mice, and Aqp1 is upregulated in the CPF group
Figure S2. Prenatal CPF exposure mouse model does not affect the development of other nephron structures
Figure S3. ADAM10 localisation and the effect of Adam10‐MO on kidney development
Figure S4. Adam10‐MO resulted in inhibition of kidney development in ex vivo E12.5 kidneys of normal ICR mice
Figure S5. There was no obvious epithelial‐to‐mesenchymal transition in 4‐ or 8‐week kidneys
Figure S6. Effects of overexpression and knockdown of ADAM10 in 293T cells
Figure S7. Verification of the overexpression and knockdown efficiency in 293T cells
Figure S8. The degree of renal interstitial fibrosis is positively correlated with ADAM10 and NOTCH2 expression in patients with IgANs
Figure S9. ADAM10 and NOTCH2 expression both increase as the degree of renal interstitial fibrosis increases in various renal biopsy tissues
Table S1. Detailed information of differentially expressed genes between the two groups
Table S2. Detailed information of differentially expressed genes in the Notch signalling pathway (GeneChip data)
Table S3. General information for the 81 IgAN patients
Table S4. Primers used in this study
Acknowledgements
We would like to thank the Department of Pathology, Kidney Disease Center, the First Affiliated Hospital, College of Medicine, Zhejiang University for assistance throughout this project. Thanks are due for the technical support by the Core Facilities, Zhejiang University School of Medicine. This work was supported by the National Natural Science Foundation of China (Nos 81470938 and 81770697) to HJ, and the National Basic Research Program of China (973 Program) (No 2011CB944002) to JC.
No conflicts of interest were declared.
Contributor Information
Jianghua Chen, Email: chenjianghua@zju.edu.cn.
Hong Jiang, Email: jianghong961106@zju.edu.cn.
References
- 1. Eckardt KU, Coresh J, Devuyst O, et al Evolving importance of kidney disease: from subspecialty to global health burden. Lancet 2013; 382 : 158–169. [DOI] [PubMed] [Google Scholar]
- 2. Bello AK, Levin A, Tonelli M, et al Assessment of global kidney health care status. JAMA 2017; 317 : 1864–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jha V, Garcia‐Garcia G, Iseki K, et al Chronic kidney disease: global dimension and perspectives. Lancet 2013; 382 : 260–272. [DOI] [PubMed] [Google Scholar]
- 4. GBD 2015 Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet 2016; 388 : 1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luyckx VA, Bertram JF, Brenner BM, et al Effect of fetal and child health on kidney development and long‐term risk of hypertension and kidney disease. Lancet 2013; 382 : 273–283. [DOI] [PubMed] [Google Scholar]
- 6. Hanson M, Gluckman P. Developmental origins of noncommunicable disease: population and public health implications. Am J Clin Nutr 2011; 94 : 1754s–1758s. [DOI] [PubMed] [Google Scholar]
- 7. Paixão AD, Alexander BT. How the kidney is impacted by the perinatal maternal environment to develop hypertension. Biol Reprod 2013; 89 : 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luyckx VA, Brenner BM. Birth weight, malnutrition and kidney‐associated outcomes – a global concern. Nat Rev Nephrol 2015; 11 : 135–149. [DOI] [PubMed] [Google Scholar]
- 9. Xu X, Nie S, Ding H, et al Environmental pollution and kidney diseases. Nat Rev Nephrol 2018; 14 : 313–324. [DOI] [PubMed] [Google Scholar]
- 10. Mehta AJ, Zanobetti A, Bind MA, et al Long‐term exposure to ambient fine particulate matter and renal function in older men: the Veterans Administration Normative Aging Study. Environ Health Perspect 2016; 124 : 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bowe B, Xie Y, Li T, et al Particulate matter air pollution and the risk of incident CKD and progression to ESRD. J Am Soc Nephrol 2018; 29 : 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu W, Zhang K, Jiang S, et al Association of co‐exposure to heavy metals with renal function in a hypertensive population. Environ Int 2018; 112 : 198–206. [DOI] [PubMed] [Google Scholar]
- 13. Valcke M, Levasseur ME, Soares da Silva A, et al Pesticide exposures and chronic kidney disease of unknown etiology: an epidemiologic review. Environ Health 2017; 16 : 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanvir EM, Afroz R, Chowdhury M, et al A model of chlorpyrifos distribution and its biochemical effects on the liver and kidneys of rats. Hum Exp Toxicol 2016; 35 : 991–1004. [DOI] [PubMed] [Google Scholar]
- 15. Sirin Y, Susztak K. Notch in the kidney: development and disease. J Pathol 2012; 226 : 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCright B. Notch signaling in kidney development. Curr Opin Nephrol Hypertens 2003; 12 : 5–10. [DOI] [PubMed] [Google Scholar]
- 17. Desgrange A, Cereghini S. Nephron patterning: lessons from Xenopus, zebrafish, and mouse studies. Cell 2015; 4 : 483–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopan R, Chen S, Little M. Nephron progenitor cells: shifting the balance of self‐renewal and differentiation. Curr Top Dev Biol 2014; 107 : 293–331. [DOI] [PubMed] [Google Scholar]
- 19. Surendran K, Boyle S, Barak H, et al The contribution of Notch1 to nephron segmentation in the developing kidney is revealed in a sensitized Notch2 background and can be augmented by reducing Mint dosage. Dev Biol 2010; 337 : 386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng H, Kim M, Valerius M, et al Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development 2007; 134 : 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chung E, Deacon P, Park JS. Notch is required for the formation of all nephron segments and primes nephron progenitors for differentiation. Development 2017; 144 : 4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chung E, Deacon P, Marable S, et al Notch signaling promotes nephrogenesis by downregulating Six2. Development 2016; 143 : 3907–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujimura S, Jiang Q, Kobayashi C, et al Notch2 activation in the embryonic kidney depletes nephron progenitors. J Am Soc Nephrol 2010; 21 : 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murea M, Park JK, Sharma S, et al Expression of Notch pathway proteins correlates with albuminuria, glomerulosclerosis, and renal function. Kidney Int 2010; 78 : 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bielesz B, Sirin Y, Si H, et al Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest 2010; 120 : 4040–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seegar TCM, Killingsworth LB, Saha N, et al Structural basis for regulated proteolysis by the α‐secretase ADAM10. Cell 2017; 171 : 1638–1648.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aster JC, Pear WS, Blacklow SC. The varied roles of Notch in cancer. Annu Rev Pathol 2017; 12 : 245–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol 2016; 17 : 722–735. [DOI] [PubMed] [Google Scholar]
- 29. Hou L, Du Y, Zhao C, et al PAX2 may induce ADAM10 expression in renal tubular epithelial cells and contribute to epithelial‐to‐mesenchymal transition. Int Urol Nephrol 2018; 50 : 1729–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lagares D, Ghassemi‐Kakroodi P, Tremblay C, et al ADAM10‐mediated ephrin‐B2 shedding promotes myofibroblast activation and organ fibrosis. Nat Med 2017; 23 : 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen W, Jiang H, Wang M, et al Effects of chlorpyrifos exposure on kidney Notch2–Jagged1 pathway of early prenatal embryo. Birth Defects Res B Dev Reprod Toxicol 2011; 92 : 97–101. [DOI] [PubMed] [Google Scholar]
- 32. Gschwind A, Hart S, Fischer OM, et al TACE cleavage of proamphiregulin regulates GPCR‐induced proliferation and motility of cancer cells. EMBO J 2003; 22 : 2411–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ricceri L, Venerosi A, Capone F, et al Developmental neurotoxicity of organophosphorous pesticides: fetal and neonatal exposure to chlorpyrifos alters sex‐specific behaviors at adulthood in mice. Toxicol Sci 2006; 93 : 105–113. [DOI] [PubMed] [Google Scholar]
- 34. Levin ED, Addy N, Nakajima A, et al Persistent behavioral consequences of neonatal chlorpyrifos exposure in rats. Brain Res Dev Brain Res 2001; 130 : 83–89. [DOI] [PubMed] [Google Scholar]
- 35. Barak H, Boyle SC. Organ culture and immunostaining of mouse embryonic kidneys. Cold Spring Harb Protoc 2011: pdb.prot 5558. [DOI] [PubMed] [Google Scholar]
- 36. Dobin A, Davis CA, Schlesinger F, et al STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 2013; 29 : 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trapnell C, Hendrickson DG, Sauvageau M, et al Differential analysis of gene regulation at transcript resolution with RNA‐seq. Nat Biotechnol 2013; 31 : 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subramanian A, Tamayo P, Mootha VK, et al Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A 2005; 102 : 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Combes AN, Phipson B, Lawlor KT, et al Single cell analysis of the developing mouse kidney provides deeper insight into marker gene expression and ligand–receptor crosstalk. Development 2019; 146 : dev178673. [DOI] [PubMed] [Google Scholar]
- 40. Yang G, Zhou R, Zhou Q, et al Structural basis of Notch recognition by human γ‐secretase. Nature 2019; 565 : 192–197. [DOI] [PubMed] [Google Scholar]
- 41. Weng CH, Li YJ, Wu HH, et al Interleukin‐17A induces renal fibrosis through the ERK and Smad signaling pathways. Biomed Pharmacother 2020; 123 : 109741. [DOI] [PubMed] [Google Scholar]
- 42. Miller MA, Meyer AS, Beste MT, et al ADAM‐10 and ‐17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc Natl Acad Sci U S A 2013; 110 : E2074–E2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tain YL, Hsieh CS, Lin IC, et al Effects of maternal l‐citrulline supplementation on renal function and blood pressure in offspring exposed to maternal caloric restriction: the impact of nitric oxide pathway. Nitric Oxide 2010; 23 : 34–41. [DOI] [PubMed] [Google Scholar]
- 44. Gray SP, Denton KM, Cullen‐McEwen L, et al Prenatal exposure to alcohol reduces nephron number and raises blood pressure in progeny. J Am Soc Nephrol 2010; 21 : 1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Slabiak‐Blaz N, Adamczak M, Gut N, et al Administration of cyclosporine A in pregnant rats – the effect on blood pressure and on the glomerular number in their offspring. Blood Press Res 2015; 40 : 413–423. [DOI] [PubMed] [Google Scholar]
- 46. Ortiz LA, Quan A, et al Effect of prenatal dexamethasone on rat renal development. Kidney Int 2001; 59 : 1663–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hertz‐Picciotto I, Sass JB, Engel S, et al Organophosphate exposures during pregnancy and child neurodevelopment: recommendations for essential policy reforms. PLoS Med 2018; 15 : e1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Self M, Lagutin OV, Bowling B, et al Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J 2006; 25 : 5214–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Combes AN, Wilson S, Phipson B, et al Haploinsufficiency for the Six2 gene increases nephron progenitor proliferation promoting branching and nephron number. Kidney Int 2018; 93 : 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int 2006; 69 : 213–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Prenatal CPF exposure mouse model has no effect on kidney function of pregnant mice, and Aqp1 is upregulated in the CPF group
Figure S2. Prenatal CPF exposure mouse model does not affect the development of other nephron structures
Figure S3. ADAM10 localisation and the effect of Adam10‐MO on kidney development
Figure S4. Adam10‐MO resulted in inhibition of kidney development in ex vivo E12.5 kidneys of normal ICR mice
Figure S5. There was no obvious epithelial‐to‐mesenchymal transition in 4‐ or 8‐week kidneys
Figure S6. Effects of overexpression and knockdown of ADAM10 in 293T cells
Figure S7. Verification of the overexpression and knockdown efficiency in 293T cells
Figure S8. The degree of renal interstitial fibrosis is positively correlated with ADAM10 and NOTCH2 expression in patients with IgANs
Figure S9. ADAM10 and NOTCH2 expression both increase as the degree of renal interstitial fibrosis increases in various renal biopsy tissues
Table S1. Detailed information of differentially expressed genes between the two groups
Table S2. Detailed information of differentially expressed genes in the Notch signalling pathway (GeneChip data)
Table S3. General information for the 81 IgAN patients
Table S4. Primers used in this study


