Abstract
Background
COVID-19 has become a major global threat. The present study aimed to develop a nomogram model to predict the survival of COVID-19 patients based on their clinical and laboratory data at admission.
Methods
COVID-19 patients who were admitted at Hankou Hospital and Huoshenshan Hospital in Wuhan, China from January 12, 2020 to March 20, 2020, whose outcome during the hospitalization was known, were retrospectively reviewed. The categorical variables were compared using Pearson’s χ2-test or Fisher’s exact test, and continuous variables were analyzed using Student’s t-test or Mann Whitney U-test, as appropriate. Then, variables with a P-value of ≤0.1 were included in the log-binomial model, and merely these independent risk factors were used to establish the nomogram model. The discrimination of the nomogram was evaluated using the area under the receiver operating characteristic curve (AUC), and internally verified using the Bootstrap method.
Results
A total of 262 patients (134 surviving and 128 non-surviving patients) were included in the analysis. Seven variables, which included age (relative risk [RR]: 0.905, 95% confidence interval [CI]: 0.868–0.944; P < 0.001), chronic heart disease (CHD, RR: 0.045, 95% CI: 0.0097–0.205; P < 0.001, the percentage of lymphocytes (Lym%, RR: 1.125, 95% CI: 1.041–1.216; P = 0.0029), platelets (RR: 1.008, 95% CI: 1.003–1.012; P = 0.001), C-reaction protein (RR: 0.982, 95% CI: 0.973–0.991; P < 0.001), lactate dehydrogenase (LDH, RR: 0.993, 95% CI: 0.990–0.997; P < 0.001) and D-dimer (RR: 0.734, 95% CI: 0.617–0.879; P < 0.001), were identified as the independent risk factors. The nomogram model based on these factors exhibited a good discrimination, with an AUC of 0.948 (95% CI: 0.923–0.973).
Conclusions
A nomogram based on age, CHD, Lym%, platelets, C-reaction protein, LDH and D-dimer was established to accurately predict the prognosis of COVID-19 patients. This can be used as an alerting tool for clinicians to take early intervention measures, when necessary.
Keywords: SARS-CoV-2, COVID-19, Risk factor, Prediction model, Nomogram
Background
Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a pandemic and major threat to global health [1–4]. As of June 2, 2020, more than 6.2 million cases have been reported worldwide, with a death toll approaching 370,000 (https://www.who.int/docs/default-source/coronaviruse/situation-reports). In clinic, 10–15% of patients with COVID-19 develop severe or critical outcomes, such as acute respiratory distress syndrome (ARDS), and eventually, multiple organ dysfunction or even death [5]. At present, there are no specific drugs for SARS-CoV-2, and the treatment for COVID-19 depends on the severity of the manifestations developed during the disease course. Theoretically, the early prediction of the subsequent development of severe or critical outcomes, including death, with early intervention would help greatly prevent severe, critical, or fatal consequences. Therefore, there is an urgent need to establish and validate models to accurately predict patient outcomes, in order to implement early intervention, and thereby optimize patient care and resource allocation during the pandemic.
A nomogram is known to be a commonly used visual tool that integrates clinical data and laboratory variables for predicting the outcome of a disease. It was invented in the nineteenth century and has been widely used as a visualized, intuitive and appreciable tool in clinical practice [6–8]. Therefore, the present study aimed to develop a predictive nomogram model to predict the survival of COVID-19 patients based on their baseline clinical and laboratory data at admission.
Methods
Patients
Patients who were diagnosed with COVID-19, and admitted at Hankou Hospital and Huoshenshan Hospital in Wuhan, China from January 12, 2020 to March 20, 2020, whose outcome (survived [recovered and discharged from the hospital] or did not survive [died] during the hospitalization) was known by April 24, 2020, were retrospectively reviewed. Their demographic, epidemiological, clinical, imaging and laboratory data, and other data were collected and analyzed. Patients with incomplete clinical data, or those who died of causes unrelated to COVID-19 were excluded from the analysis. Hankou Hospital is a grade III class B hospital designated for COVID-19 treatment, while Huoshenshan Hospital is a hospital specifically built for COVID-19 treatment during the early stage of the epidemic. These patients were divided into two groups for analysis, according to the final outcome: survival group and non-survival group.
Diagnostic criteria and clinical classification
A patient was diagnosed with COVID-19 based on the following: (1) when the patient had an etiological evidence of SARS-CoV-2 infection, and/or (2) when the patient had an epidemiological history, with any two of the clinical manifestations with or without any of the pulmonary imaging changes described below, according to the 5th or 7th edition of the COVID-19 Diagnosis and Treatment Program of China [9, 10].
An etiological evidence included: (1) positive SARS-CoV-2 nucleic acid detected by real-time fluorescence polymerase chain reaction (PCR) in the respiratory or blood samples; (2) viral gene sequencing highly homologous to the known SARS-CoV-2 in the respiratory or blood samples; or (3) positive serum SARS-CoV-2 specific IgM antibody and IgG antibody.
An epidemiological history included: (1) a history of travel or residence in Wuhan and its surrounding areas, or in other reported epidemic areas within 14 days before the onset of the disease; (2) a history of contact with a confirmed COVID-19 patient within 14 days before the onset of the disease; and (3) a history of contact with a patient from Wuhan and its surrounding areas, or a patient with fever or respiratory symptoms from the reported epidemic community within 14 days before the onset of the disease. The clinical manifestations included fever and/or respiratory symptoms with normal or decreased white blood cell count, or decreased lymphocyte count in the early stage of the disease. The pulmonary imaging changes included multiple small patch shadows and stromal changes in the early stage, which were obvious in the lung periphery, and developed into ground glass shadows, and infiltrating shadows or lung consolidation in severe cases, but rarely with pleural effusion.
The COVID-19 cases were clinically classified as mild, common, severe, or critical type [11]. Mild type was defined when the clinical symptoms were mild, without any imaging changes. Common type was defined when patients had clinical symptoms, such as fever and respiratory infection, with pulmonary imaging changes. Severe type was defined when patients presented with any of the following: (1) a respiratory rate of > 30 rate/min, (2) a pulse oxygen saturation (SpO2) of ≤93%, and (3) an arterial oxygen partial pressure to fractional inspired oxygen ratio (the ratio of partial pressure of oxygen to fraction of inspiration O2: PaO2/FiO2) of ≤300 mmHg. Critical type was defined when patients presented with any of the following: (1) respiratory failure requiring mechanical ventilation, (2) shock, and (3) other organ failure requiring intensive care unit (ICU) care. In the present study, patients diagnosed with mild and common types were assigned to the non-severe group, while patients diagnosed with severe and critical types were assigned to the severe group.
Data collection and statistical analysis
The demographic, epidemiological, clinical, imaging and laboratory data were collected from the electronic medical record system of the hospitals. The categorical variables were expressed in frequency (percentage), and continuous variables were expressed in median (interquartile range) or mean ± standard deviation, when appropriate. The Shapiro–Wilk test was used to judge the distribution of continuous variables before the statistical analysis. The categorical variables were compared using Pearson’s χ2-test or Fisher’s exact test, while continuous variables were analyzed by Student t-test or Mann Whitney U-test, as appropriate. Variables with a P-value of ≤0.1 in the univariate analyses were included in the log-binomial model using the backward stepwise method, and merely these independent risk factors were used to establish the nomogram model. The relative risk (RR) and 95% confidence interval (CI) were calculated. The bootstrap self-sampling method was used to internally verify the prediction effect of the model, and the verification result was visually presented in the calibration curve. The discrimination of the nomogram was evaluated using the area under the receiver operating characteristic (ROC) curve (AUC), and a calibration curve was constructed to determine whether the prediction result was consistent with the observation result. All statistical analyses were performed using IBM SPSS statistics 20.0 (SPSS Inc., IL, USA) and R v3.5.2 (R Foundation for Statistical Computing, Vienna, Austria, http://www.Rproject.org) through RStudio v1.0.136 (RStudio Inc., MA, USA). The nomogram model and calibrating curve were analyzed using the R software package called rms and cali, respectively. All tests were two sided with an alpha level of 0.05.
Results
Characteristics of patients with non-survival and survival COVID-19
A total of 269 COVID-19 patients were hospitalized in Wuhan Hankou Hospital (n = 215) and Huoshenshan Hospital (n = 54) from January 12, 2020 to March 20, 2020. Among them, 141 (52.4%) patients survived, while 128 (47.6%) patients did not survive. The number of patients (rates) were 125 (58.1%) and 90 (41.9%), respectively, for Hankou Central Hospital, and 16 (29.6%) and 38 (71.4%), respectively, for Huoshenshan Hospital. Seven survived patients in survival group were excluded due to incomplete information. Finally, 262 patients were included in the analysis. Among these patients, 143 patients were male and 119 were female, and their median age was 67 years old (range: 31–96 years old) (Table 1).
Table 1.
Clinical characteristics and laboratory parameters of patients in the survival and non-survival groups at admission
| Clinical features | Total (n = 262) | Survival (n = 134) | Non-survival (n = 128) | P-value | Adjusted RR (95%CI)† | P-value |
|---|---|---|---|---|---|---|
| Age, years | 67 (59, 73.25) | 62.5 (51.75, 68.25) | 70.5 (65, 78) | < 0.001* | 0.905 (0.868, 0.944) | < 0.001* |
| Gender | 0.202 | |||||
| Female% | 119 (45.4%) | 66 (49.3%) | 53 (41.4%) | |||
| Male% | 143 (54.6%) | 68 (50.7%) | 75 (58.6%) | |||
| Smoking | 14 (5.3%) | 3 (2.2%) | 11 (8.6%) | 0.022* | ||
| Alcohol drinking | 7 (2.7%) | 2 (1.5%) | 5 (3.9%) | 0.226 | ||
| Illness to length of stay, days | 10 (5, 14) | 10 (5, 14) | 10 (7, 13) | 0.962 | ||
| Comorbidities | ||||||
| Hypertension | 121 (46.2%) | 50 (37.3%) | 71 (55.5%) | 0.003* | ||
| Diabetes | 56 (21.4%) | 24 (17.9%) | 32 (25%) | 0.162 | ||
| Cardiovascular disease | 45 (17.2%) | 12 (9%) | 33 (25.8%) | < 0.001* | 0.045 (0.010, 0.205) | < 0.001* |
| COPD | 7 (2.7%) | 1 (0.7%) | 6 (4.7%) | 0.061 | ||
| Chronic kidney disease | 10 (3.8%) | 4 (3%) | 6 (4.7%) | 0.533 | ||
| Tumor | 11 (4.2%) | 3 (2.2%) | 8 (6.2%) | 0.106 | ||
| Nervous system disease | 15 (6.5%) | 5 (3.7%) | 12 (9.4%) | 0.064 | ||
| Other | 42 (16%) | 25 (18.7%) | 17 (13.3%) | 0.236 | ||
| Temperature at admission, oC | 38.5 (38, 39) | 38.5 (38, 39) | 38.6 (38, 39) | 0.430 | ||
| Duration of fever, day | 10 (5, 14) | 10 (5, 14) | 10 (6.5, 12) | 0.867 | ||
| Respiratory rate at admission, rate/min | 20 (20, 25) | 20 (20, 23) | 22 (20, 30) | < 0.001* | ||
| SpO2 at admission,% | 90 (80, 96) | 96 (92, 98) | 83 (70, 91) | < 0.001* | ||
| Symptoms | ||||||
| Fever | 208 (79.4%) | 108 (80.6%) | 100 (78.1%) | 0.621 | ||
| Cough | 190 (72.5%) | 98 (73.1%) | 92 (71.9%) | 0.819 | ||
| Expectoration | 79 (30.2%) | 47 (35.1%) | 32 (25%) | 0.076 | ||
| Pharyngalgia | 10 (3.8%) | 7 (5.2%) | 3 (2.3%) | 0.336 | ||
| Vomit | 10 (3.8%) | 4 (3%) | 6 (4.7%) | 0.533 | ||
| Poor appetite | 115 (43.9%) | 65 (48.5%) | 50 (39.1%) | 0.124 | ||
| Muscular soreness | 24 (9.1%) | 12 (8.8%) | 12 (9.4%) | 0.876 | ||
| Diarrhea | 17 (6.5%) | 11 (8.2%) | 6 (4.7%) | 0.247 | ||
| Headache | 7 (2.7%) | 5 (3.7%) | 2 (1.6%) | 0.448 | ||
| Shortness of breath | 143 (54.6%) | 60 (44.8%) | 79 (61.7%) | 0.001* | ||
| Weakness | 155 (57%) | 82 (61.2%) | 73 (57%) | 0.493 | ||
| Severity of disease at admission | 0.191 | |||||
| Non-severe | 111 (42.4%) | 62 (46.3%) | 49 (38.3%) | |||
| Severe | 151 (57.6%) | 72 (53.7%) | 79 (61.7%) | |||
| Positive nucleic acid before admission | 73 (29%) | 36 (26.9%) | 37 (31.4%) | 0.433 | ||
| Positive nucleic acid at admission | 99 (37.8%) | 49 (49.5%) | 50 (39.1%) | 0.677 | ||
| Laboratory parameter | ||||||
| White blood cell, ×109/L | 6.5 (4.5, 10.2) | 5.6 (3.9, 7.42) | 8.85 (5.67, 12.9) | < 0.001* | ||
| Lymphocyte, ×109/L | 0.7 (0.41, 1.0) | 0.8 (0.6, 1.15) | 0.66 (0.3, 0.8) | < 0.001* | ||
| LYM percentage, % | 10 (5.4, 20.525) | 16.5 (8.8, 26.2) | 6.4 (3.45, 10.25) | < 0.001* | 1.125 (1.041, 1.216) | 0.0029* |
| Blood platelet, ×109/L | 188 (137, 246) | 216.5 (157.5, 265.7) | 169 (111, 229) | 0.001* | 1.008 (1.003, 1.012) | 0.001* |
| Hemoglobin, g/L | 126 (117, 137) | 126 (117.75, 137.25) | 124 (115.5, 137) | 0.436 | ||
| CRP, mg/L | 36.45 (17.54, 101.5) | 31.98 (7.52, 37.93) | 61.76 (35.52, 148.31) | < 0.001* | 0.982 (0.973, 0.991) | < 0.001* |
| PCT, ng/ml | 0.083 (0.05, 0.20) | 0.068 (0.039, 0.09) | 0.237 (0.09, 0.60) | < 0.001* | ||
| APTT, s | 34.5 (30.4, 40.6) | 34.3 (31.32, 41.1) | 34.85 (29.97, 39.7) | 0.603 | ||
| Fibrinogen, g/L | 3.825 (2.85, 4.67) | 3.75 (2.89, 4.73) | 3.97 (2.81, 4.67) | 0.841 | ||
| TT, s | 15.3 (14.6, 16.29) | 15.3 (14.5, 15.9) | 15.3 (14.6, 16.6) | 0.287 | ||
| PT, s | 13.9 (12.85, 15.45) | 13.9 (13.0, 15.05) | 13.95 (11.95, 15.97) | 0.932 | ||
| INR | 1.14 (1.05, 1.3) | 1.12 (1.04, 1.21) | 1.2 (1.08, 1.61) | 0.002* | ||
| D-dimer, μg/mL | 0.75 (0.205, 4.61) | 0.31 (0.12, 1.3) | 1.22 (0.51, 7.39) | < 0.001* | 0.734 (0.617, 0.879) | < 0.001* |
| CK, U/L | 99.5 (59, 201.75) | 87 (43, 173) | 108.3 (72, 348) | 0.065 | ||
| CK-MB, U/L | 15.7 (11, 27) | 14 (9, 19) | 19 (13.72, 35.45) | < 0.001* | ||
| a-HBDH, U/L | 329.7 (223.1, 461.8) | 268.3 (185.85, 364.2) | 483.3 (279.75, 609.77) | < 0.001* | ||
| cTnT, ng/mL | 0.03 (0.014, 1.395) | 0.014 (0.011, 0.022) | 0.17 (0.041, 8.13) | < 0.001* | ||
| BNP, pg/mL | 199.4 (81.34, 874.3) | 118.35 (67.88,280.6) | 388.6 (145.2, 2262) | < 0.001* | ||
| TBil, μmol/L | 9.7 (7.1, 13.9) | 8.7 (6.9, 11.3) | 11.1 (7.8, 17.12) | < 0.001* | ||
| DBil, μmol/L | 3.9 (2.6, 6.47) | 3.3 (2.4, 4.3) | 5.05 (3.47, 9.3) | < 0.001* | ||
| IBil, μmol/L | 5.5 (3.8, 7.3) | 5.3 (3.6, 7.1) | 5.85 (3.87, 7.96) | 0.186 | ||
| ALT, U/L | 26.1 (17, 40) | 25 (17, 36) | 27.1 (18.15, 43.6) | 0.151 | ||
| AST, U/L | 32 (24, 49) | 28 (22, 40) | 38 (27.4, 61) | < 0.001* | ||
| LDH, U/L | 334 (236.9, 476) | 269 (199, 382) | 422 (294, 584.9) | < 0.001* | 0.993 (0.990, 0.997) | < 0.001* |
| Cr, μmol/l | 75 (57.25, 94) | 67 (56.4, 84.5) | 83 (60, 108) | 0.001* | ||
| BUN, mmol/L | 5.52 (4.14, 8.29) | 4.78 (3.8, 6.51) | 7.42 (4.48, 13.24) | < 0.001* | ||
| Alb, g/L | 31.7 (29.3, 35.7) | 32.8 (30, 36.9) | 31 (28.7, 33.95) | 0.001* | ||
The data are expressed in median (interquartile range) or number (%), when appropriate
Abbreviations: COPD Chronic obstructive pulmonary disease, CRP C-reactive protein, PCT Procalcitonin, APTT Activated partial thromboplastin time, TT Thrombin time, PT Prothrombin time, INR International normalized ratio, CK Creatine kinase, CK-MB Creatine kinase isoenzyme, CTnT Troponin T, α-HBDH α-Hydroxybutyrate dehydrogenase, TBil Total bilirubin, IBil Indirect bilirubin, DBil Direct bilirubin, ALT Alanine aminotransferase, AST Aspartate aminotransferase, LDH Lactate dehydrogenase, Cr Creatinine, BUN Urea nitrogen, Alb Albumin, RR Relative risk
*P < 0.05 was considered statistically significant
†, Variables with a P value of ≤0.1 in the univariate analyses were included in the log-binomial model and statistical data are not shown for variables that were not significant (i.e. P > 0.05) in the log-binomial model
There were significant differences in smoking, coronary heart disease (CHD), hypertension, shortness of breath, respiratory rate and SpO2 at admission between the survival and non-survival groups (P < 0.05) (Table 1). Furthermore, there were significant differences in white blood cell count (WBC), lymphocyte, lymphocyte percentage (Lym%), platelet (PLT), D-dimer, ɑ-hydroxybutyrate dehydrogenase (ɑ-HBDH), international normalized ratio (INR), CK-MB, cTnT, BNP, total bilirubin (TBil), direct bilirubin (DBIL), aspartate aminotransferase (AST), C-reactive protein (CRP), procalcitonin (PCT), lactate dehydrogenase (LDH), creatinine (Cr), urea nitrogen (BUN) and albumin (Alb) between these two groups within 24 h after admission (Table 1).
Independent risk factors for survival as determined by the multivariate logistic regression analysis
After the univariate analyses, 26 potential risk factors were included in the log-binomial model analysis. Finally, seven variables, including age (RR: 0.905, 95% CI: 0.868–0.944; P < 0.001), CHD (RR: 0.045, 95% CI: 0.0097–0.205; P < 0.001), Lym% (RR: 1.125, 95% CI: 1.041–1.216; P = 0.003), PLT (RR: 1.008, 95% CI: 1.003–1.012; P = 0.001), CRP (RR: 0.982, 95% CI: 0.973–0.991; P < 0.001), LDH (RR: 0.993, 95% CI: 0.990–0.997; P < 0.001), and D-dimer (RR: 0.734, 95% CI: 0.617–0.879; P < 0.001), were identified as independent risk factors. These were used to build the nomogram for predicting the survival probability.
Establishment and verification of the nomogram model
The foreign package of the R language was used to read the original data, and the rms package was used to bring age, CHD, Lym%, PLT, CRP, LDH and D-dimer into the functional model, and form the nomogram model for predicting the outcome of COVID-19 patients. The identified seven risk factors were incorporated into the prediction model, and presented as the nomogram in Fig. 1.
Fig. 1.
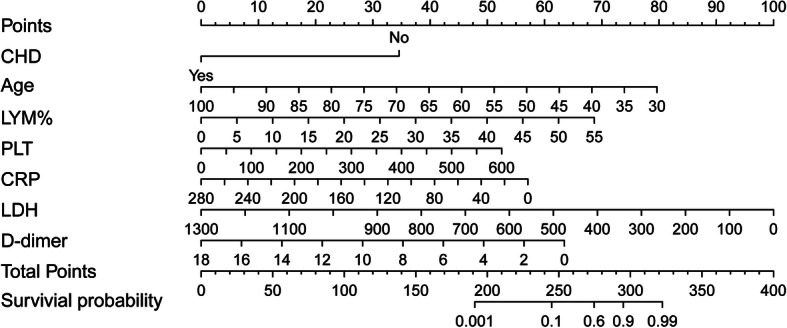
A nomogram for predicting the survival probability of patients with COVID-19. Abbreviations: Age, years; CHD: coronary heart disease; Lym%: lymphocyte percentage; PLT: platelet, × 109/L; CRP: C-reactive protein, mg/L; LDH: lactate dehydrogenase, U/L. Instructions for using the nomogram: (1) Draw a vertical line based on the value of each variable to obtain the corresponding point; (2) Add all seven points to obtain the total point; (3) Draw a vertical line based on the total point to determine the estimated survival probability
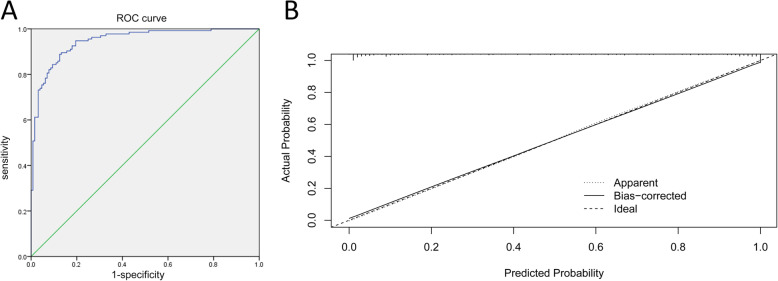
The ROC curve was generated for the nomogram, with an AUC of 0.948 (95% CI: 0.923–0.973, Fig. 2a). The internal verification using the bootstrap method was visually presented in the calibration curve (Fig. 2b). This revealed that the prediction probability of the nomogram was consistent with the actual situation. The calibration curve was slightly nonlinear. Furthermore, the consistency was better in the range of predicting the survival probability of patients < 50%, while the inconsistency slightly increased in the range of predicting the survival probability of patients ≥50%.
Fig. 2.
The receiver operating characteristic (ROC) curve (a) and calibration curve (b) for the established nomogram. OR: odds ratios; AUC: area under the curve; 95% CI: 95% confidence interval. Calibration curve reflects the extent to which the model correctly estimates the absolute probability or agreement between the predicted probability and observed outcomes. The Y-axis represents the actual survival probability. The X-axis represents the predicted survival probability. The black dot at the top represents the prediction probability corresponding to the actual observation, the black dotted line represents the ideal predicted value, and the solid line represents the actual predicted value
Discussion
In the present study, 262 patients with COVID-19, who were admitted in two designated hospitals, and either survived or died during the hospitalization, were analyzed. The multivariate logistic regression analysis identified age, CHD, and the five laboratory variables, which included Lym%, PLT, CRP, LDH and D-dimer, as independent factors for the survival of COVID-19 patients. Based on this, a nomogram for predicting in-hospital survival was established, with excellent performance.
Advanced age has been reported as an important independent predictor of SARS and MERS mortality [12]. Several studies have revealed that elderly patients and patients with underlying comorbidities tend to have poor outcomes [13–16]. The patients infected with SARS-CoV-2 were analyzed, and it was found that there was a significant difference in age between the survival and non-survival groups. Furthermore, the multivariate analysis also revealed that age was an independent factor that affected the prognosis of COVID-19 patients. In the nomogram established by our team, it was found that the survival rate of COVID-19 patients decreased with the increase in age. According to the present analysis of the clinical data, the proportions of hypertension and CHD were significantly higher in the non-survival group, when compared to the survival group, and CHD was an independent risk factor for the prognosis of COVID-19 patients. This suggests that patients with hypertension and CHD are susceptible to SARS-CoV-2 infection, and have a high mortality rate. Although the underlying mechanism remains unclear, this may have been contributed by the following causes. First, Angiotensin converting enzyme 2 (ACE2) is widely expressed in myocardial cells, myocardial fibroblasts and coronary endothelial cells. It has been reported that the Spike (S) protein of SARS-CoV-2 binds with high affinity to human ACE2, and before SARS-CoV-2’s entry into the cells [17–19], the S protein is subjected to a priming process via serine protease TMPRSS2 in order to permit the attachment of viral particles to ACE2 and thus on cell surface [20, 21]. Therefore, SARS-CoV-2 can directly attack cardiac muscle cells through this pathway [22–24]. This entry mechanism is confirmed by the fact that TMPRSS2 inhibition or TMPRSS2-KO mice show both decreased, though not abolished, S protein priming, and reduced viral entry, spread, as well as, inflammatory chemokine and cytokine release [20]. Second, there may be an imbalance between Th1 and Th2 responses in COVID-19 patients, and the cytokine storm triggered by this imbalance may be another mechanism of myocardial injury [25–27].
Interestingly, it was found PLT and D-dimer were independent risk factors for predicting the outcome of COVID-19 patients. Thrombocytopenia has been reported to be present in up to 55% of patients with SARS, and has been identified as an important risk factor for mortality [28]. In a study conducted by Zou et al., merely two variables (i.e. platelet count and hypoxemia) were used to establish a SARS prognostic model, and this was used to predict the survival rate, with an accuracy of 96.2% [5]. It has been demonstrated that the pathological features of COVID-19 are similar to those of SARS and MERS [29]. In the early stage of infection, patients present with inflammation, edema, protein exudation, focal hyperplasia of alveolar epithelial cells and patchy inflammatory infiltration, as well as multi-nucleated giant cells in the lung. In the late stage, in addition to hemorrhage and some areas of interstitial fibrosis, diffuse alveolar damage was also observed [30]. Furthermore, fibrous clots and gelatinous mucus in the small airway and diffuse intravascular coagulation were observed [31]. In a previous study, it was found that lung injury was the main cause, followed by heart, liver and kidney injuries, and coagulation system abnormality [32]. In the present study, there were abnormal coagulation functions in both the survival and non-survival groups, which mainly manifested with the increase in D-dimer. At the same time, it was found that there was a significant increase in fibrin degradation products and thrombocytopenia, which was consistent with the presence of hyper-fibrinolysis, in patients with severe COVID-19 [33]. Therefore, it is not difficult to understand that platelets and D-dimers are independent risk factors that affect the outcome of patients. Noteworthy, patients with severe SARS-CoV-2 infection often possess coagulation dysfunction at admission [34]. A recent study by Zhang et al demonstrated that INR was a prognostic factor for clinical outcomes in patients with severe COVID-19 [34]. In the present study, we observed that there was also significant in the INR between survival and non-survival group in univariate analysis. However, it was not an independent risk factor for the survival of COVID-19 patients in multivariate analysis. Thus, further investigation is required to reveal the association between coagulation disorder and adverse clinical outcome in severe COVID-19 patients.
The present study revealed that Lym% and CRP were also independent risk factors that affected the prognosis of patients. The decrease in Lym% indicates that the immune system of a patient with SARS-CoV-2 is more likely to be suppressed, and that the host loses the immune function to fight against the pathogen, leading to the persistence of the infection and deterioration of COVID-19. In addition, the elevated levels of CRP observed in the present study, as well as in previous studies [32, 35], also suggest that there is a persistent inflammatory response in COVID-19 patients. Finally, it was found that LDH was another independent risk factor to predict the outcome of COVID-19 patients. It has been shown that COVID-19 patients first present with lung injury, which subsequently leads to hypoxemia and multiple organ damage [36]. Consequently, LDH in cells is released, resulting in increased LDH levels. Indeed, LDH was used to predict the severity of tissue damage in the early stage of diseases as an auxiliary marker, and for the early identification of cases at high risk of progression to severe COVID-19 [37].
The nomogram, as a visual form of predictive models, has been used in the diagnosis, treatment and prognosis of various diseases [6–8, 38]. It is visual and intuitive, and does not need to substitute the numbers into the equation calculation. Furthermore, the user only needs to draw one or more lines to quickly and reliably obtain the prediction. In the present study, a nomogram was established, which can be used to accurately predict the prognosis of COVID-19 patients. The calibration curve revealed the accuracy of the nomogramic model for predicting the prognosis of COVID-19 patients (“dominant state”) and the bootstrap model (“bias correction state”), which can explain the relationship between the prediction probability and actual observation probability in the original data set.
There were a few limitations in the present study. First, this was a retrospective study with relatively a small sample size. Second, the nomogram in the present study was not externally verified through another cohort of COVID-19 patients. Thus, the predictive performance remains to be further confirmed. Third, patients included in the present study were all over 18 years old. Thus, the nomogram is not suitable for children and pregnant women.
Conclusions
A nomogram based on age, CHD, Lym%, PLT, CRP, LDH and D-dimer was established to accurately predict the prognosis of COVID-19 patients. In clinical practice, the survival probability of COVID-19 patients can be obtained by simple calculation. This can be used as an alerting tool for clinicians to take early intervention measures, when necessary.
Acknowledgements
The authors would like to thank Medjaden Bioscience Limited for assisting in the preparation of the manuscript.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- Alb
Albumin
- ACE2
Angiotensin converting enzyme 2
- AST
Aspartate aminotransferase
- ɑ-HBDH
ɑ-hydroxybutyrate dehydrogenase
- BUN
Urea nitrogen
- COVID-19
Coronavirus disease 2019
- CHD
Coronary heart disease
- CRP
C-reactive protein
- Cr
Creatinine
- DBIL
Direct bilirubin
- ICU
Intensive care unit
- INR
International normalized ratio
- LDH
Lactate dehydrogenase
- Lym%
Lymphocyte percentage
- PCR
Polymerase chain reaction
- PLT
Platelet
- PCT
Procalcitonin
- PaO2/FiO2
The ratio of partial pressure of oxygen to fraction of inspiration O2
- WBC
White blood cell count
- RR
Relative risk
- ROC
Receiver operating characteristic curve
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SpO2
Pulse oxygen saturation
- S protein
Spike protein
- TBil
Total bilirubin
Authors’ contributions
YD designed the study. XM, JH, YG, YD, NP, HH and WL provided the data of the COVID-19 patients. HW, SJ, QZ and XC participated in the data entry. QZ and SJ checked the data. HW and QZ were responsible for the data analysis. HW wrote the draft, and XG, CZ, NP and YD critically revised the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by National Natural Science Foundation of Guangdong, China (grant no. 2019A1515010206), Science and Technology Program of Guangzhou, China (grant no. 202002030094), Outstanding Young Medical Talents in Guangdong Province (grant no. KJ012019435) and High-level Hospital Construction Project of Guangdong Provincial People’s Hospital (grant no. DFJH2020020).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Hankou Hospital in Wuhan (HKYY-2020-028), and granted the study an exemption from informed consent due to its retrospective nature. The data used in this study was anonymised before its use.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaojun Ma, Huifang Wang, Junwei Huang and Yan Geng contributed equally to this work.
Contributor Information
Na Peng, Email: pnatz@163.com.
Yiyu Deng, Email: yiyudeng666@163.com.
References
- 1.Hoseinpour Dehkordi A, Alizadeh M, Derakhshan P, Babazadeh P, Jahandideh A. Understanding epidemic data and statistics: a case study of COVID-19. J Med Virol. 2020;92(7):868–882. doi: 10.1002/jmv.25885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onyeaka HK, Zahid S, Patel RS. The unaddressed behavioral health aspect during the coronavirus pandemic. Cureus. 2020;12(3):e7351. doi: 10.7759/cureus.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun T, Weng D. Estimating the effects of asymptomatic and imported patients on COVID-19 epidemic using mathematical modeling. J Med Virol. 2020. 10.1002/jmv.25939. [DOI] [PMC free article] [PubMed]
- 4.Ohannessian R, Duong TA, Odone A. Global telemedicine implementation and integration within health systems to fight the COVID-19 pandemic: a call to action. JMIR Public Health Surveill. 2020;6(2):e18810. doi: 10.2196/18810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou Z, Yang Y, Chen J, Xin S, Zhang W, Zhou X, Mao Y, Hu L, Liu D, Chang B, et al. Prognostic factors for severe acute respiratory syndrome: a clinical analysis of 165 cases. Clin Infect Dis. 2004;38(4):483–489. doi: 10.1086/380973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao Y, Fu X, Wang Y, Wu Y, Wang W, Zhang Q. Development and validation of risk Nomogram model predicting coronary microvascular obstruction in patients with ST-segment elevation myocardial infarction (STEMI) undergoing primary percutaneous catheterization. Sci Rep. 2019;25:5864–5877. doi: 10.12659/MSM.915960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and validation of a Radiomics Nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Q, Sun Z, Zhao G, Xie L. Nomogram for the individualized prediction of survival among patients with H7N9 infection. Risk Manag Healthc Policy. 2020;13:255–269. doi: 10.2147/RMHP.S242168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding YQ, Bian XW. Interpretation of pathological changes for “Guidelines for the Diagnosis and Treatment of COVID-19 by the National Health Commission (Trial Version 7)”. Zhonghua Bing Li Xue Za Zhi. 2020;49:E011. doi: 10.3760/cma.j.cn112151-20200318-00221. [DOI] [PubMed] [Google Scholar]
- 10.National Health Commission of the People’s Republic of China . Diagnosis and treatment scheme for COVID-19 (5th edition, China) (in Chinese) 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang WM, Kim UN, Jang DH, Jung H, Cho S, Eun SJ, Lee JY. Influence of trust on two different risk perceptions as an affective and cognitive dimension during Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak in South Korea: serial cross-sectional surveys. BMJ Open. 2020;10(3):e033026. doi: 10.1136/bmjopen-2019-033026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lippi G, Wong J, Henry BM. Hypertension and its severity or mortality in coronavirus disease 2019 (COVID-19): a pooled analysis. Pol Arch Intern Med. 2020;130(4):304–330. doi: 10.20452/pamw.15272. [DOI] [PubMed] [Google Scholar]
- 16.Ren H, Wang Q, Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, Ji R, Wang H, et al. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: a systematic review and meta-analysis. J Med Virol. 2020;127:104371. [Google Scholar]
- 17.Kumar V, Dhanjal JK, Bhargava P, Kaul A, Wang J, Zhang H, Kaul SC, Wadhwa R, Sundar D. Withanone and Withaferin-a are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J Biomol Struct Dyn. 2020:1–13. 10.1080/07391102.2020.1775704. [DOI] [PMC free article] [PubMed]
- 18.Rahman N, Basharat Z, Yousuf M, Castaldo G, Rastrelli L, Khan H. Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2) Molecules. 2020;25(10):2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Ye D, Mullick AE, Li Z, Danser AHJ, Daugherty A, Lu HS. Effects of Renin-Angiotensin Inhibition on ACE2 and TMPRSS2 Expression: Insights into COVID-19. bioRxiv. 2020. 10.1101/2020.06.08.137331. [DOI] [PMC free article] [PubMed]
- 20.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bestle D, Heindl MR, Limburg H, van Van Lam T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3(9):e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kai H, Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens Res. 2020;43(7):648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astuti I, Ysrafil Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aleksova A, Ferro F, Gagno G, Cappelletto C, Santon D, Rossi M, Ippolito G, Zumla A, Beltrami AP, Sinagra G. COVID-19 and renin-angiotensin system inhibition: role of angiotensin converting enzyme 2 (ACE2) - is there any scientific evidence for controversy? J Intern Med. 2020;288(4):410–421. doi: 10.1111/joim.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano T, Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52(5):731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long B, Brady WJ, Koyfman A, Gottlieb M. Cardiovascular complications in COVID-19. Am J Emerg Med. 2020;38(7):1504–1507. doi: 10.1016/j.ajem.2020.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Rhee JW, Cheng P, Waliany S, Chang A, Witteles RM, Maecker H, Davis MM, Nguyen PK, Wu SM. Cardiovascular complications in patients with COVID-19: consequences of viral toxicities and host immune response. Curr Cardiol Rep. 2020;22(5):32. doi: 10.1007/s11886-020-01292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang HJ, Du SH, Yue X, Chen CX. Review and Prospect of pathological features of Corona virus disease. Fa yi xue za zhi. 2020;36(1):16–20. doi: 10.12116/j.issn.1004-5619.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Park SE. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; coronavirus Disease-19) Clin Exp Pediatr. 2020;63(4):119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Li T, Han M, Li X, Wu D, Xu Y, Zhu Y, Liu Y, Wang X, Wang L. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu R, Ling Y, Zhang YH, Wei LY, Chen X, Li XM, Liu XY, Liu HM, Guo Z. Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease-19. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji HL, Zhao R, Matalon S, Matthay MA. Elevated plasmin (ogen) as a common risk factor for COVID-19 susceptibility. Physiol Rev. 2020;100(3):1065–1075. doi: 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, He L, Chen H, Lu S, Xiong Y, Liu J, Zheng Y, Wang S, Liu L. Manifestations of blood coagulation and its relation to clinical outcomes in severe COVID-19 patients: retrospective analysis. Int J Lab Hematol. 2020;42:766–72. [DOI] [PMC free article] [PubMed]
- 35.Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID-19 patients: a retrospective analysis of 115 cases from a single center in Wuhan city, China. Liver Int. 2020;40(9):2095–2103. doi: 10.1111/liv.14455. [DOI] [PubMed] [Google Scholar]
- 36.Yang F, Shi S, Zhu J, Shi J, Dai K, Chen X. Analysis of 92 deceased patients with COVID-19. J Med Virol. 2020. 10.1002/jmv.25891. [DOI] [PMC free article] [PubMed]
- 37.Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, Shi J, Zhou M, Wu B, Yang Z, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146(1):110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology – more than meets the eye. Lancet Oncology. 2015;16(4):e173–e180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.