Abstract
Coronavirus disease 2019 (COVID-19), caused by the SARS-CoV-2 virus, has developed into a pandemic causing major disruptions and hundreds of thousands of deaths in wide parts of the world. As of July 3, 2020, neither vaccines nor approved drugs for effective treatment are available. In this article, we showcase how to individuate drug targets and potentially repurposable drugs in silico using CoVex a recently presented systems medicine platform for COVID-19 drug repurposing. Starting from initial hypotheses, CoVex leverages network algorithms to individuate host proteins involved in COVID-19 disease mechanisms, as well as existing drugs targeting these potential drug targets. Our analysis reveals GLA, PLAT, and GGCX as potential drug targets, and urokinase, argatroban, dabigatran etexilate, betrixaban, ximelagatran and anisindione as potentially repurposable drugs.
Keywords: systems medicine, network-based drug repurposing, SARS-CoV-2
Introduction
In December 2019, the novel coronavirus SARS-CoV-2 was discovered in Wuhan, China. It causes coronavirus disease 2019 (COVID-19), which since then has developed into a pandemic with more than 10.5 million confirmed infections and more than 500 thousand confirmed deaths. As of July 3, 2020, neither vaccines nor effective drugs are available, and, consequently, researchers all over the world are working on new opportunities for treatment.
In this article, we show how to use our recently proposed computational system's medicine platform, CoVex, to identify new drug targets and possibly repurposable drugs for effective treatment.1 CoVex provides an easy-to-use web interface, which allows experts from pharmacology, biomedicine, and other fields to mine the COVID-19 virus–host–drug interactome in an intuitive, hypothesis-driven way. In particular, CoVex leverages network algorithms to unravel host proteins involved in key pathways of SARS-CoV-2 infection and points the user to existing drugs targeting these proteins. CoVex is available at https://exbio.wzw.tum.de/covex/.
Using the interaction data and network analysis algorithms implemented in CoVex (cf. “Results and Discussion” for details), we obtained two interesting results, which we will use to exemplify how to employ CoVex for identifying potential drug targets and possibly repurposable drugs: First, we observed that the enzyme α-galactosidase (α-GAL), encoded by the GLA gene, is not only a direct interactor of the viral protein NSP14, but is also involved in an important pathway of angiotensin-converting enzyme 2 (ACE2), which serves as the main entry point for SARS-CoV-2.2 Deficiency of α-GAL causes Fabry disease, a rare genetic disorder.3 Investigating further, we found that GLA is strongly expressed in lung cells and conferred by email with Fabry disease centers across Europe (in Germany, Switzerland, Slovenia, Norway, and Denmark). None of them report a single confirmed COVID-19 case. We hence formulate the hypothesis that α-GAL might play an important role in the life cycle of SARS-CoV-2 and, consequently, α-GAL inhibitors might be effective for COVID-19 treatment.
Second, we found that 10 host proteins that directly interact with the viral proteins NSP7, NSP13, E, M, N, ORF9C, and ORF8, are also involved in coagulation pathways. Using these proteins as starting points for the analysis in CoVex, we obtained the subnetwork connecting them and identified the vitamin K-dependent gamma-glutamyl-carboxylase, encoded by GGCX, and tissue plasminogen activator, encoded by PLAT, as potential drug targets (cf. “Results and Discussion” for details). They can be targeted by existing drugs, such as argatroban, dabigatran etexilate, betrixaban, ximelagatran, and anisindione.
Several systems medicine-based approaches for COVID-19 drug repurposing have been presented recently. Zhou et al. constructed a coronavirus–human interactome by integrating publicly available virus–human protein–protein interaction (PPI) data for SARS-CoV, MERS-CoV, HCoV-229E, and HCoV-NL with a human PPI network and known drug–target interactions.4 Subsequently, they used a network proximity algorithm to compile a list of potentially repurposable drugs. Gordon et al. used affinity purification mass spectrometry to generate the SARS-CoV-2-specific virus–host interactome and identified potentially promising drug candidates targeting the direct virus interactors.5 Gysi et al. used a similar approach as Zhou et al., but worked with the SARS-CoV-2-specific virus–host interactome generated by Gordon et al. instead of using a generic coronavirus–host interactome.6
A major advantage of CoVex w.r.t. the works of Zhou et al., Gordon et al., and Gysi et al. is that CoVex allows to explore the virus–drug–host interactome in an interactive, hypothesis-driven way. CoVex hence allows experts in the field to leverage their medical knowledge and browse the interactome in a more specific and directed fashion than by simply running generic algorithms on the data. How exactly this can be done is explained and exemplified in this article.
The remainder of this article is organized as follows: In the Methods section, we explain the systems medicine approach implemented in CoVex and provide a concise overview of how to use the main features. In the Results and Discussion section, we discuss GLA as a potential drug target and PLAT and GGCX as targets of possibly repurposable drugs, such as argatroban, dabigatran etexilate, betrixaban, ximelagatran, and anisindione. We conclude the article by pointing out possibilities for future work.
Methods
Leveraging Systems Medicine for Drug Repurposing and Drug Target Individuation
The general paradigm of systems medicine-based drug development and repurposing is to individuate a molecular disease mechanism and to target proteins involved in the extracted mechanism through new or existing drugs.7,8 In the case of SARS-CoV-2 infection, this paradigm translates into the task to unravel key virus–host pathways, which SARS-CoV-2 requires for replication or which are involved in the pathogenesis of severe COVID-19 cases.9 Host proteins involved in the extracted pathways are potential drug targets and existing drugs targeting them are potentially repurposable drugs. In comparison to drug development or repurposing strategies that aim at directly targeting the viral proteins,10 this approach has the advantage that drug resistances due to viral evolution are less probable.9
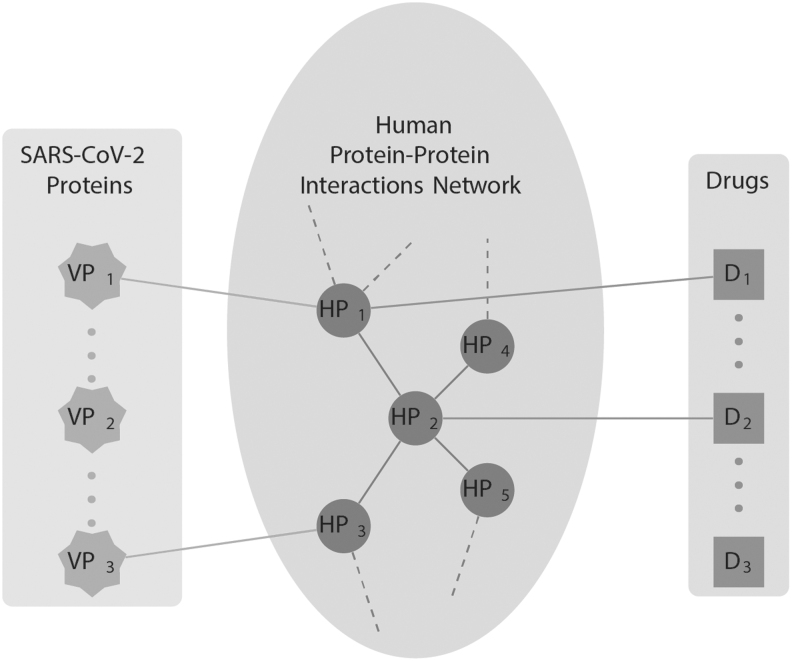
Figure 1 visualizes how CoVex can be used to mine the virus–host–drug interactome of SARS-CoV-2 infection in a hypothesis-driven way. Assume, for instance, that the host proteins HP4 and HP5 are known to be involved in a pathway related to a phenotype observed in many critical COVID-19 cases. This knowledge can be leveraged in CoVex by seeding the available network algorithms with HP4, HP5, and the viral proteins. CoVex then returns a subnetwork whose nodes correspond to key viral proteins for the phenotype of interest, potential drug targets, and potentially repurposable drugs. In the toy example shown in Figure 1, the viral proteins VP1 and VP3 are connected to the phenotype-related pathway through the direct interactors HP1 and HP3, as well as the indirect interactor HP2. Moreover, there are existing drugs D1 and D2 targeting HP1 and HP2, respectively. Hence, HP1, HP2, and HP3 are potential drug targets and D1 and D2 are potentially repurposable drugs.
Fig. 1.
Systems medicine approach employed by CoVex. CoVex integrates experimentally validated data about interaction between SARS-CoV-2 proteins (star nodes) with a human PPI network (round nodes) and known drug–target interactions (square nodes). Interactions between SARS-CoV-2 and human proteins are obtained from Gordon et al.,5 the human PPI network is taken from IID,11 and the drug–host links are obtained from ChEMBL,12 DrugBank,13 DrugCentral,14 Therapeutic Target Database,15 Guide to Pharmacology,16 PharmGKB,17 and BindingDB.18 PPI, protein–protein interaction.
Using the Functionality Available in CoVex
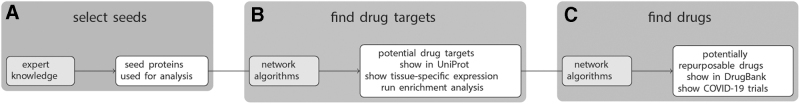
Figure 2 provides a schematic overview of CoVex’ main features and the intended workflow. The analysis always starts with a set of seed proteins, which have to be selected by the user (Fig. 2A). There are several ways to select the seeds. The easiest option is to graphically select some or all of the viral proteins and/or direct host interactors. Alternatively, the user can select proteins with high expression in a tissue of interest (e.g., lung), or provide a custom list of seed proteins. This latter option is especially interesting, because it allows to use CoVex in a hypothesis-driven way. For instance, the user-provided list of seed proteins could contain host proteins involved in symptom-related pathways or host proteins, which are differentially expressed in COVID-19 patients.
Fig. 2.
The CoVex workflow. In the first step, the user has to select a set of seed proteins from which the analysis should be run (A). Subsequently, network algorithms are employed to find proteins that are putative drug targets (B) and potentially repurposable drugs targeting them (C).
After selecting the seeds, the user can choose among six network algorithms to individuate potential drug targets, that is, host proteins involved in virus–host pathways, which are relevant to the selected seeds (Fig. 2B). The following algorithms are available at this stage: degree, closeness, and betweenness centrality,19 TrustRank,20 KeyPathwayMiner,21 and a novel multi-Steiner tree algorithm. While these algorithms differ w.r.t. their mathematical underpinnings (cf. Sadegh et al.1 for details), they all aim at extracting pathways that connect the selected seeds. The returned host proteins can then be displayed in UniProt22 and be annotated with or filtered by tissue-specific expression values. Moreover, g:Profiler23 can be called to run enrichment analysis on the found drug targets.
The next step is to individuate drugs targeting the discovered drug targets (Fig. 2C). For this, CoVex provides four different network algorithms: degree and closeness centrality,19 TrustRank,20 and network proximity.24 All of them extract ranked lists of drugs that hit (in the close vicinity of) the found drug targets and are hence candidates for drug repurposing. The drugs can be displayed in DrugBank and it is shown which of them are currently in clinical trial for COVID-19 treatment. Note that this step (Fig. 2C) of the CoVex workflow can also be run from drug targets provided by the user. That is, the automated individuation of drug targets (Fig. 2B) can be substituted by expert knowledge. Note that CoVex—by design—is dependent on expert input, as it follows the expert-in-the-loop concept. Consequently, the final results always depend on user input at different stages of the analyses.
Results and Discussion
In this study, we show two application examples to find drug targets and drug repurposing candidates. The first example demonstrates the potential relevance of the α-GAL protein as a drug target in SARS-CoV-2 infection, and the second example shows potential drug targets and possibly repurposable drugs for thromboembolic complications frequently observed in COVID-19 patients. Note that the obtained results are hypotheses that have not been validated through further genetic or chemical approaches and, consequently, should be treated with caution. In the present article, their main purpose is to exemplify the CoVex workflow.
Potential Drug Target: GLA
The α-GAL protein, encoded by the GLA gene, was identified by Gordon et al., as a direct interactor of the viral protein NSP14 of SARS-CoV-2 virus.5 The α-GAL protein is a lysosomal enzyme, highly expressed in the lung25 that degrades glycolipids, particularly globotriaosylceramide.26 Deficiencies of this enzyme cause Fabry disease, a rare genetic disorder characterized by the accumulation of globotriaosylceramide in endothelial cells, leading to severe symptoms.3,26 Fabry patients are often treated with blockers of the renin–angiotensin system, particularly angiotensin-converting enzyme inhibitors and angiotensin II receptor antagonists. Importantly, these proteins are expressed in vascular endothelial cells of the lung, kidney, and gastrointestinal tract, which are among the main tissues affected in Fabry disease and in COVID-19 patients.26,27
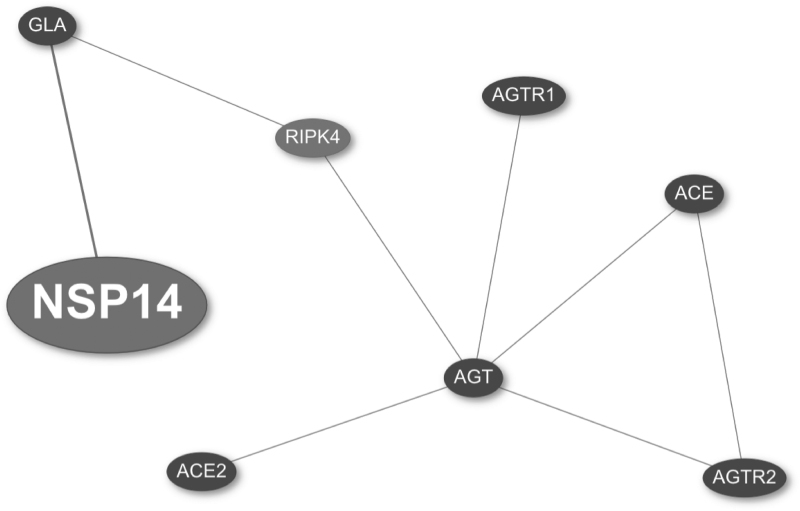
The points mentioned above raise the question whether the α-GAL protein could be involved with the renin–angiotensin system in the context of SARS-CoV-2 infection. Using CoVex, we aimed at extracting a subnetwork that could link α-GAL with this pathway. Thus, we used AGT, ACE, ACE2, AGTR1, and AGTR2 (main players of the renin–angiotensin system), as well as NSP14 and GLA as seeds. Next, we used the multi-Steiner algorithm to identify the connected subnetwork relevant to the seed proteins, and then ran closeness centrality to find potentially repurposable drugs.
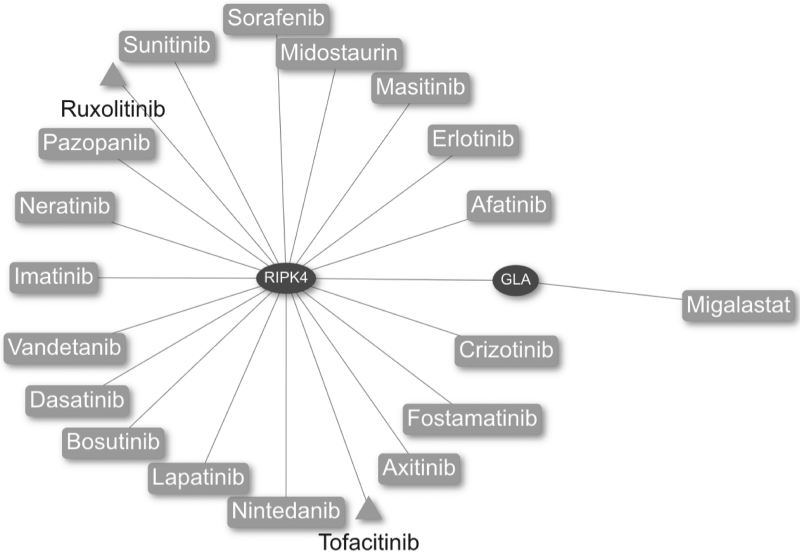
The results shown in Figure 3 reveal that GLA is closely related to important components of the renin–angiotensin system through the receptor-interacting serine/threonine-protein kinase 4 protein, encoded by the RIPK4 gene. Interestingly, the drugs ruxolitinib and tofacitinib, which are currently used in clinical trials in COVID-19 patients, also target this protein (Fig. 4).
Fig. 3.
Virus–host interaction network obtained with the multi-Steiner algorithm in CoVex. The viral protein NSP14 (large node) and the host proteins GLA, ACE2, AGT, ACE, AGTR2, and AGTR1 were used as seeds (small black nodes). The resulting subnetwork contains a new potential drug target RIPK4 (small gray node). ACE2, angiotensin-converting enzyme 2; RIPK4, receptor-interacting serine/threonine protein kinase 4 protein.
Fig. 4.
Drug–PPI network obtained with the closeness centrality algorithm in CoVex. The host proteins RIPK4 and GLA were used as seeds (elliptical nodes). The resulting network contains approved drugs (rectangular nodes) and drugs which are in clinical trials for COVID-19 treatment (triangular nodes). COVID-19, coronavirus disease 2019.
To further elucidate the role of α-GAL in SARS-CoV-2 infection, we contacted several Fabry disease centers across Europe by email. We received numbers of Fabry disease patients and confirmed COVID-19 cases among them from Germany, Switzerland, Slovenia, Norway, and Denmark. Interestingly, all centers reported zero confirmed COVID-19 cases among their patients. Hence, we formulate the hypothesis that α-GAL might play a role in COVID-19 pathogenesis and could hence be an attractive drug target for treatment. However, one should note that Fabry disease is a rare disease and these centers follow only a few thousand patients such that the evidence of not having a single COVID-19 case among them is rather weak. We followed up this thread because of the observed molecular interaction between α-GAL and the viral protein NSP14.
Potential Drug Targets: PLAT and GGCX
Several studies reported an increased incidence of cardiovascular complications and venous thrombotic events in hospitalized COVID-19 patients irrespectively of disease severity as well as abnormal coagulation parameters.28,29 While the mechanism of COVID-19 coagulopathy remains unclear, the cytokine storm syndrome30,31 and the endothelial dysfunction occurring in COVID-19 have been suggested as a possible main driver.32 In this second application example of CoVex, we hypothesize that the virus is directly involved in the disruption of the coagulation process, which, to some extent, could contribute to the observed hypercoagulable states.
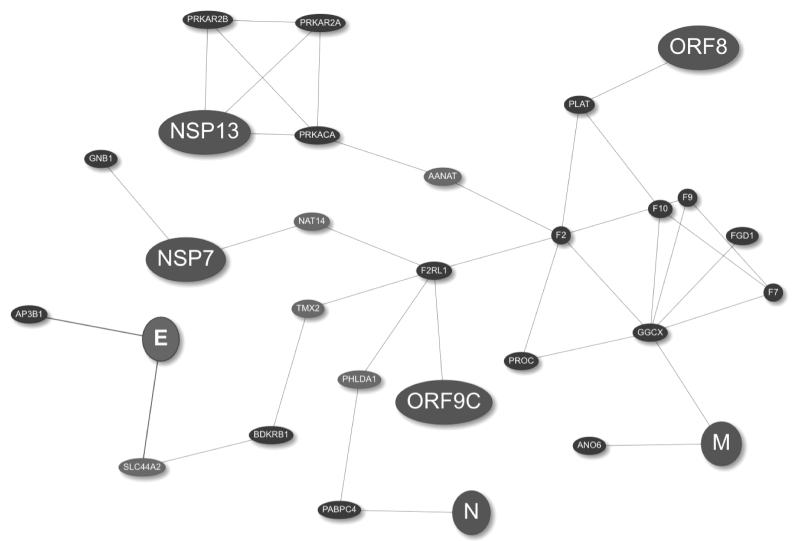
To evaluate this hypothesis, we used SARS-CoV-2 host interactors and viral proteins involved in coagulation as seeds (proteins mapping to the Gene Ontology (GO) term coagulation GO:0050817, cf. Table 1). Next we ran the “Find drug target” analysis using TrustRank and the multi-Steiner algorithms to extract the corresponding subnetwork (Fig. 5), which is enriched with genes involved in coagulation (Fig. 6). This means that CoVex was capable of identifying new genes in a network, which are also involved with the biological process of interest.
Table 1.
Virus–Host Interactions Related to Coagulation in Gordon et al.a Dataset
| Viral proteins | Host proteins |
|---|---|
| NSP7 | GNB1 |
| NSP13 | PRKAR2B, PRKACA, PRKAR2A |
| E | AP3B1 |
| M | GGCX, ANO6 |
| N | PABPC4 |
| ORF9C | F2RL1 |
| ORF8 | PLAT |
Reference.5
Fig. 5.
Virus–host interaction network obtained with the TrustRank and multi-Steiner algorithms in CoVex. The viral protein (large nodes) and the host proteins shown in Table 1 were used for the initial analysis with TrustRank, the resulting network (small black nodes) was further analyzed with the multi-Steiner algorithm, which identified additional connector proteins (small gray nodes).
Fig. 6.
Functional enrichment analysis of the coagulation-related interaction network in CoVex. The nonseed host proteins from the coagulation-related network (Fig. 5) were analyzed with g:Profiler using GO biological process, KEGG pathways, and WikiPathways as data sources. The upper panel shows the significantly enriched terms for each data source, and the lower panel shows the terms with the smallest p-values. BP, biological process; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; WP, Wikipathways.
Additionally, the resulting network shows new identified target proteins connected to the viral proteins, M, ORF8, and ORF9C, and to the host interactors, PLAT and GGCX. These new targets are of special interest because they are among the main proteins in the coagulation cascade (F2, F7, F9, F10, and protein C [PROC]).33 Proteins F2, F7, F9, and F10 correspond to the clotting factors thrombin (II), proconvertin (VII), Christmas factor (IX), and Stuart-Prower factor (X), respectively; and they are necessary to induce thrombus formation.33 On the other hand, PROC plays an important role in the inhibition of thrombi formation; similarly, PLAT is involved in fibrinolysis (degradation of thrombi).33 Finally, GGCX is expressed in the liver and catalyzes a vitamin K-dependent post-translational modification required by several coagulation proteins to properly function.34 We selected PLAT and GGCX as potential drug targets from the network analysis because they directly interact with viral proteins and are key regulators of coagulation. A dysfunction of these proteins could explain, at least in part, the thromboembolic complications observed during SARS-CoV-2 infection.
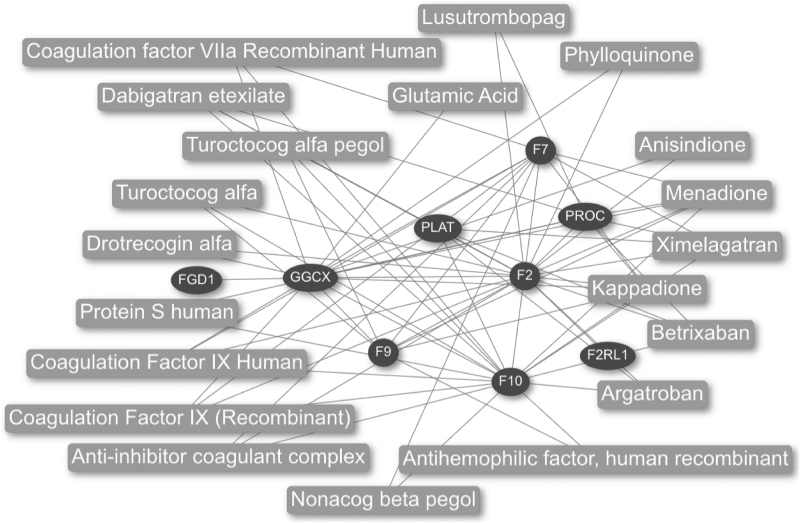
Next, we obtained approved drugs with the “Find drugs” analysis using closeness centrality (Fig. 7). The analysis shows the top 20 drugs identified by the network analysis. Attractive drugs are argatroban, dabigatran etexilate, betrixaban, ximelagatran, and anisindione, because they exhibit anticoagulant effects and additionally target the proteins PLAT and GGCX; thus these drugs could be considered as additional treatment in COVID-19. Of note, dabigatran is an approved anticoagulant drug routinely used in different settings. Our data provide pathophysiological insights concerning the benefit of anticoagulant treatment during SARS-CoV-2 infection, which may reduce the impact of COVID-19 coagulopathy. Clinical trials investigating different antiplatelet therapies are ongoing worldwide and may validate our in silico findings.
Fig. 7.
Drug–PPI network obtained with the closeness centrality algorithm in CoVex. The resulting network contains host seed proteins (elliptical nodes) and approved drugs (rectangular nodes).
In summary, we showed that CoVex is capable of extracting biologically relevant networks useful to put the interactors of SARS-CoV-2 into molecular context by integrating drug–PPI information and network-based drug repurposing methods.
Conclusions and Outlook
Systems medicine is a powerful emerging paradigm for drug discovery and drug repurposing, which aims at targeting the molecular disease mechanisms. In this article, we showcased how to leverage systems medicine for individuating putative drug targets and potentially repurposable drugs for COVID-19 treatment using the SARS-CoV-2 systems medicine platform CoVex. We individuated GLA, PLAT, and GGCX as putative drug targets and argatroban, dabigatran etexilate, betrixaban, ximelagatran, and anisindione as potentially repurposable drugs. Since CoVex is designed to allow easy incorporation of expert knowledge, we hope that more and more researchers from pharmacology and related fields will use it to unravel further promising strategies for COVID-19 treatment.
Acknowledgments
The authors are grateful for support by Harald H.H.W. Schmidt and Frank Weidemann for establishing the contacts to Fabry disease experts across Europe.
Abbreviations Used
- α-GAL
α-galactosidase
- ACE2
angiotensin-converting enzyme 2
- BP
biological process
- COVID-19
coronavirus disease 2019
- GGCX
gamma-glutamyl carboxylase
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PLAT
tissue plasminogen activator
- PPI
protein–protein interaction
- PROC
protein C
- RIPK4
receptor-interacting serine/threonine-protein kinase 4 protein
- WP
Wikipathways
Disclosure Statement
No competing financial interests exist.
Funding Information
J.B. and S.S. are grateful for financial support from H2020 grant REPO-TRIAL (no. 777111). M.S.-A. is a doctoral student from “Programa de Maestría y Doctorado en Ciencias Bioquímicas, UNAM” and is grateful for a PhD fellowship funding from CONACYT (CVU659273 and 312021) and the German Academic Exchange Service, DAAD (ref. 91693321). J.B. and J.M. acknowledge financial support from H2020 grant FeatureCloud (no. 826078) and BMBF grant Sys_CARE (no. 5103388). D.B.B. gratefully acknowledges financial support from the German Center for Cardiovascular Research, DZHK (no. 81X3600606).
References
- 1. Sadegh S, Matschinske J, Blumenthal DB, et al. : Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat Commun 2020;11:3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS: Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 2020;46:586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karen JK, Hale EK, Ma L: Angiokeratoma corporis diffusum (Fabry disease). Dermatol Online J 2005;11:8. [PubMed] [Google Scholar]
- 4. Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F: Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gordon DE, Jang GM, Bouhaddou M, et al. : A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gysi DM, Do Valle Í, Zitnik M, et al. : Network medicine framework for identifying drug repurposing opportunities for COVID-19. arXiv.org 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baumbach J, Schmidt HHHW: The end of medicine as we know it: introduction to the new journal, Systems Medicine. Syst Med 2018;1:1–2 [Google Scholar]
- 8. Casas AI, Hassan AA, Larsen SJ, et al. : From single drug targets to synergistic network pharmacology in ischemic stroke. Proc Natl Acad Sci U S A 2019;116:7129–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen K-Y: Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov 2016;15:327–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Senanayake SL: Drug repurposing strategies for COVID-19. Future Drug Discov 2020;2. DOI: 10..4155/fdd-2020-0010. [Google Scholar]
- 11. Kotlyar M, Pastrello C, Malik Z, Jurisica I: IID 2018 update: context-specific physical protein–protein interactions in human, model organisms and domesticated species. Nucleic Acids Res 2019;47:D581–D589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mendez D, Gaulton A, Bento AP, et al. : ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 2019;47:D930–D940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wishart DS, Feunang YD, Guo AC, et al. : DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074–D1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ursu O, Holmes J, Bologa CG, et al. : DrugCentral 2018: an update. Nucleic Acids Res 2019;47:D963–D970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Zhang S, Li F, et al. : Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res 2020;48:D1031–D1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Armstrong JF, Faccenda E, Harding SD, et al. : The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res 2020;48:D1006–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barbarino JM, Whirl-Carrillo M, Altman RB, Klein TE: PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med 2018;10:e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J: BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 2016;44:D1045–D1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kacprowski T, Doncheva NT, Albrecht M: NetworkPrioritizer: a versatile tool for network-based prioritization of candidate disease genes or other molecules. Bioinformatics 2013;29:1471–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gyöngyi Z, Garcia-Molina H, Pedersen J: Combating web spam with TrustRank. In: Proceedings 2004 VLDB Conference, pp. 576–87. Toronto, Canada: Elsevier, 2004 [Google Scholar]
- 21. Alcaraz N, List M, Dissing-Hansen M, et al. : Robust de novo pathway enrichment with KeyPathwayMiner 5. F1000Res 2016;5:1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. UniProt Consortium T, The UniProt consortium: UniProt: the universal protein knowledgebase. Nucleic Acids Res 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raudvere U, Kolberg L, Kuzmin I, et al. : g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191–W198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guney E, Menche J, Vidal M, Barábasi A-L: Network-based in silico drug efficacy screening. Nat Commun 2016;7:10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. GLA gene. GTEx Portal. Available at: https://www.gtexportal.org/home/gene/GLA Last accessed on June9, 2020
- 26. Germain DP: Fabry disease. Orphanet J Rare Dis 2010;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Geng X, Tan Y, et al. : New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed Pharmacother 2020;127:110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oudkerk M, Büller HR, Kuijpers D, et al. : Diagnosis, prevention, and treatment of thromboembolic complications in COVID-19: report of the National Institute for Public Health of the Netherlands. Radiology 2020;297:E216–E222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Atallah B, Mallah SI, AlMahmeed W: Anticoagulation in COVID-19. Eur Heart J Cardiovasc Pharmacother 2020;6:260–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jose RJ, Manuel A: COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med 2020;8:e46–e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Budnik I, Brill A: Immune factors in deep vein thrombosis initiation. Trends Immunol 2018;39:610–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Varga Z, Flammer AJ, Steiger P, et al. : Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020;395:1417–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palta S, Saroa R, Palta A: Overview of the coagulation system. Indian J Anaesth 2014;58:515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Presnell SR, Stafford DW: The vitamin K-dependent carboxylase. Thromb Haemost 2002;87:937–946 [PubMed] [Google Scholar]