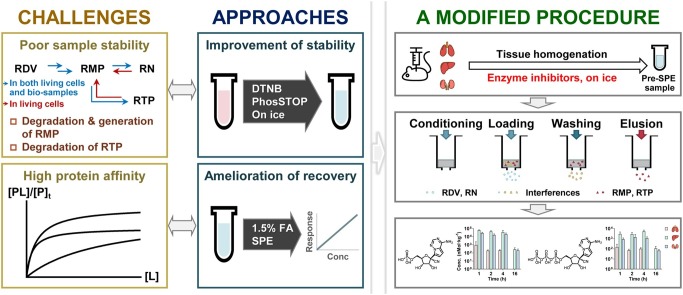
Graphical abstract
Abbreviations: ACN, acetonitrile; AgNO3, silver nitrate; ATNs, active triphosphate nucleotide metabolites; ATP, adenosine triphosphate; BNPP, bis (p-nitrophenyl) phosphate; COVID 19, coronavirus disease 2019; DTNB, 55′ dithiobis (2-nitrobenzoic acid); ESI, electrospray ionization; FA, formic acid; FDA, United States Federal Drug Administration; FT-ICR MS, fourier transform ion cyclotron resonance mass spectrometry; HSA, human serum albumin; IS, internal standard; KD, equilibrium dissociation constant; LLE, liquid-liquid extraction; LLOQ, lower limit of quantitation; MeOH, methanol; NH4Ac, ammonium acetate; PPT, protein precipitation; QC, quality control; RdRp, RNA-dependent RNA polymerases; RDV, remdesivir; RMP, remdesivir monophosphate; RN, remdesivir nucleoside; SPE, solid-phase extraction; RSDs, relative standard deviations; RTP, remdesivir nucleotide triphosphate; SARS CoV 2, severe acute respiratory coronavirus 2; TCEP, tris (2 carboxyethyl) phosphine hydrochloride; UPLC-MS/MS, ultra-performance liquid chromatography coupled triple-quadrupole mass spectrometry; UTP, uridine triphosphate
Keywords: Remdesivir, Nucleotide monophosphate, Nucleotide triphosphate, Stability, Recovery, LC–MS/MS
Highlights
-
•
A robust and reliable LC–MS/MS method for the quantification of RMP and RTP was optimized and validated.
-
•
The novel method solved the major challenges of direct determination of RMP and RTP in biological matrix through improvement of LC retention, stability and recovery.
-
•
The method was validated and successfully applied to mouse tissue distribution study.
-
•
This method provides useful information for further study of remdesivir as well as extends the approach for phosphate determination.
-
•
The method solved the major challenges for determining RMP and RTP in biological matrix such as LC retention, stability and recovery.
Abstract
Remdesivir is a prodrug of the nucleotide analogue and used for COVID-19 treatment. However, the bioanalysis of the active metabolites remdesivir nucleotide triphosphate (RTP) and its precursor remdesivir nucleotide monophosphate (RMP) is very challenging. Herein, we established a novel method to separate RTP and RMP on a BioBasic AX column and quantified them by high-performance liquid chromatography-tandem mass spectrometry in positive electrospray ionization mode. Stepwise, we optimized chromatographic retention on an anion exchange column, improved stability in matrix through the addition of 5,5′-dithiobis-(2nitrobenzoic acid) and PhosSTOP EASYpack, and increased recovery by dissociation of tight protein binding with 2 % formic acid aqueous solution. The method allowed lower limit of quantification of 20 nM for RMP and 10 nM for RTP. Method validation demonstrated acceptable accuracy (93.6%–103% for RMP, 94.5%–107% for RTP) and precision (RSD < 11.9 % for RMP, RSD < 11.4 % for RTP), suggesting that it was sensitive and robust for simultaneous quantification of RMP and RTP. The method was successfully applied to analyze RMP and RTP in mouse tissues. In general, the developed method is suitable to monitor RMP and RTP, and provides a useful approach for exploring more detailed effects of remdesivir in treating diseases.
1. Introduction
Currently, the novel coronavirus disease 2019 (COVID19) is a global pandemic and has severely affected the normal life and work of human beings in the world [1]. Since the severe acute respiratory coronavirus 2 (SARS-CoV2) is well identified as the pathogen [2,3], the development of antivirus drugs and vaccines towards SARS-CoV2 is extremely urgent and important for COVID-19 treatment [4].
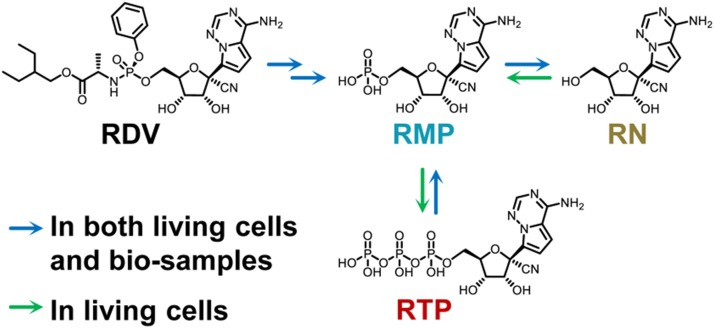
Remdesivir (RDV) is a prodrug of a 1′-cyano-substituted adenine C-nucleoside ribose analogue. It undergoes metabolic activation to exert potent inhibitory activity against SARS-CoV-2 [5]. In brief, RDV is first completely hydrolysed by various esterases, such as carboxylesterase, phosphoramidase, acetylesterase and nucleotidase, to form monophosphate (RMP) and nucleoside (RN). Then, RMP is further phosphorylated to the pharmacologically active form, remdesivir nucleotide triphosphate (RTP), which interferes with the activity of viral RNA-dependent RNA polymerases (RdRp) of SARS-CoV-2 [6].
RDV has been granted emergency use authorization for patients with severe COVID19 by the United States Federal Drug Administration (FDA), and its widespread clinical use is on the horizon [7,8]. However, there still exist some issues regarding efficacy and safety that need to further evaluated and confirmed, such as dose judgements for special populations and potential drug-drug interactions. [[9], [10], [11]]. The exposure of RDV and its metabolites can help to address these considerations. Several studies have revealed that both RDV and RN could be effectively determined in human and animal samples after intravenous injection or intragastric administration of RDV [12,13]. To date, there are few methods reported in detail for direct determination of the more important active compound RTP and its precursor form RMP in biological matrix, which might be attributed to methodological challenges of nucleotides, such as poor stability, high protein affinity, and weak chromatographic retention [[14], [15], [16], [17]].
Therefore, in this study, we aimed to solve the aforementioned challenges step by step and establish a robust LC–MS/MS method for simultaneous quantification of both RMP and RTP in different mouse tissues. Moreover, we successfully applied it to measure the exposure of several tissues to RMP and RTP after a single oral administration of RDV in mice.
2. Materials and methods
2.1. Chemicals and reagents
RDV (purity ≥ 98 %), RMP (purity of 93 %), and RTP (purity of 98 %) were synthesized as previously reported [6,18]. The structure and purity of each compound were identified by NMR and HPLC-UV. RN (purity of 98 %) was purchased from Target Molecule Corp. (Boston, MA, USA). Adenosine triphosphate (ATP, purity of 99 %), uridine triphosphate (UTP, purity of 96 %), 13C,15N-ATP (purity of 98 %), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, purity ≥ 98 %), bis (p-nitrophenyl) phosphate (BNPP, purity ≥ 99 %), tris-(2-carboxyethyl)phosphine hydrochloride (TCEP, purity ≥ 98 %), Boc-Lys(Boc)-OH, ammonium hydroxide (NH3∙H2O, 28 % NH3 in H2O), and ammonium acetate (NH4Ac) were obtained from Sigma-Aldrich (St. Louis, MO, USA). PhosSTOP™ EASYpack (PhosSTOP) was purchased from Roche (Basel, Switzerland). The Invitrosol™ LC/MS Protein Solubilizer Kit was purchased from ThermoFisher (MA, USA). Phosphoric acid (H3PO4, purity ≥ 85 %) and silver nitrate (AgNO3) were obtained from Sinopharm Chemical Regent Co., Ltd. (Shanghai, China). Acetonitrile (ACN), formic acid (FA), and methanol (MeOH) were obtained from Merck (NJ, USA). Ultrapure water was obtained using a Millipore Milli-Q® Advantage A10 Purification System (Millipore, Molsheim, France). All other regents were of the highest quality commercially available.
2.2. Instrumentation
The development and validation processes for quantification of RMP and RTP were conducted in an ultra-performance liquid chromatography coupled triple-quadrupole mass spectrometry (UPLC-MS/MS) instrument, which comprised an AQUITY UPLC system with a thermostatted autosampler and an ultrahigh performance binary pump (I-class, Waters, MA, USA), and a triple quadrupole mass spectrometer with electrospray ionization (ESI) source (Xevo TQ-S, Waters, MA, USA). The chromatographic separation was performed on a BioBasic AX column (2.1 × 50 mm, 4.6 μm; ThermoFisher) at a temperature of 45 °C, using ACN-H2O (3:7, v/v) with 10 mM NH4Ac (pH 6.0) and ACN-H2O (3:7, v/v) with 1 mM NH4Ac (pH 10.5) as mobile phases A and B, respectively. The flow rate was 0.6 mL min−1 under a gradient elution: 0–0.5 min, 5 % B; 0.5–0.8 min, 5%–10% B; 0.8–4.5 min, 10%–90% B; 4.5–5.5 min, 90 % B; 5.5–6.5 min, 90 % – 5 % B. The autosampler was set at 15 °C and the injection volume was 5 μL. The following transitions were monitored: m/z 372.0 → m/z 202.1 for RMP, m/z 532.1 → m/z 202.1 for RTP, and m/z 523.1 → m/z 146.0 for 13C,15N-ATP (IS). Mass spectrometry settings were as follows: capillary voltage 3.0 kV, source offset 50 V, desolvation gas flow 1000 L∙h-1, cone gas flow 150 L∙h-1, nebulizer pressure 7.0 bar, collision gas (argon) flow 0.14 mL min-1, and desolvation temperature 500 °C. Data were captured and analyzed using MassLynx software (version 4.1, Waters, MA, USA).
2.3. Optimization of tissue stability
The stabilization procedure was optimized in liver homogenates under different conditions at 15 and 30 min. A 50 μL liver homogenate was immediately added to a 1.5 mL polyethylene tube containing 5 μL RMP or RTP solution (10 μM in water) and different stabilizing agents as follows: (a) 10 μL 100 mM BNPP; (b) 10 μL 500 mM DTNB; (c) 5 μL PhosSTOP solution (the solution was prepared by dissolving one piece of PhosStop tablet into 1 mL waters, and the spiked sample concentration of PhosSTOP was referred to as 10 % PhosSTOP); or (d) 10 μL 500 mM DTNB and 5 μL PhosSTOP solution. Then a 150 μL 2 % FA aqueous solution was added to the prepared tube for further solid-phase extraction (SPE). For comparison, another 50 μL liver homogenate was added to a 1.5 mL polyethylene tube with 500 μL of ACN-MeOH (1:1, v/v) to precipitate protein and inactivate enzymes thoroughly, and then RMP or RTP solution was added into the supernatant. Next, after SPE, the samples were analyzed.
2.4. Evaluation of protein affinity of RTP
Binding studies between human serum albumin (HSA, purity ≥ 98 %, Sigma-Aldrich) and RTP, ATP, or UTP were conducted under native conditions using an electrospray ionization-Fourier transform ion cyclotron resonance mass spectrometry (ESI-FT-ICR MS) method as previously reported [19]. The commercial HSA was purified five times by buffer exchange with 10 mM NH4Ac (pH 6.0) buffer in an Amicon Ultra 30k cellulose membrane filter (Merck). Purified HSA (3 μM) and different concentrations of RTP, ATP, and UTP were prepared in 10 mM NH4Ac as spray solutions to form differential HSA-nucleotide ratios. The instrument conditions for the scimaX MRMS system (Bruker Daltonics, Bremen, Germany) were optimized to simultaneously maximize the relative ionization efficiency of the HSA-nucleotide complex over the free HSA and minimize the HSA-nucleotide complex dissociation during the ionization and transference processes as follows: Skimmer 1 50 V, Funnel 1 180 V, Funnel RF Amplitude 200 V, collision cell frequency 1.4 MHz, ion transfer frequency 1 MHz, time of flight to analyzer cell 2.0 ms with radio frequency amplitude of 4500 V, source temperature 120 °C. The ion accumulation time was set to 400 ms for the optimal signal-to-noise ratio of the mass spectrum. Two hundred single scans were combined for the final mass spectrum. Finally, the spray solutions were infused into the FT-ICR MS at a flow rate of 5 μL∙min−1 in positive ion spray mode. Data were acquired in the m/z range 1000–5000 with a resolving power of 54.000 at m/z 3240. The charges stated 17+ to 19+ were isolated and used to calculate the intensity ratios of the free HSA to the HSA-nucleotide complex. Mass spectra data were deconvoluted using the SNAP2 function in DataAnalysis 5.2 software (Bruker Daltonics, Germany). The equilibrium dissociation constant (KD) was calculated using a nonlinear regression in GraphPad Prism 8.2.1.
2.5. Optimization of recovery
The recovery was optimized using liver homogenate under several different conditions, including 2 % FA in H2O, 3 % FA in H2O, H3PO4, 10 M NH4Ac, 20 mg mL−1 AgNO3, 1 M Boc-Lys(Boc)-OH, 10 mM TCEP, 20 % Invitrosol, 50 μL liver homogenate, which was immediately added to a 1.5 mL polyethylene tube containing 5 μL RMP or RTP solution and prepared by 150 μL aforementioned regents, respectively.
2.6. Optimization of sample preparation
Tissue samples were individually homogenized with four equivalent volumes of H2O containing PhosSTOP and DTNB. Then, 50 μL homogenate was immediately added to a 1.5 mL polyethylene tube containing 150 μL 2 % FA aqueous solution and 5 μL of IS working solution (10 μM 13C,15N-ATP in water), then vortexed thoroughly and centrifuged at 4 °C, 11,363 × g for 3 min. Then, 160 μL supernatant was mixed with an equivalent volume of water to carry out the SPE with a weak anion exchange plate (Waters Oasis WAX μ-Elution plate). Prior to SPE, the cartridge was preconditioned with 200 μL of MeOH and then further conditioned with 200 μL of H2O. After sample loading, the cartridge was washed with 200 μL 5 mM NH4Ac aqueous solution (pH 4.5) and 200 μL MeOH, then eluted with 200 μL 10 % NH3·H2O in ACN-MeOH (6:4, v/v). Finally, the eluate was gently evaporated to dryness under a gentle stream of air, and the residue was reconstituted with 100 μL H2O for analysis. All SPE extractions were performed on a Waters Positive Pressure-96 processor (Waters, MA, USA).
2.7. Method validation
To minimize experimental errors resulting from instability, the RMP and RTP working solutions were freshly prepared before use. For the establishment of standard curves, RMP and RTP working solutions were serially diluted with liver homogenate to final concentrations of 20, 50, 100, 200, 500, 1,000, 4,000, and 10,000 nM for RMP and 10, 20, 50, 100, 200, 500, 1,000, 4,000, and 10,000 nM for RTP, spiked with 5 μL of IS (10 μM in water), and prepared as described above in the sample preparation section. The calibration curves were evaluated by plotting the absolute peak-area ratios of the analyte to the IS area against the corresponding RMP and RTP concentrations. The linearity of RMP and RTP to IS peak area ratio versus the theoretical concentration was verified using 1/x2-weighted linear regression. The precision and accuracy for intra- and inter-day analyses of the RMP and RTP were performed by quality control (QC) samples at concentrations of 30, 300 and 7,500 nM. The QC samples were prepared and extracted as described in standard samples. To assess intra-day precision and accuracy, six replicates were prepared and analyzed on the same day. To assess inter-day precision, samples were analyzed for three consecutive days. The precision was evaluated by recoveries and intra- and inter-day relative standard deviations (RSDs). The accuracy was calculated from the detected concentrations divided by the theoretical concentration of RMP and RTP. Sensitivity was assessed by the lower limit of quantitation (LLOQ). Matrix effects are analyzed by comparison of extracted blank matrix spiked with analyte and a dilution of analytes at the same concentration in solvent used for injection. In addition, partial validation including accuracy, precision, recovery, matrix, and etc., were conducted in kidney and lung homogenates with respective QC samples as the same concentrations as those of liver homogenates.
The tissue concentrations were calculated by the equation as follows: Ctissue = Chomogenate × V/M, where Ctissue is the tissue concentration (nmol·kg−1), Chomogenate is the tissue homogenate concentration (nM), V is the tissue homogenate volume (mL), and M is the weight of the tissue sample (g).
2.8. Animal study
Caesarean Derived-1 (CD-1) mice (male, 7–8 weeks, 18–22 g) were purchased from Shanghai Super-B&K Laboratory Animal Co., Ltd. (Shanghai, China). Twenty-four CD-1 mice were randomly divided into four groups (n = 6) and allowed free access to water but fasted for 12 h prior to the experiment. All mice received a single oral administration of 100 mg kg−1 RDV, which was dissolved in DMSO-0.5 % hydroxypropyl methylcellulose (HPMC) (5/95, v/v). Mice were anaesthetized at 1, 2, 4, and 16 h post-dosing. Tissue samples, including liver, kidney, and lung were harvested after perfusion with saline and immediately homogenized for further analysis. All animal experiment procedures were conducted in accordance with the guidelines of the National Research Council. The experimental protocol was approved by the Animal Care and Use Committee of the Shanghai Institute of Materia Medica (No.: 2020-02-YY-10).
3. Results
3.1. Instrumentation optimization
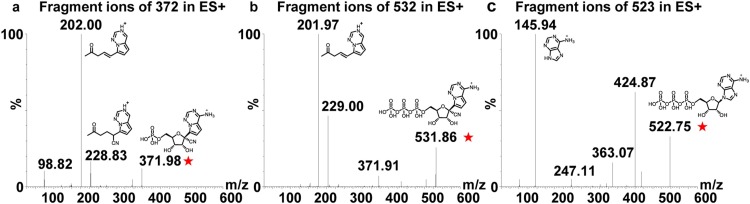
RMP, RTP, and 13C,15N-ATP (IS) contained both amino and phosphate groups, which could be ionized under ESI in both positive and negative modes. Since both analytes and IS showed higher signal responses in positive mode and less fragmentation ion interference, their [M+H]+ ions were chosen as parent ions and transitions of m/z 372 → m/z 202, m/z 532 → m/z 202, and m/z 523 → m/z 146 were chosen for monitoring RMP, RTP, and IS, respectively (Fig. 2 ).
Fig. 2.
MS2 spectra of RMP (a), RTP (b) and 13C,15N-ATP (c).
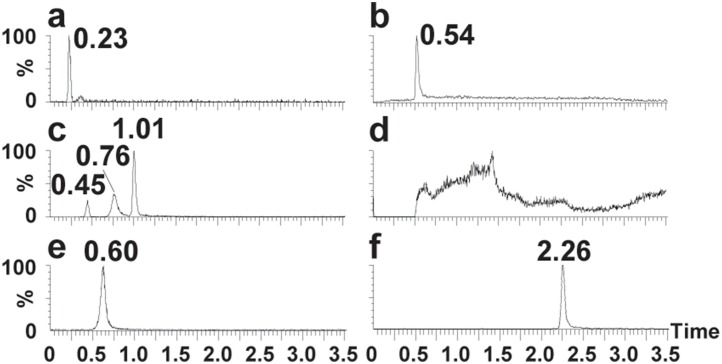
The optimization of chromatographic retention for RMP and RTP was conducted on several LC columns with different stationary phases. First, HSS T3 (Waters) columns were evaluated. Due to their compatibility with the 100 % aqueous phase, these columns are good choices for highly polar analytes. However, RTP could not be retained in these columns with MS compatible mobile phases (Fig. 3 a and b). Then, hydrophilic interaction LC columns, including BEH Amide column (Waters) and Kinetex Polar C18 column (Phenomenex), were tried using common mobile phase, but failed to obtain satisfactory retention for RTP (Fig. 3c and d). In addition to the above tested methods, ion-pairing chromatography is another approach to improve the retention of highly hydrophilic compounds [15,20]. However, the ion-pairing reagents in the mobile phase could cause high ion suppression for MS response. Alternatively, we further chose an anion exchange column (Biobasic AX, 2.1 × 50 mm, 4.6 μm, Thermo Fisher) with MS compatible mobile phase [21], which could retain highly polar compounds by adjusting the pH conditions. Using solvents of 10 mM NH4Ac in ACN-H2O (3:7, v/v) at different pH values, several LC conditions were compared (Fig. 3e and f). We finally achieved the desired retention and peak shape using a gradient program, with which the retention times of RMP and RTP were 1.38 min and 2.26 min, respectively.
Fig. 3.
Chromatograms of RTP under different LC conditions. (a) Column: HSS T3 (2.1 × 50 mm, 1.8 μm, Waters); Solvent: 5 mM NH4Ac in H2O; (b) Column: HSS T3 (2.1 × 100 mm, 1.8 μm, Waters); Solvent: 5 mM NH4Ac in H2O; (c) Column: BEH Amide (2.1 × 100 mm, 1.7 μm, Waters); Solvent: 5 mM NH4Ac in H2O – MeOH, gradient; (d) Column: Kinetex Polar C18 (2.1 × 100 mm, 1.6 μm, Phenomenex); Solvent: 5 mM NH4Ac in H2O – MeOH, gradient; (e) Column: Biobasic AX (2.1 × 50 mm, 4.6 μm, Thermo); Solvent: 10 mM NH4Ac in ACN-H2O (3:7, v/v), pH 6.0–10 mM NH4Ac in ACN-H2O (3:7, v/v), pH 10.0 = 7:3, isocratic; (f) Column: Biobasic AX (2.1 × 50 mm, 4.6 μm, Thermo); Solvent: 10 mM NH4Ac in ACN-H2O (3:7, v/v), pH 6.0–10 mM NH4Ac in ACN-H2O (3:7, v/v), pH 10.0, gradient.
3.2. Initial optimization for sample preparation
Regarding biological sample preparation, protein precipitation (PPT), liquid-liquid extraction (LLE) and SPE were the most commonly used approaches. Owing to the extremely high hydrophilicity of phosphates, water-immiscible organic solvents could not extract RMP and RTP from tissue homogenate. Therefore, LLE could not be applied in sample preparation for phosphate bioanalysis. PPT using ACN, MeOH, acetone, or the other organic solvents is convenient and efficient for removing large amounts of protein from biological matrices. We attempted this approach, however, serious ion suppression was observed due to the co-elution of endogenous components (data not shown). Thus, SPE was used for further development.
Due to the high polarity of RMP and RTP, reversed C18 or C8 packing materials could not be used for the separation of analytes and other endogenous interferences. On account of the strong acidity of the phosphate group, the ion-exchange resin was evaluated. Oasis WAX (Waters) SPE cartridge was packed with anion-exchange sorbent, which could sufficiently absorb analytes under low pH and release them under high pH [14,22,23]. We carefully optimized the balancing, loading and elution procedures. The pH of the balancing solvent was optimized as 4.5 with 5 mM NH4Ac buffer. The elution solvent was optimized as 10 % NH3·H2O in ACN-MeOH (6:4, v/v). With the optimized balancing and elution procedures, RMP and RTP in solution could achieve good recovery in any acid loading solvents. Meanwhile, using this SPE procedure, the sample was purified such that under the HPLC conditions used, there were no observed matrix effects.
3.3. Optimization for stability challenge
Generally, the nucleotide analogue drugs have very poor stability in biological samples [16,21]. As shown in Fig. 1, RDV could degrade into RMP, while RMP could further degrade into RN in biological matrices. Meanwhile, RTP could also degrade into RMP. To determine accurate concentrations in bio-samples, degradation of all RDV, RMP and RTP should be prevented.
Fig. 1.
Transformation pathways of RDV.
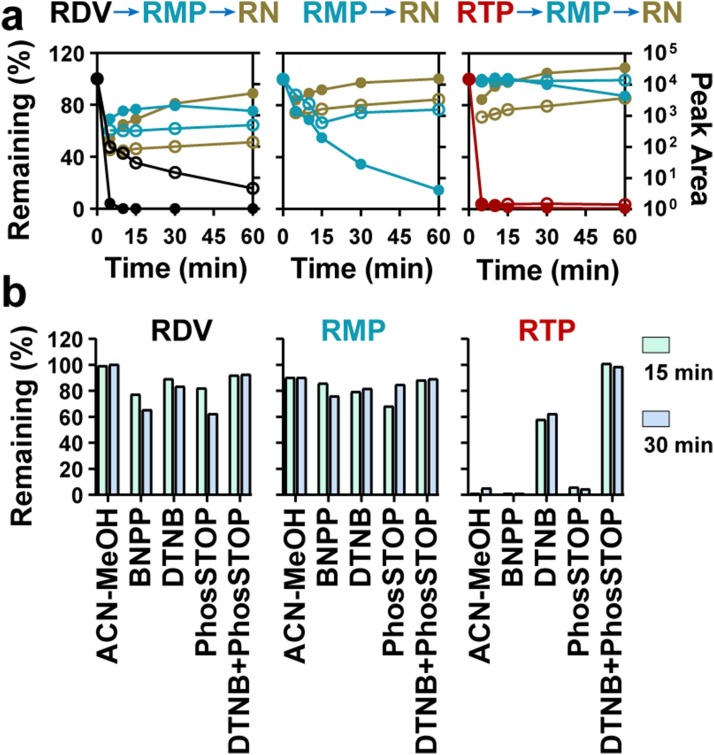
Throughout the pretreatment process, reaction on ice is a normal approach for unstable compounds. Unfortunately, we observed minor resistance to degradation for RDV, RDP, and RTP when placed on ice (Fig. 4 a). Specifically, the ice condition had little effect on both RDV and RTP degradation, which almost completely degraded in 5 min both on ice, as well as at room temperature. Then, several stabilizing agents, including organic solvent (ACN-MeOH, 1:1, v/v), carboxylesterase inhibitors (BNPP and DTNB), and phosphatase inhibitor cocktail (phosSTOP), were investigated alone or in combination (Fig. 4b). Among them, the combination of DTNB and phosSTOP exhibited the best suppression effect for degradation of RDV, RMP and RTP. Thus, 0.5 M DTNB and 10 % phosSTOP mixture was used as the stabilizer during sample preparation.
Fig. 4.
Transformation of RMP and RTP in bio-samples: (a) Degradation/generation curves of RDV (black), RMP (blue) and RTP (red) at room temperature (●) and on ice (○); (b) Inhibition of transformation by different methods.
However, a confusing phenomenon was also observed that RTP showed very low concentrations in the ACN-MeOH treated samples. Theoretically, organic solvent should thoroughly inhibit the activities of metabolic enzymes. After further investigation, we found it was caused by the extremely low recovery of RTP in organic solvent treated samples.
3.4. Optimization for protein binding challenge
Because extremely low recovery of RTP was noticed during method development, we suspected the coprecipitation of RTP and protein. To further investigate the hypothesis and discover the solution, we evaluated the affinity between RTP and albumin and optimized the sample preparation procedure accordingly.
3.4.1. Confirmation of the high affinity between RTP and albumin
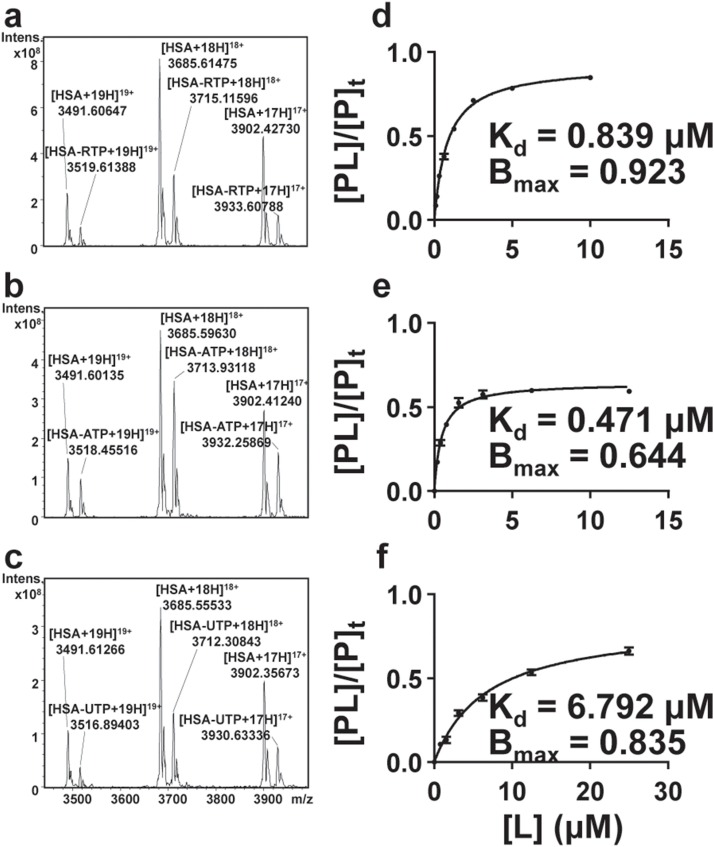
ATP was reported to have a very tight serum albumin binding [24], while RTP, as its analogue, could have similar properties. The affinity between RTP and HSA was evaluated by the FT-ICR MS method (Table 1 and Fig. 5 ). The KD of RTP with HSA was 0.839 μM and close to that of ATP (0.471 μM), which confirmed a high serum albumin binding affinity for RTP. UTP with a much different nucleotide showed a much lower binding affinity. The tight protein binding of RTP could cause coprecipitation when biological samples were precipitated using organic solvent and then a consequent low recovery.
Table 1.
The equilibrium dissociation constants (Kd) of RTP, ATP, and UTP with human serum albumin (HSA), respectively.
| Name | Kd (μM) | Bmax ([PL]/[P]t) |
|---|---|---|
| RTP | 0.8388 ± 0.0268 | 0.9227 ± 0.0067 |
| ATP | 0.4710 ± 0.0238 | 0.6443 ± 0.0138 |
| UTP | 6.7920 ± 0.3326 | 0.8357 ± 0.0180 |
Fig. 5.
Native MS spectra of HSA with RTP (a), ATP (b), and UTP (c), and their saturation curves (d, e, f). n = 3 for each concentration.
3.4.2. Approaches to improve recovery
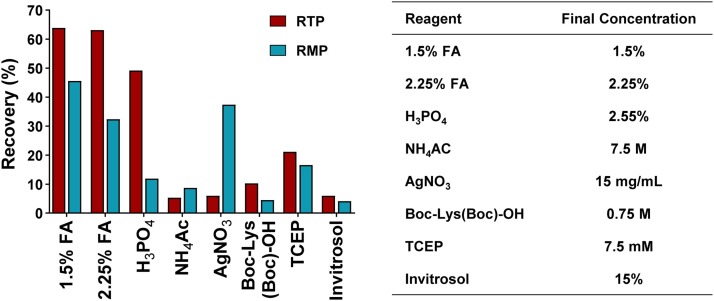
To overcome the issue of tight protein binding, several protein dissociation approaches were carefully evaluated, including acid dissociation (FA and phosphoric acid), salting-out (NH4Ac), disulfide bond reduction (TCEP), thioester cleaving (AgNO3), tyrosine residue blocking (Boc-LYS(Boc)-OH), and protein solubilizing (Invitrosol). The details and results are shown in Fig. 6 . Eventually, 1.5 % FA aqueous solution was used as the loading solvent, with which we achieved relatively high recoveries for RMP and RTP.
Fig. 6.
Recovery of RTP and RMP with different preparation methods.
3.5. Method validation
3.5.1. Linearity of calibration standards
The calibration curves were linear over the concentration ranges of 20 to 10,000 nM and 10 to 10,000 nM for RMP and RTP, respectively. In the assessment, the accuracy values of the back-calculated concentrations of calibration standards including the LLOQs from three different runs ranging from 94.4%–101% and 95.3%–113% for RMP and RTP, respectively. For all concentrations, RMP and RTP showed excellent precision with RSDs less than 2.6 % and 6.2 %, respectively. The determination coefficients (r2) were more than 0.99 across all the analytes in 3 runs. The typical equations of RMP and RTP in tissue homogenate were y = 0.000319 X + 0.00219 (r2 = 0.9903) and y = 0.000678 X - 0.00450 (r2 = 0.9929), respectively.
3.5.2. Specificity
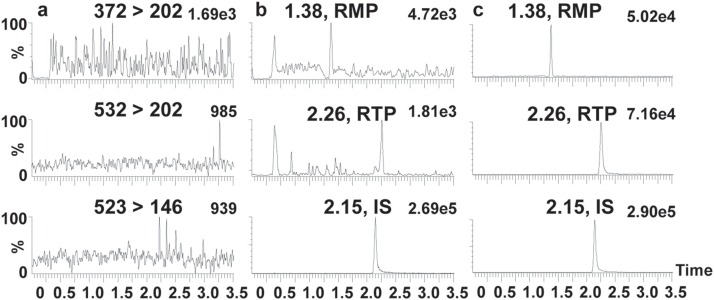
MRM chromatograms of blank sample were compared with that at the LLOQ. The typical chromatograms are shown in Fig. 7 . Under the optimized LC–MS/MS condition, no significant interferences were observed at the retention times of the analytes.
Fig. 7.
Representative MRM chromatograms for RMP, RTP, and IS in mouse liver homogenates: (a) blank mouse liver homogenates; (b) mouse liver homogenates spiked with RMP (20 nM), RTP (10 nM), and IS (1,000 nM); (c) mouse liver homogenates spiked with RMP (500 nM), RTP (500 nM), and IS (1,000 nM).
3.5.3. Precision and accuracy
Accuracy and precision were evaluated by six replicate analyses of the LLOQ (10 nM and 20 nM for RMP and RTP, respectively), low (30 nM for both RMP and RTP), medium (300 nM for both RMP and RTP) and high QC (7,500 nM for both RMP and RTP) levels in three separate analytical runs. The acceptance criteria of accuracy and precision were 85 %–115 % and less than 15 % for all concentration levels except for LLOQ. The acceptance criteria of accuracy and precision for LLOQ were 80%–120% and less than 20 %.
The results of intra- and inter-day accuracy and precision based on LLOQ and QC samples are summarized in Table 2 . The accuracy for RMP ranged from 93.6%–103%. The RSD of intra-assay precision for the RMP was < 6.3 %, while the RSD of inter-assay precision was < 11.9 %. The accuracy for RTP ranged from 94.5%–107%. The RSD of intra-assay precision for the RTP was < 6.2 %, while the RSD of inter-assay precision was < 11.4 %. The method was determined to be reliable and reproducible as all the accuracy and the intra- and inter-day precision values were within acceptable ranges.
Table 2.
Accuracy and precision for the analysis of RMP and RTP in mouse liver homogenate (3 days with 6 replicates for each concentration per day).
| Analyte | Nominal conc. (nM) |
Mean conc. (nM) |
Accuracy (%) |
Inter-day RSD (%) |
Intra-day RSD (%) |
|---|---|---|---|---|---|
| RMP | 20.0 | 20.6 | 103.0 | 5.7 | 5.8 |
| 30.0 | 29.4 | 98.2 | 3.2 | 6.3 | |
| 300 | 285 | 95.0 | 9.7 | 5.3 | |
| 7,500 | 7,022 | 93.6 | 11.9 | 4.2 | |
| RTP | 10.0 | 10.7 | 106.7 | 11.4 | 6.2 |
| 30.0 | 28.3 | 94.5 | 8.7 | 5.8 | |
| 300 | 290 | 96.8 | 8.1 | 5.1 | |
| 7,500 | 7,416 | 98.9 | 8.2 | 4.8 |
3.5.4. Matrix effect
The matrix effect ranged from 97.0–97.1% for RMP and 94.9–103% for RTP (Table 3 ). The matrix effect of IS at 1,000 nM was 101 %. The variation (RSD) values of the matrix effects for all the analytes and IS were <11.7 %. Thus, ionization enhancement or suppression was negligible after sample extraction under the current conditions.
Table 3.
Matrix effect of RMP, RTP, and IS in mouse liver homogenate (n = 6 for each concentration).
| Matrix factor (%) | Conc. of RMP (nM) |
Conc. of RTP (nM) |
Conc. of IS (nM) | ||
|---|---|---|---|---|---|
| 30 | 7,500 | 30 | 7,500 | 1,000 | |
| 1 | 112.8 | 97.3 | 90.6 | 93.5 | 96.7 |
| 2 | 100.4 | 97.4 | 92.8 | 95.9 | 99.9 |
| 3 | 98.1 | 103.4 | 97.1 | 96.0 | 102.8 |
| 4 | 88.0 | 106.3 | 103.6 | 95.8 | 101.2 |
| 5 | 82.3 | 115.1 | 97.1 | 107.9 | 102.0 |
| 6 | 88.0 | 100.1 | 101.4 | 92.8 | 103.6 |
| Mean (%) | 94.9 | 103.3 | 97.1 | 97.0 | 101.0 |
| RSD (%) | 11.7 | 6.5 | 5.1 | 5.7 | 2.5 |
3.6. Application to mouse pharmacokinetics and tissue distribution study
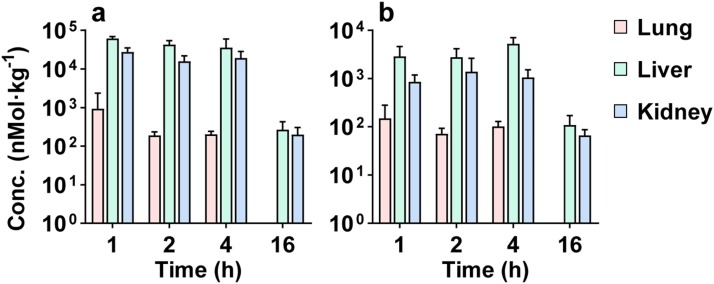
The validated method was applied to the tissue distribution study of RMP and RTP after a single oral dose of remdesivir at 100 mg kg−1 in mice (Fig. 8 ). The main pharmacokinetic parameters are listed in Table 4 . The Cmax and AUC0-t of RMP in the liver, kidney and lung were 61,582, 27,365, and 937 nmol kg−1, and 247,045, 119,349, and 1,329 h∙nmol∙kg−1, respectively. Meanwhile, the Cmax and AUC0-t of RTP in the liver, kidney and lung were 5,251, 1,394, and 151 nmol kg−1, and 28,222, 8,706, and 355 h∙nmol∙kg−1, respectively. RMP could be detected up to 16 h post-dosing in the three tissues, while RTP could be detected up to 16 h post-dosing in liver and kidney and up to 4 h post-dosing in lung (Fig. 8). In the three tissues RMP showed a much higher exposure than RTP. Among the three tissues liver was most abundant and followed by kidney for both RMP and RTP.
Fig. 8.
Tissue concentrations of RMP (a) and RTP (b) in mouse lung, liver and kidney after oral administration of RDV 100 mg kg−1 in mice (n = 6, Mean ± SD).
Table 4.
Main pharmacokinetic parameters of RMP and RTP in mouse tissues after po administration of RDV 100 mg kg−1 (n = 6).
| Tissue | RMP |
RTP |
||||
|---|---|---|---|---|---|---|
| Cmax nmol∙kg−1 |
tmax h |
AUC0-t h∙nmol∙kg−1 |
Cmax nmol∙kg−1 |
tmax h |
AUC0-t h∙nmol∙kg−1 |
|
| Lung | 937 | 1 | 1,329 | 151 | 1 | 355 |
| Liver | 61,582 | 1 | 247,045 | 5,251 | 4 | 28,222 |
| Kidney | 27,365 | 1 | 119,349 | 1,394 | 2 | 8,706 |
4. Discussion
Nucleoside analogues are normally prodrugs that require intracellular phosphorylation to active triphosphate nucleotide metabolites (ATNs) for their therapeutic effects. Numerous assays have been described for the determination of those nucleotides or nucleotide analogues, such as electrochemical method [25], fluorescence spectrometry [26,27] and LC–MS [[14], [15], [16], [17],[20], [21], [22],[28], [29], [30]]. Although reversed phase LC–MS/MS is considered to be the most commonly used tool in bioanalysis, it is still very challenging to measure ATNs and related mono/di-phosphate nucleotide metabolites due to their instability, high hydrophilicity, confusing endogenous interferences, as well as very low concentrations in blood and tissues [31]. To date, there is still a lack of detailed methods for direct determination of RTP and its precursor form RMP in biological matrices, which might hamper therapeutic studies of RDV in COVID-19.
In the current study, facing these vital issues, we optimized the LC–MS/MS method step by step. Initially, similar to the general process for method development of nucleotide compounds, we modified the MS and LC conditions and finally achieved a desired MS response (positive ion mode) and a satisfactory LC retention (separation on an ion-exchange chromatography column). Then, regarding two key challenges of the current method, i.e., stability and recovery, we performed a careful investigation and fit-to-purpose optimization.
Considering stability, degradation is normally the most serious issue for most unstable analytes. However, for the analytes in the current study, especially for RMP, the situation was more complicated. On one hand, RMP could degrade into RN; on the other hand, it could be generated from RDV, as well as RTP [32]. Regarding tissue samples, the preparation procedure contained several steps including homogenization and SPE and thus took some time. Consequently, to obtain accurate concentrations, degradation of all RDV, RMP and RTP should be prevented in tissue sample preparation. We optimized the pretreatment processes in terms of temperature and stabilizing agents, which included organic solvent (ACN-MeOH, 1:1, v/v), carboxylesterase inhibitors (BNPP and DTNB), and phosphatase inhibitor cocktail (phosSTOP). Only low temperature could not prevent degradation, while combined esterase and phosphatase inhibitors could significantly improve stability. Finally, using the modified conditions, the samples were stable for 60 min, which provided an adequate period for sample preparation.
In the experiment, we found very low recovery of RTP by protein precipitation (MeOH or ACN), which is normally used in pretreatment of ATNs in biological matrix [33]. Regarding the issue of RTP’s low recovery, the main reason was investigated and confirmed to be protein-analyte coprecipitation based on the results of protein binding affinity. To resolve this problem, various protein denaturing reagents, such as acid dissociation (FA and phosphoric acid), salting-out (NH4Ac), disulfide bond reduction (TCEP), thioester cleaving (AgNO3), tyrosine residue blocking (Boc-LYS(Boc)-OH), and protein solubilizing (Invitrosol), were tested and compared. Among them, FA treatment showed the highest recovery and was chosen for sample preparation. Meanwhile, RDV is an ester and both RMP and RTP are phosphates, all of which are unstable in either acidic or basic conditions. Thus, their stabilities were monitored during the evaluation of protein denaturing. Using this optimized protein denaturing coupled with the SPE procedure, we achieved a desired recovery for both RMP and RTP.
In summary, targeting several challenges, we performed a stepwise optimization, set up several approaches to overcome the key issues, and eventually achieved a modified strategy. Furthermore, this method was validated and applied to the study of tissue distribution of RMP and RTP in mice after RDV administration.
5. Conclusion
In the current study, we developed a novel sensitive method based on LC–MS/MS for the determination of the mono- and triphosphate types of RDV in mouse tissues. The challenges of weak chromatographic retention, poor stability and high protein affinity were overcome through stepwise fit-for-purpose optimization. The established method was fully validated in terms of linearity, specificity, sensitivity, precision and accuracy and successfully applied to a tissue distribution study of RDV in mice. This method can provide useful information for further study of RDV and may be extended for the determination of a relatively wide group of metabolites that contain mono- and triphosphate groups.
CRediT authorship contribution statement
Wenjuan Hu: Methodology, Software, Writing - original draft. Lu Chang: Data curation, Investigation. Changqiang Ke: Data curation, Investigation. Yuanchao Xie: Resources. Jingshan Shen: Resources. Bo Tan: Conceptualization, Writing - review & editing. Jia Liu: Conceptualization, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This study was supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2016263) and Shanghai Shuguang Hospital (SGXZ-201907).
References
- 1.World Health Organization . 2020. WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/ (Accessed 1 September 2020) [Google Scholar]
- 2.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D., van der Voort P.H., Mulder D.J., van Goor H. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., I. China Novel Coronavirus, T. Research A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., Chen Y.S., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wei X., Gaggar A., Brainard D.M., Towner W.J., Munoz J., Mullane K.M., Marty F.M., Tashima K.T., Diaz G., Subramanian A. GS-US-540-5773 investigators, remdesivir for 5 or 10 days in patients with severe covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.U.S. Food and Drug Administration . 2020. Remdesivir for Certain Hospitalized COVID-19 Patients.https://www.fda.gov/media/137564/download (Accessed 1 September 2020) [Google Scholar]
- 9.Gotzinger F., Santiago-Garcia B., Noguera-Julian A., Lanaspa M., Lancella L., Calo Carducci F.I., Gabrovska N., Velizarova S., Prunk P., Osterman V., Krivec U., Lo Vecchio A., Shingadia D., Soriano-Arandes A., Melendo S., Lanari M., Pierantoni L., Wagner N., L’Huillier A.G., Heininger U., Ritz N., Bandi S., Krajcar N., Roglic S., Santos M., Christiaens C., Creuven M., Buonsenso D., Welch S.B., Bogyi M., Brinkmann F., Tebruegge M., ptbnet C.-S.G. COVID-19 in children and adolescents in Europe: a multinational, multicentre cohort study. Lancet Child Adolesc. Health. 2020;4:653–661. doi: 10.1016/S2352-4642(20)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maharaj A.R., Wu H., Hornik C.P., Balevic S.J., Hornik C.D., Smith P.B., Gonzalez D., Zimmerman K.O., Benjamin D.K., Jr., Cohen-Wolkowiez M., C. Best Pharmaceuticals for Children Act-Pediatric Trials Network Steering Simulated assessment of pharmacokinetically guided dosing for investigational treatments of pediatric patients with coronavirus disease 2019. JAMA Pediatr. 2020 doi: 10.1001/jamapediatrics.2020.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leegwater E., Strik A., Wilms E.B., Bosma L.B.E., Burger D.M., Ottens T.H., van Nieuwkoop C. Drug-induced liver injury in a COVID-19 patient: potential interaction of remdesivir with P-glycoprotein inhibitors. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humeniuk R., Mathias A., Cao H., Osinusi A., Shen G., Chng E., Ling J., Vu A., German P. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin. Transl. Sci. 2020 doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puy J.Y., Jordheim L.P., Cros-Perrial E., Dumontet C., Peyrottes S., Lefebvre-Tournier I. Determination and quantification of intracellular fludarabine triphosphate, cladribine triphosphate and clofarabine triphosphate by LC-MS/MS in human cancer cells. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1053:101–110. doi: 10.1016/j.jchromb.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Mateos-Vivas M., Rodriguez-Gonzalo E., Garcia-Gomez D., Carabias-Martinez R. Hydrophilic interaction chromatography coupled to tandem mass spectrometry in the presence of hydrophilic ion-pairing reagents for the separation of nucleosides and nucleotide mono-, di- and triphosphates. J. Chromatogr. A. 2015;1414:129–137. doi: 10.1016/j.chroma.2015.08.040. [DOI] [PubMed] [Google Scholar]
- 16.Liu A., Lute J., Gu H., Wang B., Trouba K.J., Arnold M.E., Aubry A.F., Wang J. Challenges and solutions in the bioanalysis of BMS-986094 and its metabolites including a highly polar, active nucleoside triphosphate in plasma and tissues using LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015;1000:29–40. doi: 10.1016/j.jchromb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Jimmerson L.C., Ray M.L., Bushman L.R., Anderson P.L., Klein B., Rower J.E., Zheng J.H., Kiser J.J. Measurement of intracellular ribavirin mono-, di- and triphosphate using solid phase extraction and LC-MS/MS quantification. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015;978–979:163–172. doi: 10.1016/j.jchromb.2014.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 19.Pedro L., Van Voorhis W.C., Quinn R.J. Optimization of electrospray ionization by statistical design of experiments and response surface methodology: protein-ligand equilibrium dissociation constant determinations. J. Am. Soc. Mass Spectrom. 2016;27:1520–1530. doi: 10.1007/s13361-016-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnsen E., Wilson S.R., Odsbu I., Krapp A., Malerod H., Skarstad K., Lundanes E. Hydrophilic interaction chromatography of nucleoside triphosphates with temperature as a separation parameter. J. Chromatogr. A. 2011;1218:5981–5986. doi: 10.1016/j.chroma.2011.01.066. [DOI] [PubMed] [Google Scholar]
- 21.Gautam N., Lin Z., Banoub M.G., Smith N.A., Maayah A., McMillan J., Gendelman H.E., Alnouti Y. Simultaneous quantification of intracellular lamivudine and abacavir triphosphate metabolites by LC-MS/MS. J. Pharm. Biomed. Anal. 2018;153:248–259. doi: 10.1016/j.jpba.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen R.S., Rosing H., Schellens J.H., Beijnen J.H. Mass spectrometry in the quantitative analysis of therapeutic intracellular nucleotide analogs. Mass Spectrom. Rev. 2011;30:321–343. doi: 10.1002/mas.20280. [DOI] [PubMed] [Google Scholar]
- 23.Cohen S., Jordheim L.P., Megherbi M., Dumontet C., Guitton J. Liquid chromatographic methods for the determination of endogenous nucleotides and nucleotide analogs used in cancer therapy: a review. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010;878:1912–1928. doi: 10.1016/j.jchromb.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Takeda S., Miyauchi S., Nakayama H., Kamo N. Adenosine 5’-triphosphate binding to bovine serum albumin. Biophys. Chem. 1997;69:175–183. doi: 10.1016/s0301-4622(97)00084-7. [DOI] [PubMed] [Google Scholar]
- 25.Goyal R.N., Oyama M., Tyagi A. Simultaneous determination of guanosine and guanosine-5’-triphosphate in biological sample using gold nanoparticles modified indium tin oxide electrode. Anal. Chim. Acta. 2007;581:32–36. doi: 10.1016/j.aca.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Hai X.M., Li N., Wang K., Zhang Z.Q., Zhang J., Dang F.Q. A fluorescence aptasensor based on two-dimensional sheet metal-organic frameworks for monitoring adenosine triphosphate. Anal. Chim. Acta. 2018;998:60–66. doi: 10.1016/j.aca.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 27.Yu F., Li L., Chen F. Determination of adenosine disodium triphosphate using prulifloxacin-terbium(III) as a fluorescence probe by spectrofluorimetry. Anal. Chim. Acta. 2008;610:257–262. doi: 10.1016/j.aca.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Cohen S., Megherbi M., Jordheim L.P., Lefebvre I., Perigaud C., Dumontet C., Guitton J. Simultaneous analysis of eight nucleoside triphosphates in cell lines by liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877:3831–3840. doi: 10.1016/j.jchromb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 29.Tu X., Lu Y., Zhong D., Zhang Y., Chen X. A sensitive LC-MS/MS method for quantifying clofarabine triphosphate concentrations in human peripheral blood mononuclear cells. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014;964:202–207. doi: 10.1016/j.jchromb.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Philibert G.S., Olesik S.V. Characterization of enhanced-fluidity liquid hydrophilic interaction chromatography for the separation of nucleosides and nucleotides. J. Chromatogr. A. 2011;1218:8222–8230. doi: 10.1016/j.chroma.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 31.Gautam N., Alamoudi J.A., Kumar S., Alnouti Y. Direct and indirect quantification of phosphate metabolites of nucleoside analogs in biological samples. J. Pharm. Biomed. Anal. 2020;178 doi: 10.1016/j.jpba.2019.112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao D., Ling K.H.J., Custodio J., Majeed S.R., Tarnowski T. Quantitation of intracellular triphosphate metabolites of antiretroviral agents in peripheral blood mononuclear cells (PBMCs) and corresponding cell count determinations: review of current methods and challenges. Expert Opin. Drug Metab. Toxicol. 2018;14:781–802. doi: 10.1080/17425255.2018.1500552. [DOI] [PubMed] [Google Scholar]