ABSTRACT
Congenital heart disease (CHD) is one of the major debilitating birth defects resulting in significant impact on neonatal and child mortality globally. The etiology of CHD is complex and multifactorial. Many causative genes responsible for CHDs have been identified from the familial forms previously. Still, the non-Mendelian inheritance and predominant sporadic cases have stimulated research to understand the epigenetic basis and environmental impact on the incidence of CHD. The fetal epigenetic programming affecting cardiac development is susceptible to the availability of key dietary factors during the crucial periconceptional period. This article highlights the need and importance of in-depth research in the new emerging area of maternal nutritional epigenetics and CHD. It summarizes the current research and underlines the limitations in these types of studies. This review will benefit the future research on nutrition as a modifiable environmental factor to decrease the incidence of CHD.
Keywords: congenital heart disease, embryonic development, epigenetic programming, maternal nutrition, DNA methylation
Introduction
Congenital heart diseases (CHDs) are the most common birth defects encountered among the congenital anomalies. They are due to the abnormal formation of heart and great blood vessels during embryonic development. CHDs vary in complexity, from simple shunt lesions to complex critical heart diseases such as hypoplastic left heart syndrome. Some of the congenital heart anomalies are as follows: atrial septal defect (ASD), coarctation of aorta, transposition of great arteries, Ebstein's anomaly, patent ductus arteriosus, single ventricle defects, tetrology of Fallot (TOF), total anomalous pulmonary venous connection, truncus arteriosus, and ventricular septal defect.
CHD contributes to nearly one-third of all the congenital defects. With advances in health care and detection, the incidence of CHDs is currently ∼9.410 per 1000 live births, with a rapid increase in the past 15 y indicating a steep rise in the global burden of CHD (1). The highest prevalence of CHD is reported in Asia, with 9.3 live cases per 1000 live births (2). The mortality and morbidity of CHD can vary with the severity of the disease. There might be a need for multiple surgeries to correct a defect, leading to compromised life quality (3). This highlights the need to study the molecular basis of CHD and formulate preventive strategies to decrease the incidence.
CHDs are classified based on different criteria including, cyanotic or acyanotic, position of defect (in septa, atria, ventricles, veins, and/or great arteries) and more (4). Pathophysiological classification presents these diseases in the following categories (4):
With increased pulmonary blood flow (septal defects with left to right shunt and without pulmonary obstruction).
With decreased pulmonary blood flow (septal defects with right to left shunt and pulmonary obstruction).
No septal defects and obstructive lesions.
Severe forms with multiple lesions and hypoplastic ventricles.
Asymptomatic until adulthood.
The etiology of most CHDs remains elusive. In most cases, genetic as well as environmental factors play key roles contributing to these multifactorial structural malformations (5). Genetic variations contributing to the development of CHD have been explored extensively and present from complex polygenic to simple monogenic high-impact models (6). The genetic mutations responsible for CHD can be sporadic or familial (7). Chromosomal abnormalities also contribute to significant cases of CHD and these are mostly syndromic forms including numeric or structural chromosomal aberrations (8). The genetics of CHD is heterogenous (7) and has been explored for decades and is now developing swiftly with advances in the genetic technologies. Along with the multifactorial etiology of CHD, partial correlation between genotype and phenotype further adds to the complexity (9). The role of genetic factors in CHD development was conceptualized with the finding of recurrence of the disease in families. Familial CHDs can present as autosomal recessive, autosomal dominant, or X-linked traits, with the latter 2 mostly attributed to de novo mutations (7). The study on familial forms has many limitations, including the rare availability of large families with many CHD-affected members. Probable pathogenic mutation in BMPR1A(bone morphogenetic protein receptor type 1A) was recorded in 12 members in a family of 19 individuals (10). More challenging in this type of research is the occurrence of various anatomic forms of CHD in different members of the family, making the interpretation even more complex. Different anatomical expressions of the same contributing genetic factors also highlight the potential impact of environmental factors in the disease etiology (7).
Progress in research in the past few years has uncovered evidence that many genes are responsible for inherited and sporadic CHD. The products of these genes are mostly the transcription factors that have key roles in the ventricular septation or the outflow tract morphogenesis (3).
Mutations in certain transcription factor genes like NKX2–5 (NK2 Homeobox 5), GATA4 (GATA Binding Protein 4), TBX5 (T-Box Transcription Factor 5), and TBX1 (T-Box Transcription Factor 1) alter the normal development of the heart (3, 11–13). In addition, genes encoding signaling molecules and cellular structural components are shown to have a Mendelian inheritance pattern in CHD development. The key examples of this are NOTCH1 (Notch Receptor 1) and JAG1 (Jagged Canonical Notch Ligand 1) (9). Over 440 de novo mutations potentially contributing to CHD were reported recently in a single cohort of 2871 probands (14).
Despite this evidence, the identification of causal genes responsible for several isolated CHDs is still far from completion considering their complex etiology (15). Apart from known causes of 20% of the CHD cases, the cause remains elusive for almost 80% of cases (2). With this hurdle it is essential to explore the environmental and epigenetic causes of these defects.
Since CHD is a structural anomaly, the period of cardiogenesis during embryonic development is a major focus of research. Maternal and fetal environmental factors can deeply influence this developmental process. Research conducted to date in this area unravels the most common potentially causative factors, such as infections during pregnancy (e.g., rubella), certain drugs, maternal diseases, and maternal stress, reflected in increased corticotropin-releasing hormone (CRH) (5). Maternal diet during the periconceptional period is now evolving as an extremely important target for intervention due to its potential role as the protective factor against development of CHD in the offspring (16, 17).
The impact of maternal nutrition on fetal development and birth outcome is well studied in animal studies, but replication in humans has faced many limitations. Baseline maternal nutrition, maternal nutritional variables, time and assessment methods, and socioeconomic factors may skew the study model (18). Another hurdle in such studies is the selection of a nutrient(s). Nutritional deficiencies are more common in low socioeconomic strata and, hence, there is commonly the occurrence of multiple nutritional deficiencies at the same time (18).
The present review provides insights into this relatively new intriguing area of research—that is, maternal nutritional epigenetics and aberrant cardiogenesis during early embryonic development. The term “epigenetics” was first used by Conrad Waddington in the 1940s: “An epigenetic trait is a stably inherited phenotype resulting from changes in a chromosome without alterations in the DNA sequence”(15). The genotype is more or less conserved throughout the life of an organism. The genotype is the same for all cells of the body, with the exception of certain cells such as gametes and cells of the immune system with exclusive phenotypic and functional variability among different cell types. The phenotypic differences found despite the identical genotype are attributed to epigenetic regulation. Epigenetics is an extra-genomic gene regulator (19) and epigenotype is mitotically inheritable and is a potentially reversible molecular alteration of the DNA sequence and chromatin (20). This inheritance plays an important role in maintaining stable gene expression in differentiated cells defining their identity (21). The epigenetic regulation during embryonic development that shapes the life of the individual is summarized in Figure 1.
FIGURE 1.
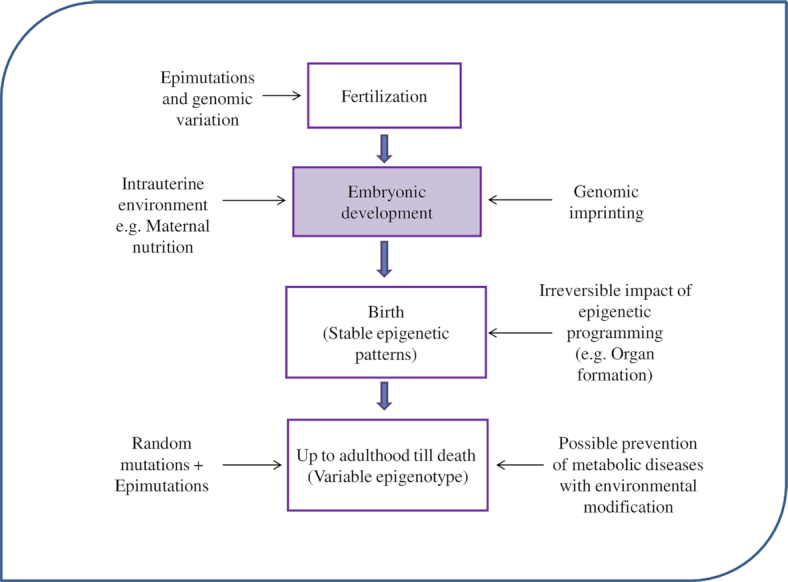
Embryonic programming.
Polygenic diseases such as CHD have been proven to have altered epigenetic patterns through the experimental and clinical research until today (19). The epigenetic set of modifications is diverse in nature and the epigenetic mechanisms include DNA methylation, noncoding RNA (ncRNA)–mediated gene regulation, and histone modifications with consequent chromatin remodeling. The epigenetic regulation is executed by a complex interplay of all these mechanisms. The few studies that have explored the epigenetic basis of CHD are mentioned in Table 1.
TABLE 1.
Studies on the epigenetic basis of CHD1
| Study, year (reference) | Study population/sample type | Sample size, n | Methylation measures/other analyses | Epigenetic change |
|---|---|---|---|---|
| Human | ||||
| Chowdhury et al. 2011 (28) | Women <30 y, maternal blood | Cases: 180Controls: 187 | Genome-wide | Lower global DNA methylation and higher risk of CHD-affected pregnancy |
| Chowdhury et al. 2011 (29) | Women <30 y, maternal blood | Cases: 180Controls: 187 | Gene-specific | Significant differential methylation, but not up to genome-wide statistical significance |
| Hypermethylated CpG sites in cases located in CpG islands | ||||
| Sheng et al., 2013 (26) | Children <5 y, heart tissue | Cases: 30Controls: 6 | Gene-specificMass array, RT-PCR | Aberrant methylation statuses of the NKX2–5 gene body and HAND1 promoter regions in TOF |
| Bahado-Singh et al., 2016 (30) | Newborns, blood spots | Cases: 60Controls: 32 | Genome-wide | Differential methylation in various genes in different CHDs |
| Li et al., 2014 (31) | Toddlers, plasma | Cases: 3; Controls: 3; and Cases: 20Controls: 15 | miRNA microarray analysis and RT-PCR for verification | 36 differentially expressed miRNAs with VSD and upregulation of 1 miRNA and downregulation of 7 miRNAs |
| Serra-Juhe et al., 2015 (25) | Fetuses, heart tissues | Cases: 18Controls: 4 | Genome-wide | 3 aberrant methylation regions per sample and 18 differentially methylated regions; hypermethylation of MSX1 in isolated heart malformation and GATA4 in cases and certain controls |
| Radhakrishna et al. 2018 (32) | Newborns, blood spots | Cases: 24Controls: 24 | Genome-wide | 64 differentially methylated CpG sites with 25 having high predictive accuracy for TOF and CpG methylation variation >10% in 51 CpG islands |
| Radhakrishna et al. 2019 (33) | Newborns, placental tissue | Cases: 8Controls: 10 | Genome-wide | Differential methylation patterns in several genes with certain genes essential in cardiac development and disease: HEY2, ISL1, SRF, ACTC1, and HEYL and 8 miRNA species-potential biomarkers for VSD |
| Rat | ||||
| Feng et al., 2013 (34) | Maternal blood samples, fetal heart tissues | Cases: 20 ratsControls: 10 rats | Gene-specific, gene expression analysis by real-time PCR, GATA-4 gene product estimation analysis by Western blot, vitamin A estimation in serum | Reduction in vitamin A concentrations; higher proportion of cardiac malformations including VSD, single ventricle, valves malformation, and myocardial hypertrophy; hypermethylation of CpG loci of GATA4 and reduction in GATA4 gene expression; upregulation of DNMT1 and downregulation of DNMT3A and 3B expression |
ACTC1, Actin Alpha Cardiac Muscle 1; CHD, congenital heart disease; DNMT, DNA methyltransferase; DNMT1, DNA Methyltransferase 1; DNMT3A, DNA Methyltransferase 3 Alpha; DNMT3B, DNA Methyltransferase 3 Beta;HAND1, Heart And Neural Crest Derivatives Expressed 1; HEYL, Hes Related Family BHLH Transcription Factor With YRPW Motif Like;HEY2, Hes Related Family BHLH Transcription Factor With YRPW Motif 2; ISL1, ISL LIM Homeobox 1; miRNA, micro-RNA; PCR, polymerase chain reaction; RT-PCR, reverse transcript–polymerase chain reaction; SRF, Serum Response Factor; TOF, tetrology of Fallot; VSD, ventricular septal defect.
Epigenetic regulation of cardiac-specific genes
Stages of development of the human heart and heart defects arising at each stage were well summarized in a recent review (19). Pathogenic cardiac remodeling that raises the chances of defects during heart development is also attributed to epigenetic alterations and mechanobiological events (19). Diversified gene expression profiles in different cells and tissues or maintaining the state of tissue differentiation are facilitated by epigenetic mechanisms (22).
DNA methylation
DNA methylation has been largely explored as one of the crucial epigenetic events, and distinct methylation patterns have been observed in patients with CHD (19). DNA methylation patterns are dynamically established as maintenance methylation by the enzyme DNA (cytosine-5)-methyltransferase (DNMT) 1 (DNMT1 product). Products of DNMT3A (DNA Methyltransferase 3 Alpha) and DNMT3B (DNA Methyltransferase 3 Beta) are involved in the establishment of de novo methylation patterns during gametogenesis and embryogenesis. Correct DNA methylation patterns are important for normal embryonic development. Appropriate DNA methylation is vital for normal embryonic growth and development, and altered DNA methylation processes may significantly increase the chances of adverse birth outcomes (23).
Dinucleotide CpG sites are predominantly targeted for DNA methylation in vertebrate genomes. This global methylation at CpG sites spares the “CpG islands,” which are ∼1000 bp long and with higher GC composition (24). CpG islands are associated with ∼70% of the annotated promoters in the vertebrate genome (19). These regions have been conserved throughout evolution in terms of location and this highlights their functional importance (22). CpG islands are rich in relatively stable GC content (24). These regions undergo differential methylation during stages of gametogenesis and early embryonic development. DNA methylation at these islands regulates gene expression during differentiation and embryonic development beyond the imprinted genes (22). The various hypotheses explaining the possible mechanism by which CpG islands escape the wave of global methylation in the blastocyst stage of embryonic development have been put forth by researchers to date (24). Binding of transcription factors has been addressed to be one of the key mechanisms protecting the unmethylated state of the CpG islands (22, 24). These conserved regions are dispersed throughout the genome and are immune to DNA methylation. This explains the clustering of DNA methylation events at distinct genomic loci.
Mutations in transcription binding sites of certain promoters associated with CpG islands may expose these CpG islands to methylation (22). Such methylation events at CpG islands located in gene promoters result in gene silencing and, in certain cases where CpG islands are located outside the promoter regions, the methylation event increases the transcription (19).
In a global methylation analysis, 3 regions of abnormal methylation per sample and 18 differentially methylated genes between study groups were reported. These DNA methylation errors are implicated in the occurrence of CHD (25). Epimutations in the genes involved in apoptosis, the folate pathway, and growth regulation were recorded. Possible pathogenic epimutations were present in MSX1(Msh Homeobox 1) (gene involved in outflow tract morphogenesis) in a fetus affected with an isolated heart defect were found. Similar epimutations were also seen in GATA4 in fetuses with Down syndrome with or without CHD and fetuses with isolated heart defects. All the aberrant DNA methylation patterns detected were found to have a positive correlation with CHD (25).
A recent study has reported that there are aberrant DNA methylation pattens in NKX2–5, GATA4, and HAND1 (Heart And Neural Crest Derivatives Expressed 1) in patients with TOF (26). Also, a negative correlation was found between mRNA expression of these genes in the cardiac tissues and altered methylation status (26). Seventy-nine genes were found to be differentially methylated in the heart tissues from embryonic day (E) 11.5–14.5 embryos and these patterns showed correlation with altered gene expression in a genome-wide global DNA methylation analysis (27). HAS2 (Hyaluronan Synthase 2) is essential for heart-valve formation and the expression of this gene is downregulated at E14.5 with hypermethylation of its enhancer. This research provided strong evidence for the epigenetic regulation of the genes required for heart development and confirmed that an aberrant DNA methylation is one of the mechanisms for the development of CHD.
The key enzymes that regulate the DNA methylation process are DNMT1 and DNMT3A and 3B. DNMT1 acts on a hemi-methylated substrate and is specialized for maintenance methylation. DNMT3A and 3B are known as de novo DNMTs as they are capable of introducing the methyl tags into a naked DNA. DNMT3A is required for normal cellular differentiation, whereas DNMT3B is essential during early development (22). DNMT3L is a noncatalytic component that is mainly expressed in germ cells and the thymus in adults and is essential during early embryonic development. It works with DNMT3A and 3B to stimulate their methyltransferase function. Genomic imprinting and compaction of X chromosomes are mainly mediated by DNMT3L. Although the mechanism of de novo methylation process targeting distinct genomic loci still remains to be elucidated, transcription factors are most commonly thought to be the mediators either to recruit DNMTs for transcription or to protect the genome sequences from methylation (22).
Brief introduction to ncRNAs
Another important epigenetic regulation is executed by micro-RNA (miRNA)–mediated silencing. mi-RNAs of 21–25 nucleotides are small RNA molecules that mediate the process of post-transcriptional gene silencing. miRNAs take part in RNA-directed DNA methylation where the process of transcriptional gene silencing regulates gene expression (35). A single mRNA can be regulated by multiple miRNAs and 1 miRNA can regulate many target mRNAs. miRNAs also have important roles in mesoderm formation and differentiation of embryonic stem cells (36). These ncRNA molecules have diverse roles in inducing epigenetic modifications by post-transcriptional gene regulation in a complex multilineage process of cardiac development (36).
Several miRNAs have been proven to be implicated in CHD (37). miRNAs have been reported to control fundamental cardiac transcription factors, such as GATA4, MEF2C (Myocyte Enhancer Factor 2C), SRF (Serum Response Factor), and TBX1. Cardiomyocyte proliferation is regulated by miR-1/-133. In mouse experiments, miR-1 overexpression was found to inhibit ventricular myocyte proliferation (36). miRNA biogenesis and functional significance during embryonic development in the context of heart formation are summarized in Figure 2.
FIGURE 2.
miRNA biogenesis and significance in heart development. DGCR8, microprocessor complex subunit DGCR8 [DiGeorge syndrome chromosomal (or critical) region 8], dsRNA, double-stranded RNA; GTP, Guanosine-5′-triphosphate; miRNA, micro-RNA; Pre-miRNA, precursor-microRNA; polyA, polyadenylic acid; Pri-miRNA, primary microRNA; RanGTP, Ras-related nuclear protein (GTP binding protein); RISC, RNA-induced silencing complex; RNA Pol, RNA polymerase; TRBP, trans-activation response RNA-binding protein.
Long noncoding RNAs (lncRNAs) are also components of epigenetic regulation. They can be generated from intronic, exonic, or intergenic sites (38). Their feature is stability in plasma, urine, and tissue and disease specificity (39). The first cardioprotective lncRNA that prevents cardiac hypertrophy was identified in a recent study (40). lncRNAs can act as the guide molecules for binding to transcription factors or protein subunits of chromatin remodeling complexes (39). Scaffold lncRNAs are the complexes that facilitate the former role of complex formation and are themselves the functional components. In the role of enhancer lncRNAs these molecules act as enhancers for transcription by maintaining accessible 3D conformation of the genome essential for the transcription machinery to access the promoter region. In the regulatory role, lncRNAs compete with transcription factors and chromatin-remodeling complexes for the targets (39). Previous studies have shown that several lncRNAs, namely Braveheart, Carmen, Fendrr, Upperhand, Terminator,Alien, and Punisher, are implicated in heart development (39).
Histones: the “epiregulatory” blocks?
Histones are the structural core of the basic unit of chromatin (i.e., nucleosome). H1, H2A, H2B, H3, and H4 are the histone molecules that also form the regulatory core of chromosomes (41). Any covalent modifications in the structure of these proteins change the structural dynamics of the nucleosome core. As a result, the accessibility of genes to transcription machinery is also modulated, resulting in altered expression of genes (23).
Histones are the important regulatory proteins, and their structural modification by acetylation, phosphorylation, methylation, and ubiquitination becomes a part of epigenetic modifications. The histone modification process is substantially influenced by histone acetyltransferases and deacetylases. All histone changes grossly influence RNA expression (i.e., transcription process and hence the further downstream expression of proteins) (41).
BAF60c is the subunit of SWI/SNF-like BAF chromatin remodeling complex (SMARCD3) and is expressed in high amounts in the developing heart. The interaction of transcription factors (e.g., GATA4, NKX2–5, and TBX5) and BAF60c is yet to be completely understood (15).
The mechanism of establishment, maintenance, and re-establishment of DNA methylation marks and histone structure patterns has still not been clearly elucidated. Factors affecting these structural proteins during fetal growth and development can potentially give rise to adverse birth outcomes. Histone deacetylase (HDAC) enzymes have critical roles in the development of the heart (15). In a recent study, overactivity of HDAC was found to be correlated with the single ventricular CHD (42). The research also points to exploration of HDAC inhibitors for the treatment of CHD (42).
Epigenetics during embryonic development
For normal embryonic development, accurate methylation pattern establishment is a must (43). The period of early embryonic development is crucial in the establishment of the epigenome. Any error in establishment of the epigenomic program during this time can have an irreversible and permanent impact throughout the life of the organism. This underlines the influence of critical environmental factors during pregnancy, and maternal nutrition is one such factor (44). Inadequate maternal nutrition during pregnancy was proven to alter the DNA methylation pattern and expression of certain imprinted genes such as Insulin Like Growth Factor 2 Receptor (IGF2R) and H19 Imprinted Maternally Expressed Transcript (H19) in ovine fetuses (45).
The hypothesis of the Developmental Origins of Health and Disease proposes that there is a crucial role of exposure to environmental factors during early development in the onset of disease in childhood or adulthood. Maternal nutrition is among the most important environmental variables in deciding the fate of the developing embryo (46).
Sperm and egg genomes are transcriptionally inactive as they are hypermethylated before fertilization (47). Active demethylation of the paternal genome and passive demethylation of the maternal genome occurs at the early stages of embryogenesis (48). First, the removal of established DNA methylation patterns occurs at the blastocyst stage. Second, this event occurs in primordial germ cells (PGCs) advancing towards development of gametocytes (49). The imprint control regions (ICRs) escaping this global wave of demethylation undergo comprehensive programming at the second episode in PGCs (49).
Irrespective of the global DNA hypomethylation, some imprinted genes retain their methylation pattern in the early stage of embryonic development. All genome-encompassing de novo DNA methylation is initiated following the implantation of the inner cell mass (43). This indicates that early embryogenesis is the crucial step to establish epigenotypes (23).
The heart is among the first organs to be developed during embryogenesis (50). There are intricate signaling events, active transcription factor machinery, and ultimate regulation of gene expression in differentiating cardiomyocytes (51). Multiplication, migration, differentiation, and apoptosis are the key cellular events in the process of heart development along with the structural modifications involving looping and septation (49).
Potentially pathogenic epimutations and aberrant methylation patterns were found in global methylation analysis of individuals affected with CHD (25). Unlike the repression effect of methylation at the promoter regions, the deviant methylation patterns in any other gene regions have totally different regulatory outcomes (25). The specific role of DNA methylation in heart development has yet to be explored. Although this fact persists, certain adverse environmental triggers can accelerate premature termination of the differentiation of cardiomyocytes by promoting hypermethylation. This can be one of the mechanisms underlying the pathogenic remodeling of the heart (49). This effect of DNA methylation in the developing heart varies according to genomic loci, cell type, and stage of development. Equally critical is the role of several factors important in cardiac development, including products of genes namely; histone modifying HAT p300 (histone acetyltransferase p300), HDAC1 (Histone Deacetylase 1) and HDAC2 (Histone Deacetylase 2), SMYD1 (SET And MYND Domain Containing 1), histone demethylase–Jumonji family, LSD-1 (Lysine-specific histone demethylase 1A), miR-499, miR-206, miR-208 and more (49).
Impact of maternal nutrition on fetal growth
In mammals, epigenetic inheritance is either intergenerational or transgenerational (52). Intergenerational inheritance will pass on the epimutations established in gametes that survive the epigenetic reprogramming events during the preimplantation stage. If epimutations persist throughout the additional reprogramming during germ cell development in the later part of embryonic growth, it is referred to as transgenerational inheritance (52). The transgenerational epigenetic effect of nutrition has been explored by various researchers. Many animal studies have been reported in this field (53). Nongenomic transmission of induced phenotypes may prove to be one of the vital mechanisms for the origin of human disease (54). On similar lines, the origin of CHD needs to be explored in depth with respect to modifiable, causal environmental risk factors, maternal nutrition being one of them (15).
It is known that maternal dietary factors (e.g., micronutrients and proteins) have maximum influence on fetal growth during the period of peri-implantation and rapid placental development (55). For several decades, evidence from various studies stated that maternal nutrition at the time of conception affects the pregnancy and birth outcome. External phenomena that are potential stimuli for epigenetic alterations involve prenatal malnutrition, cigarette smoking, and UV radiation, and the resulting epigenetic changes are fairly stable and ensure adaptation of the unaltered cellular genetic makeup to the environmental variations (41). Thus, environmental factors during embryonic development affect the epigenome due to rapid DNA synthesis during this period and establishment of new methylation patterns (15).
The impact of various nutrients during embryonic development has been largely studied using mouse models. Various nutrients supplemented to mice at different stages of development affect different genes, thereby grossly influencing the phenotype (56). In a study to observe the effect of protein supplementation on fetal growth, a diet with limited protein fed to mothers resulted in reduced numbers of cells in inner cell mass and trophoectoderm, ultimately resulting in impaired development of the organism. Organ development was grossly irregular and abnormal (23).
The Dutch Hunger Winter is a classic example showing the association between nutrition and birth outcome. Acute starvation (400–800 kcal/d from 1800 kcal/d) occurred for a period of 6 mo in west Netherlands, in the winter season of 1944–1945; this starvation was more severe than the inadequate dietary supply received by many populations today. This starvation in the first trimester of pregnancies resulted in increased rates of preterm births, stillbirths, and deaths in the first week of life. In case of acute starvation after the third trimester, the birth weight of the newborns was less and there was increased mortality in the first 3 mo of life. This was an example of a “natural experiment” that demonstrated the long-term adverse impact of periconceptional dietary inadequacies (57). Hypomethylation was recorded in differentially methylated regions of imprinted gene insulin-like growth factor 2 (IGF2) in the individuals exposed to famine in utero. This was the first study to provide evidence pointing to an association between periconceptional exposure to environmental factors and DNA methylation (58). There was a higher risk of cardiovascular defects present at birth in the affected population. The exposure period in the course of conception was of profound importance, as reflected in the birth outcomes (54, 57).
It is known that, although rogue methylation is a reversible process, it has a major role in the development of health conditions like cardiovascular diseases, Alzheimer disease, and Parkinson disease in later life, and the process is affected by dietary factors. Apart from the lifetime of the organism, the development of the embryo is affected by the nutrient supply. The crucial stage of rapid growth and cellular differentiation during pregnancy largely depends on the availability of the correct nutrients. The nutrient supply obtainable for the child depends on multiple factors, including maternal metabolism and nutrition partitioning between maternal storage and availability for the fetus. This marks the indirect relation between maternal and fetal nutrition. Pregnancy and lactation are the crucial periods that demand higher nutrient intake and an adequate supply of methyl donors from the diet (59).
DNA methylation is very dependent on the availability of methyl group donors and cofactors obtained from food that are components in folate and methionine metabolism (23). The maternal diet during the periconceptional period is seen to alter the epigenetic regulation by alteration of the DNA methylation status globally or at specific loci. This process is mediated either by direct provision of methyl donors or by providing cofactors for proper activity of the enzyme DNMT, or by providing cofactors to enzymes that regulate the one-carbon cycle (44).
Methyl group donors and cofactors from the ingested food impact the DNA methylation process in various ways. The critical amino acid in this context is methionine, which is provided from the diet, after the protein degradation and synthesis from homocysteine by the process of methylation. The primary donor for DNA methylation is S-adenosylmethionine (SAM). Being the methyl donor, demethylated SAM becomes S-adenosylhomocysteine (SAH). The SAM to SAH ratio in the cell decides the methylation capability in the tissue. SAH is hydrolyzed to homocysteine and adenosine reversibly. Homocysteine metabolism is invariably linked to the methionine cycle and folate cycle and all of these together contribute to one-carbon metabolism (23). This pathway is cyclical and needs a frequent indispensable supply of dietary micronutrients (20).
All methyl group donors (lipotropes) are derived from diet. Folic acid (vitamin B-9) is vital for all DNA repair and biosynthesis. Similarly important is dietary choline, which has limited synthesis capacity in the body. Approximately 10–20% of methyl donors are derived from folate, 20% are from methionine, and 60% are from choline (59).
Research showed that periconceptional low choline, methionine, and folate intake alters the gene expression by some indirect mechanism rather than by direct alteration of global methylation (60). In another pilot study it was established that inadequate folate supplementation during pregnancy led to more methylation of long interspersed nuclear element (LINE) regions (61).
Inadequate or excess dietary nutrients that participate in the methylation process may affect the epigenotype. The point to be noted here is that this epigenotype can be preserved throughout the life of the organism to influence phenotypic traits by influencing gene expression (23). This suggests that establishment of epigenetic programming shapes the phenotype broadly with defined structural formation. The epigenotype can undergo certain limited variations over the lifespan of the organism, which is the reflection of altered DNMT expression in response to certain external factors. Inherited genotype of the organism remains relatively static in comparison to the epigenotype, which is subjected to constant flux leading to regulation of gene expression (62).
Maternal nutrition in early life can provide clues when investigating the current health condition in the offspring. There might be a direct impact of nutrients on the epigenetic regulatory enzymes, namely DNMT, HDAC, or HAT. In another way, they might alter the availability of substrates essential for the reactions catalyzed by these enzymes (63). This process may lead to permanent changes in the phenotype of the offspring.
Role of micronutrients in embryonic development
Micronutrients are the chemical elements required in trace amounts for the normal development and growth of living organisms. The supply of 1 micronutrient in the diet is not straightforward to visualize its positive effects considering the complex interaction between several micronutrients. Vitamin A is indispensable for normal embryogenesis, which is proven in several animal studies. It is proposed that vitamin A interacts with certain minerals such as iron, copper, and zinc, but the significance of this interaction during pregnancy is yet unclear (64). During embryonic development and growth, fetal organs develop at different times, marking the “critical windows” where the organ is more susceptible to alteration in a particular macronutrient's or micronutrients’ supply. Maternal micronutrient concentrations potentially determine fetal phenotype to a great extent (65). Like the components of one-carbon metabolism, the vital enzyme cofactors—namely, riboflavin, vitamin B-6, and vitamin B-12—are essentially derived from the diet. Deficiencies of these nutrients in pregnancy and the resultant visible impact on pregnancy outcomes need to be further explored. Maternal micronutrient deficiency is one of the major causes of intrauterine growth retardation (66).
Although fetal nutrition and growth are completely derived and dependent on the maternal diet, multiple factors control the transfer of nutrition from the mother to the developing fetus and, hence, maternal nutrition is not equivalent to the nutrient supply to the fetus (65). The studies in human populations have provided evidence to prove the correlation between periconceptional dietary supplementation and reduced risk of deleterious birth outcomes. The Pune Maternal Nutrition Study provided crucial evidence to prove the importance of one-carbon metabolism in embryonic development and programming. This study also established the relationship between maternal nutrition, fetal growth, and size at birth and postnatal growth (67). An investigation established that women who consume multivitamins during the periconceptional period have a 30–35% lower risk of conotruncal or limb defects in their children (68). One previous study also reported a similar finding that periconceptional vitamin use reduces the occurrence of neural tube defects (69). Since it is essential that the fetus should be provided just enough nutrients for correct development and growth, any nutrient excess can also lead to unfavorable effects (23).
Maternal nutrition and epigenetic mechanism of the development of CHD
Maternal nutrition is thus an important topic in itself, which demands more targeted research. Although structural heart defects are proposed to arise due to improper maternal periconceptional diet, intervention-based studies can establish this correlation in a better way. The most crucial period of heart development is the initial 2–7 wk of gestation, and hence exposure to environmental risk factors during the periconceptional period (3 mo before pregnancy through first trimester) can significantly affect the cardiac development (15).
During the development of the heart, folate and its derivative homocysteine have key roles in cell migration of the cardiac neural crest cells from the cranial region of the embryonic neural tube to the thoracic area to develop the great blood vessels, other cardiac sections, and the outflow tract (30). Folate deficiency during the periconceptional period can thus potentially give rise to CHDs in newborns.
Certain animal studies have provided evidence towards the causal link between the availability of micronutrients during critical periods of pregnancy and CHD in offspring. A recent study demonstrated the role of vitamin A deficiency in the development of cardiac defects in rat offspring (34). A vitamin A–fortified diet was supplemented to 1 group of female rats before mating and this diet was continued throughout the study, whereas a vitamin A–deficient normal diet was provided to another group before mating. There was further supplementation of vitamin A during the pregnancy cycle to a few rats of the second group fed the vitamin A–deficient diet. Aberrant methylation with repression of GATA4 was observed in the tissue samples from the hearts of the offspring of rats fed the vitamin A–deficient diet. Supplementation during pregnancy reduced this aberrant methylation, showing an intermediate effect. Also, DNMT1 was significantly upregulated and DNMT3A and DNMT3B were downregulated in these offspring. There was a high incidence of heart defects in the hearts of the embryos born from the study group fed the vitamin A–deficient diet. The study thus provided evidence to highlight the importance of the periconceptional period in shaping the epigenotype for life. Also, this study provided strong, supportive evidence to mark the importance of consequent abnormal methylation following nutrition deprivation as a crucial epigenetic mechanism leading to adverse outcome of CHD (34).
The influence of diet quality on the incidence of conotruncal and septal heart defects was recently evaluated in a population-based study. Maternal DNA hypomethylation with a history of inadequate nutrition was reflected in the pregnancies affected with CHDs (16). The differential methylation patterns were observed in women with CHD-affected and -unaffected pregnancies, with maternal undernutrition as an influential variable under consideration (28, 29).
As stated before, maternal overnutrition also adversely affects fetal growth. In a recent study, increased prevalence rate ratios measured for aortic branch defects and transposition of great arteries, ASD, and PDA were found to be increased with severity of maternal obesity (70). Another population-based study showed that obese women are at higher risk of giving birth to a child affected with CHD and this risk increases with increased maternal BMI (71). It is proposed that this causal association has an underlying epigenetic mechanism (72).
From the analyses presented in this paper it can be inferred that dietary interventions, if provided during critical periods of embryonic development, have a profound impact on the epigenome of the organism and are also responsible for shaping the phenotype of the offspring. Based on the availability of sufficient periconceptional nutrition to the mother, embryonic heart development may progress towards a healthy heart or structural malformation and this has been summarized in Figure 3. The regular, calculated, and balanced intake of bioactive dietary components (referred to as an “epigenetics diet”) that can induce epigenetic modifications can provide preventive solutions against the development of metabolic and structural diseases such as CHD when provided around the early critical period of life (53, 73).
FIGURE 3.
Periconceptional nutrition and impact on cardiogenesis. ncRNA, noncoding RNA.
Conclusions
Genetic and epigenetic mechanisms regulate the complex morphogenetic process of cardiac development. The severity of the phenotype and relatively minor variations in gene expression have potential epigenetic triggers (74).
The multifactorial etiology of the isolated CHDs demands a more systematic, integrated research to establish the causal associations. Various environmental factors need to be investigated as potential epigenetic triggers leading to CHD as the adverse birth outcome. Maternal nutrition is one such important environmental factor. Inadequate nutrition at different key stages of gamete formation and embryonic development and intrauterine environment can have diverse outcomes in determining the fate of the newborn (Figure 3). Hence, defining the exact phase for the dietary intervention to the mother to improve the birth outcome is of utmost importance.
Apart from the growing evidence to establish the association between periconceptional nutrition and in utero onset of developmental defects, these studies in humans face certain limitations. Maternal dietary patterns vary across geographical locations, across and within populations, and even at individual levels. While planning any diet-supplementation study, problems occur in the accuracy of food recall, history-based evaluation of diet during conception, compliance with the study design, and practical follow-ups for the requisite number of times (75). Although studies with larger sample sizes are more difficult to execute in all aspects, the study outcomes of small-scale studies cannot be directly correlated with the maternal periconceptional diet.
A causal relationship between or among the variables of interest may be established from well-organized randomized controlled trials. These trials can derive the recommended dosage of essential dietary components and adequate period of administration. However, such trials, in this case from nutritional baseline to the endpoint of disease under investigation, are extremely difficult to organize and execute because the pregnancy outcome can be variable and may range from a healthy to diseased state not limited to the disease under investigation.
Studies in humans are complex to conduct, require considerable time for the generation of interpretable results, and may face difficulty in maintaining uniformity in the sample composition due to inherent variations in diet patterns.
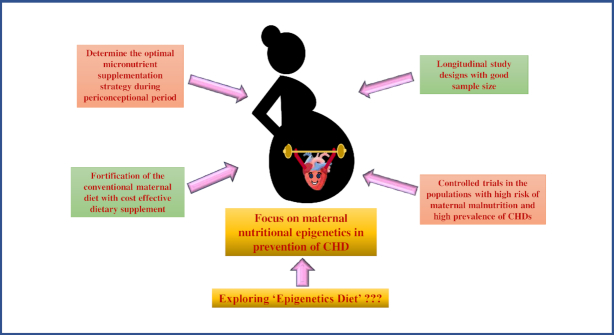
In addition to DNA methylation, which is explored the most to discover its correlation with the consequence of pregnancy, more research is needed to understand the role of other epigenetic regulators that are possibly influenced by the maternal diet. Possible future research approaches to demonstrate the role of maternal nutritional epigenetics in the prevention of CHD are summarized in Figure 4.
FIGURE 4.
Role of maternal nutritional epigenetics in CHD. CHD, congenital heart disease.
Future Perspectives
Nutrition shapes the lifetime of the individual with its impact at the epigenetic level. There needs to be further research in this direction with the following:
Well-organized, good-quality, controlled trials in the populations with high risk of malnutrition and prevalence of CHDs.
Longitudinal study designs with a good sample size to better illustrate the causal link between maternal nutrition and incidence of CHD.
An intervention that tests the scientifically justified and measured levels of micronutrients, with different combinations of them, to deduce the synergistic and antagonistic interactions to derive the recommended dosage to be consumed to reduce the incidence of CHD.
Fortification of the existing diet with cost-effective dietary supplements, which can produce a sustainable solution for possible prevention of the problem.
Thus, because diet is a modifiable behavior, the research in this direction to put forth measurable effects of an intervention on genotype and phenotype can justify the link between maternal nutrition and the development of CHD with the evolution of practical preventive strategies that can potentially be used in the population to decrease the incidence of CHD.
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—ROJ: designed and wrote the paper; SC and PK: contributed to revision of the final manuscript; PK: has primary responsibility for final content; and all authors: read and approved the final manuscript.
Notes
This study was supported by Sri Sathya Sai Health and Education Trust. The authors reported no external funding received.
Author disclosures: The authors report no conflicts of interest.
Abbreviations used: ASD, atrial septal defect; CHD, congenital heart disease; DNMT, DNA methyltransferase; E, embryonic day; HDAC, histone deacetylase; lncRNA, long noncoding RNA; miRNA, micro-RNA; ncRNA, noncoding RNA; PGC, primordial germ cells; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; TOF, tetrology of Fallot.
Contributor Information
Radha O Joshi, Department of Genomics Research, Sri Sathya Sai Sanjeevani Research Foundation, Palwal, Haryana, India.
Subramanian Chellappan, Department of Anesthesia, Sri Sathya Sai Sanjeevani International Centre for Child Heart Care and Research, Palwal, Haryana, India.
Prachi Kukshal, Email: drprachi.kukshal@srisathyasaisanjeevani.com, Department of Genomics Research, Sri Sathya Sai Sanjeevani Research Foundation, Palwal, Haryana, India.
References
- 1. Liu Y, Chen S, Zühlke L, Black GC, Choy M, Li N, Keavney BD. Global birth prevalence of congenital heart defects 1970–2017: updated systematic review and meta-analysis of 260 studies. Int J Epidemiol. 2019;48:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Li P, Chen S, Xi L, Guo Y, Guo A, Sun K. Influence of genes and the environment in familial congenital heart defects. Mol Med Rep. 2014;9:695–700. [DOI] [PubMed] [Google Scholar]
- 3. Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8. [DOI] [PubMed] [Google Scholar]
- 4. Thiene G, Frescura C. Anatomical and pathophysiological classification of congenital heart disease. Cardiovasc Pathol. 2010;19:259–74. [DOI] [PubMed] [Google Scholar]
- 5. Setty HS, Setty N, Patil SSG, Ramegowda RT, Vijaykumar V IB, Manjunath CN. Comprehensive approach to congenital heart defects. J Cardiovasc Disease Res. 2017;8:01. [Google Scholar]
- 6. Edwards JJ, Gelb BD. Genetics of congenital heart disease. Curr Opin Cardiol. 2016;31:235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung I, Rajakumar G. Genetics of congenital heart defects: the NKX2-5 gene, a key player. Genes. 2016;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trevisan P, Zen TD, Rosa RFM, Silva JN, Koshiyama DB, Paskulin GA, Zen PRG. Chromosomal abnormalities in patients with congenital heart disease. Arq Bras Cardiol. 2013;101:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res. 2017;120:923–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Demal TJ, Heise M, Reiz B, Dogra D, Brænne I, Reichenspurner H, Männer J, Aherrahrou Z, Schunkert H, Erdmann J et al. A familial congenital heart disease with a possible multigenic origin involving a mutation in BMPR1A. Sci Rep. 2019;9:2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kern MJ, Argao EA, Potter SS. Homeobox genes and heart development. Trends Cardiovasc Med. 1995;5:47–54. [DOI] [PubMed] [Google Scholar]
- 12. Chen J, Fishman M. Genetics of heart development. Trends Genet. 2000;16:383–8. [DOI] [PubMed] [Google Scholar]
- 13. Blue GM, Kirk EP, Giannoulatou E, Sholler GF, Dunwoodie SL, Harvey RP, Winlaw DS. Advances in the genetics of congenital heart disease: a clinician's guide. J Am Coll Cardiol. 2017;69:859–70. [DOI] [PubMed] [Google Scholar]
- 14. Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vecoli C, Pulignani S, Foffa I, Andreassi MG. Congenital heart disease: the crossroads of genetics, epigenetics and environment. Curr Genomics. 2014;15:390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Botto LD, Krikov S, Carmichael SL, Munger RG, Shaw GM, Feldkamp ML. Lower rate of selected congenital heart defects with better maternal diet quality: a population-based study. Arch Dis Child Fetal Neonatal Ed. 2016;101:43. [DOI] [PubMed] [Google Scholar]
- 17. Yang J, Kang Y, Cheng Y, Zeng L, Yan H, Dang S. Maternal dietary patterns during pregnancy and congenital heart defects: a case-control study. Int J Environ Res Public Health. 2019;16:2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abu-Saad K, Fraser D. Maternal nutrition and birth outcomes. Epidemiol Rev. 2010;32:5–25. [DOI] [PubMed] [Google Scholar]
- 19. Jarrell DK, Lennon ML, Jacot JG. Epigenetics and mechanobiology in heart development and congenital heart disease. Diseases. 2019;7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism, and DNA methylation. J Nutr Biochem. 2012;23:853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spearman AD. Epigenetics for paediatric cardiologist. Congenit Heart Dis. 2017;12:(6):828–33. [DOI] [PubMed] [Google Scholar]
- 22. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacol. 2013;38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chmurzynska A. Fetal programming: link between early nutrition, DNA methylation, and complex diseases. Nutr Rev. 2010;68:87–98. [DOI] [PubMed] [Google Scholar]
- 24. Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serra-Juhe C, Cusco I, Homs A, Flores R, Toran N, Perez-Jurado LA. DNA methylation abnormalities in congenital heart disease. Epigenetics. 2015;10:167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sheng W, Qian Y, Wang H, Ma X, Zhang P, Diao L, An Q, Chen L, Ma D, Huang G. DNA methylation status of NKX2-5, GATA4 and HAND1 in patients with tetralogy of Fallot. BMC Med Genomics. 2013;6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chamberlain AA, Lin M, Lister RL, Maslov AA, Wang Y, Suzuki M, Wu B, Greally JM, Zheng D, Zhou B. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J Am Heart Assoc. 2014;3:e000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chowdhury S, Cleves MA, MacLeod SL, James SJ, Zhao W, Hobbs CA. Maternal DNA hypomethylation and congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chowdhury S, Erickson SW, MacLeod SL, Cleves MA, Hu P, Karim MA, Hobbs CA. Maternal genome-wide DNA methylation patterns and congenital heart defects. PLoS One. 2011;6:e16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bahado-Singh RO, Zaffra R, Albayarak S, Chelliah A, Bolinjkar R, Turkoglu O, Radhakrishna U. Epigenetic markers for newborn congenital heart defect (CHD). J Matern Fetal Neonatal Med. 2016;29:1881–7. [DOI] [PubMed] [Google Scholar]
- 31. Li D, Ji L, Liu L, Liu Y, Hou H, Yu K, Sun Q, Zhao Z. Characterization of circulating microRNA expression in patients with a ventricular septal defect. PLoS One. 2014;9:e106318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Radhakrishna U, Vishweswaraiah S, Veerappa AM, Zafra R, Albayrak S, Sitharam PH, Saiyed NM, Mishra NK, Guda C, Bahado-Singh R. Newborn blood DNA epigenetic variations and signaling pathway genes associated with tetralogy of Fallot (TOF). PLoS One. 2018;13:e0203893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radhakrishna U, Albayrak S, Zafra R, Baraa A, Vishweswaraiah S, Veerappa AM, Mahishi D, Saiyed N, Mishra NK, Guda C et al. Placental epigenetics for evaluation of fetal congenital heart defects: ventricular septal defect (VSD). PLoS One. 2019;14:e0200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng Y, Zhao LZ, Hong L, Shan C, Shi W, Cai W. Alteration in methylation pattern of GATA4 promoter region in vitamin A-deficient offspring's heart. J Nutr Biochem. 2013;24:1373–80. [DOI] [PubMed] [Google Scholar]
- 35. Pickford AS, Cogoni C. RNA-mediated gene silencing. Cell Mol Life Sci. 2003;60:871–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagy O, Baráth S, Ujfalusi A. The role of microRNAs in congenital heart disease. Clin Chem Lab Med. 2019;30:165–78. [PMC free article] [PubMed] [Google Scholar]
- 37. Smith T, Rajakaruna C, Caputo M, Emanueli C. MicroRNAs in congenital heart disease. Ann Transl Med. 2015;3:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen J, Xue Y. Emerging roles of non-coding RNAs in epigenetic regulation. Sci China Life Sci. 2016;59:227–35. [DOI] [PubMed] [Google Scholar]
- 39. Dueñas A, Expósito A, Aranega A, Franco D. The role of non-coding RNA in congenital heart diseases. J Cardiovasc Dev Dis. 2019;6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ et al. A long non-coding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Majo FD, Calore M. Chromatin remodeling and epigenetic state regulation by non-coding RNAs in the diseased heart. Non-coding RNA Research. 2018;3:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blakeslee WW, Demos-Davies KM, Lemon DD, Lutter KM, Cavasin MA, Payne S, Nunley K, Long CS, McKinsey TA, Miyamoto SD. Histone deacetylase adaptation in single ventricle heart disease and a young animal model of right ventricular hypertrophy. Pediatr Res. 2017;82:642–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kierszenbaum AL. Genomic imprinting and epigenetic reprogramming: unearthing the garden of forking paths. Mol Reprod Dev. 2002;63:269–72. [DOI] [PubMed] [Google Scholar]
- 44. Zhang N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Animal Nutrition. 2015;1:144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Duan J, Zhang M, Flocka K, Seesic SA, Mandoi I, Jones A, Johnsona E, Pillaia S, Hoffmana M, McFaddena K et al. Effects of maternal nutrition on the expression of genomic imprinted genes in ovine fetuses. Epigenetics. 2018;13:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng J, Xiao X, Zhang Q, Yu M. DNA methylation: the pivotal interaction between early-life nutrition and glucose metabolism in later life. Br J Nutr. 2014;112:1850–57. [DOI] [PubMed] [Google Scholar]
- 47. Fraser R, Lin CJ. Epigenetic reprogramming of the zygote in mice and men: on your marks, get set, go!. Reproduction. 2016;152:R211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006;310:13–22. [DOI] [PubMed] [Google Scholar]
- 49. Martinez SR, Gay MS, Zhang L. Epigenetic mechanisms in heart development and disease. Drug Discov Today. 2015;20:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vallaster M, Vallaster CD, Wu SM. Epigenetic mechanisms in cardiac development and disease. Acta Biochim Biophys Sin. 2012;44:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gilsbach R, Schwaderer M, Preissl S, Grüning BA, Kranzhöfer D, Schneider P, Nührenberg TG, Mulero-Navarro S, Weichenhan D, Braun C et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat Commun. 2018;9:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaspar D, Hastreiter S, Irmler M, Angelis MH, Beckers J. Nutrition and its role in epigenetic inheritance of obesity and diabetes across generations. Mamm Genome. 2020;31:119–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park JH, Kim SH, Lee MS, Kim MS. Epigenetic modification by dietary factors: implications in metabolic syndrome. Mol Aspects Med. 2017;54:58–70. [DOI] [PubMed] [Google Scholar]
- 54. Burdge GC, Lillycrop KA. Nutrition, epigenetics, and developmental plasticity: implications for understanding human disease. Annu Rev Nutr. 201030:315–39. [DOI] [PubMed] [Google Scholar]
- 55. Nafee TM, Farrell WE, Carroll WD, Fryer AA, Ismaila KMK. Epigenetic control of fetal gene expression. BJOG. 2007;115:158–68. [DOI] [PubMed] [Google Scholar]
- 56. Niculescu MD. Nutritional epigenetics. ILAR J. 2012;53:270–8. [DOI] [PubMed] [Google Scholar]
- 57. King JC. A summary of pathways or mechanisms linking preconception maternal nutrition with birth outcomes. J Nutr. 2016;146:1437S–44S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Heijmans BT, Tobia EW, Stein AD, Putter H, Blauwd GJ, Sussere ES, Slagbooma PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci. 2008;105:17046–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C1 metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc Nutr Soc. 2012;71:154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maloney CA, Hay SM, Rees WD. Folate deficiency during pregnancy impacts on methyl metabolism without affecting global DNA methylation in the rat fetus. Br J Nutr. 2007;97:1090–98. [DOI] [PubMed] [Google Scholar]
- 61. Khan MFJ, Little J, Mossey PA, Steegers-Theunissen RPM, Autelitano L, Lombardo I, Andreasi RB, Rubini M. Evaluating LINE-1 methylation in cleft lip tissues and its association with early pregnancy exposures. Epigenomics. 2018;10:105–13. [DOI] [PubMed] [Google Scholar]
- 62. Whitelaw NC, Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum Mol Genet. 2006;15:R131–7. [DOI] [PubMed] [Google Scholar]
- 63. Tiffon C. The impact of nutrition and environmental epigenetics on human health and disease. Int J Mol Sci. 2018;19:3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McArdle HJ, Ashworth CJ. Micronutrients in fetal growth and development. Br Med Bull. 1999;55:499–510. [DOI] [PubMed] [Google Scholar]
- 65. Yajnik C. Nutritional control of fetal growth. Nut Rev. 2006;64:50. [DOI] [PubMed] [Google Scholar]
- 66. Fall CHD, Yajnik CS, Rao S, Davies AA, Brown N, Farrant HJW. Micronutrients and fetal growth. J Nutr. 2003;133:1747S–56S. [DOI] [PubMed] [Google Scholar]
- 67. Yajnik CS, Deshmukh US. Fetal programming: maternal nutrition and role of one-carbon metabolism. Rev Endocr Metab Disord. 2012;13:121–7. [DOI] [PubMed] [Google Scholar]
- 68. Shaw GM, O'Malley CD, Wasserman CR, Tolarova MM, Lamer EJ. Maternal periconceptional use of multivitamins and reduced risk for conotruncal heart defects and limb deficiencies among offspring. Am J Med Genet. 1995;59:536–45. [DOI] [PubMed] [Google Scholar]
- 69. Czeizel AE, Dobo M, Vargha P. Hungarian cohort-controlled trial of periconceptional multivitamin supplementation shows a reduction in certain congenital abnormalities. Birth Defects Res A Clin Mol Teratol. 2004;70:853–61. [DOI] [PubMed] [Google Scholar]
- 70. Persson M, Razaz N, Bonmay AKE, Villamor E, Cnattinguis S. Maternal overweight and obesity and risk of congenital heart defects. J Am Coll Cardiol. 2019;73:44–53. [DOI] [PubMed] [Google Scholar]
- 71. Mills JL, Troendle J, Conley MR, Carter T, Druschel CM. Maternal obesity and congenital heart defects: a population-based study. Am J Clin Nutr. 2010;91:1543–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Catalano PM, Shankar K. Obesity and pregnancy: mechanisms of short term and long-term adverse consequences for mother and child. BMJ. 2017;356:j1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li S, Chen M, Li Y, Tollefsbol TO. Prenatal epigenetics diets play protective roles against environmental pollution. Clin Epigenet. 2019;11:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Moore-Morris T, Vliet PP, Andelfinger G, Puceat M. Role of epigenetics in cardiac development and congenital diseases. Physiol Rev. 2018;98:2453–75. [DOI] [PubMed] [Google Scholar]
- 75. Aiken CE, Tarry-Adkins JL, Ozanne SE. Transgenerational effects of maternal diet on metabolic and reproductive ageing. Mamm Genome. 2016;27:430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]