Abstract
Coronavirus disease 2019 (COVID-19) is a newly emerged human infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In a global pandemic, development of a cheap, rapid, accurate, and easy-to-use diagnostic test is necessary if we are to mount an immediate response to this emerging threat. Here, we report the development of a specific lateral flow immunoassay (LFIA)-based biosensor for COVID-19. We used phage display technology to generate four SARS-CoV-2 nucleocapsid protein (NP)-specific single-chain variable fragment-crystallizable fragment (scFv-Fc) fusion antibodies. The scFv-Fc antibodies bind specifically and with high affinity to the SARS-CoV-2 NP antigen, but not to NPs of other coronaviruses. Using these scFv-Fc antibodies, we screened three diagnostic antibody pairs for use on a cellulose nanobead (CNB)-based LFIA platform. The detection limits of the best scFv-Fc antibody pair, 12H1 as the capture probe and 12H8 as the CNB-conjugated detection probe, were 2 ng antigen protein and 2.5 × 104 pfu cultured virus. This LFIA platform detected only SARS-CoV-2 NP, not NPs from MERS-CoV, SARS-CoV, or influenza H1N1. Thus, we have successfully developed a SARS-CoV-2 NP-specific rapid diagnostic test, which is expected to be a simple and rapid diagnostic test for COVID-19.
Keywords: COVID-19, SARS-CoV-2, Lateral flow immunoassay, Rapid diagnostic test, Antibody
Highlights
-
•
SARS-CoV-2 NP-specific scFv-Fc antibodies were generated.
-
•
Antibody pairs for the specific and sensitive detection of SARS-CoV-2 were found.
-
•
Lateral flow immunoassay-based rapid diagnostic tests sensitively detected NP antigen and SARS-CoV-2 virus.
-
•
The COVID-19 biosensor showed no cross-reactivity with other coronaviruses, SARS-CoV and MERS-CoV.
1. Introduction
Coronavirus disease 2019 (COVID-19), which was first reported in Wuhan city, Hubei province, China, in December 2019 (Wang et al., 2020; WHO 2020c), is caused by the novel virus acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu et al., 2020; Zhou et al., 2020; Zhu et al., 2020). Infection with SARS-CoV-2 primarily causes pneumonia-like symptoms, including fever, cough, and fatigue (Le Bert et al., 2020; Long et al., 2020). Human-to-human transmission is rapid; therefore, the World Health Organization declared the COVID-19 outbreak a global pandemic (WHO 2020a). As of August 2020, more than 24,000,000 cases of COVID-19 have been confirmed world-wide, and 838,924 people have died (WHO 2020b). No COVID-19-specific drugs or vaccines are available at present; therefore, accurate diagnosis is crucial if we are to manage the COVID-19 pandemic. Rapid screening and isolation of COVID-19 patients prevent super-spreading events and enables patients to receive treatment at the early stage of the illness.
Detection of SARS-CoV-2 RNAs using real-time polymerase chain reaction (RT-PCR) was approved by the US Food and Drug Administration (FDA) for emergency use authorization (EUA); this test is used word-wide (FDA 2020). Nucleic acid amplification tests are the primary method of diagnosis for emerging pathogens due to their high accuracy and low limits of detection. However, molecular diagnosis requires specific instruments, expensive reagents, and skilled technicians. The steps are also complicated and time-consuming (Seo et al., 2020). Thus, molecular diagnostic tests are not suitable for point-of-care testing (POCT) for COVID-19. Recently, antibody-based detection tests (i.e., lateral flow immunoassays (LFIA)) were developed and approved for EUA (FDA 2020). These serological tests detect IgM and IgG antibodies specific for COVID-19 virus in patient blood samples. SARS-CoV-2-specific antibodies usually appear between 1 and 2 weeks after symptom onset (Younes et al., 2020); therefore, antibody-based methods for COVID-19 diagnosis may not be that useful in the early stages of infection, even though LFIA is rapid and suitable for POCT. Hence, immunological diagnostic tests that detect viral antigens directly are necessary for rapid POC diagnosis of COVID-19.
Coronaviruses (CoV)s are positive-sense single-stranded RNA viruses with a genome of approximately 30 kbp, which encodes four structural proteins, such as spike, envelop, matrix, and nucleocapsid. CoVs are divided into four genera (alpha, beta, gamma, and delta) and cause mild to moderate upper respiratory tract illnesses in both human and animals (Li et al., 2020). SARS-CoV-2 is a betacoronavirus (beta-CoV) (subgenus sarbecovirus), as is severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (Park et al., 2020). Thus, specific detection of SARS-CoV-2 without cross-reactivity with SARS-CoV or MERS-CoV is essential for diagnosis of COVID-19. Currently, researchers are trying to develop specific immunoassays to detect SARS-CoV-2 antigens. The LFIA-based rapid diagnostic test (RDT), which is reliable, cheap, and easy-to-use, is used widely for diagnosis of acute infection (Kubina and Dziedzic 2020; Younes et al., 2020). For specific detection of antigen proteins, matched antibody pairs (one capture antibody and one detection antibody) are necessary. Gold nanoparticles, cellulose nanobeads (CNBs), and fluorophores are used widely for labeling detection antibodies (Bishop et al., 2019).
Here, we used phage display technology (Ledsgaard et al., 2018) to generate single-chain variable fragment (scFv)-crystallizable fragment (Fc) fusion proteins (scFv-Fcs) for use as antibodies for specific detection of SARS-CoV-2 nucleocapsid protein (NP). The interaction between SARS-CoV-2 NP and scFv-Fc antibodies was examined by western blotting, enzyme-linked immunosorbent assay (ELISA), and biolayer interferometry (BLI) to measure affinity (KD). Extensive LFIA screening identified three SARS-CoV-2-specific diagnostic antibody pairs. The LFIA-based biosensors based on these sc-Fv-Fc antibody pairs were sensitive for SARS-CoV-2 NP antigen and virus, without cross-reacting with NP from other CoVs or influenza virus. Thus, we have developed a SARS-CoV-2 NP-specific RDT, which is expected to be a simple and rapid diagnostic test for COVID-19.
2. Materials and methods
2.1. Cloning, expression, and purification of SARS-CoV-2 NP
The full-length SARS-CoV-2 NP gene was synthesized and cloned into a pET28a vector (Bionics, South Korea) with an N-terminal 6 × His tag. This was then transformed into E. coli BL21(DE3). Expression of recombinant NP was induced by adding 0.5 mM isopropyl -D-1-thiogalactopyronoside to the bacterial culture. Bacterial cells were lysed by sonication, and soluble NP was purified using Ni2+ affinity chromatography (HisTrap FF; GE Healthcare, IL, USA) and a size-exclusion column (HiLoad 16/60 Superdex 200 PG, GE Healthcare). The purity and homogeneity of the recombinant protein were purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Finally, the protein was concentrated to 2 mg/ml in phosphate-buffered saline (PBS) using a 30 kDa molecular-weight cutoff centrifugal filter (Millipore, MA, USA).
2.2. Development of phage display-derived SARS-CoV-2-specific antibodies
The scFv clones were isolated from a chicken naïve phage library (YntoAb, South Korea) by biopanning using purified recombinant SARS-CoV-2 NP. Monoclonal binders were selected using an ELISA based on SARS-CoV-2 NP (40588-V08B; Sino Biological, Inc., China); clones that were cross-reactive for SARS-CoV NP (40143-V08B, Sino Biological, Inc.) and MERS-CoV NP (40068-V08B, Sino Biological, Inc.) were eliminated. SARS-CoV-2-specific scFv binders were inserted to scFv-Fc vectors and expressed in 293F cells (YntoAb). Monoclonal scFv-Fc antibodies were purified by protein A affinity chromatography. After dialysis with PBS, the antibody concentration was measured using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, MA, USA).
2.3. ELISA
A 96-well plate was coated with each NP antigen (100 ng) for 1 h at 37 °C and then blocked with 5% BSA. Each scFv-Fc antibody (1:100 dilution) was added to the blocked well and the sample was incubated for 1 h at room temperature. After washing with TBST (Tris-buffered saline, 0.1% Tween 20), an HRP-conjugated anti-human IgG antibody (1:2000 dilution; ThermoFisher Scientific) was added and the sample was incubated for 1 h at room temperature. After extensive washing with TBST, an ELISA substrate (1-Step Ultra TMB-ELISA; Thermo Fisher Scientific) was applied, and signals were obtained using a Synergy HTX plate reader (BioTek Instruments, Winooski, VT).
2.4. Western blot analysis and dot blot assay
For Western blotting, SARS-CoV-2 NP was separated by SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane. For the dot blot, SARS-CoV-2 NP was spotted directly onto a nitrocellulose (NC) membrane, which was then dried for 1 h at 37 °C. The membrane was blocked for 1 h at room temperature with 5% skim milk in TBST, followed by incubation with purified scFv-Fc antibodies (1:1000 dilution) for 2 h at 37 °C. Afterward, membranes were washed with TBST and incubated for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-human IgG (1:5000 dilution, ThermoFisher Scientific). Next, the membranes were washed in TBST, and blotted proteins were visualized using ECL western blot substrate reagents (Millipore). Finally, the chemiluminescent signals were analyzed using a ChemiDoc MP imaging system (Bio-Rad, CA, USA).
2.5. BLI
The binding kinetics of the scFv-Fc antibodies and SARS-CoV-2 NP antigen were analyzed by BLI using an Octet QK384 instrument (ForteBio, CA, USA). First, Ni-NTA biosensors were hydrated for at least 10 min prior to measurement. Next, SARS-CoV-2 NP antigen (10 μg/ml) and four different antibodies (31.3, 15.6, 7.8, or 3.9 nM) were prepared in 1×PBS and 0.09% Tween 20. The association and dissociation steps were adjusted to 900 and 1200 s, respectively. After each step, the biosensor tip was equilibrated in 1×PBS and 0.09% Tween 20 for 60 s. KD values were calculated by ForteBio data analysis software using a 1:1 binding model.
2.6. CNB conjugation
Protein A and scFv-Fc antibodies were conjugated to Nano Act™ CNBs (Asahi Kasei, Japan) using a CNB conjugation kit (DCN Diagnostics, CA, USA). Briefly, protein A and antibodies (0.5 mg/ml in 120 μl conjugation buffer) were mixed with CNB (0.5% CNB in 120 μl conjugation buffer) and incubated for 2 h at 37 °C. Then, 7.2 ml blocking buffer were added to block the CNB surface. After blocking for 2 h at 37 °C, unconjugated CNB was removed and the protein A and antibody-CNB conjugants were washed with 7.5 ml wash buffer by centrifugation at 14,400 g for 20 min at 4 °C. Finally, the pellets were suspended gently in 300 μl wash buffer. The concentration of the protein A and antibody-CNB conjugants was measured using UV–vis spectrophotometry (BioTek Instruments, Inc., VT, USA) at an absorption intensity of 554 nm.
2.7. LFIA
An LFIA strip comprised a sample pad, a conjugate pad, an NC membrane, and an absorbent pad. The control and test lines were dispensed onto the NC membrane using a line dispenser (BTM Inc., South Korea) at a dispensing speed of 50 mm/s and a dispensing rate of 1 μl/cm. The membrane was dried at 37 °C for 1 h, blocked for 1 h at 37 °C with blocking solution, and then dried again at 37 °C for 1 h in a vacuum drying oven. Prior to conjugation, the conjugate pad was soaked in 0.1% triton X-100 and dried for 1 h at 37 °C. The three parts (sample and conjugate pads, and the adsorbent pad) were pasted onto a backing card comprising a nitrocellulose membrane, cut into uniform 38 mm strips, and housed in plastic cassettes for the lateral flow tests.
To construct the indirect LFIA used for antibody screening, NP antigens (50 μg/ml in sample diluent buffer) were dispensed onto the test line and anti-human IgG antibodies (50 μg/ml in sample diluent buffer) were applied to the control line. Protein A-CNB conjugants were sprayed onto the conjugate pad and dried for 1 h at 37 °C in a vacuum drying oven. For the test, 100 μl of each antibody-CNB conjugant (100 ng in dilution buffer) was applied to the sample pad of the assembled LFIA test kit. After 20 min, the density of the test and control lines was measured using the LFIA reader (careSTART™ Light-G; Wells bio, Seoul, South Korea).
To detect SARS-CoV-2 using LFIA, each scFv-Fc antibody (1 mg/ml) was dispensed onto the test line, and the anti-human IgG antibodies (50 ng/ml in sample diluent buffer) were applied to the control line. The conjugate pad was treated with each scFv-Fc antibody-CNB conjugate (final concentration of CNB, 0.5%) in stabilizing buffer and dried for 1 h at 37 °C in a vacuum drying oven. For the test, 100 μl of analyte (antigen proteins and cultured virus in lysis buffer; Wells bio) were applied to the sample pad of the assembled LFIA test kit. After 20 min, the density of the test and control lines was measured using the LFIA reader.
2.8. SARS-CoV-2 culture
Virus culture was conducted in a biosafety level 3 laboratory. African green monkey kidney Vero E6 cells were infected with a clinical isolate of SARS-CoV-2 (BetaCoV/Korea/KCDC03/2020, provided by Korea CDC). After 48 h, the culture medium containing mature infectious virions (virus medium) was collected, and the viral titer was measured in a plaque assay. Live virus was inactivated by heating to 100 °C for 15 min and then stored at −80 °C until required.
3. Results and discussion
3.1. Overall process for development of the SARS-CoV-2 biosensor using scFv-Fc antibodies
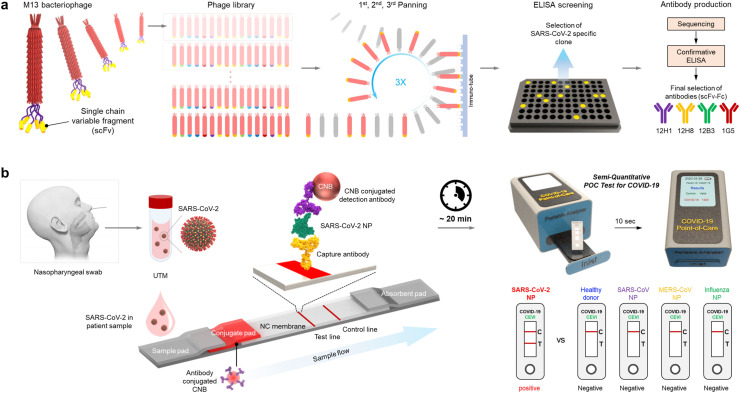
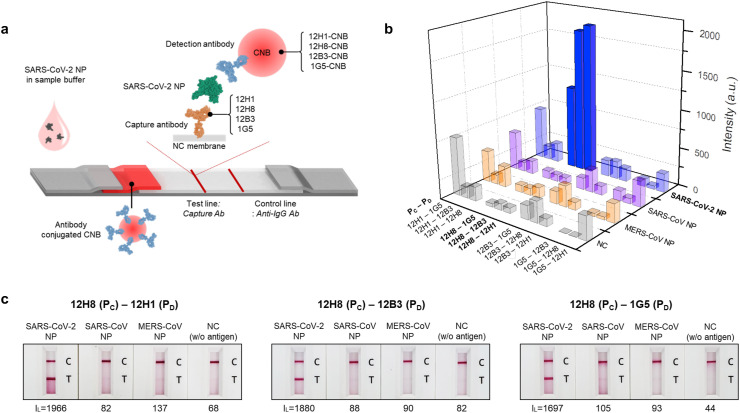
To develop the SARS-CoV-2 NP-specific LFIA-based biosensor, we first used phage display technology. The phage library contains a considerable number (>1012) of phages displaying different single-chain variable fragments (scFv). Through three rounds of biopanning and ELISA screening, the specific clones for the SARS-CoV-2 NP were primarily selected. To generate scFv-Fc, the sequences of selected clones were confirmed, and scFv-Fc fusion proteins were generated based on this sequence information. Finally, SARS-CoV-2 NP-specific scFv-Fc fusion proteins (12H1, 12H8, 12B3, and 1G5) were screened by confirmative ELISA (Scheme 1 a). The LFIA strip consisted of a sample pad, a conjugate pad, a nitrocellulose membrane, and an absorbent pad. Newly developed scFv-Fc antibodies specific for SARS-CoV-2 NP were used as capture and detection antibodies. When samples containing SARS-CoV-2 were loaded onto the LFIA biosensor, NP antigens of SARS-CoV-2 are first recognized by the detection antibodies conjugated to CNB. Finally, the sandwich complex (capture antibody-NP antigen-detection antibody) was formed at the test line, allowing the presence of SARS-CoV-2 to be confirmed directly with the naked eye within 20 min. Furthermore, the quantitative and more accurate analysis was achieved by using a portable LFIA reader. When the LFIA biosensor is mounted onto the portable LFIA reader, the image obtained from the LFIA is analyzed automatically using LED and CMOS sensors, and a quantitative result is obtained within 10 s (Scheme 1b).
Scheme 1.
A SARS-CoV-2-specific LFIA-based biosensor using scFv-Fc fusion proteins. (a) Schematic illustration of the development processes of SARS-CoV-2-specific scFv-Fc fusion proteins based on phage display technology. First, the SARS-CoV-2-specific scFv-Fc fusion proteins were screened, and four different scFv-Fcs with high affinity and specificity for the SARS-CoV-2 NP were selected using ELISAs. (b) Schematic showing the scFv-Fc-based LFIA consisting of a sample pad, a conjugate pad, a nitrocellulose (NC) membrane, and an absorbent pad. The selected scFv-Fc pairs were used as capture and detection antibodies. A test line placed on the NC membrane contains the capture scFv-Fc, and the CNB-conjugated detection scFv-Fc was immobilized on the conjugate pad. In the presence of SARS-CoV-2 NP, a sandwich complex (capture scFv-Fc-SARS-CoV-2 NP-detection scFv-Fc) was formed, and a clear red line appeared on the test line within 20 min. A handheld LFIA reader was used to analyze the line intensity semi-quantitatively. The proposed LFIA has is both sensitive and specific for SARS-CoV-2 NP, with no cross-reactivity with NPs from other coronaviruses such as MERS-CoV and SARS-CoV.
3.2. Development of SARS-CoV-2 NP-specific scFv-Fc antibodies
The NP was selected as the target antigen for diagnosis. The SARS-CoV-2 NP is an RNA-binding protein that forms a helical ribonucleoprotein necessary for viral RNA transcription and replication. It is a highly abundant protein expressed at the early stage of viral infection and is the most sensitive target for rapid CoV diagnosis (Das et al., 2010). Recombinant SARS-CoV-2 NP was expressed in E. coli and purified as an antigen to generate antibodies specific for SARS-CoV-2 (Fig. S1a).
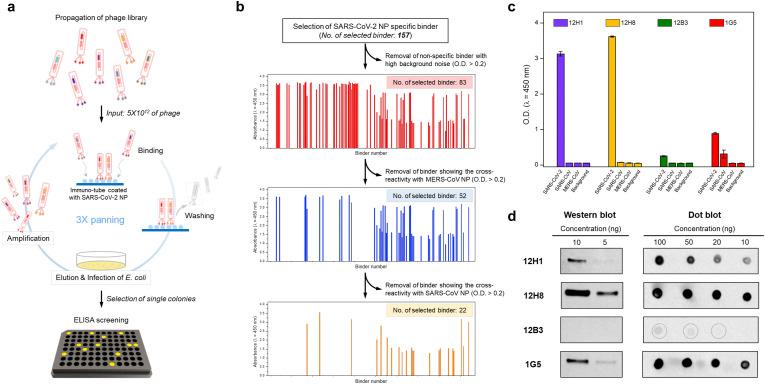
For antibody development, we used phage display technology. Generally, the phage disaply technology is faster than traditional hybridoma technology to discover antibodies and identify sequences of paratope, the antigen binding site (Winter et al., 1994). To screen scFv binders that interact specifically with SARS-CoV-2 NP, we performed phage display screening using a chicken naïve scFv antibody library (Fig. 1 a). After three rounds of biopanning, we isolated 157 positive clones with strong positive binding signals. Next, to isolate SARS-CoV-2 NP-specific scFv binders, non-specific scFv binders showing high background signals and cross-reactivity with NPs from MERS-CoV and SARS-CoV were eliminated. After removing non-specific scFv binders, 22 clones specific for SARS-CoV-2 NP were isolated (Fig. 1b), and four unique clones (12H1, 12H8, 12H3, and 1G5) with different complementary-determining region sequences of heavy and light chains were identified by DNA sequencing (Fig. S2). To generate specific antibodies for diagnosis, the four scFv binders were cloned into a scFv-Fc plasmid, and scFv-Fc antibodies were expressed and purified (Fig. S1b). The scFv-Fc format allows for characterization of scFvs before conversion into a full-length IgG and also can be used itself as antibody.
Fig. 1.
Screening of SARS-CoV-2 NP-specific scFv-Fc fusion proteins. (a) Schematic illustration of the phage display screening process using a chicken naïve scFv antibody library. After three rounds of biopanning, positive clones were isolated by ELISA screening. (b) Selection of SARS-CoV-2 NP-specific binders. Of the 157 positive clones obtained from phage display screening, non-specific scFv binders showing a high background signal (n = 74), or cross-reactivity with MERS-CoV NP (n = 31) and SARS-CoV NP (n = 30), were eliminated; ultimately, 22 specific binders were isolated. (c) ELISA results for specific interactions between the scFv-Fc antibodies and the SARS-CoV-2 NP antigen. The purified scFv-Fc antibody bound specifically to SARS-CoV-2 NPs. (d) Western and dot blot assay results. 12H1, 12H8, and IG5 scFv-Fc antibodies bound strongly to SARS-CoV-2 NP. The scFv-Fc antibody bound weakly to SARS-CoV-2 NP in the dot blot assay.
3.3. Interaction between scFv-Fc antibodies and the SARS-CoV-2 NP antigen
The virus that causes COVID-19, SARS-CoV-2, is a beta-CoV, which has a genome similar to that of SARS-CoV and MERS-CoV. Thus, specific detection of SARS-CoV-2 is essential for accurate diagnosis of COVID-19. To determine whether the scFv-Fc antibodies are specific for SARS-CoV-2, the binding of purified scFv-Fc antibodies to NP antigens from three known pathogenic beta-CoVs was investigated by biochemical analyses. ELISA experiments revealed that the four scFv-Fc antibodies bound to SARS-CoV-2 NP, but not to NPs from SARS-CoV or MERS-CoV (Fig. 1c). This indicates that the scFvs isolated from phage display screening are specific for SARS-CoV-2. The interaction between scFv-Fc antibodies and SARS-CoV-2 NP was also analyzed in western and dot blot experiments (Fig. 1d). The results showed that 12H1, 12H8, and 1G5 scFv-Fcs bound strongly to SARS-CoV-2 NP on both western and dot blots. However, binding of 12B3 scFv-Fc was weaker in the dot blot experiment, and it did not detect the antigen on the western blot. This suggests that 12H1, 12H8, and 1G5 scFv-Fc antibodies bind linear epitopes, whereas the 12B3 scFv-Fc antibody may recognize a conformational epitope.
3.4. Binding kinetics of the SARS-CoV-2 NP-specific scFv-Fc antibodies
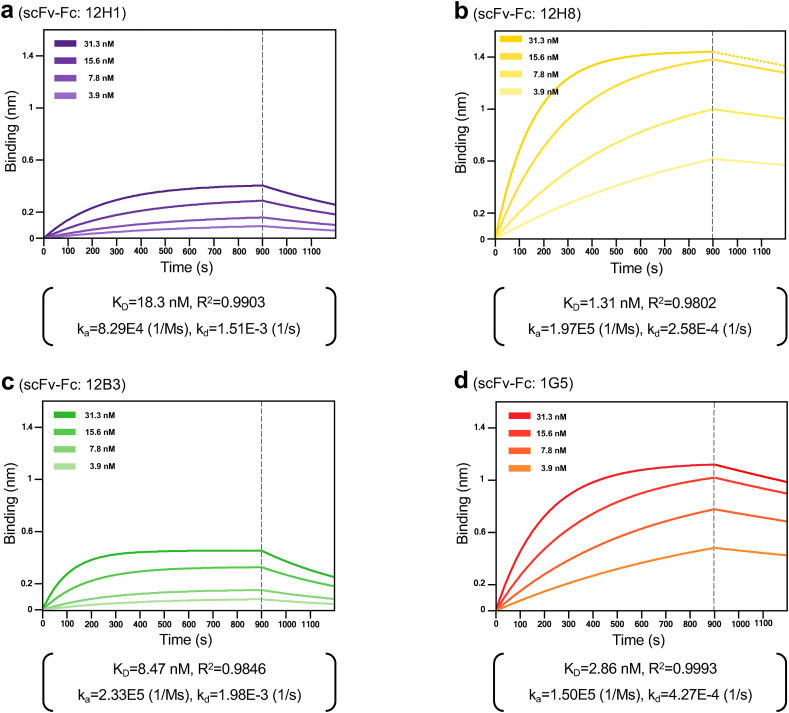
Next, to determine whether the scFv-Fc antibodies are sensitive enough for use as diagnostic antibodies, we measured their binding affinity of scFv-Fc antibodies for SARS-CoV-2 NP using a real-time label-free BLI biosensor. SARS-CoV-2 NP with a 6 × His tag (ligand) was immobilized onto a Ni-NTA biosensor, and the binding affinity of the scFv-Fc antibodies (analyte) was measured. Fig. 2 shows the kinetic interaction between SARS-CoV-2 NP and each scFv-Fc antibody. The KD value for the interaction of 12H1, 12H8, 12B3, and 1G5 with SARS-CoV-2 NP was 18.3, 1.31, 8.47, and 2.86 nM, respectively. It suggests that scFv-Fc antibodies with KD value in the nanomolar range are suitable for rapid detection of SARS-CoV-2 antigen.
Fig. 2.
The biolayer interferometry (BLI) results of scFv-Fc antibodies against the SARS-CoV-2 NP antigen. (a) 12H1, (b) 12H8, (c) 12B3, and (d) 1G5. Dotted lines represent the response curves of BLI measurement, and solid lines represent the fitting curves based on the 1:1 binding model. Binding kinetics were measured for four different concentrates of each scFv-Fc antibody. KD: equilibrium dissociation constant; R2: coefficient of determination; ka: association rate constant; kd: dissociation rate constant.
3.5. Application of scFv-Fc antibodies to the LFIA platform
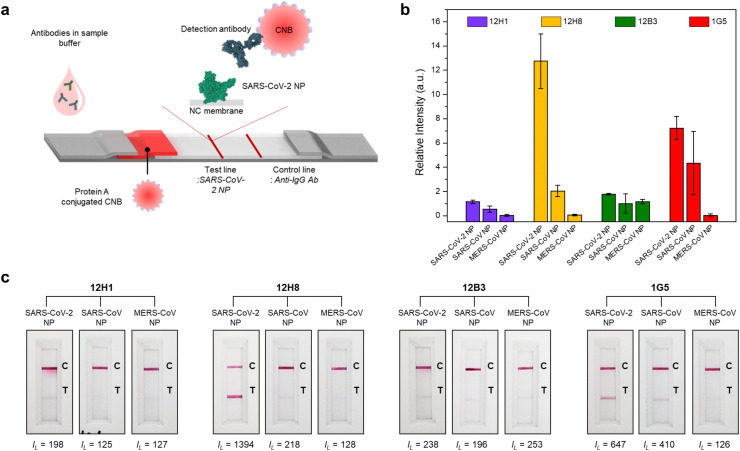
To confirm whether the scFv-Fc antibodies are suitable for use in the LFIA platform, we performed an indirect LFIA. We used CNB as a detection probe because it is more stable and sensitive than gold nanoparticles (Choi et al., 2020; Sakurai et al., 2014). The working principle of the indirect LFIA system is shown in Fig. 3 a. Briefly, 50 μg/ml NP antigen derived from three different CoVs were dispensed onto the test line of the NC membrane. For the control line, an anti-human IgG antibody was applied to confirm correct flow of the sample along the LFIA strip. In general, protein A has a high affinity for the Fc region of IgG-type antibodies. Thus, protein A-CNB conjugates were sprayed onto the conjugate pad. If the CNB-conjugated protein A complex and the antibody bind to the target SARS-CoV NP, the test line turns red. When the scFv-Fc antibodies were applied to the indirect LFIA test strip, all the four scFv-Fc antibodies detected SARS-CoV-2 NP antigen, with minor cross-reactivity with NPs from SARS-CoV and MERS-CoV (Fig. 3b and c). 12H8 showed the highest sensitivity to SARS-CoV-2 NP, with minor signals generated for NPs of SARS-CoV and MERS-CoV. 12H1 and 12B3 showed relatively weak positive binding to SARS-CoV-2 NP, but the signals were clearly distinguishable from those of NPs from other CoVs. In the case of 1G5, it showed a weak cross-reactivity with SARS-CoV NP (Fig. 3b and c). This indicates that the scFv-Fc antibodies are suitable for the LFIA diagnostic platform. With regard to specificity, considering that the concentration of antigens coated on test line was high, the weak positive signals for NPs from SARS-CoV and MERS-CoV could be managed in the sandwich LFIA.
Fig. 3.
Indirect LFIA results showing specific interactions between the scFv-Fc antibodies and SARS-CoV-2 NP. (a) Schematic diagram of the indirect LFIA using SARS-CoV-2 NP as a capture probe. Protein A-conjugated CNB c bound strongly to all antibodies in the sample buffer. The antibody-protein A-conjugated CNB complex binds to the pre-immobilized SARS-CoV-2 NP on the test line in a manner dependent upon the affinity between the antibody and SARS-CoV-2 NP. (b) Bar graph showing the intensities of the test lines. After 20 min of sample flow, the intensity of the test line was measured using a portable LFIA reader, and the line intensities were normalized using the following equation: Relative intensity = (IL– I0)/I0, IL: Line intensity in the presence of antibodies, I0: Line intensity in the absence of antibodies. (c) Results of the indirect LFIA. Three different NPs (derived from SARS-CoV-2, SARS-CoV, and MERS-CoV) were used for the test lines, and each scFv-Fc antibody was introduced onto the LFIA strip. After 20 min, the line intensities were measured using the portable LFIA reader (IL: line intensity) and the LFIA strips were photographed with a smartphone.
3.6. Screening of a scFv-Fc antibody pair for rapid diagnosis
To develop the LFIA-based RDT, two different antibodies (one for capture and the other for detection) are needed for a sandwich immunoassay. To select the optimum antibody pair for RDT, all possible combinations of the four different antibodies were tested. Each scFv-Fc antibody was immobilized onto the test line of the NC membrane, and each CNB-labeled antibody was added to the conjugation pad (Fig. 4 a). To test the RDTs, the same concentration of NP antigen from different CoVs was applied. The results are shown in Fig. S3. Among the 12 candidate pairs, three (12H8 as capture probe and 12H1, 12B3, and 1G5 as the CNB-conjugated detection probe) generated the strongest signals for SARS-CoV-2 NP, with no positive signal for NP from other CoVs (Fig. 4b and c). The 12H1-1G5 (capture-detection) pair also showed a strong positive signal for SARS-CoV-2 NP, but the background signal was even higher. With the results of 1G5-12H1 pair, it is suggested that 12H1 and 1G5 scFv-Fc antibodies may interact with each other (Fig. 4b and Fig. S3). Therefore, the 12H8–12H1, 12H8–12B3, 12H8-1G5 pairs were selected for specific detection of SARS-CoV-2. The common feature of the selected three diagnostic antibody pairs is that they share 12H8 as the capture antibody. The 12H8 scFv-Fc shows the highest affinity for the target antigen (Fig. 3b and c). When 12H8 was used as a detection antibody, however, the antibody pairs showed very weak positive signals (Fig. 4b and Fig. S3). This may suggest that for the development of a sandwich diagnostic pair, the antibody with higher affinity is better employed as the capture antibody rather than the detection antibody. In fact, the duration of antigen contact is much shorter for the capture antibody than that for the detection antibody. Thus, using an antibody with higher affinity as the capture antibody would improve the sensitivity of the LFIA.
Fig. 4.
Identification of the sandwich pair that best detects the SARS-CoV-2 NP antigen. (a) Schematic diagram of the sandwich LFIA. A total of 12 pairs obtained from four different scFv-Fc antibodies were analyzed using a positive control (SARS-CoV-2 NP) and negative controls (SARS-CoV NP, MERS-CoV NP, and background). (b) Bar graph showing line intensity according to the target antigen (SARS-CoV-2 NP, SARS-CoV NP, MERS-CoV NP, and background) and sandwich pair (PC: capture probe; PD: detection probe). Among the 12 pairs, three optimal pairs [12H8(PC)/12H1(PD); 12H8(PC)/12B3(PD); and 12H8(PC)/1G5(PD)] were sensitive and specific for the SARS-CoV-2 NP. (c) Sandwich LFIA results for the three selected pairs. SARS-CoV-2 NP, SARS-CoV NP, MERS NP, and running buffer (negative control, NC) were introduced onto the LFIA strips. After 20 min of sample flow, the line intensities were measured using the portable LFIA reader (IL: line intensity) and the LFIA strips were photographed using a smartphone.
3.7. Virus detection using the scFv-Fc antibody pairs
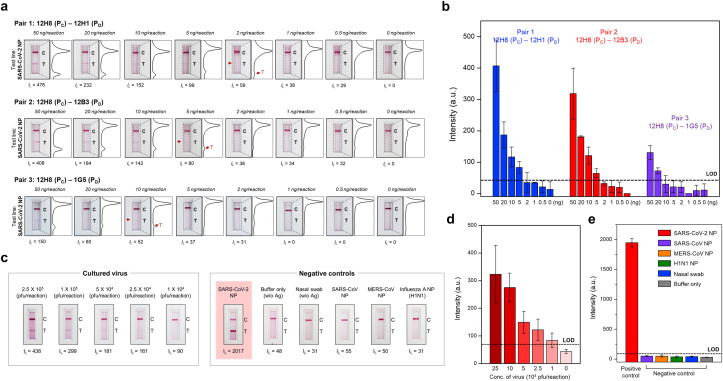
Finally, we used the three optimal combinations of scFv-Fc antibodies to ascertain the detection limit for antigen protein and cultured virus. The results showed that the detection limit of the 12H8–12H1, 12H8–12B3, 12H8-1G5 pairs for NP antigen was 2 ng, 5 ng, and 10 ng, respectively (Fig. 5 a and b). Using the best pair of scFv-Fc antibodies, we then analyzed the limit of detection for SARS-CoV-2 virus. The results showed that LFIA using the 12H8–12H1 scFv-Fc antibody pair could detect SARS-CoV-2 virus at levels as low as 2.5 × 104 pfu/reaction (Fig. 5c and d). Moreover, there was no cross-reactivity with NPs of SARS-Co-V, MERS-CoV, influenza virus, or negative control nasal swab specimens (Fig. 5c and e). These results indicate that the LFIA biosensor developed herein can successfully distinguish between SARS-CoV-2-positive and -negative samples. Moreover, the detection limit of LFIA biosensor would meet the conditions for the clinical use through optimization.
Fig. 5.
Sensitive and specific detection of the SARS-CoV-2 NP antigen. (a) Sensitivity of the sandwich LFIA for SARS-CoV-2 NP. Serially diluted SARS-CoV-2 NP (concentration range: 50 ng/reaction to 0.5 ng/reaction) was tested. After 20 min, the line intensities were analyzed using the portable LFIA reader (IL: line intensity) and the LFIA strips were photographed using a smartphone. In addition, the intensity of the test and control lines was converted to a peak histogram by an image analyzer. (b) Intensity of the test lines measured by the portable LFIA reader. The limit of detection was calculated as the mean value of the negative controls plus three times the standard deviation. The 12H8(PC)/12H1(PD) pair was the most sensitive for SARS-CoV-2 NP (as low as 2 ng of target antigen). (c) Sensitivity and selectivity of the best pair [12H8(PC)/12H1(PD)] for SARS-CoV-2 NP. Serially diluted cultured SARS-CoV-2 virus samples (concentration ranges: 2.5 × 105 pfu to 1 × 104 pfu) were used to evaluate the diagnostic performance of the LFIA biosensor. Moreover, NP from SARS-CoV, MERS-CoV, and influenza virus, or nasal swab specimens, were tested to investigate cross-reactivity (antigen concentration of all controls: 100 ng/reaction). After 20 min of sample flow, the intensities (IL) of test lines were measured using the portable LFIA reader and each LFIA strip was photographed using a smartphone. (d) Ability of the LFIA biosensor to detect cultured SARS-CoV-2. Using the best pair [12H8(PC)/12H1(PD)], the LFIA biosensor successfully detected cultured SARS-CoV-2 levels as low as 1 × 104 pfu. (e) Cross-reactivity of the proposed LFIA biosensor with various negative controls. There was no cross-reactivity with SARS-CoV, MERS, influenza NP, or nasal swab specimens (antigen concentration of all controls: 100 ng/reaction).
4. Conclusion
In summary, we developed a SARS-CoV-2-specific LFIA-based biosensor for rapid diagnosis of COVID-19 using scFv-Fc antibodies. The SARS-CoV-2 NP-specific scFv-Fc antibodies were generated by phage display technology, and the best diagnostic antibody pair for LFIA was screened. This COVID-19 biosensor detected SARS-CoV-2 virus within 20 min and distinguished SARS-CoV-2 from other similar CoVs, such as SARS-CoV and MERS-CoV. Further studies to enable more sensitive diagnosis are necessary. This may require changing the CNB detection dye to a fluorescent dye, and use of multiple scFv-Fc antibody pairs.
CRediT authorship contribution statement
Hye-Yeon Kim: Conceptualization, Methodology, Investigation, Writing - original draft. Jong-Hwan Lee: Conceptualization, Methodology, Investigation, Data curation, Writing - original draft. Mi Jeong Kim: Methodology, Investigation, Data curation, Writing - original draft. Sun Cheol Park: Investigation. Minsuk Choi: Investigation. Wonbin Lee: Investigation. Keun Bon Ku: Investigation. Bum Tae Kim: Funding acquisition, Supervision. Edmond Changkyun Park: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Hong Gi Kim: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Seung Il Kim: Project administration, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the National Culture Collection for Pathogens of the Korean CDC for providing the clinical SARS-CoV-2 isolates. We also thank Mr. S.Y. Lee (YntoAb) for the use of chicken naïve phage library. This work was supported by National Research Council of Science and Technology (CRC-16-01-KRICT) and National Research Foundation (NRF-2020M3E9A1043749) funded by the Ministry of Science and ICT, Republic of Korea and 2020 RIGHT Fund Grant – Technical Accelerator Award (PI2020112).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112868.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bishop J.D., Hsieh H.V., Gasperino D.J., Weigl B.H. Sensitivity enhancement in lateral flow assays: a systems perspective. Lab Chip. 2019;19(15):2486–2499. doi: 10.1039/c9lc00104b. [DOI] [PubMed] [Google Scholar]
- Choi E.S., Al Faruque H., Kim J.H., Cho J.H., Park K.M., Kim E. Immunochromatographic assay to detect alpha-tubulin in urine for the diagnosis of kidney injury. J. Clin. Lab. Anal. 2020;34(1) doi: 10.1002/jcla.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D., Kammila S., Suresh M.R. Development, characterization, and application of monoclonal antibodies against severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Vaccine Immunol. 2010;17(12):2033–2036. doi: 10.1128/CVI.00293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2020. Emergency Use Authorization (EUA) Information, and List of All Current EUAs. [Google Scholar]
- Kubina R., Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6) doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I., Wang L.F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G., Tan Y.J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Ledsgaard L., Kilstrup M., Karatt-Vellatt A., McCafferty J., Laustsen A.H. Basics of antibody phage display technology. Toxins. 2018;10(6) doi: 10.3390/toxins10060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Park W.B., Kwon N.J., Choi S.J., Kang C.K., Choe P.G., Kim J.Y., Yun J., Lee G.W., Seong M.W., Kim N.J., Seo J.S., Oh M.D. Virus isolation from the first patient with SARS-CoV-2 in Korea. J. Kor. Med. Sci. 2020;35(7):e84. doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A., Takayama K., Nomura N., Yamamoto N., Sakoda Y., Kobayashi Y., Kida H., Shibasaki F. Multi-colored immunochromatography using nanobeads for rapid and sensitive typing of seasonal influenza viruses. J. Virol. Methods. 2014;209:62–68. doi: 10.1016/j.jviromet.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang Z., Dong Y., Chang R., Xu C., Yu X., Zhang S., Tsamlag L., Shang M., Huang J., Wang Y., Xu G., Shen T., Zhang X., Cai Y. Phase-adjusted estimation of the number of coronavirus disease 2019 cases in wuhan, China. Cell. Discov. 2020;6:10. doi: 10.1038/s41421-020-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus Disease 2019 (COVID-19) Situation Report – 52. [Google Scholar]
- WHO . 2020. Coronavirus Disease (COVID-19) Weekly Epidemiological Update. [Google Scholar]
- WHO . 2020. Pneumonia of Unknown Cause – China. [Google Scholar]
- Winter G., Griffiths A.D., Hawkins R.E., Hoogenboom H.R. Making antibodies by phage display technology. Annu. Rev. Immunol. 1994;12(1):433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12(6) doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.