Abstract
Increased cytokine levels, acute phase reactants and immune checkpoint expression changes have been described in patients with Coronavirus Disease 2019 (COVID-19). Here, we have reported a monocyte polarization towards a low HLA-DR and high PD-L1 expression after long exposure to proteins from SARS-CoV-2. Moreover, CD86 expression was also reduced over SARS-CoV-2 proteins exposure. Additionally, T-cells proliferation was significantly reduced after stimulation with these proteins. Eventually, patients with long-term SARS-CoV-2 infection also exhibited a significant blockade of T-cells proliferation.
Keywords: Inflammation, Infectious disease, Pathology, Virology, Immunology, COVID19, Sepsis, T cell proliferation, Immune checkpoints, PD-L1/PD-1, T cell exhaustion
Inflammation; Infectious disease; Pathology; Infectious Disease; Virology; Immunology; COVID19, sepsis, T cell proliferation, immune checkpoints, PD-L1/PD-1, T cell exhaustion
1. Introduction
The emergence of SARS-CoV-2 is a global issue that has triggered significant political and social changes. Structurally, SARS-CoV-2 has four main structural proteins including spike (S) glycoprotein, small envelope (E) glycoprotein, membrane (M) glycoprotein, and nucleocapsid (N) protein, and also several accessory proteins such as the Papain-like protease (P) [1]. The infection caused by SARS-CoV-2 leads to a primary viral pneumonia, which resembles the Severe Acute Respiratory Syndrome (SARS) [2] and, eventually, a sepsis figure [3, 4, 5, 6].
Some innate immune system (IIS) cells, principally monocytes and macrophages, have been identified as key players in orchestrating the inflammatory response by activating a number of crucial pathways including the nuclear factor kappa B (NF-κB) and interferon regulatory factor (IRF) pathways in several clinical contexts [7, 8]. As previously described for sepsis, patients with severe COVID-19 infection show inflammation and cytokine storms. In these patients the overexpression of interleukin (IL)-1β, IL-6, and tumor necrosis factor alpha (TNFα), in the early phase of the disease have been reported [9]. All these hallmarks of sepsis have been explained by a monocyte/macrophage activation [10]. Numerous studies have indicated that the inflammatory phase for severe COVID-19 patients is restricted to the initial phase of the disease [1]. The subsequent chronic basal inflammation, which may last for several days, leads the immune system towards a patent refractory state, as it is also described in prolonged sepsis.
Many authors have indicated that not only the IIS but also the adaptive system become deregulated in patients with severe COVID-19 infection [4, 9]. While monocytes/macrophages appear to play a crucial role in the early phase of SARS-CoV-2 infection, the adaptive immune system emerges as a critical factor in the late phase of the disease. The total number of lymphocytes is significantly reduced in those patients with a poor prognosis [9]. In line, a multicenter retrospective study indicated that the lymphocyte count was an independent high-risk factor connected with COVID-19 progression [11]. Other studies have revealed an inverse correlation between lymphocyte count and time to symptom reappearance in a cohort of COVID-19 patients discharged from the hospital [12]. It should be noted that these observations have been reported only in patients who have required hospitalization.
Likewise, lymphocytes depletion has been described during sepsis, that which compromises the adaptive response in the latter phase of this disease [13]. We and others have studied this phenomenon, demonstrating the implication of the immune checkpoint (IC) programmed death-1 (PD-1) and its ligand PD-L1 [14]. Moreover, PD-1/PD-L1 axis activation during sepsis induced a patent “lymphocyte exhaustion”. This last effect was reverted by blocking monoclonal antibodies against either PD-1 or PD-L1 [15,16]. Along these lines, SARS-CoV-2 induced apoptosis of peripheral blood lymphocytes through P53 activation [17] and PD-1 was found upregulated in patients who were in the last phase of COVID19 infection [18].
Here, we established an in vitro model using a cocktail of proteins from SARS-CoV-2 and blood cells from healthy volunteers. S glycoprotein, a transmembrane protein localized in the outer portion of the virus that facilitates binding to the host cells through angiotensin-converting enzyme 2 (ACE2); N protein, a structural component bound to the RNA of the virus; and the Papain-like protease (P), which is required to process the viral polyprotein into functional subunits; were used to mimic the presence of the virus. Our data demonstrated the induction of a specific monocyte profile (HLA-DRlowPD-L1high) due to the presence of the cocktail of proteins from SARS-CoV-2, which in turn caused a patent T-cells exhaustion.
2. Materials and methods
2.1. Reagents
Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen), supplemented with Fetal Bovine Serum (FBS) to 10% and 1% Penicillin and Streptomycin mix (Gibco), was used for cells culture. SARS-CoV-2 proteins Spike (S) and Nucleoprotein (N) were purchased from Sino Biological and the Papain-like protease (P) from R&D Systems. Carboxyfluorescein succinimidyl ester (CFSE) was purchased from ThermoFisher Scientific, to assess T-cells proliferation using Dynabeads ® Human T-Activator CD3/CD28 (ThermoFisher Scientific) and Lectin from Phytolacca americana (pokeweed, PWD) from Sigma-Aldrich as stimuli. For PD-L1/PD-1 crosstalk inhibition, fully human anti-PD-L1 (pembrolizumab, Merck Sharp & Dohme) and anti-PD-1 (nivolumab, Bristol Myers Squibb) blocking antibodies were used. Human Th1/Th2/Th17 Cytometric Bead Array (CBA) from BD Biosciences was utilized for cytokine quantitation.
2.2. PBMCs isolation and SARS-CoV-2 infection in vitro model
Peripheral blood mononuclear cells (PBMCs) from healthy volunteers and COVID-19 patients from La Paz University Hospital were isolated by Ficoll-Plus (GE Healthcare Bio-Sciences) gradient. PBMCs were cultured in RPMI 1640 medium, supplemented with FBS to 10% and 1% Penicillin and Streptomycin mix. For the in vitro model, we performed 3 conditions: i)PBMCs were treated once with S, N and P proteins of SARS-CoV-2 (250 ng/mL for S and N and 25 nM for P protein) simulating an acute infection (dark square). ii) PBMCs treated every 48 h with S, N and P proteins (250 ng/mL for S and N and 25 nM for P protein) simulating a steady infection without resolution (red triangle); and iii) PBMCs treated with PBS as negative control stimulus (empty circle).
A scheme is shown in Figure 1A. After for each condition, supernatants were collected and stored at -80 °C until cytokine measurement, and cells were labelled with proper antibodies for flow cytometry analysis.
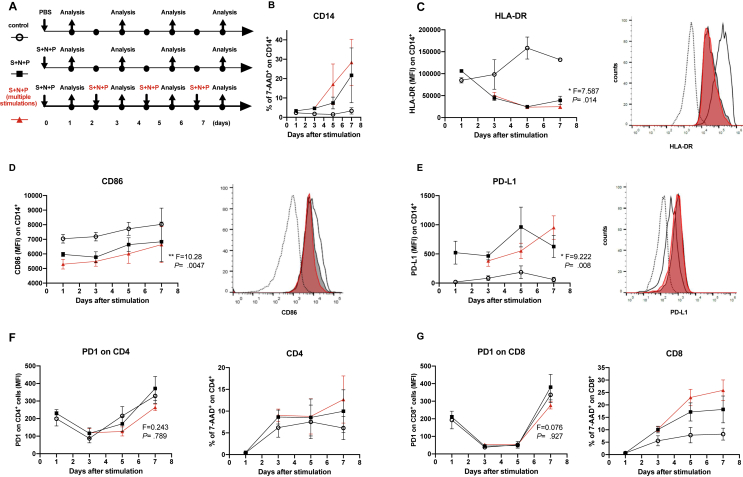
Figure 1.
Proteins from SARS-CoV-2 polarize human monocytes towards an HLA-DRlowPD-L1high phenotype. (A) Experimental design. Peripheral blood mononuclear cells (PBMCs) from healthy volunteers were cultured for 7 days and stimulated once or every 48 h with a cocktail of proteins from the virus including S glycoprotein (250 ng/mL), N protein (250 ng/mL) and the Papain-like protease (P, 25 nM). (B) Viability of CD14+ cells were measured by 7-AAD staining of dead cells cultured following the experimental design (A) is shown. (C) Mean Fluorescence Intensity (MFI) expressions of HLA-DR on CD14+ cells are shown (left panel) (∗, p < 0.05, one-way ANOVA test), n = 5. A representative histogram overlay is shown, fluorescent minus one (FMO, dotted line), unstimulated control (empty), S + N + P (solid black), S + N + P multiple stimulations (solid red), right panel. (D) MFI expressions of CD86 on CD14+ cells are shown (left panel) (∗, p < 0.05, one-way ANOVA test), n = 5. A representative overlay is shown, fluorescent minus one (FMO, dotted line), unstimulated control (empty), S + N + P (solid black), S + N + P multiple stimulations (solid red), right panel. (E) MFI expressions of PD-L1 on CD14+ cells are shown (left panel) (∗, p < 0.05, one-way ANOVA test), n = 5. A representative overlay is shown, fluorescent minus one (FMO, dotted line), unstimulated control (empty), S + N + P (solid black), S + N + P multiple stimulations (solid red), right panel. (F and G, left panels) Expressions of PD-1 on CD4+ (F) and CD8+ (G) cells are shown. Viability of CD4+ (F) and CD8+ (G) cells are shown (right panels).
2.3. Flow cytometry analysis
Fluorescence-activated cell sorting (FACS) analysis were developed using specific human antibodies (Abs) to the following surface molecules: CD8-Allophycocianin (APC), CD4-Peridinin-chlorophyll-A protein (PerCP) and HLA-DR-Fluorescein-isothiocyanate (FITC) (all three from ImmunoStep), 7-Amino-Actinomycin D (7-AAD), CD8-BV510, CD86-BUV737, CD14-BV395, PD-L1-Phycoerythrin (PE) and PD-1 FITC (all five from BD Biosciences). Cells were stained with proper antibodies for 30 min at 4 °C in the dark and washed once with Phosphate Buffer Saline (PBS). Not stained cells were used as negative controls. For all assays, samples were run in FACS Calibur or FACS Celesta (BD Biosciences) flow cytometers and data were analyzed with FlowJo (TreeStar) v. 10.6.2 software or BD Diva v10.
2.4. T cell proliferation assay
PBMCs were labelled with CFSE according to the manufacturer's protocol and seeded into round bottom 96-wells plates (2x105 cells per well) in RPMI 1640 medium supplemented with FBS to 10% and 1% Penicillin and Streptomycin. CFSE-labelled PBMCs were exposed to S, N and P proteins at the above-mentioned concentrations. Immediately after, they were stimulated or not with the PWD (2.5 μg/mL) mitogen or Dynabeads Human T-activator CD3/CD28 (0.5 μL/well) and treated or not with fully human anti-PD-L1 antibody or fully human anti-PD-1 antibody (both at 50 μg/mL) for 5 days. PBS was used as negative control stimulus.
2.5. Cytokine measurement
The cytokine levels in the culture supernatants were determined using the Human Th1/Th2/Th17 Cytometric Bead Array (CBA), following the manufacturer's protocol. Briefly, culture supernatants were labelled by incubating them with specific beads, washed, acquired using a FACS Calibur flow cytometer and finally analyzed with FCAP Array v3.0 Software (both from BD Biosciences).
2.6. Patients and healthy volunteers
This study was conducted in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Committee for Human Subjects of La Paz University Hospital. All the participants provided written consent for the study. Patients older than 18 years old who fulfilled the diagnostic criteria for COVID-19 (Symptoms of acute respiratory infection and positive RT-PCR for SARS-CoV-2 by nasopharyngeal or oropharyngeal swab) were included in the study. These were classified into three groups according to the length of hospital stay at the time of sample collection as group 1 (from 0 to 2 days), group 2 (from 5-7 days), group 3 (8–20 days) and group 4 (more than 20 days). Note that patients included in the last group were SARS-CoV-2 RT-PCR positive at admission, but at the time of sample collections they were SARS-CoV-2 negative by the same technique. Healthy volunteers were subjects without relevant pathologies and with a negative RT-Q-PCR for SARS-CoV-2 (see Table 1 for details).
Table 1.
Patient and healthy volunteers’ characteristics.
| 0-2 (n = 5) | 5-7 (n = 5) | 8-20 (n = 6) | >20 (n = 9) | HVs (n = 6) ∗∗ | |
|---|---|---|---|---|---|
| Age, years | 60.75 ± 16.58 | 46.25 ± 8.95 | 57.71 ± 19.26 | 56.42 ± 12.89 | 37 ± 9.55 |
| Sex, male, n (%) | 3 (50) | 3 (60) | 4 (66.66) | 9 (100) | 3 (50) |
| Symptoms onset, days | 6.67 ± 4.04 | 12.4 ± 2.97 | 23.3 ± 2.86 | 65.4 ± 8.17 | |
| q-SOFA, n (%)∗ | |||||
| 0 | 2 (40) | 2 (40) | 2 (33.33) | 0 (0) | - |
| 1 | 2 (40) | 2 (40) | 3 (50) | 2 (22.22) | - |
| 2 | 1 (20) | 1 (20) | 1 (16.66) | 6 (66.66) | - |
| 3 | 0 (0) | 0 (0) | 0 (0) | 1 (11.11) | - |
Data are mean ± SD, or number (%).
qSOFA: quick Sequential Organ Failure Assessment.
At admission.
Healthy volunteers.
2.7. Statistical analysis
The number of experiments analyzed is indicated in each figure. The statistical significance was calculated using a one-way ANOVA or paired t-test, depending on the specific assay. The statistical significance was set at p < 0.05, and the statistical analyses were conducted using GraphPad Prism 8.0 software.
3. Results
3.1. Proteins from SARS-CoV-2 polarize human monocytes toward a profile with low HLA-DR and high PD-L1 expression
To evaluate the potential impact of SARS-CoV-2 on monocytes and its interaction with lymphocytes, we followed the experimental approach showed in Figure 1A. PBMCs were cultured for 7 days and stimulated once or every 48 h with a cocktail of S, N and P proteins from the virus. Note that multiple stimulation with the cocktail simulated a steady viral presence without resolution, while the single stimulation mimicked an acute infection.
The major histocompatibility complex (MHC) class II cell surface receptor HLA-DR and the IC ligand PD-L1 were evaluated on CD14+ cells. Both single and multiple stimulations with the SARS-CoV-2 proteins did not affected viability of monocytes (Figure 1B) but reduced HLA-DR expression on monocytes in a time dependent manner (Figure 1C). In the same line, the antigen presentation co-stimulatory molecule CD86, showed a decreased expression in monocytes when these become stimulated with the viral proteins (Figure 1D). In contrast, PD-L1 was up-regulated by the cocktail (Figure 1E). On the other hand, no differences of PD-1 expression on lymphocytes were found between the conditions tested (Figure 1 F and G, left panels). Additionally, viability changes on CD4+ and CD8+ were not statistically significant when associated with the presence of the virus proteins cocktail (Figure 1 F and G, right panels). For all experiments, no differences between single and multiple stimulations were found.
3.2. Long exposure to SARS-CoV-2 proteins reduces the T lymphocytes proliferative ability in vitro
Induction of an HLA-DRlowCD86lowPD-L1high phenotype in monocytes stimulated with S, N and P proteins prompted us to study the proliferative ability of T lymphocytes in this context.
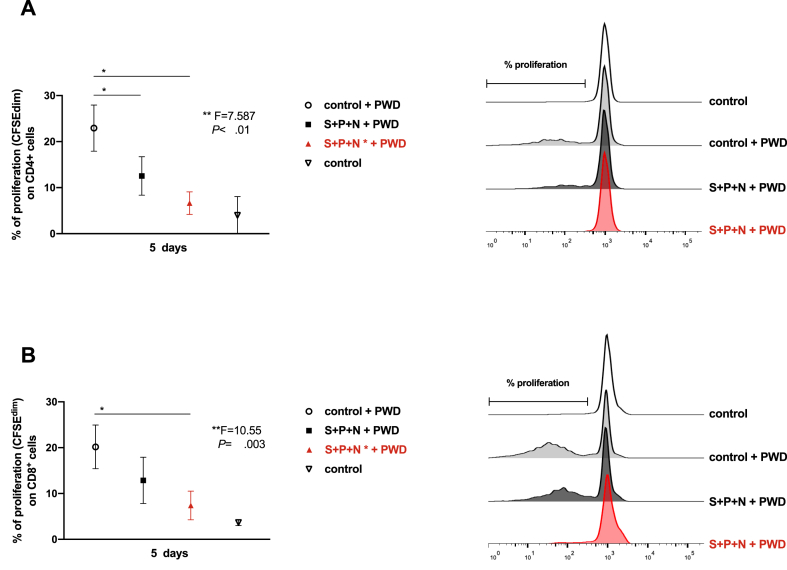
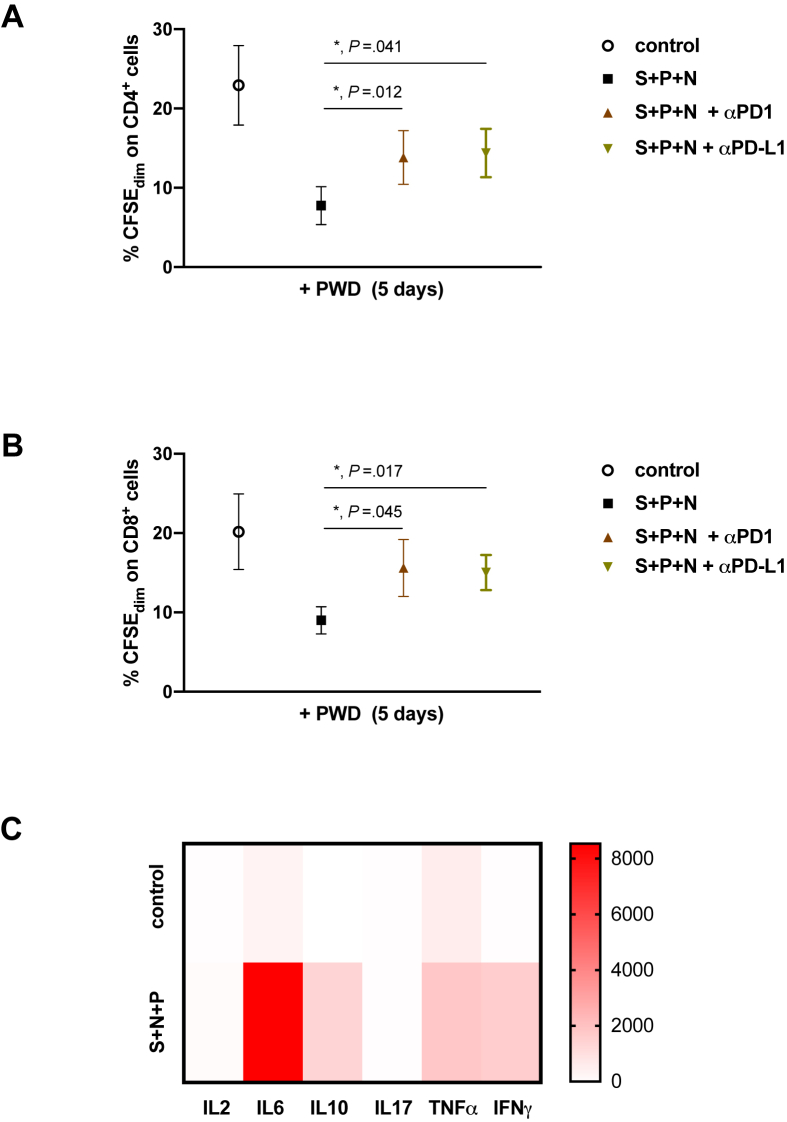
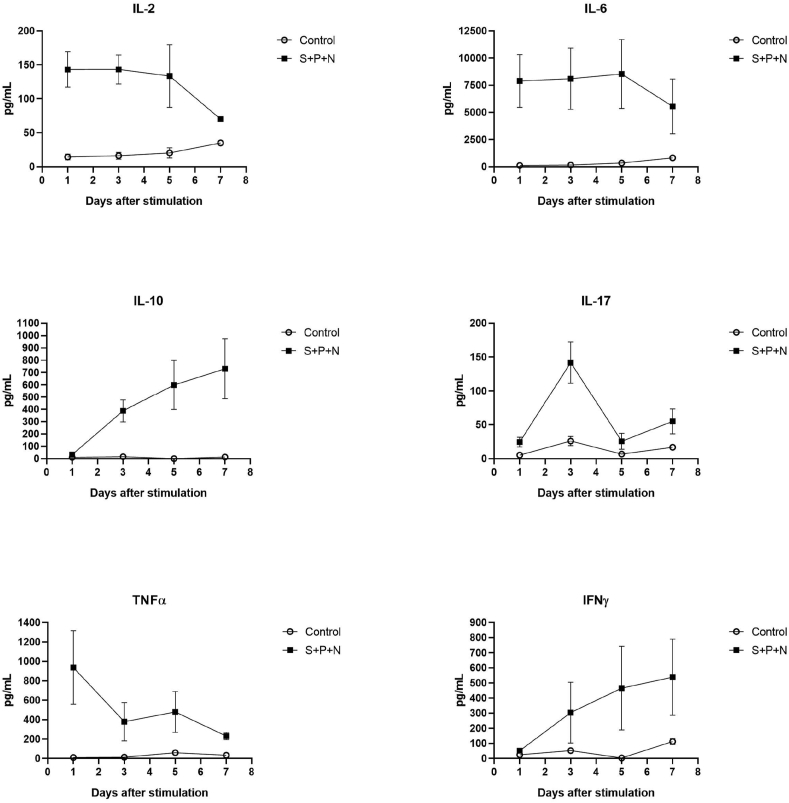
According to our data, after 5 days of challenge with the protein cocktail, monocytes achieved the lowest HLA-DR and highest PD-L1 expression. Therefore, we decided to test the T lymphocytes proliferation in this experimental point. As Figure 2 shows, presence of the SARS-CoV-2 proteins impaired both CD4 and CD8 proliferation when cultures were treated with the classical mitogen PWD. Next, we evaluated the potential role of the PD-L1/PD-1 axis in this phenomenon. A slight but significant proliferative recovery was observed in both CD4 and CD8 populations when blocking anti-PD-L1 or anti-PD-1 antibodies were added independently (Figure 3 A and B). Interestingly, high levels of proinflammatory cytokines as IL-6 and TNFα were detected in the supernatants of cultures in the early response (24 h post-stimulation) and were maintained after 5 days of stimulation with the proteins cocktail (Figure 3C and Figure 4). Other cytokines, such as IL-10 and IFNγ were increased over time.
Figure 2.
Long exposure to SARS-CoV-2 proteins reduces the T lymphocytes proliferative ability in vitro.PBMCs from healthy volunteers were labelled with CFSE, then cells were exposed or not (open circle) to S, N and P proteins for 5 days (solid black square) or multiple stimulations (Figure 1A) until 5 days (solid red triangle). Cells were also stimulated or not (open triangle) with Pokeweed (PWD, 2.5 μg/mL). (A) Percentage of proliferative CD4+ populations (CFSEdim), measured by flow cytometry, are shown (left panel) (∗, p < 0.05, Mann-Whitney t-test; ∗∗, p < 0.01, one-way ANOVA test), n = 5. A representative histogram overlay is shown (right panel). (B) Percentage of proliferative CD8+ populations, measured by flow cytometry, are shown (left panel) (∗, p < 0.05, Mann-Whitney t test; ∗∗, p < 0.01, one-way ANOVA test), n = 5. A representative histogram overlay is shown (right panel).
Figure 3.
T-cells proliferation is recovered by PD-L1/PD-1 axis blockage. PBMCs from healthy volunteers were labelled with CFSE, then cells were exposed or not (open circle) to S, N and P proteins for 5 days (solid black square). Anti-PD-L1 (solid black triangle) or anti-PD-1 (solid grey inverted triangle) were added. All cultures were also stimulated with Pokeweed (PWD, 2.5 μg/mL). Percentage of proliferative (CFSEdim) CD4+ (A) and CD8+ (B) populations, measured by flow cytometry, are shown (∗, p < 0.05, paired t-test), n = 5. (C) An array analysis is shown for indicated cytokines, that were analyzed in the supernatant by Cytometric Bead Array (CBA) after stimulation with S, N and P proteins from SARS-CoV-2 (S + N + P) or not (control) of PBMCs from healthy volunteers for 5 days; n = 5.
Figure 4.
SARS-CoV-2 proteins induce cytokine production. Peripheral blood mononuclear cells (PBMCs) from healthy volunteers were stimulated with SARS-CoV-2 proteins S glycoprotein (250 ng/mL), N protein (250 ng/mL) and the Papain-like protease (P, 25 nM). Levels of IL-2, IL-6, IL-10, IL-17, TNFα and INFγ in cell culture supernatants after 24, 72, 120 and 168 h are shown, n = 5.
3.3. Patients exposed to SARS-CoV-2 exhibit a low rate of T-cells proliferation
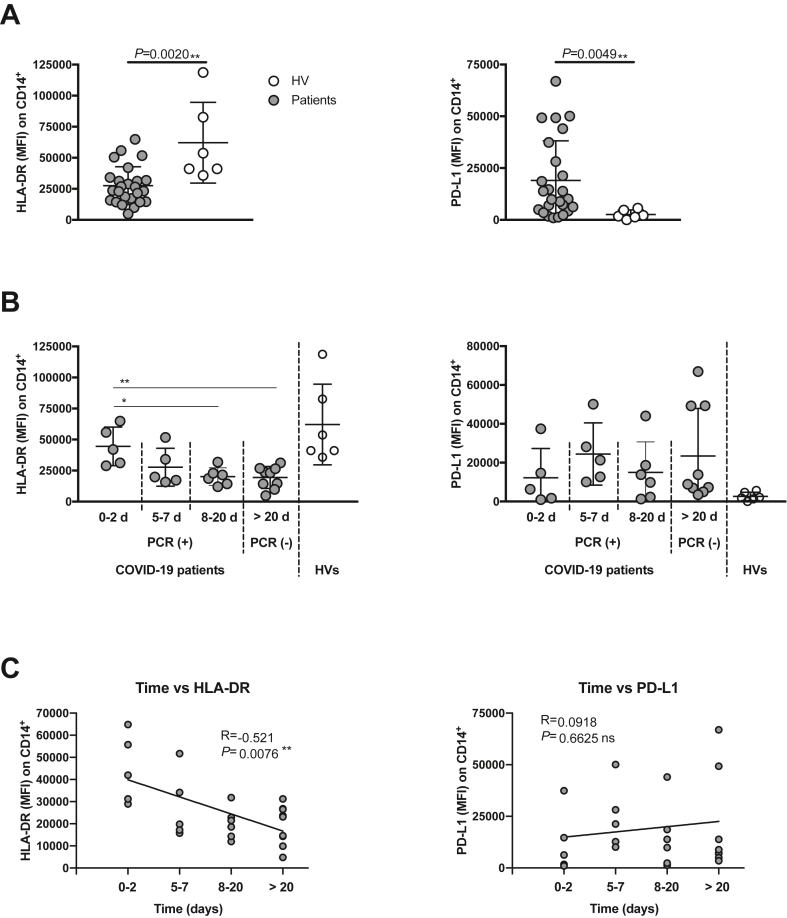
To corroborate our in vitro data, we analyzed the HLA-DR and PD-L1 expression on monocytes and the proliferative ability of circulating T-cells from COVID-19 patients. Blood samples were obtained from four groups of patients. The first group had been hospitalized for 0–2 days, the second group for 5–7 days, the third group for 8–20 days, and the fourth one, for more than 20 days (Table 1). Note that the latter patient group, after being found RT-PCR positive for SARS-CoV-2 on admission, had already cleared the virus when sampled since they were RT-PCR negative at this point, but nonetheless, their clinical condition persisted.
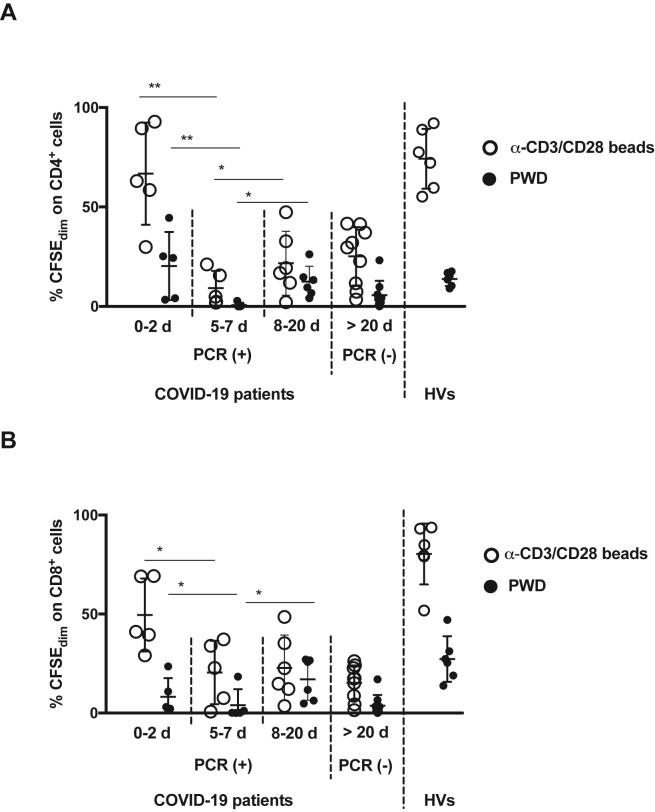
In line with the data obtained in the in vitro model, we found an HLA-DRlowPD-L1high phenotype in COVID-19 patients monocytes compared with HVs (Figure 5). Next, cells were labelled and cultured under two different proliferation induction conditions using either anti-CD3/CD28 beads or PWD for 5 days. Both CD4 and CD8 cells exhibited a significant proliferation reduction after long exposure to the virus (Figure 6). Interestingly, the third group maintained low T-cells proliferation, even though the virus was yet undetectable by RT-PCR.
Figure 5.
Monocytes from COVID-19 patients have decreased HLA-DR and increased PD-L1 expression. (A) HLA-DR (left) and PD-L1 (right) expression on CD14+ cells from COVID-19 patients (n = 25) and HVs (n = 6). (B) COVID-19 patients were classified according their admission time. Their HLA-DR (left) and PD-L1 (right) expression on CD14+ cells are shown. (C) Spearman correlation between sample collection time collection from patient admission time and the expression of HLA-DR (left) and PD-L1 (right). ∗, p < 0.05; ∗∗, p < 0.01 Mann-Whitney t-test for (A) and (B) and Spearman correlation test in (C); MFI, Mean Fluorescence Intensity; R = Spearman r; vs, versus.
Figure 6.
Patients exposed to SARS-CoV-2 exhibit a low rate of T-cells proliferation. Four groups of COVID-19 patients were recruited according to their length of hospital stay: 0–2 days (n = 5), 5–7 days (n = 5), 8–19 days (n = 6), or more than 20 days (n = 9). PBMCs were isolated from these and from healthy volunteers (HVs, n = 6), labelled with CFSE and stimulated with Pokeweed (PWD, 2.5 μg/mL, solid black circle) or anti CD3/CD28 beads (0.5 μL/well, open black circle) for 5 days. Percentage of proliferative CD4+ (A) and CD8+ (B) populations, measured by flow cytometry, are shown (∗, p < 0.05; ∗∗, p < 0.01 Mann-Whitney t-test). The RT-PCR status for SARS-CoV-2 infection at moment of blood recruitment for each group of patients are also shown.
4. Discussion
Data generated denote that SARS-CoV-2 infection is a complex disease that might show two potentially overlapping phases. Whereas, the first phase is described by a lopsided innate response with a huge cytokine storm, the second stage generates both a significant damage to the body and an acute lymphopenia, a condition also observed during sepsis.
We and others have described a T-cell exhaustion in septic patients associated with poor prognosis [13, 14, 15, 19, 20, 21, 22]. This phenomenon was governed by an activation of the PD-L1/PD-1 axis in these patients [16]. Here we demonstrate that blood cells from healthy volunteers, incubated with a SARS-CoV-2 protein cocktail, develop a phenotype that mimic the COVID-19 condition. While monocytes show a negative regulation of HLA-DR, an important component of the antigen presentation pathway, together with high levels of the IC ligand PD-L1, lymphocytes experience an exhaustion. Remarkably, T-cell exhaustion is partially but significantly reverted by either anti-PD-L1 or anti-PD-1. These data suggest a potential role of the PD-L1/PD-1 axis in this clinical situation and support the use of antibodies against ICs in patients with COVID-19. Along these lines, camrelizumab (a PD-1 monoclonal antibody) and thymosin have been included in a clinical trial (NCT04268537) for COVID-19 treatment [23].
We highlight that the observed effects have taken place using viral recombinant proteins rather than the virus itself. We have selected spike (S) glycoprotein, nucleocapsid (N) protein and papain-like protease (P) because they are described as essential players in the structure and pathogenicity of SARS-CoV-2 [24, 25, 26]. Therefore, the data indicate that with the sole presence of some viral proteins, a response can be triggered in such a way that compromises the health of patients. Hence, therapeutic decisions in patients with negative RT-PCR for SARS-CoV-2 should be considered on this basis.
Our data herein though from a limited cohort of patients with COVID-19 have eventually revealed an acute T-cells exhaustion effect associated to the time of hospitalization. While patients in the first days of the disease showed a discrete T-cells proliferation, an acute down-regulation of the proliferation was observed in those within 5–7 days, 8–19 days and more than 20 days of hospitalization. Interestingly, those long-term patients who cleared the virus kept the low T-cells proliferation profile. Accordingly, a painstaking study of the interaction between the virus and the immune system is necessary in order to identify suitable pharmaceutical targets. We have to reach a precise balance between regulating the first wave of cytokines and reactivating an appropriate adaptive response; a fine balance struck between simultaneously blocking and unblocking the host's immune response. Therefore, biomarkers that plainly show the proper approach must be identified.
Declarations
Author contribution statement
E. Lopez-Collazo: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
J. Avendaño-Ortiz: Conceived and designed the experiments; Analyzed and interpreted the data.
R. Lozano-Rodríguez: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
K. Montalbán-Hernández, V. Terrón, F. de la Bastida, C. Cubillos-Zapata and J. Valentín: Performed the experiments.
L. Aguirre: Analyzed and interpreted the data; Wrote the paper.
A. Martín-Quirós, C. Maroun-Eid, M. García-Garrido and A. del Balzo-Castillo: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by Fundación Mutua Madrileña, Alonso Family Foundation, Fundación Banco Santander, Real Seguros, Despacho Uria and Ayuntamiento de Madrid. K. Montalbán-Hernández was supported by “la Caixa” Foundation.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank the Alonso Family Fundación, Santander Bank, Real Seguros, Fundación Mutua Madrileña, Fundación Uria, Fundación Caixa and Ayuntamiento de Madrid for their support.
Contributor Information
Luis A. Aguirre, Email: luis.augusto.aguirre@idipaz.es.
Eduardo López-Collazo, Email: elopezc@salud.madrid.org.
References
- 1.Astuti I. Null Ysrafil, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borobia A.M., Carcas A.J., Arnalich F., Álvarez-Sala R., Monserrat-Villatoro J., Quintana M., Figueira J.C., Torres Santos-Olmo R.M., García-Rodríguez J., Martín-Vega A., Buño A., Ramírez E., Martínez-Alés G., García-Arenzana N., Núñez M.C., Martí-de-Gracia M., Moreno Ramos F., Reinoso-Barbero F., Martin-Quiros A., Rivera Núñez A., Mingorance J., Carpio Segura C.J., Prieto Arribas D., Rey Cuevas E., Prados Sánchez C., Rios J.J., Hernán M.A., Frías J., Arribas J.R. Null on behalf of the covid hulp working group, A cohort of patients with COVID-19 in a major teaching hospital in Europe. J. Clin. Med. 2020;9 doi: 10.3390/jcm9061733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S. China medical treatment Expert group for covid-19, clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao J.Y., Derespina K.R., Herold B.C., Goldman D.L., Aldrich M., Weingarten J., Ushay H.M., Cabana M.D., Medar S.S. Clinical characteristics and outcomes of hospitalized and critically ill children and adolescents with coronavirus disease 2019 (COVID-19) at a tertiary care medical center in New York city. J. Pediatr. 2020 doi: 10.1016/j.jpeds.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas S.K., Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 8.López-Collazo E., del Fresno C. Pathophysiology of endotoxin tolerance: mechanisms and clinical consequences. Crit. Care. 2013;17:242. doi: 10.1186/cc13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L., Xiong L., Guo C., Tian J., Luo J., Yao J., Pang R., Shen H., Peng C., Liu T., Zhang Q., Wu J., Xu L., Lu S., Wang B., Weng Z., Han C., Zhu H., Zhou R., Zhou H., Chen X., Ye P., Zhu B., Wang L., Zhou W., He S., He Y., Jie S., Wei P., Zhang J., Lu Y., Wang W., Zhang L., Li L., Zhou F., Wang J., Dittmer U., Lu M., Hu Y., Yang D., Zheng X. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalova I.N., Lim J.Y., Chittezhath M., Zinkernagel A.S., Beasley F., Hernández-Jiménez E., Toledano V., Cubillos-Zapata C., Rapisarda A., Chen J., Duan K., Yang H., Poidinger M., Melillo G., Nizet V., Arnalich F., López-Collazo E., Biswas S.K. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Wu J., Liu J., Zhao X., Liu C., Wang W., Wang D., Xu W., Zhang C., Yu J., Jiang B., Cao H., Li L. Clinical characteristics of imported cases of COVID-19 in Jiangsu province: a multicenter descriptive study. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan J., Kou S., Liang Y., Zeng J., Pan Y., Liu L. PCR assays turned positive in 25 discharged COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotchkiss R.S., Tinsley K.W., Swanson P.E., Schmieg R.E., Hui J.J., Chang K.C., Osborne D.F., Freeman B.D., Cobb J.P., Buchman T.G., Karl I.E. Sepsis-induced apoptosis causes progressive profound depletion of B and CD4+ T lymphocytes in humans. J. Immunol. 2001;166:6952–6963. doi: 10.4049/jimmunol.166.11.6952. [DOI] [PubMed] [Google Scholar]
- 14.Chang K., Svabek C., Vazquez-Guillamet C., Sato B., Rasche D., Wilson S., Robbins P., Ulbrandt N., Suzich J., Green J., Patera A.C., Blair W., Krishnan S., Hotchkiss R. Targeting the programmed cell death 1: programmed cell death ligand 1 pathway reverses T cell exhaustion in patients with sepsis. Crit. Care. 2014;18:R3. doi: 10.1186/cc13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avendaño-Ortiz J., Maroun-Eid C., Martín-Quirós A., Lozano-Rodríguez R., Llanos-González E., Toledano V., Gómez-Campelo P., Montalbán-Hernández K., Carballo-Cardona C., Aguirre L.A., López-Collazo E. Oxygen saturation on admission is a predictive biomarker for PD-L1 expression on circulating monocytes and impaired immune response in patients with sepsis. Front. Immunol. 2018;9:2008. doi: 10.3389/fimmu.2018.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avendaño-Ortiz J., Maroun-Eid C., Martín-Quirós A., Toledano V., Cubillos-Zapata C., Gómez-Campelo P., Varela-Serrano A., Casas-Martin J., Llanos-González E., Alvarez E., García-Río F., Aguirre L.A., Hernández-Jiménez E., López-Collazo E. PD-L1 overexpression during endotoxin tolerance impairs the adaptive immune response in septic patients via HIF1α. J. Infect. Dis. 2018;217:393–404. doi: 10.1093/infdis/jix279. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., Tong S.Y.C., Lewin S.R., Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boomer J.S., To K., Chang K.C., Takasu O., Osborne D.F., Walton A.H., Bricker T.L., Jarman S.D., Kreisel D., Krupnick A.S., Srivastava A., Swanson P.E., Green J.M., Hotchkiss R.S. Immunosuppression in patients who die of sepsis and multiple organ failure. J. Am. Med. Assoc. 2011;306:2594–2605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shindo Y., McDonough J.S., Chang K.C., Ramachandra M., Sasikumar P.G., Hotchkiss R.S. Anti-PD-L1 peptide improves survival in sepsis. J. Surg. Res. 2017;208:33–39. doi: 10.1016/j.jss.2016.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao R., Fang Y., Yu H., Zhao L., Jiang Z., Li C.-S. Monocyte programmed death ligand-1 expression after 3-4 days of sepsis is associated with risk stratification and mortality in septic patients: a prospective cohort study. Crit. Care. 2016;20:124. doi: 10.1186/s13054-016-1301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue S., Suzuki K., Komori Y., Morishita Y., Suzuki-Utsunomiya K., Hozumi K., Inokuchi S., Sato T. Persistent inflammation and T cell exhaustion in severe sepsis in the elderly. Crit. Care. 2014;18:R130. doi: 10.1186/cc13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41:363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Astuti I., Ysrafil, Astuti I., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diab. Metab. Sydr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., Van Der Heden Van Noort G.J., Ovaa H., Müller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.