Abstract
The SARS-Cov2 infection triggers a multisystem inflammatory disorder, knowing as COVID-19, a pandemic disease. This disease is characterized by acute respiratory distress syndrome, cytokine-driven hyperinflammation, and leukocytes count changes. The innate immune response has been linked to COVID-19 immunopathogenesis (e.g., dysfunctional IFN response and myeloid inflammation). In this regard, neutrophils have been highlighted as essential effector cells in the development of COVID-19. This review summarized the significant finds about neutrophils and its effector mechanisms (e.g., neutrophils enzymes and cytokines, neutrophil extracellular traps) in COVID-19 so far.
Keywords: Inflammation, Chemokines, NETs, SARS-CoV2
1. Introduction: COVID-19
COVID-19 (Coronavirus disease 2019) is an infectious inflammatory disease caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) [1], a new type of coronavirus identified in China in December 2019 after several patients were diagnosed with nonspecific pneumonia [2]. The coronavirus outbreak began in Wuhan, the capital of Hubei province, and quickly spread across continental dimensions, turning Covid-19 into a pandemic disease [3].
Coronaviruses are single-stranded RNA viruses that are characterized by having corona-like projections on their surface. There are four main proteins in the structure of these microorganisms, including the spike protein (S), which is related to the host cell mechanism of invasion [4]. The SARS-CoV-2 is the third virus of the β coronavirus group to demonstrate the capacity to infect humans with pandemic potential [5]. SARS-CoV and the MERS-CoV (Middle Eastern respiratory syndrome coronavirus) were responsible for previous relevant outbreaks of respiratory disease in 2003 [6], [7] and 2012 [8], [9], respectively.
Human-to-human transmission occurs through direct contact or respiratory droplets from infected individuals, whether symptomatic or asymptomatic [10], [11], [12]. Several reports have suggested that other forms of transmission, such as the fecal-oral route [13], [14], [15], [16] and intrauterine vertical transmission, may also happen [17], [18]. However, more studies need to be carried out to confirm this form of transmission.
The clinical features of COVID-19 may appear after an incubation period of around 5–14 days [19]. Some early symptoms resemble those of other viral respiratory infections, such as those caused by influenza viruses. However, dyspnea and high fever define the main clinical difference between COVID-19 and common cold [20]. Additionally, when compared to the influenza virus, SARS-CoV-2 infection presents greater chances of progressing to severe and critical infections, which require oxygen therapy and ventilatory support [21]. Elderly patients and those with chronic conditions have higher risks of rapid progression to acute respiratory distress syndrome (ARDS) and multiple organ failure, often resulting in death. These features demonstrate a systemic aspect of this infection, which is accompanied by an intense inflammatory process [22], [23], [24].
2. COVID-19 and inflammation
The COVID-19 infection starts by exposure to microdroplets present in the exhalations of infected individuals. Then, the SARS-CoV-2 spreads to the bronchioles and alveolar spaces [25], entrancing into the host cells (e.g., endothelial, epithelial, and smooth muscle cells) by binding the angiotensin-converting enzyme (ACE)-2, a metallopeptidase present on the cell surface [26], [27], [28], [29].
In the lung, SARS-CoV-2 infects the alveolar cells (type I and II pneumocytes and alveolar macrophages) and then starts intracellular replication in pulmonary tissues. Type I and III interferons (IFN) production is an early defense mechanism in the alveolar cells [25]. However, recent researchers have found deficient expression of these cytokines, besides the upregulated expression of chemokines and interleukins [30], [31]. In normal human bronchial epithelial (NHBE) cells culture, the cytokine profile includes the IFNs deficiency and elevated expression of CCL20, CXC-type chemokines, IL-1β, IL-6, and tumor necrosis factor (TNF) [31]. The type I and III IFN absence shows that, although SARS-CoV-2 is sensitive to IFN antiviral effect, the virus can inhibit its induction [31], [32], [33], [34]. This ability may come from, at least, one mechanism of blocking the activation of the IFN signaling pathway at an early step following the nuclear transport of interferon regulatory factors (IRF) [35]. Furthermore, the recruitment of leukocytes, a hallmark of inflammation, is strongly related to the chemokine profile. For example, CCL2 and CCL8 recruit monocytes/macrophages, CXCL16 is a chemoattractant of NK cells, CXCL8 is the principal neutrophil chemoattractant, and CXCL9 and CXCL10 chemoattract T cells. Thus, the chemokine profile may be a driver of the signature pathology of SARS-CoV-2 [36].
The immune features between moderate and severe disease are modified after ten days of infection when severely ill patients remain with high proinflammatory cytokines [37]. Furthermore, deregulated inflammatory response to an infection may result in the cytokine storm syndrome, which is associated with severe COVID-19 [38], [39]. This syndrome is characterized by high levels of interleukins, TNF-α, G-CSF, MCP-1, and MIP-1α, which are higher in intensive care unit patients than non-intensive care unit patients [37], [40], [41]. Additionally, the inflammasome NLRP3, a multiprotein complex crucial to the host defense, is highly activated in COVID-19 patients. Inflammasome-induced cytokines IL-1β and IL-18 also contribute to cytokine storm, and sustained NLRP3 inflammasome activation is directly associated with the disease's severity [42], [43], [44]. The cytokines milieu recruits immune cells and activate T helper type 1 (Th1) response, which is related to the activation of a specific immune response. Moreover, Th1 cells stimulate IL-6 production by inflammatory monocytes in severe COVID-19 and contribute to the cytokine storm [45]. However, Th2 cytokines are also presented in COVID-19 serum patients and may impair the Th1-inflammatory response [40]. Thereby, chemokines/cytokines milieu comprises a possible therapeutic target for COVID-19 [46].
Peripheral blood immune cells (PBMCs) of COVID-19 patients present low T cell number and frequency in both CD4+ and CD8+ populations, which are more activated. On the order hand, monocytes are increased, but they present a reduction in HLA-DR expression compared with the control group (non-infected) [37]. Additionally, in severe COVID-19, patients present a reduced number of B cells and natural killer (NK) cells associated with severe T cell depletion, and a high neutrophil population [37], [40], [47], [48], [49]. This neutrophilia occurs after seven days symptoms onset [50].
3. Neutrophils in COVID-19
Neutrophils are the most abundant immune cells in human blood. They account for approximately 50–70% of all leukocytes. Besides serving as first responders to many infections, neutrophils have critical homeostatic functions being also implicated in chronic inflammatory diseases [51]. These polymorphonuclear cells play a protective role during bacterial or fungal infections; however, their role in viral infections is not fully understood [52], [53]. Although the evidence is limited, it has been suggested that neutrophils enhance antiviral defenses by interaction with other immune cell populations, virus internalization and killing mechanism, cytokines release, degranulation, oxidative burst, and neutrophil extracellular traps (NETs) [53], [54].
Neutrophils are present in many lung diseases associated with ARDS, as reported in infections by influenza virus and SARS-CoV-1 [55]. A bioinformatic study presented data indicating that neutrophil activation and degranulation are highly activated processes in the SARS infection [56]. Recently, the recruitment of this polymorphonuclear (PMN) was observed in the immune response triggered by SARS-CoV-2. Furthermore, neutrophilia has been described as an indicator of severe respiratory symptoms and a poor outcome in patients with COVID-19 [57], [58], [59].
Several studies have reported that neutrophil-to-lymphocyte ratio (NLR), a clinical inflammation biomarker, is increased and predicts severe illness in the early stage of SARS-CoV-2 infection [59], [60], [61], [62]. Higher D-dimer and C-reactive protein (CRP) levels follow NLR's increase in these patients [63], [64]. Also, increased NLR has been considered an independent risk factor for mortality in hospitalized patients [41], [65], [66], related to some comorbidities (e.g., diabetes and cardiovascular disease) [67]. A study observed that COVID-19 diabetes patients with higher NLR had heavier severity and more extended hospital stay [68]. This fact supports the idea that pre-existing chronic inflammation contributes to COVID-19 severity [65], [69].
In addition to the NLR, neutrophil to CD4+ lymphocyte ratio (NCD4LR) has been associated with the negative conversion time (NCT) of SARS-CoV-2. A study found that high NCD4LR indicates worse immune function and prolonged virus clearance [70]. Another biomarker involving this PMN, the neutrophil count to albumin ratio (NAR), has been described as a new predictor of mortality in COVID-19 patients [71]. Therefore, the NCD4LR and NAR values also could be used as clinical markers for COVID-19 progression in addition to the NLR [41].
Besides, the increase of neutrophils is not reported only in the bloodstream but also in the lungs [72]. PMN infiltration in pulmonary capillaries with extravasation to alveolar space and neutrophilic mucositis was observed in lung autopsies obtained from patients who died from COVID-19, indicating inflammation in the entire lower respiratory tract [73], [74]. Moreover, immature phenotype and/or dysfunctional mature neutrophils have been described in severe COVID-19 patients [75], [76]. These studies indicate that the increased infiltration of immature and/or dysfunctional neutrophil contributes to the imbalance of the lungs' immune response in severe cases.
Respiratory epithelium infection by SARS-CoV-2 leads to cell secretion of multiple cytokines, chemokines, and DAMPs, as previously described [31], [77]. Transcriptional analysis of bronchoalveolar lavage fluid (BALF) from COVID-19 patients reported high levels of CXCL-2 and CXCL-8, chemokines that facilitate the PMN recruitment to the site of infection [78], [79], [80], [81], [82]. Although the neutrophils could present a protective role, extensive and prolonged activation of these leukocytes can lead to detrimental effects in the lungs and result in pneumonia and/or ARDS [83], [84]. Wang and colleagues [50] also demonstrated that neutrophilia coincides with lung injury in severe COVID-19 patients.
It has been described that neutrophils play a pivotal role in the development of ARDS caused by influenza infection [55]. In COVID-19, neutrophils accumulation generates toxic molecules that might contribute to ARDS's physiopathology [85]. Respiratory burst from activated neutrophils induces ROS release, such as superoxide radicals and H2O2, leading to oxidative stress that contributes to the cytokine storm and blood clots formation in SARS-CoV-2 infection [86], [87]. Moreover, decreased expression of the antioxidant enzyme superoxide dismutase 3 (SOD3) in the lung tissue of old patients with COVID-19 was also reported [88]. Therefore, excessive oxidative stress induced by PMN infiltration is related to the alveolar damage, thrombosis, and severity in COVID-19 [87]. In addition to ROS formation, neutrophil elastase has been implicated in COVID-19 pathogenesis [89], [90], [91]. This proteolytic enzyme, which is stored in azurophil granules, is secreted to degrade antigens. Nevertheless, an imbalance of the elastase and other proteinases induces damage in the alveolar-capillary barrier, resulting in tissue injury and edema formation [92].
Furthermore, persistently activated neutrophils contribute to maintaining the inflammatory state in the lungs by cytokine release, as observed in MERS and SARS-CoV-1 infections [93]. Similar findings were described in SARS-CoV-2 infection by Parackova and colleagues [76] that reported the neutrophils as drivers of hyperinflammation by enhanced degranulation of primary granules and pro-inflammatory cytokines release. Taken together, these molecules secreted by PMN can cause severe damage in alveolar tissue, independently of the virus cytopathic effect.
Additionally, Meizlish and colleagues [94] identified neutrophil activators (IL-8 and G-CSF) and effectors (resistin, lipocalin-2, and hepatocyte growth factor) as early biomarkers of severe COVID-19 patients. The authors also demonstrated a positive association between high levels in immature granulocytes and neutrophil counts with increased mortality [94]. These data highlight the neutrophil role in the severity of COVID-19 disease.
Viral infection can also induce the release of neutrophils extracellular traps (NETs) by neutrophils [95]. The NETs mechanism was first described by Brinkmann and colleagues in 2004 [96]. These traps consist of chromatin fibers associated with enzymes such as neutrophil elastase, cathepsin G, and myeloperoxidase [97], [98]. NETs are known to immobilize and degrade bacteria, fungi, viruses, being a critical effector mechanism to contain infections [99]. However, NETs can act as a double-edged sword of immunity [98], having a pro- or anti-inflammatory effect [100], [101]. Schauer and colleagues [102] reported that an aggregate of NETs can degrade cytokines and chemokines, reducing inflammation. This anti-inflammatory effect has also been demonstrated in the ocular microenvironment [103]. On the other hand, NETs can promote tissue damage, having already been shown that NETs and platelets' interaction can cause endothelial damage in infections by Escherichia coli [104]. NETs can also participate in the pathogenesis of autoimmune diseases, such as systemic lupus erythematosus and rheumatoid arthritis, where elevated levels of NETs have been seen in serum and synovial fluid, respectively, in patients with these diseases [105], [106].
Studies have been reported an elevated level of NETs in patients with COVID-19 [107], [108], [109], and an increased plasma NETs is correlated with increased COVID-19 severity [109], besides contributing to lung injury and microvascular thrombosis [107]. The vascular occlusion caused by NETs is not only reported in lung tissue [110] but also in kidney and liver [111], which suggests that NETs thrombotic effects may be related to systemic and harmful effects of COVID-19. This relationship between NETs and thrombosis may also be related to complement system activation. Indeed, C3 [112] and C5 [113] inhibition dampen NET release in COVID-19 patients. Since coagulation disorders are a worse prognosis to COVID-19 [114], [115], [116], and both NETs and complement proteins are associated with these thrombotic events [113], therapies that focus on this triple complement-NETs-coagulation axis may be a therapeutic opportunity.
At the transcriptional level, Wang and collaborators [50] demonstrated activation of several NETs-associated in COVID-19 patients. They hypothesized that some of them could be related to negative regulation of NK and T cell, dampening antiviral response [50]. In severe COVID-19, Veras and colleagues [109] demonstrated that neutrophils, both circulating and lung-infiltrating, release high levels of NETs. The authors also present data that demonstrate a NETs release directly induced by SARS-CoV-2 [117]. This SARS-CoV-2-induced NETs release is PAD-4-dependent [109]. PAD4 is critical to NET formation because it promotes a process of hypercitrulination of histones, resulting in chromatin decondensation [118]. The SARS-CoV-2-activated neutrophils can also induce apoptosis in lung epithelial (A549 cells), reinforcing neutrophil role in COVID-19 immunopathology and other coronavirus infections [109].
4. Conclusions
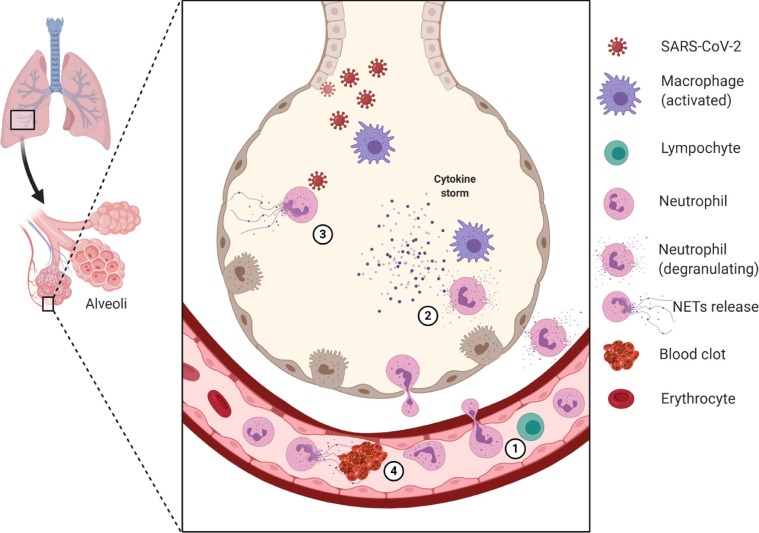
The literature related to neutrophil and COVID-19 so far demonstrated a crucial role of these polymorphonuclear cells in the pathogenesis of COVID-19 (Fig. 1 ). Despite the immune system modulation needs being tightly controlled to avoid immunosuppression, the different neutrophil mechanisms (e.g., neutrophils enzymes and cytokines, NETs) are potential targets to treat COVID-19, mainly the severe cases.
Fig 1.
The neutrophil role in the lung tissue during infection by SARS-CoV-2. (1) The neutrophil-to-lymphocyte ratio (NLR) is elevated in the bloodstream. (2) The migrated neutrophils contribute to storm cytokines formation and release other mediators (e.g., elastase neutrophilic). (3) SARS-CoV-2 infection promotes neutrophil extracellular traps release, which can contribute to lung damage and (4) immunothrombosis. These many steps may be potential therapeutic targets. Several other cells and mediators are involved in COVID-19 immunopathology, but they are suppressed in this figure to highlight the neutrophil role. The figure was created with BioRender.com [119].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to CAPES (PROCAD) and CNPq (“Conselho Nacional de Desenvolvimento Científico e Tecnológico”) to fellowships support (154801/2019-4 and 141304/2017-0).
References
- 1.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop. Med. Int. Heal. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morens D.M., Breman J.G., Calisher C.H., Doherty P.C., Hahn B.H., Keusch G.T., Kramer L.D., LeDuc J.W., Monath T.P., Taubenberger J.K. The origin of COVID-19 and why it matters. Am. J. Trop. Med. Hyg. 2020;103:955–959. doi: 10.4269/ajtmh.20-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong N.S., Zheng B.J., Li Y.M., Poon L.L.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris J.S.M., Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February. Lancet. 2003;362(2003):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 8.Reina J., Reina N. The middle east respiratory syndrome coronavirus. Med. Clínica (English Ed. 2015;145:529–531. doi: 10.1016/j.medcle.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X., Ma H., Chen W., Lin Y., Zheng Y., Wang J., Hu Z., Yi Y., Shen H. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vella P., Senia M., Ceccarelli E., Vitale H., Maltezou R., Taibi A., Lleshi E.V., Rullo G.F., Pellicano V.R., Nunnari C.L. Transmission mode associated with coronavirus disease, A review. Eur. Rev. Med. Pharmacol. Sci. 2019;24(2020):7889–7904. doi: 10.26355/eurrev_202007_22296. [DOI] [PubMed] [Google Scholar]
- 12.Yang X., Chen F. Letter to the editor: “asymptomatic carrier transmission of coronavirus disease 2019 (COVID-19) and multipoint aerosol sampling to assess risks in the operating room during a pandemic”. World Neurosurg. 2020 doi: 10.1016/j.wneu.2020.07.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal A., Chen A., Ravindran N., To C., Thuluvath P.J. Gastrointestinal and liver manifestations of COVID-19. J. Clin. Exp. Hepatol. 2020;10:263–265. doi: 10.1016/j.jceh.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohan N., Deswal S. Corona Virus Disease (COVID-19) fecal-oral transmission: Is it a potential risk for Indians?, Indian. J. Gastroenterol. 2020:1–2. doi: 10.1007/s12664-020-01072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuicchi D., Lazzarotto T., Poggioli G. Fecal-oral transmission of SARS-CoV-2: review of laboratory-confirmed virus in gastrointestinal system. Int. J. Colorectal Dis. 2020 doi: 10.1007/s00384-020-03785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vigil-De Gracia P., Luo C., Epifanio Malpassi R. Perinatal transmission with SARS-CoV-2 and route of pregnancy termination: a narrative review. J. Matern. Neonatal Med. 2020 doi: 10.1080/14767058.2020.1788533. [DOI] [PubMed] [Google Scholar]
- 18.Bwire G.M., Njiro B.J., Mwakawanga D.L., Sabas D., Sunguya B.F. Possible vertical transmission and antibodies against SARS-CoV-2 among infants born to mothers with COVID-19: A living systematic review. J. Med. Virol. 2020:jmv.26622. doi: 10.1002/jmv.26622. [DOI] [PubMed] [Google Scholar]
- 19.Wang W., Tang J., Wei F. Updated understanding of the outbreak of novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2019;92(2020):441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He D., Gao D., Li Y., Zhuang Z., Cao P., Lou Y., Yang L. An updated comparison of COVID-19 and influenza. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3573503. [DOI] [Google Scholar]
- 21.Yi Y., Lagniton P.N.P., Ye S., Li E., Xu R.H. COVID-19: What has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical Characteristics of 138 Hospitalized Patients with, Novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 2019;323(2020):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J.T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P.M., Cowling B.J., Lipsitch M., Leung G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020;26:506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct. Target. Ther. 2020;5:1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García L.F. Immune response, inflammation, and the clinical spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu H., Chan J.F.W., Wang Y., Yuen T.T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X., Zhang X., Cai J.P., Zhou J., Yuan S., Kok K.H., To K.K.W., Chan I.H.Y., Zhang A.J., Sit K.Y., Au W.K., Yuen K.Y. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: Implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi C.-S., Qi H.-Y., Boularan C., Huang N.-N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., Weber F. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/jvi.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot A.E.I. Chemokine receptors: Multifaceted therapeutic targets. Nat. Rev. Immunol. 2002;2:106–115. doi: 10.1038/nri722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Obaid A., Lu-Culligan A., Nelson A., Brito A., Nunez A., Martin A., Watkins A., Geng B., Kalinich C., Harden C., Todeasa C., Jensen C., Kim D., McDonald D., Shepard D., Courchaine E., White E.B., Song E., Silva E., Kudo E., DeIuliis G., Rahming H., Park H.J., Matos I., Nouws J., Valdez J., Fauver J., Lim J., Rose K.A., Anastasio K., Brower K., Glick L., Sharma L., Sewanan L., Knaggs L., Minasyan M., Batsu M., Petrone M., Kuang M., Nakahata M., Campbell M., Linehan M., Askenase M.H., Simonov M., Smolgovsky M., Sonnert N., Naushad N., Vijayakumar P., Martinello R., Datta R., Handoko R., Bermejo S., Prophet S., Bickerton S., Velazquez S., Alpert T., Rice T., Khoury-Hanold W., Peng X., Yang Y., Cao Y., Strong Y., Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., Li G., Li Y., Xing L., Peng L., Wei J., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Liu L., Liu Y. Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. MedRxiv. 2020;2020(03) doi: 10.1101/2020.03.02.20029975. [DOI] [Google Scholar]
- 39.Vaninov N. In the eye of the COVID-19 cytokine storm. Nat. Rev. Immunol. 2020;20:277. doi: 10.1038/s41577-020-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with, novel coronavirus in Wuhan. China, Lancet. 2019;395(2020):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Du X., Chen J., Jin Y., Peng L., Wang H.H.X., Luo M., Chen L., Zhao Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeman T.L., Swartz T.H. Targeting the NLRP3 inflammasome in severe COVID-19. Front. Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigues T.S., de Sá K.S., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Gonçalves A.V., Perucello D.B., Andrade W.A., Castro R., Veras F.P., Toller- J.E., Nascimento D.C., de Lima M.H., Silva C.M.S., Caetite D.B., Martins R.B., Castro I.A., Pontelli M.C., de Barros C., do Amaral N.B., Giannini M.C., Bonjorno L.P., Isabel Lopes M.F., Benatti M.N., Santana R.C., Vilar F.C., Auxiliadora-Martins M., Luppino-Assad R., de Almeida S.C., de Oliveira F.R., Batah S.S., Siyuan L., Benatti M.N., Alves-Filho J.C., Cunha F.Q., Cunha L.D., Kohlsdorf T., Fabro A.T., Arruda E., De R.D., Louzada-Junior P., Zamboni D.S. Inflammasome activation in COVID-19 patients. MedRxiv. 2020;2020(08) doi: 10.1101/2020.08.05.20168872. [DOI] [Google Scholar]
- 44.van den Berg D.F., te Velde A.A. Severe COVID-19: NLRP3 inflammasome dysregulated. Front. Immunol. 2020;11:1580. doi: 10.3389/fimmu.2020.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodarzi P., Mahdavi F., Mirzaei R., Hasanvand H., Sholeh M., Zamani F., Sohrabi M., Tabibzadeh A., Jeda A.S., Niya M.H.K., Keyvani H., Karampoor S. Coronavirus disease 2019 (COVID-19): Immunological approaches and emerging pharmacologic treatments. Int. Immunopharmacol. 2020;88 doi: 10.1016/j.intimp.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus, (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2019;71(2020):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., Shi Y. Immunopathological characteristics of coronavirus disease, cases in Guangzhou China. Immunology. 2019;160(2020):261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J., Li Q., Yin Y., Zhang Y., Cao Y., Lin X., Huang L., Hoffmann D., Lu M., Qiu Y. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soehnlein O., Steffens S., Hidalgo A., Weber C. Neutrophils as protagonists and targets in chronic inflammation. Nat. Rev. Immunol. 2017;17:248–261. doi: 10.1038/nri.2017.10. [DOI] [PubMed] [Google Scholar]
- 52.Tomar B., Anders H.-J., Desai J., Mulay S.R. Neutrophils and neutrophil extracellular traps drive necroinflammation in COVID-19. Cells. 2020;9:1383. doi: 10.3390/cells9061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galani I.E., Andreakos E. Neutrophils in viral infections: Current concepts and caveats. J. Leukoc. Biol. 2015;98:557–564. doi: 10.1189/jlb.4vmr1114-555r. [DOI] [PubMed] [Google Scholar]
- 54.Naumenko V., Turk M., Jenne C.N., Kim S.J. Neutrophils in viral infection. Cell Tissue Res. 2018;371:505–516. doi: 10.1007/s00441-017-2763-0. [DOI] [PubMed] [Google Scholar]
- 55.Camp J.V., Jonsson C.B. A role for neutrophils in viral respiratory disease. Front. Immunol. 2017;8:1. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemmat N., Derakhshani A., Bannazadeh Baghi H., Silvestris N., Baradaran B., De Summa S. Neutrophils, crucial, or harmful immune cells involved in coronavirus infection: A bioinformatics study. Front. Genet. 2020;11:641. doi: 10.3389/fgene.2020.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh K., Mittal S., Gollapudi S., Butzmann A., Kumar J., Ohgami R.S. A meta-analysis of SARS-CoV-2 patients identifies the combinatorial significance of D-dimer, C-reactive protein, lymphocyte, and neutrophil values as a predictor of disease severity. Int. J. Lab. Hematol. 2020 doi: 10.1111/ijlh.13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guan J., Wei X., Qin S., Liu X., Jiang Y., Chen Y., Chen Y., Lu H., Qian J., Wang Z., Lin X. Continuous tracking of COVID-19 patients’ immune status. Int. Immunopharmacol. 2020;89 doi: 10.1016/j.intimp.2020.107034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., Song M., Wang L., Zhang W., Han B., Yang L., Wang X., Zhou G., Zhang T., Li B., Wang Y., Chen Z., Wang X. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu J., Kong J., Wang W., Wu M., Yao L., Wang Z., Jin J., Wu D., Yu X. The clinical implication of dynamic neutrophil to lymphocyte ratio and D-dimer in COVID-19: A retrospective study in Suzhou China. Thromb. Res. 2020;192:3–8. doi: 10.1016/j.thromres.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb B.J., Peltan I.D., Jensen P., Hoda D., Hunter B., Silver A., Starr N., Buckel W., Grisel N., Hummel E., Snow G., Morris D., Stenehjem E., Srivastava R., Brown S.M. Clinical criteria for COVID-19-associated hyperinflammatory syndrome: a cohort study. Lancet Rheumatol. 2020 doi: 10.1016/s2665-9913(20)30343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye W., Chen G., Li X., Lan X., Ji C., Hou M., Zhang D., Zeng G., Wang Y., Xu C., Lu W., Cui R., Cai Y., Huang H., Yang L. Dynamic changes of D-dimer and neutrophil-lymphocyte count ratio as prognostic biomarkers in COVID-19. Respir. Res. 2020;21:169. doi: 10.1186/s12931-020-01428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tatum D., Taghavi S., Houghton A., Stover J., Toraih E., Duchesne J. Neutrophil-to-Lymphocyte ratio and outcomes in Louisiana COVID-19 Patients. Shock. 2020;54:652–658. doi: 10.1097/shk.0000000000001585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang A.P., ping Liu J., qiang Tao W., ming Li H. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020;84 doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azab B., Camacho-Rivera M., Taioli E. Average values and racial differences of neutrophil lymphocyte ratio among a nationally representative sample of United States subjects. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu G., Zhang S., Hu H., Liu T.T., Huang J. The role of neutrophil-lymphocyte ratio and lymphocyte–monocyte ratio in the prognosis of type 2 diabetics with COVID-19. Scott. Med. J. 2020;65:154–160. doi: 10.1177/0036933020953516. [DOI] [PubMed] [Google Scholar]
- 69.Yang P., Wang N., Wang J., Luo A., Gao F., Tu Y. Admission fasting plasma glucose is an independent risk factor for 28-day mortality in patients with COVID-19. J. Med. Virol. 2020:jmv.26608. doi: 10.1002/jmv.26608. [DOI] [PubMed] [Google Scholar]
- 70.Wang H., Zhang Y., Mo P., Liu J., Wang H., Wang F., Zhao Q. Neutrophil to CD4+ lymphocyte ratio as a potential biomarker in predicting virus negative conversion time in COVID-19. Int. Immunopharmacol. 2020;85 doi: 10.1016/j.intimp.2020.106683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.C. Varim, S. Yaylaci, T. Demirci, T. Kaya, A. Nalbant, H. Dheir, D. Senocak, R. Kurt, H. Cengiz, C. Karacaer, Neutrophil count to albumin ratio as a new predictor of mortality in patients with COVID-19 ınfection, Rev. Assoc. Med. Bras. 66Suppl 2 (2020) 77–81. https://doi.org/10.1590/1806-9282.66.S2.77. [DOI] [PubMed]
- 72.Buja L.M., Wolf D., Zhao B., Akkanti B., McDonald M., Lelenwa L., Reilly N., Ottaviani G., Elghetany M.T., Trujillo D.O., Aisenberg G.M., Madjid M., Kar B. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): Report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc. Pathol. 2020;48 doi: 10.1016/j.carpath.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S., Loda M., Looney M.R., McAllister F., Rayes R., Renaud S., Rousseau S., Salvatore S., Schwartz R.E., Spicer J.D., Yost C.C., Weber A., Zuo Y., Egeblad M. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.X.H. Yao, T.Y. Li, Z.C. He, Y.F. Ping, H.W. Liu, S.C. Yu, H.M. Mou, L.H. Wang, H.R. Zhang, W.J. Fu, T. Luo, F. Liu, C. Chen, H.L. Xiao, H.T. Guo, S. Lin, D.F. Xiang, Y. Shi, Q.R. Li, X. Huang, Y. Cui, X.Z. Li, W. Tang, P.F. Pan, X.Q. Huang, Y.Q. Ding, X.W. Bian, [A pathological report of three COVID-19 cases by minimally invasive autopsies]., Zhonghua Bing Li Xue Za Zhi = Chinese J. Pathol. 49 (2020) E009. https://doi.org/10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed]
- 75.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., De Domenico E., Wendisch D., Grasshoff M., Kapellos T.S., Beckstette M., Pecht T., Saglam A., Dietrich O., Mei H.E., Schulz A.R., Conrad C., Kunkel D., Vafadarnejad E., Xu C.J., Horne A., Herbert M., Drews A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., Müller-Redetzky H., Heim K.M., Machleidt F., Uhrig A., Bosquillon de Jarcy L., Jürgens L., Stegemann M., Glösenkamp C.R., Volk H.D., Goffinet C., Landthaler M., Wyler E., Georg P., Schneider M., Dang-Heine C., Neuwinger N., Kappert K., Tauber R., Corman V., Raabe J., Kaiser K.M., Vinh M.T., Rieke G., Meisel C., Ulas T., Becker M., Geffers R., Witzenrath M., Drosten C., Suttorp N., von Kalle C., Kurth F., Händler K., Schultze J.L., Aschenbrenner A.C., Li Y., Nattermann J., Sawitzki B., Saliba A.E., Sander L.E., Angelov A., Bals R., Bartholomäus A., Becker A., Bezdan D., Bonifacio E., Bork P., Clavel T., Colome-Tatche M., Diefenbach A., Dilthey A., Fischer N., Förstner K., Frick J.S., Gagneur J., Goesmann A., Hain T., Hummel M., Janssen S., Kalinowski J., Kallies R., Kehr B., Keller A., Kim-Hellmuth S., Klein C., Kohlbacher O., Korbel J.O., Kurth I., Ludwig K., Makarewicz O., Marz M., McHardy A., Mertes C., Nöthen M., Nürnberg P., Ohler U., Ossowski S., Overmann J., Peter S., Pfeffer K., Poetsch A.R., Pühler A., Rajewsky N., Ralser M., Rieß O., Ripke S., Nunes da Rocha U., Rosenstiel P., Schiffer P., Schulte E.C., Sczyrba A., Stegle O., Stoye J., Theis F., Vehreschild J., Vogel J., von Kleist M., Walker A., Walter J., Wieczorek D., Ziebuhr J. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parackova Z., Zentsova I., Bloomfield M., Vrabcova P., Smetanova J., Klocperk A., Mesežnikov G., Casas Mendez L.F., Vymazal T., Sediva A. Disharmonic inflammatory signatures in COVID-19 Augmented neutrophils’ but impaired monocytes’ and dendritic cells’ responsiveness. Cells. 2020;9:2206. doi: 10.3390/cells9102206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cicco S., Cicco G., Racanelli V., Vacca A. Neutrophil Extracellular Traps (NETs) and Damage-Associated Molecular Patterns (DAMPs): Two potential targets for COVID-19 treatment. Mediators Inflamm. 2020;2020 doi: 10.1155/2020/7527953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Didangelos A. COVID-19 hyperinflammation: What about neutrophils? MSphere. 2020;5 doi: 10.1128/msphere.00367-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ronit A., Berg R.M.G., Bay J.T., Haugaard A.K., Ahlström M.G., Burgdorf K.S., Ullum H., Rørvig S.B., Tjelle K., Foss N.B., Benfield T., Marquart H.V., Plovsing R.R. Compartmental immunophenotyping in COVID-19 ARDS: a case series. J. Allergy Clin. Immunol. 2020 doi: 10.1016/j.jaci.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X., Liu Y., Li J., Sun L., Yang J., Xu F., Zhou J., Wan L., Xu X., Le A., Zhang W. Immune characteristics distinguish patients with severe disease associated with SARS-CoV-2. Immunol. Res. 2020 doi: 10.1007/s12026-020-09156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abraham E. Neutrophils and acute lung injury. Crit. Care Med. 2003;31 doi: 10.1097/01.ccm.0000057843.47705.e8. [DOI] [PubMed] [Google Scholar]
- 84.Miyazawa M. Immunopathogenesis of SARS-CoV-2-induced pneumonia: lessons from influenza virus infection. Inflamm. Regen. 2020;40:39. doi: 10.1186/s41232-020-00148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang S.-C., Tsai Y.-F., Pan Y.-L., Hwang T.-L. Understanding the role of neutrophils in acute respiratory distress syndrome. Biomed. J. 2020 doi: 10.1016/j.bj.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cecchini R., Cecchini A.L. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laforge M., Elbim C., Frère C., Hémadi M., Massaad C., Nuss P., Benoliel J.J., Becker C. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020;20:515–516. doi: 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abouhashem A.S., Singh K., Azzazy H.M.E., Sen C.K. Is low alveolar type II Cell SOD3 in the lungs of elderly linked to the observed severity of COVID-19? Antioxidants Redox Signal. 2020;33:59–65. doi: 10.1089/ars.2020.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mohamed M.M.A., El-Shimy I.A., El-Shimy I.A., Hadi M.A. Neutrophil elastase inhibitors: A potential prophylactic treatment option for SARS-CoV-2-induced respiratory complications? Crit. Care. 2020;24:311. doi: 10.1186/s13054-020-03023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thierry A.R. Anti-protease treatments targeting plasmin(Ogen) and neutrophil elastase may be beneficial in fighting covid-19. Physiol. Rev. 2020;100:1597–1598. doi: 10.1152/physrev.00019.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sahebnasagh A., Saghafi F., Safdari M., Khataminia M., Sadremomtaz A., Talaei Z., Rezai Ghaleno H., Bagheri M., Habtemariam S., Avan R. Neutrophil elastase inhibitor (sivelestat) may be a promising therapeutic option for management of acute lung injury/acute respiratory distress syndrome or disseminated intravascular coagulation in COVID-19. J. Clin. Pharm. Ther. 2020 doi: 10.1111/jcpt.13251. [DOI] [PubMed] [Google Scholar]
- 92.Aikawa N., Kawasaki Y. Clinical utility of the neutrophil elastase inhibitor sivelestat for the treatment of acute respiratory distress syndrome. Ther. Clin. Risk Manag. 2014;10:621–629. doi: 10.2147/TCRM.S65066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’’ in COVID-19’. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meizlish M.L., Pine A.B., Bishai J.D., Goshua G., Nadelmann E.R., Simonov M., Chang C.-H., Zhang H., Shallow M., Bahel P., Owusu K., Yamamoto Y., Arora T., Atri D.S., Patel A., Gbyli R., Kwan J., Won C.H., Dela Cruz C., Price C., Koff J., King B.A., Rinder H.M., Perry Wilson F., Hwa J., Halene S., Damsky W., van Dijk D., Lee A.I., Chun H.J. A neutrophil activation signature predicts critical illness and mortality in COVID-19. MedRxiv. 2020;2020(09) doi: 10.1101/2020.09.01.20183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schönrich G., Raftery M.J. Neutrophil extracellular traps go viral. Front. Immunol. 2016;7:366. doi: 10.3389/fimmu.2016.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.V. Brinkmann, U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D.S. Weiss, Y. Weinrauch, A. Zychlinsky, Neutrophil Extracellular Traps Kill Bacteria, Science (80-.). 303 (2004) 1532–1535. https://doi.org/10.1126/science.1092385. [DOI] [PubMed]
- 97.Delgado-Rizo V., Martínez-Guzmán M.A., Iñiguez-Gutierrez L., García-Orozco A., Alvarado-Navarro A., Fafutis-Morris M. Neutrophil extracellular traps and its implications in inflammation: An overview. Front. Immunol. 2017;8:81. doi: 10.3389/fimmu.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaplan M.J., Radic M. Neutrophil extracellular traps: double-edged swords of innate immunity. J. Immunol. 2012;189:2689–2695. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 2018;18:134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 100.Hahn J., Knopf J., Maueröder C., Kienhöfer D., Leppkes M., Herrmann M. Neutrophils and neutrophil extracellular traps orchestrate initiation and resolution of inflammation. Clin. Exp. Rheumatol. 2016;34:6–8. [PubMed] [Google Scholar]
- 101.Muñoz L.E., Bilyy R., Biermann M.H.C., Kienhöfer D., Maueröder C., Hahn J., Brauner J.M., Weidner D., Chen J., Scharin-Mehlmann M., Janko C., Friedrich R.P., Mielenz D., Dumych T., Lootsik M.D., Schauer C., Schett G., Hoffmann M., Zhao Y., Herrmann M. Nanoparticles size-dependently initiate self-limiting NETosis-driven inflammation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5856–E5865. doi: 10.1073/pnas.1602230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schauer C., Janko C., Munoz L.E., Zhao Y., Kienhöfer D., Frey B., Lell M., Manger B., Rech J., Naschberger E., Holmdahl R., Krenn V., Harrer T., Jeremic I., Bilyy R., Schett G., Hoffmann M., Herrmann M. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat. Med. 2014;20:511–517. doi: 10.1038/nm.3547. [DOI] [PubMed] [Google Scholar]
- 103.Mahajan A., Grüneboom A., Petru L., Podolska M.J., Kling L., Maueröder C., Dahms F., Christiansen S., Günter L., Krenn V., Jünemann A., Bock F., Schauer C., Schett G., Hohberger B., Herrmann M., Muñoz L.E. Frontline science: Aggregated neutrophil extracellular traps prevent inflammation on the neutrophil-rich ocular surface. J. Leukoc. Biol. 2019;105:1087–1098. doi: 10.1002/JLB.HI0718-249RR. [DOI] [PubMed] [Google Scholar]
- 104.Clark S.R., Ma A.C., Tavener S.A., McDonald B., Goodarzi Z., Kelly M.M., Patel K.D., Chakrabarti S., McAvoy E., Sinclair G.D., Keys E.M., Allen-Vercoe E., DeVinney R., Doig C.J., Green F.H.Y., Kubes P. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 105.Gupta S., Kaplan M.J. The role of neutrophils and NETosis in autoimmune and renal diseases. Nat. Rev. Nephrol. 2016;12:402–413. doi: 10.1038/nrneph.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee K.H., Kronbichler A., Park D.D.Y., Park Y.M., Moon H., Kim H., Choi J.H., Choi Y.S., Shim S., Lyu I.S., Yun B.H., Han Y., Lee D., Lee S.Y., Yoo B.H., Lee K.H., Kim T.L., Kim H., Shim J.S., Nam W., So H., Choi S.Y., Lee S., Il Shin J. Neutrophil extracellular traps (NETs) in autoimmune diseases: A comprehensive review. Autoimmun. Rev. 2017;16:1160–1173. doi: 10.1016/j.autrev.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 107.Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M., Cody M.J., Manne B.K., Portier I., Harris E., Petrey A.C., Beswick E.J., Caulin A.F., Iovino A., Abegglen L.M., Weyrich A.S., Rondina M.T., Egeblad M., Schiffman J.D., Yost C.C. Neutrophil Extracellular Traps (NETs) contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., Woods R.J., Kanthi Y., Knight J.S. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Veras F.P., Pontelli M.C., Silva C.M., Toller-Kawahisa J.E., de Lima M., Nascimento D.C., Schneider A.H., Caetité D., Tavares L.A., Paiva I.M., Rosales R., Colón D., Martins R., Castro I.A., Almeida G.M., Lopes M.I.F., Benatti M.N., Bonjorno L.P., Giannini M.C., Luppino-Assad R., Almeida S.L., Vilar F., Santana R., Bollela V.R., Auxiliadora-Martins M., Borges M., Miranda C.H., Pazin-Filho A., da Silva L.L.P., Cunha L., Zamboni D.S., Dal-Pizzol F., Leiria L.O., Siyuan L., Batah S., Fabro A., Mauad T., Dolhnikoff M., Duarte-Neto A., Saldiva P., Cunha T.M., Alves-Filho J.C., Arruda E., Louzada-Junior P., Oliveira R.D., Cunha F.Q. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Radermecker C., Detrembleur N., Guiot J., Cavalier E., Henket M., d’Emal C., Vanwinge C., Cataldo D., Oury C., Delvenne P., Marichal T. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., Stürzl M., Staats L., Mahajan A., Schauer C., Kremer A.N., Völkl S., Amann K., Evert K., Falkeis C., Wehrfritz A., Rieker R.J., Hartmann A., Kremer A.E., Neurath M.F., Muñoz L.E., Schett G., Herrmann M. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mastellos D.C., Pires da Silva B.G.P., Fonseca B.A.L., Fonseca N.P., Auxiliadora-Martins M., Mastaglio S., Ruggeri A., Sironi M., Radermacher P., Chrysanthopoulou A., Skendros P., Ritis K., Manfra I., Iacobelli S., Huber-Lang M., Nilsson B., Yancopoulou D., Connolly E.S., Garlanda C., Ciceri F., Risitano A.M., Calado R.T., Lambris J.D. Complement C3 vs C5 inhibition in severe COVID-19: Early clinical findings reveal differential biological efficacy. Clin. Immunol. 2020;220 doi: 10.1016/j.clim.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C., Tektonidou M.G., Konstantinidis T., Papagoras C., Mitroulis I., Germanidis G., Lambris J.D., Ritis K. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020 doi: 10.1172/jci141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Macciò A., Madeddu C., Caocci G., La Nasa G. Multifactorial pathogenesis of COVID-19-related coagulopathy: Can defibrotide have a role in the early phases of coagulation disorders? J. Thromb. Haemost. 2020;18:3106–3108. doi: 10.1111/jth.15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Price L.C., McCabe C., Garfield B., Wort S.J. Thrombosis and COVID-19 pneumonia: The clot thickens! Eur. Respir. J. 2020;56 doi: 10.1183/13993003.01608-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pujhari S., Paul S., Ahluwalia J., Rasgon J.L. Clotting disorder in severe acute respiratory syndrome coronavirus 2. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Veras F.P., Pontelli M., Silva C., Toller-Kawahisa J., de Lima M., Nascimento D., Schneider A., Caetite D., Rosales R., Colon D., Martins R., Castro I., Almeida G., Lopes M.I., Benatti M., Bonjorno L., Giannini M., Luppino-Assad R., Almeida S., Vilar F., Santana R., Bollela V., Martins M., Miranda C., Borges M., Pazin-Filho A., Cunha L., Zamboni D., Dal-Pizzol F., Leiria L., Siyuan L., Batah S., Fabro A., Mauad T., Dolhnikoff M., Duarte-Neto A., Saldiva P., Cunha T., Alves-Filho J.C., Arruda E., Louzada-Junior P., Oliveira R., Cunha F. SARS-CoV-2 triggered neutrophil extracellular traps (NETs) mediate COVID-19 pathology. MedRxiv. 2020;2020(06) doi: 10.1101/2020.06.08.20125823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jorch S.K., Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 2017;23:279–287. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- 119.Adapted from “Cytokine Storm”, by BioRender.com (2020). Retrieved from https://app.biorender.com/biorender-templates.