Abstract
Background
Small studies have correlated hypertension with pneumonia risk; whether this is recapitulated in larger prospective studies, and represents a causal association, is unclear.
Methods
We estimated the risk for prevalent hypertension with incident respiratory diseases over mean follow-up of 8 years among 377,143 British participants in the UK Biobank. Mendelian randomization of blood pressure on pneumonia was implemented using 75 independent, genome-wide significant variants associated with systolic and diastolic blood pressures among 299,024 individuals not in the UK Biobank. Secondary analyses with pulmonary function tests were performed.
Findings
In total, 107,310 participants (30%) had hypertension at UK Biobank enrollment, and 9,969 (3%) developed pneumonia during follow-up. Prevalent hypertension was independently associated with increased risk for incident pneumonia (HR: 1.36; 95% CI: 1.29–1.43; p < 0.001), as well as other incident respiratory diseases. Genetic predisposition to a 5 mm Hg increase in blood pressure was associated with increased risk for incident pneumonia for systolic blood pressure (HR: 1.08; 95% CI: 1.04–1.13; p < 0.001) and diastolic blood pressure (HR: 1.11; 95% CI: 1.03–1.20; p = 0.005). Additionally, consistent with epidemiologic associations, increased blood pressure genetic risk was significantly associated with reduced performance on pulmonary function tests (p < 0.001).
Conclusions
These results suggest that elevated blood pressure increases risk for pneumonia. Maintaining adequate blood pressure control, in addition to other measures, may reduce risk for pneumonia.
Funding
S.M.Z. (1F30HL149180-01), M.H. (T32HL094301-07), and P.N. (R01HL1427, R01HL148565, and R01HL148050) are supported by the National Institutes of Health. J.P. is supported by the John S. LaDue Memorial Fellowship.
Keywords: hypertension, high blood pressure, pulmonary, pneumonia, population genetics, Mendelian randomization, epidemiology
Graphical Abstract
Context and Significance
Epidemiologic analyses have correlated hypertension with pneumonia risk. Whether this represents a direct consequence of hypertension or the influence of co-morbid risk factors such as age, diabetes mellitus, or smoking is unclear. Here, across 377,143 individuals from the UK Biobank, we show that hypertension is independently associated with significantly increased risk for incident pneumonia. Additionally, using Mendelian randomization, we show that a genetic predisposition to elevated blood pressure across 75 independent genetic variants is associated with increased risk for incident pneumonia and reduced pulmonary function test performance. These results suggest that elevated blood pressure may be a causal risk factor for pneumonia. Maintaining adequate blood pressure control, in addition to other measures, may reduce risk for pneumonia.
Through epidemiological and genetic association analyses in the UK Biobank (N = 377,143), Zekavat et al. link high blood pressure with increased risk for incident pneumonia and reduced performance on pulmonary function tests. These results suggest that maintaining adequate blood pressure control may reduce risk for pneumonia and improve pulmonary function.
Introduction
Hypertension is a highly prevalent, modifiable risk factor for cardiovascular disease and mortality.1 Epidemiologic analyses have correlated hypertension with pneumonia risk and more recently with pneumonia from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.2 , 3 Whether this represents a direct consequence of hypertension or influence of co-morbid risk factors such as age, diabetes mellitus, air pollution, or smoking is unclear.
Prior studies have linked hypertension with decreased performance on pulmonary function tests, which may potentially suggest a mechanism toward heightened pneumonia risk.4 , 5 Chronic obstructive pulmonary disease (COPD), diagnosed by abnormal pulmonary function test results, is a well-established risk factor for pneumonia and is co-morbid with several cardiovascular diseases and risk factors, including hypertension.6 Together, these studies and others suggest several mechanisms that may link hypertension and pulmonary obstruction: (1) both may involve physiological degradation of arterial and airway elasticity,5 (2) endothelial and vascular dysfunction may also influence pulmonary vascular endothelial cells and lead to pulmonary vascular dysfunction resulting in lung tissue destruction and airway obstruction,7 and (3) systemic inflammation associated with hypertension may additionally alter pulmonary function.8
Although epidemiologic analyses support a plausible causal relationship between blood pressure regulation and respiratory infection risk, confounding from co-morbid conditions, as indicated by the aforementioned correlations, limits such inference. Mendelian randomization is a statistical approach using genetic instruments for an exposure as opposed to the exposure itself to mitigate risks for confounding, facilitating more robust causal inference.9 Blood pressure is a highly heritable trait with several known associated genomic loci that may serve as a robust aggregated genetic proxy for Mendelian randomization.10
Here, in the UK Biobank, we (1) estimate the epidemiologic association of hypertension with incident pneumonia risk and indices of pulmonary function and (2) apply Mendelian randomization to test the hypothesis that blood pressure independently causally influences risks for pneumonia and reduced pulmonary function.
Results
Baseline Characteristics
A total of 377,143 genotyped individuals in the UK Biobank passed quality control criteria. Among these individuals, the median age was 58 years (interquartile range [IQR] 51–63 years), 202,369 (53.7%) were women, 18,943 (5.0%) had diabetes mellitus, 107,310 (29.7%) had hypertension, and 20,825 (5.7%) had coronary artery disease; 170,713 individuals (45.4%) were prior or current smokers, and 95,737 (24.5%) were prescribed antihypertensive medications. Significant differences between low (<20th percentile), intermediate (20th to 80th percentiles), and high (>80th percentile) systolic blood pressure (SBP) and diastolic blood pressure (DBP) polygenic risk scores (PRSs) were consistently observed across multiple phenotypes, including body mass index (BMI) (with high PRS being associated with lower BMI), as well as antihypertensive medications, hypertension, coronary artery disease, and hypercholesterolemia (with increased prevalence in individuals with high PRSs). No significant differences in age or sex were observed across the different PRS groups (Table 1 ).
Table 1.
Baseline Characteristics by Strata of SBP and DBP PRS
| SBP PRS Percentiles |
DBP PRS Percentiles |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 to 20th | 20th to 80th | >80th | p | 0 to 20th | 20th to 80th | >80th | p | ||
| Demographics | N | 75,283 | 225,849 | 75,284 | 75,283 | 225,849 | 75,284 | ||
| age, years, mean (SD) | 56.98 (7.92) | 56.95 (7.93) | 56.93 (7.95) | 0.504 | 57.01 (7.94) | 56.94 (7.93) | 56.94 (7.95) | 0.096 | |
| sex, male, n (%) | 35,033 (46.5) | 104,439 (46.2) | 34,947 (46.4) | 0.332 | 34,991 (46.5) | 104,503 (46.3) | 34,925 (46.4) | 0.578 | |
| smoking status, n (%) | 0.005 | 0.407 | |||||||
| Never | 41,121 (54.8) | 122,727 (54.5) | 40,883 (54.5) | 41,015 (54.7) | 122,755 (54.5) | 40,961 (54.6) | |||
| Previous | 26,352 (35.1) | 79,222 (35.2) | 26,757 (35.7) | 26,279 (35.0) | 79,522 (35.3) | 26,530 (35.4) | |||
| Current | 7,540 (10.1) | 23,115 (10.3) | 7,405 (9.9) | 7,706 (10.3) | 22,829 (10.1) | 7,525 (10.0) | |||
| BMI, kg/m2, mean (SD) | 27.51 (4.77) | 27.40 (4.74) | 27.35 (4.72) | <0.001 | 27.49 (4.78) | 27.41 (4.74) | 27.33 (4.70) | <0.001 | |
| Blood pressure traits | DBP, mm Hg, mean (SD) | 81.81 (11.10) | 83.42 (11.39) | 85.12 (11.52) | <0.001 | 81.60 (11.12) | 83.42 (11.36) | 85.32 (11.55) | <0.001 |
| SBP, mm Hg, mean (SD) | 138.81 (20.30) | 141.93 (20.82) | 145.16 (21.07) | <0.001 | 139.17 (20.43) | 141.92 (20.83) | 144.82 (21.03) | <0.001 | |
| antihypertensive medication, n (%) | <0.001 | <0.001 | |||||||
| None | 61,608 (81.9) | 172,820 (76.5) | 52,393 (69.6) | 61,709 (82.0) | 172,831 (76.5) | 52,281 (69.5) | |||
| ACEi only | 3,454 (4.6) | 13,341 (5.9) | 5,890 (7.8) | 3,428 (4.6) | 13,383 (5.9) | 5,874 (7.8) | |||
| ARB only | 1,160 (1.5) | 4,760 (2.1) | 2,020 (2.7) | 1,203 (1.6) | 4,684 (2.1) | 2,053 (2.7) | |||
| beta-blocker only | 1,999 (2.7) | 6,960 (3.1) | 2,798 (3.7) | 1,936 (2.6) | 7,036 (3.1) | 2,785 (3.7) | |||
| dihydropyridine calcium channel blocker only | 1,175 (1.6) | 4,726 (2.1) | 2,142 (2.8) | 1,169 (1.6) | 4,758 (2.1) | 2,116 (2.8) | |||
| multiple antihypertensive classes | 3,230 (4.3) | 13,125 (5.8) | 5,944 (7.9) | 3,226 (4.3) | 13,057 (5.8) | 6,016 (8.0) | |||
| other antihypertensive | 2,643 (3.5) | 10,078 (4.5) | 4,087 (5.4) | 2,596 (3.4) | 10,062 (4.5) | 4,150 (5.5) | |||
| hypertension, n (%) | 16,437 (22.7) | 63,626 (29.4) | 27,055 (37.6) | <0.001 | 16,516 (22.9) | 63,482 (29.4) | 27,120 (37.7) | <0.001 | |
| Prevalent cardiovascular risk factors | type 2 diabetes, n (%) | 1,568 (2.1) | 4,676 (2.1) | 1,695 (2.3) | 0.009 | 1,550 (2.1) | 4,741 (2.2) | 1,648 (2.2) | 0.17 |
| coronary artery disease, n (%) | 3,716 (5.1) | 12,326 (5.6) | 4,750 (6.5) | <0.001 | 3,680 (5.0) | 12,315 (5.6) | 4,797 (6.6) | <0.001 | |
| hypercholesterolemia, n (%) | 9,624 (13.2) | 31,365 (14.4) | 11,515 (15.9) | <0.001 | 9,840 (13.5) | 31,013 (14.3) | 11,651 (16.1) | <0.001 | |
| Prevalent respiratory disease history | chronic obstructive pulmonary disease, n (%) | 1,643 (2.2) | 4,987 (2.2) | 1,655 (2.2) | 0.924 | 1,715 (2.3) | 4,925 (2.2) | 1,645 (2.2) | 0.271 |
| asthma, n (%) | 9,134 (12.3) | 27,435 (12.3) | 9,077 (12.2) | 0.793 | 9,245 (12.4) | 27,430 (12.3) | 8,971 (12.0) | 0.078 | |
ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker.
Across a median follow-up time of 8 years (IQR 7–11 years), 9,969 individuals (2.6%) developed pneumonia, 11,972 (3.3%) developed influenza or pneumonia, 18,172 (5.5%) developed acute upper respiratory infections, 21,734 (6.2%) developed other lower respiratory infections (e.g., bronchitis), 12,963 (4.0%) developed chronic lower respiratory disease, 1,792 (0.5%) developed other interstitial respiratory disease, and 2,621 (0.7%) developed acute respiratory distress syndrome (ARDS) or respiratory failure.
Epidemiological Association of Prevalent Hypertension with Incident Respiratory Disease
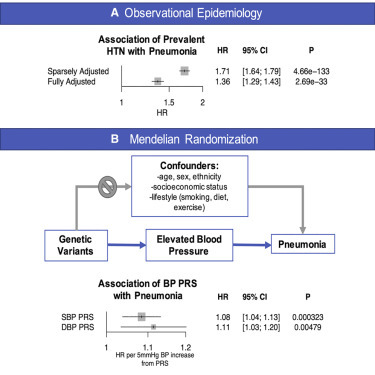
Prevalent hypertension was independently associated with risk for incident respiratory disease including pneumonia (hazard ratio [HR]: 1.36; 95% confidence interval [CI]: 1.29–1.43; p < 0.001), chronic lower respiratory disease (HR: 11.30; 95% CI: 1.25–1.36; p < 0.001), influenza or pneumonia (HR: 1.31; 95% CI: 1.25–1.37; p < 0.001), ARDS or pulmonary failure (HR: 1.43 (95% CI: 1.29–1.59), p < 0.001), other lower respiratory infections (HR: 1.15; 95% CI: 1.37–1.45; p < 0.001), other acute upper respiratory infection (HR: 1.06; 95% CI: 1.11–1.19; p = 0.002), other interstitial respiratory disease (HR: 1.16; 95% CI: 1.02–1.31; p = 0.022), and influenza or viral pneumonia (HR: 1.12; 95% CI: 1.01–1.23; p = 0.032), after adjustment for age, age2, sex, smoking status, BMI, prevalent diabetes, prevalent coronary artery disease, and the first ten principal components of population stratification (Figure 1 ).
Figure 1.
Epidemiological Association of Prevalent Hypertension with Incident Respiratory Diseases
Associations between prevalent hypertension (HTN) and incident respiratory diseases (defined in Table S2) are shown in sparsely adjusted and fully adjusted models. The sparsely adjusted model is adjusted by age, age2, sex, smoking status (current, prior, or never smoker), and the first ten principal components of population stratification. The fully adjusted model is additionally adjusted by prevalent coronary artery disease, prevalent diabetes mellitus, and body mass index. HR and 95% CI are displayed. ARDS, adult respiratory disease syndrome; HR, hazard ratio.
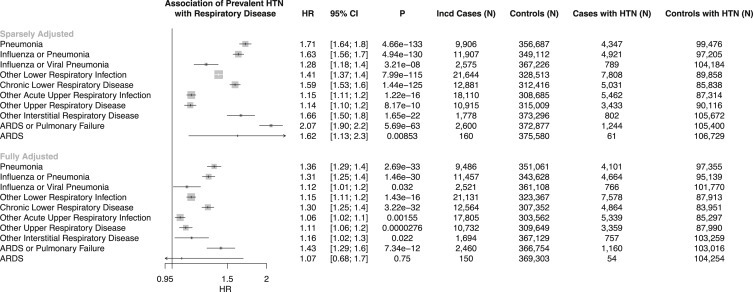
Epidemiological Association of Antihypertensive Use with Incident Pneumonia
Prescriptions for angiotensin-converting enzyme inhibitors (ACEi), angiotensin receptor blockers (ARBs), dihydropyridine calcium channel blockers, beta-blockers, combination antihypertensive medications, and other antihypertensive medications were similarly associated with increased risk for incident pneumonia (Figure 2 ). Further adjustment for prevalent hypertension status rendered all of these associations nonsignificant after multiple testing correction (Figure 2; Table 2 ).
Figure 2.
Epidemiological Association of Antihypertensive Use with Incident Pneumonia
Association of antihypertensive use with incident pneumonia, adjusted by age, age2, sex, smoking status, prevalent coronary artery disease, prevalent diabetes, body mass index, and the first ten principal components of population stratification, displayed with and without adjusting for prevalent hypertension, suggests that the effect of antihypertensives on increased risk for incident pneumonia is driven by hypertensive status. HR and 95% CI are displayed. ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; HR, hazard ratio.
Table 2.
Multivariate Associations of Antihypertensive Medications with Incident Pneumonia Adjusted for Prevalent Hypertension Status at Enrollment, in Addition to Age, Age2, Sex, Smoking Status, BMI, Prevalent Diabetes, Prevalent Coronary Artery Disease, and the First Ten Principal Components of Genetic Ancestry
| Multivariate Model Components | HR | p | Lower 95% CI | Upper 95% CI |
|---|---|---|---|---|
| Antihypertensive: ACEi only | 1.06 | 0.24 | 0.96 | 1.17 |
| Antihypertensive: ARB only | 1.03 | 0.73 | 0.89 | 1.18 |
| Antihypertensive: beta-blocker only | 0.95 | 0.42 | 0.84 | 1.08 |
| Antihypertensive: dihydropyridine calcium channel blocker only | 1.17 | 0.02 | 1.03 | 1.34 |
| Antihypertensive: multiple antihypertensive classes | 1.10 | 0.05 | 1.00 | 1.22 |
| Antihypertensive: other antihypertensive | 1.03 | 0.58 | 0.92 | 1.15 |
| Prevalent hypertension | 1.30 | 1.79E-13 | 1.22 | 1.40 |
| Age | 0.96 | 0.11 | 0.92 | 1.01 |
| Age2 | 1.00 | 6.14E-06 | 1.00 | 1.00 |
| Sex (male) | 1.24 | 1.38E-19 | 1.18 | 1.30 |
| Smoking status: previous smoker | 1.35 | 5.46E-31 | 1.29 | 1.42 |
| Smoking status: current smoker | 2.87 | 5.25E-228 | 2.69 | 3.06 |
| BMI | 1.02 | 1.74E-12 | 1.01 | 1.02 |
| Prevalent diabetes | 1.50 | 1.49E-24 | 1.39 | 1.62 |
| Prevalent coronary artery disease | 1.73 | 1.09E-47 | 1.61 | 1.86 |
| PC1 | 0.99 | 0.10 | 0.97 | 1.00 |
| PC2 | 1.00 | 0.81 | 0.98 | 1.01 |
| PC3 | 0.99 | 0.27 | 0.98 | 1.01 |
| PC4 | 1.00 | 0.53 | 0.99 | 1.01 |
| PC5 | 1.00 | 0.70 | 1.00 | 1.01 |
| PC6 | 0.99 | 0.23 | 0.98 | 1.01 |
| PC7 | 1.01 | 0.05 | 1.00 | 1.03 |
| PC8 | 1.00 | 0.71 | 0.99 | 1.02 |
| PC9 | 1.00 | 0.79 | 1.00 | 1.01 |
| PC10 | 1.00 | 0.87 | 0.99 | 1.01 |
ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; BMI, body mass index; PC, principal component of ancestry.
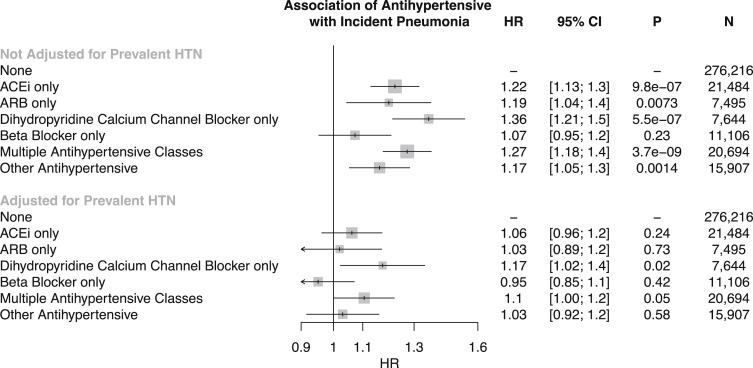
Genetic Association of Blood Pressure with Incident Respiratory Disease
We first used one-sample Mendelian randomization to determine whether a genetic predisposition to increased blood pressure is associated with increased risk for incident pneumonia as well as other respiratory diseases.
Our SBP and DBP genetic instruments each consisted of 75 independent variants (linkage disequilibrium r2 < 0.2) that were genome-wide significant among 299,024 individuals external to the UK Biobank from the International Consortium for Blood Pressure Genomics (Tables S3 and S4). The resulting PRSs for SBP and, separately, for DBP were significantly associated with their respective phenotypes in the UK Biobank, with each SD increase in the SBP PRS increasing SBP by 2.26 mm Hg (F-statistic = 4,176) and each SD increase in the DBP PRS increasing DBP by 1.32 mm Hg (F-statistic = 4,785) (Table S5). Fifty-three single-nucleotide polymorphisms (SNPs) (or SNPs in perfect linkage disequilibrium across traits) were common to both SBP and DBP PRSs. Sensitivity analyses were performed to assess for potential social and lifestyle confounders associating with the SBP and DBP PRSs and demonstrated no significant associations between the PRSs and the Townsend deprivation index for socioeconomic status estimation, smoking status, alcohol intake frequency, vegetable intake, sweet intake, significant life stressor in the past 2 years, and exercise frequency (Table S6).
Each 5 mm Hg increase in SBP conferred by the SBP PRS was associated with a significant increased risk for incident pneumonia (HR: 1.08; 95% CI: 1.04–1.13; p < 0.001), influenza or pneumonia (HR: 1.06; 95% CI: 1.02–1.11; p = 0.003), and other lower respiratory infection (HR: 1.03; 95% CI: 1.00–1.06; p = 0.029). Additionally, each 5 mm Hg increase in DBP conferred by the DBP PRS was associated with an increased risk for incident pneumonia (HR: 1.11; 95% CI: 1.03–1.20; p = 0.005), and other lower respiratory infections (HR: 1.07; 95% CI: 1.01–1.12; p = 0.012) (Figure 3 ). The UK Biobank BiLEVE project subgroup11 was designed to investigate respiratory pathology by sampling individuals from the extremes of lung function distribution and genotyped with the BiLEVE array. Although effects were stronger among those with BiLEVE genotyping, consistently significant effects were observed for both the BiLEVE and UK Biobank Axiom arrays (Figure S1).
Figure 3.
Genetic Association of Blood Pressure with Incident Respiratory Disease
Association of SBP PRS and DBP PRS with incident respiratory diseases (defined in Table S2), adjusted for age, age2, sex, smoking status, and the first ten principal components of population stratification in the UK Biobank. Effects are interpreted as HR per 5 mm Hg increase from the respective PRS. HR and 95% CI are displayed. SBP, systolic blood pressure; DBP, diastolic blood pressure; PRS, polygenic risk score; HR, hazard ratio.
Two-sample Mendelian randomization was additionally performed using pneumonia association statistics from the UK Biobank as the outcome and with blood pressure summary statistics external to the UK Biobank as above as the exposure. Both penalized, robust, inverse-variance weighted (IVW) and robust adjusted profile score (MR-RAPS)12 methods produced similarly significant results (Table S7; Figure S2). Sensitivity analyses were additionally performed to analyze the robustness of the results. First, across both SBP and DBP genetic instruments, the IVW heterogeneity test and the MR-Egger regression intercept term were both insignificant, suggesting negligible contribution of heterogeneity and directional horizontal pleiotropy (Table S7). Additionally, the Steiger directionality test13 suggests the correct causal direction of blood pressure on pneumonia. Last, leave-one-out analyses were additionally performed, suggesting that no single SNP drives the observed association (Figure S3). Variants previously described to influence ACE expression in the lung (rs145126552, rs4277405) or kidney (rs6504163) in the Genotype-Tissue Expression (GTEx) project14 were not individually associated with pneumonia risk (p > 0.05).
Epidemiological and Genetic Associations of Prevalent Hypertension with Pulmonary Function Tests
Secondary analyses identified significant associations between prevalent hypertension and reduced pulmonary function test performance (forced expiratory volume in 1 s [FEV1]: −0.07 SD [95% CI: −0.07 to −0.06; p < 0.001]; forced vital capacity [FVC]: −0.06 SD [95% CI: −0.07 to −0.06; p < 0.001]; FEV1/FVC: −0.02 SD [95% CI: −0.03 to −0.01; p < 0.001]), independent of age, age2, sex, smoking status, BMI, standing height, prevalent coronary artery disease, prevalent diabetes mellitus, and the first ten principal components of ancestry (Figure 4 A).
Figure 4.
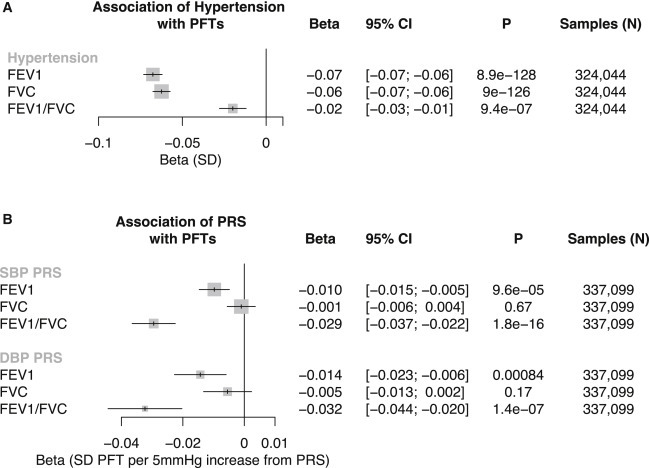
Epidemiological and Genetic Associations of Elevated Blood Pressure with Pulmonary Function Tests
(A) Association of prevalent hypertension with pulmonary function tests (PFTs), adjusted for age, age2, sex, standing height, smoking status, prevalent coronary artery disease, prevalent diabetes, body mass index, and the first ten principal components of population stratification in the UK Biobank. Beta and 95% CI are displayed.
(B) Association of SBP PRS and DBP PRS PFTs, adjusted for age, age2, sex, standing height, body mass index, smoking status, and the first ten principal components of population stratification in the UK Biobank. Beta values are interpreted as SD change in PFT per 5 mm Hg increase from the respective PRS. Beta and 95% CI are displayed.
FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; HTN, hypertension.
Each 5 mm Hg increase in SBP PRS was significantly associated with reduced pulmonary function test performance suggesting obstructive pulmonary disease (FEV1: −0.01 SD [95% CI: −0.015 to −0.005; p < 0.001]; FEV1/FVC: −0.029 SD [95% CI: −0.037 to −0.022; p < 0.001]). Similar associations were identified for between DBP PRS and pulmonary function test performance (FEV1: −0.014 SD [95% CI: −0.023 to −0.006; p < 0.001]; FEV1/FVC −0.032 SD [95% CI: −0.044 to −0.02; p < 0.001]) (Figure 4B). No significant difference was observed upon stratification by BiLEVE genotyping array (Figure S4).
Discussion
In a large, prospective, population-based cohort, we show that prevalent hypertension is a risk factor for incident pneumonia, lower respiratory infections, ARDS or respiratory failure, and many other respiratory diseases. Additionally, our epidemiological analyses also demonstrate an association between prevalent hypertension and increased pulmonary obstruction as indicated by reduced FEV1/FVC. Our Mendelian randomization studies support a causal relationship between increased blood pressure with increased risk for pneumonia as well as reduced pulmonary function.
Our findings may have important implications for the prevention of pneumonia. First, our epidemiological and genetic analyses establish that hypertension is an important independent risk factor for the development of pneumonia. Hypertension has been proposed to promote key factors that may predispose to infection via several potential mechanisms. First, hypertensive stimuli promote dysregulation of the adaptive immune response. Chronic angiotensin II infusions in mice increase markers of T lymphocyte activation and perivascular adipose infiltration.15 Additional murine studies have indicated that monocytes and neutrophils may be key factors in angiotensin II-mediated hypertension and resultant vascular dysfunction.16 A recent Mendelian randomization study supported a causal relationship between blood pressure and subsequent alteration in neutrophil, monocyte, and eosinophil indices.8 Second, endothelial dysfunction as a consequence of hypertension may promote infection. Dysregulation of nitric oxide release and signaling in murine models of pulmonary inflammation leads to exacerbated lung injury.17
Second, blood pressure elevations may result in pulmonary function alterations predisposing to the development of pneumonia. Our Mendelian randomization analyses with pulmonary function tests support a causal association between blood pressure and pulmonary obstruction as indicated by reduced FEV1/FVC. Several prior observational studies have linked hypertension with decreased performance on pulmonary function tests, reduced lung function, and increased pulmonary obstruction.4 , 5 COPD is a well-established risk factor for pneumonia and is co-morbid with several cardiovascular diseases and risk factors, including hypertension.6 Our study extends these observations to show that increased blood pressure may causally lead to increased pulmonary obstruction representing a putative mechanism toward heightened pneumonia risk. Together, these studies and others suggest several mechanisms that may link hypertension and pulmonary obstruction: (1) both may involve physiological degradation of arterial and airway elasticity,5 (2) endothelial and vascular dysfunction may also influence pulmonary vascular endothelial cells and lead to pulmonary vascular dysfunction resulting in lung tissue destruction and airway obstruction,7 and (3) systemic inflammation associated with hypertension may additionally alter pulmonary function.8
Third, as our results are consistent with a causal relationship between blood pressure and pneumonia risk, blood pressure optimization is anticipated to reduce pneumonia risk in the population orthogonal to other strategies aimed at reducing infection risk. Our observational association of antihypertensives with increased pneumonia risk was rendered nonsignificant when adjusting for hypertension consistent with confounding by indication. Furthermore, we do not observe that genetic variants influencing the expression of ACE in the lung or kidney significantly increase the risk for pneumonia. However, our results are aligned with current recommendations to maintain stable, normal blood pressure, including with ACEi as indicated during the coronavirus disease 2019 (COVID-19) pandemic.18 Consistent with our findings, the Systolic Blood Pressure Intervention Trial (SPRINT) also showed that intensive blood pressure reduction (to a mean SBP of 121 mm Hg) resulted in fewer cases of incident pneumonia (2.1% versus 2.4%) compared with standard blood pressure reduction (to a mean SBP of 136 mm Hg).19 A meta-analysis comprising randomized controlled and observational studies has also indicated that ACEi may be protective for pneumonia risk.20
This study has several strengths, including the analysis of a large, genotyped population-based cohort with high-fidelity phenotyping, including with subclinical respiratory phenotypes. Furthermore, diverse phenotyping facilitates extensive individual-level covariate adjustment in the models and sensitivity analyses to assess for pleiotropy. Our overall study design, with the incorporation of Mendelian randomization, permits more robust causal inference in humans beyond observational prospective analyses.
Limitations of Study
Although our study has several strengths, some limitations should be considered. First, the present analyses were conducted among individuals of white British ancestry residing in the United Kingdom; whether the present findings generalize to diverse ethnicities and other geographic regions remains to be tested. Second, our model assumes the association of the genetic instrument to the outcome occurs via the primary exposure and is not confounded by pleiotropy.21 We have performed a number of sensitivity analyses to assess for possible confounders; in particular, we observe that our genetic instruments for blood pressure are not associated with key socioeconomic and lifestyle factors influencing both blood pressure and pneumonia risk. We further maintain a sparsely adjusted model in our one-sample Mendelian randomization analyses to reduce the potential for collider bias.22 Additionally, secondary two-sample Mendelian randomization analyses provide consistent results with no evidence of horizontal pleiotropy, heterogeneity, or individual variants driving the association. Third, although our results do not yield an association between ACE expression quantitative trait loci (eQTLs) and pneumonia risk, we cannot rule out the possibility of reduced power for this analysis. Fourth, given the high correlation between the blood pressure PRS and antihypertensive medication use, the results cannot rule out the possibility that the association between PRS and pneumonia could be mediated in part by antihypertensive medication use. Fifth, outcomes for the present analyses occurred prior to the COVID-19 pandemic. The first release of COVID-19 phenotypes from the UK Biobank participants has few COVID-19 events (approximately 0.2%), which limits power. Furthermore, with testing data only in 0.3% of UK Biobank participants, analyses are currently limited by ascertainment bias. Whether the current findings translate to COVID-19 require further verification.
Conclusions
Our study provides evidence that hypertension directly influences the risk for pneumonia. Blood pressure optimization may reduce risk for pneumonia. Whether these results extend to the ongoing COVID-19 pandemic requires further study.
STAR★Methods
Resource Availability
Lead Contact
Further information and requests for resources should be directed to the Lead Contact, Pradeep Natarajan (pnatarajan@mgh.harvard.edu).
Materials Availability
The study did not generate any new reagents or materials.
Data and Code Availability
UK Biobank individual-level data are available for request through the UK Biobank with application (https://www.ukbiobank.ac.uk). The code used toward genome wide association of pneumonia and subsequent two-sample Mendelian randomization is available at https://github.com/mzekavat/Pneumonia_BP_CellMed. Additionally, the full pneumonia genome wide association summary statistics have been uploaded onto the LocusZoom website (https://my.locuszoom.org/gwas/842685/). The published article includes all other data generated or analyzed during this study.
Experimental Model and Subject Details
Individual-level genomic data and longitudinal phenotypic data from the UK Biobank, a large-scale population-based cohort with genotype and phenotype data in approximately 500,000 volunteer participants recruited from 2006-2010 was used.23 Baseline assessments were conducted at 22 assessment centers across the UK using touch screen questionnaire, computer assisted verbal interview, physical tests, and sample collection including for DNA (https://www.ukbiobank.ac.uk). Secondary use of the data was approved by the Massachusetts General Hospital institutional review board (protocol 2013P001840) and facilitated through UK Biobank Application 7089.
Method Details
UK Biobank
Of 488,377 individuals genotyped in the UK Biobank, we used data for 377,143 participants with white British ancestry consenting to genetic analyses, with genotypic-phenotypic sex concordance, without sex aneuploidy, and one from each pair of 1st or 2nd degree relatives selected randomly. Genome-wide genotyping was previously performed in the UK Biobank using two genotyping arrays sharing 95% of marker content: Applied Biosystems UK BiLEVE Axiom Array (807,411 markers in 49,950 participants) and Applied Biosystems UK Biobank Axiom Array (825,927 markers in 438,427 participants) both by Affymetrix (Santa Clara, CA).23 Variants used in the present analysis include those also imputed using the Haplotype Reference Consortium reference panel of up to 39 million single nucleotide polymorphisms (SNPs).24 Poor quality variants and genotypes were filtered as previously described.23
Hypertension, covariates, and medication measures
Disease definitions for hypertension and clinical disease covariates are as previously described.25 In brief, hypertension was defined by self-reported hypertension and billing codes for essential hypertension, hypertensive disease with and without heart failure, hypertensive heart and renal diseases, and secondary hypertension. There was a high concordance between self-reported and physician-diagnosed hypertension, with 91% of self-reported hypertensive individuals also having physician-diagnosed hypertension from the aforementioned billing codes.
Blood pressure medications were characterized by medication type into angiotensin converting enzyme inhibitors (ACEi), angiotensin receptor blockers (ARB), dihydropyridine calcium channel blockers, beta blockers, multiple antihypertensive classes, or other medication (Table S1). Patients who reported that they were taking a blood pressure medication (via UK Biobank Field IDs 6153 and 6177) but were not taking one of the listed antihypertensive medications were included in the ‘other’ category.
Respiratory outcomes
Clinical disease definitions for our primary outcome (pneumonia) and related respiratory outcomes are detailed in Table S2. In summary, these included respiratory diseases using the first reported occurrences of respiratory system disorders in Category 2410 as categorized by the UK Biobank (https://biobank.ndph.ox.ac.uk/showcase/label.cgi?id=2410) which maps primary care data, ICD-9 and ICD-10 codes from hospital inpatient data, ICD-10 codes in death register records, and self-reported medical conditions reported at the baseline, to ICD-10 codes. For each set of phenotypes, the time to first incident event after baseline examination in individuals free of prevalent history of respiratory system disorder was used.
Pneumonia includes viral, bacterial, and unspecified etiologies (J12-J18). Influenza or viral pneumonia includes confirmed or suspected influenza or viral pneumonia (J09-J12). The acute upper respiratory infections category includes acute nasopharyngitis, sinusitis, pharyngitis, tonsillitis, laryngitis, tracheitis, croup, epiglottitis, or upper respiratory infections of multiple and unspecified sites (J00-J06). The other acute lower respiratory infections category includes acute bronchitis, bronchiolitis, or unspecified acute lower respiratory infections (J20-J22). The other diseases of the upper respiratory tract category includes rhinitis, nasopharyngitis, pharyngitis, chronic sinusitis, nasal polyps, other disorders of the nose and nasal sinuses, chronic diseases of the tonsils and adenoids, peritonsillar abscess, chronic laryngitis, laryngotracheitis, diseases of the vocal cords and larynx, or other diseases of the upper respiratory tract (J30-J39). Chronic lower respiratory diseases include bronchitis, emphysema, chronic obstructive pulmonary disease, asthma, bronchiectasis (J40-J47). Other interstitial respiratory diseases refer to acute respiratory distress syndrome (ARDS), pulmonary edema, or other interstitial pulmonary diseases (J80, J81, J84). Respiratory failure refers to J96 (respiratory failure not elsewhere classified).
Quantitative phenotypes included best-measure pulmonary function tests from spirometry using a Vitalograph Pneumotrac 6800 (Buckingham, United Kingdom), including forced expiratory volume in the first second (FEV1), forced vital capacity (FVC), and the ratio of these two measurements (FEV1/FVC). For each individual measurement, extreme outliers were determined and filtered by adjusting the traditional box and whisker upper and lower bounds and accounting for skewness in the phenotypic data identified using the Robustbase package in R (setting range = 3) (https://cran.r-project.org/web/packages/robustbase/robustbase.pdf). Phenotypes were then inverse rank normalized for analysis.
Quantification and Statistical Analysis
Association between prevalent hypertension and respiratory conditions in the UK Biobank
Phenotypic association of prevalent hypertension with incident respiratory diseases and with pulmonary function tests was performed using Cox proportional hazards models among individuals without the corresponding prevalent condition in R-3.5. The proportional hazards assumption was assessed by Schoenfeld residuals and was satisfied. The sparsely adjusted model included age, age2 (included to fully account for age given significant associations in multivariate models), sex, smoking status (current, previous, or never smoker), and the first ten principal components of ancestry. For the pulmonary function test associations, further adjustment for body mass index and height was performed given potential biases conferred by these two physical measures on pulmonary function.The fully adjusted model additionally incorporated body-mass index (BMI), prevalent diabetes mellitus, and prevalent coronary artery disease. Alpha Bonferroni threshold for significance based on non-overlapping phenotypes was 0.05/8 = 0.00625.
Genetic instruments for blood pressure and association analyses
One-sample Mendelian randomization was performed in the UK Biobank by associating inverse-rank normalized systolic blood pressure and diastolic blood pressure polygenic risk scores (SBP PRS, DBP PRS) with incident respiratory disease phenotypes. The 75 variants comprising each genetic instrument (Tables S3 and S4) were determined by identifying genome-wide significant (p < 5x10−8), largely uncorrelated (linkage disequilibrium r2 < 0.2) variants for each phenotype from the International Consortium for Blood Pressure (ICBP) GWAS summary statistics across 299,024 individuals among 77 cohorts excluding the UK Biobank.10 Betas used to develop the polygenic risk scores were from those of European ancestry (excluding UK Biobank participants) in the ICBP GWAS.
Additive polygenic risk scores were determined as such: , where is the weight for each allele of from the ICBP GWAS summary statistics, and is the number of alleles (i.e., 0, 1, or 2) for in participant in the UK Biobank. To confirm that the SBP PRS and DBP PRS were strong instruments, we assessed their associations with SBP and DBP, adjusted for blood pressure medications by adding 15 and 10 mmHg to SBP and DBP, respectively, as previously done.26 , 27 Each genetic instrument was validated against its exposure by calculating an F-statistic derived from unadjusted linear regression of the exposure against its PRS. An F-statistic greater than 10 indicates low risk of weak-instrument bias. The SBP and DBP PRS were further scaled from standard deviation units to 5mmHg units by multiplying the SBP PRS by 2.26/5 (since each standard deviation of the SBP PRS conferred 2.26mmHg increase in SBP in the UK Biobank), and multiplying the DBP PRS by 1.32/5 (since each standard deviation of the SBP PRS conferred 1.32mmHg increase in SBP in the UK Biobank).
Additional sensitivity analyses tested associations between each PRS and potential social and lifestyle confounders including the Townsend deprivation index for socioeconomic status estimation,28 smoking status (Field ID 20116), alcohol intake frequency (Filed ID 1558), vegetable serving intake (Field ID 104060), handfuls of sweet intake (Field ID 102330), significant life stressor over the past two years (Field ID 6145), and exercise frequency (Field ID 3637).
Association analysis of the SBP and DBP PRS with incident respiratory diseases was performed using Cox proportional hazards models in R-3.5, adjusting for age, age2, sex, smoking status, and the first ten principal components of ancestry. Alpha Bonferroni threshold for significance based on non-overlapping phenotypes across both the SBP PRS and DBP PRS was 0.05/16 = 0.003125. The proportional hazards assumption was assessed by Schoenfeld residuals and was satisfied. Association analyses between the SBP and DBP PRS and pulmonary function tests was performed using a generalized linear model adjusted for age, age2, sex, smoking status, the first ten principal components of ancestry, as well as standing height and body mass index. Alpha Bonferroni threshold for significance based on non-overlapping phenotypes across both the SBP PRS and DBP PRS was 0.05/6 = 0.0083.
To verify our genetic associations with two-sample Mendelian randomization, first each of the 75 variants for each PRS were associated with both prevalent and incident pneumonia using a logistic regression Wald test in Hail-0.2,29 adjusting for age, age2, sex, smoking status, the first ten principal components of ancestry, and genotype array. Using these associations, two-sample Mendelian randomization was performed using the external ICBP-derived SBP and DBP genetic instruments as exposures and the respective effects of each variant on pneumonia in the UK Biobank as outcomes. Two-sample Mendelian randomization was performed using both the robust, penalized inverse variance weighted (IVW) method from the MendelianRandomization package in R,30 , 31 as well as robust adjusted profile score (MR-RAPS)12 for comparison. Two-sided statistical significance was assigned at 0.05 for this confirmatory procedure. IVW 2-sample Mendelian randomization uses a weighted linear regression of the ratio of the SNP effects on the outcomes to the SNP effects on the risk factor, without using an intercept term. MR-RAPS models the systematic pleiotropy using a random effects model to create a robust adjusted profile score. We additionally use the TwoSampleMR package in R (https://mrcieu.github.io/TwoSampleMR/index.html) to perform multiple sensitivity analyses including heterogeneity tests, the MR Egger intercept test for horizontal pleiotropy, and leave-one-out analyses to determine if there is a single variant driving the genetic association.
Acknowledgments
We would like to acknowledge and thank the participants and staff of the UK Biobank and of the International Consortium of Blood Pressure (ICBP) cohorts. This work was supported by UK Biobank (application number 7089).
Author Contributions
S.M.Z. and P.N. devised the project, analyzed the data, interpreted results, and wrote and revised the manuscript. M.H. contributed to analysis and critically revised the manuscript. J.P. contributed to data curation. P.N., P.K., E.W.K., C.N., and H.Z. supported the investigation, provided funding and supervision, and revised the manuscript. P.N. and S.M.Z. designed the project and are the guarantors. All authors approved final manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Declaration of Interests
P.N. has received grants and personal fees from Apple, grants from Amgen, grants from Boston Scientific, personal fees from Blackstone Life Sciences, personal fees from Novartis, personal fees from Genentech, and spouse employment and equity at Vertex. C.N.-C. has received personal fees from GE Healthcare and Novartis. P.K. reports employment and equity at Vertex, outside the submitted work. The remaining authors declare no competing interests.
Published: November 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.medj.2020.11.001.
Supplemental Information
ACEi = angiotensin converting enzyme inhibitor, ARB = angiotensin receptor blocker
ARDS = Acute respiratory distress syndrome
References
- 1.Bennett D.A., Holmes M.V. Mendelian randomisation in cardiovascular research: an introduction for clinicians. Heart. 2017;103:1400–1407. doi: 10.1136/heartjnl-2016-310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boszhurt B., Kovacs R., Harrington B. AHA Science News; 2020. HFSA/ACC/AHA statement addresses concerns re: using RAAS antagonists in COVID-19.https://www.acc.org/latest-in-cardiology/articles/2020/03/17/08/59/hfsa-acc-aha-statement-addresses-concerns-re-using-raas-antagonists-in-covid-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess S., Bowden J., Dudbridge F., Thompson S.G. Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. arXiv. 2016 https://arxiv.org/abs/1606.03729 arXiv:1606.03729. [Google Scholar]
- 4.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 2017 doi: 10.1101/166298. [DOI] [Google Scholar]
- 5.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldeira D., Alarcão J., Vaz-Carneiro A., Costa J. Risk of pneumonia associated with use of angiotensin converting enzyme inhibitors and angiotensin receptor blockers: systematic review and meta-analysis. BMJ. 2012;345:e4260. doi: 10.1136/bmj.e4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 8.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duprez D.A., Hearst M.O., Lutsey P.L., Herrington D.M., Ouyang P., Barr R.G., Bluemke D.A., McAllister D., Carr J.J., Jacobs D.R., Jr. Associations among lung function, arterial elasticity, and circulating endothelial and inflammation markers: the multiethnic study of atherosclerosis. Hypertension. 2013;61:542–548. doi: 10.1161/HYPERTENSIONAHA.111.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evangelou E., Warren H.R., Mosen-Ansorena D., Mifsud B., Pazoki R., Gao H., Ntritsos G., Dimou N., Cabrera C.P., Karaman I., et al. Million Veteran Program Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright J.T., Jr., Williamson J.D., Whelton P.K., Snyder J.K., Sink K.M., Rocco M.V., Reboussin D.M., Rahman M., Oparil S., Lewis C.E., et al. SPRINT Research Group A randomized trial of intensive versus standard blood-pressure control. N. Engl. J. Med. 2015;373:2103–2116. doi: 10.1056/NEJMoa1511939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzik T.J., Hoch N.E., Brown K.A., McCann L.A., Rahman A., Dikalov S., Goronzy J., Weyand C., Harrison D.G. Role of the T cell in the genesis of angiotensin II induced hypertension and vascular dysfunction. J. Exp. Med. 2007;204:2449–2460. doi: 10.1084/jem.20070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer K.S., Newell J.D., Jr., Jin D., Fuld M.K., Saha P.K., Hansdottir S., Hoffman E.A. Quantitative dual-energy computed tomography supports a vascular etiology of smoking-induced inflammatory lung disease. Am. J. Respir. Crit. Care Med. 2016;193:652–661. doi: 10.1164/rccm.201506-1196OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jankowich M.D., Taveira T., Wu W.C. Decreased lung function is associated with increased arterial stiffness as measured by peripheral pulse pressure: data from NHANES III. Am. J. Hypertens. 2010;23:614–619. doi: 10.1038/ajh.2010.37. [DOI] [PubMed] [Google Scholar]
- 16.Müllerova H., Agusti A., Erqou S., Mapel D.W. Cardiovascular comorbidity in COPD: systematic literature review. Chest. 2013;144:1163–1178. doi: 10.1378/chest.12-2847. [DOI] [PubMed] [Google Scholar]
- 17.Okaishi K., Morimoto S., Fukuo K., Niinobu T., Hata S., Onishi T., Ogihara T. Reduction of risk of pneumonia associated with use of angiotensin I converting enzyme inhibitors in elderly inpatients. Am. J. Hypertens. 1999;12:778–783. doi: 10.1016/s0895-7061(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 18.Olsen M.H., Angell S.Y., Asma S., Boutouyrie P., Burger D., Chirinos J.A., Damasceno A., Delles C., Gimenez-Roqueplo A.P., Hering D., et al. A call to action and a lifecourse strategy to address the global burden of raised blood pressure on current and future generations: the Lancet Commission on hypertension. Lancet. 2016;388:2665–2712. doi: 10.1016/S0140-6736(16)31134-5. [DOI] [PubMed] [Google Scholar]
- 19.Paternoster L., Tilling K., Davey Smith G. Genetic epidemiology and Mendelian randomization for informing disease therapeutics: Conceptual and methodological challenges. PLoS Genet. 2017;13:e1006944. doi: 10.1371/journal.pgen.1006944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Y., Chen Y., Huang Z., Huang J., Li X., Tian Z., Li J. Associations between untraditional risk factors, pneumonia/lung cancer, and hospital fatality among hypertensive men in Guangzhou downtown. Sci. Rep. 2020;10:1425. doi: 10.1038/s41598-020-58207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siedlinski M., Jozefczuk E., Xu X., Teumer A., Evangelou E., Schnabel R.B., Welsh P., Maffia P., Erdmann J., Tomaszewski M., et al. White blood cells and blood pressure: a Mendelian randomization study. Circulation. 2020;141:1307–1317. doi: 10.1161/CIRCULATIONAHA.119.045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speyer C.L., Neff T.A., Warner R.L., Guo R.F., Sarma J.V., Riedemann N.C., Murphy M.E., Murphy H.S., Ward P.A. Regulatory effects of iNOS on acute lung inflammatory responses in mice. Am. J. Pathol. 2003;163:2319–2328. doi: 10.1016/S0002-9440(10)63588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hail Team Hail 0.2. 2020. https://hail.is/index.html
- 24.Tobin M.D., Sheehan N.A., Scurrah K.J., Burton P.R. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat. Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 25.Townsend P., Phillimore P., Beattie A. Croom Helm; 1989. Health and Deprivation: Inequality and the North. [Google Scholar]
- 26.Wain L.V., Shrine N., Miller S., Jackson V.E., Ntalla I., Soler Artigas M., Billington C.K., Kheirallah A.K., Allen R., Cook J.P., et al. UK Brain Expression Consortium (UKBEC) OxGSK Consortium Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir. Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren H.R., Evangelou E., Cabrera C.P., Gao H., Ren M., Mifsud B., Ntalla I., Surendran P., Liu C., Cook J.P., et al. International Consortium of Blood Pressure (ICBP) 1000G Analyses. BIOS Consortium. Lifelines Cohort Study. Understanding Society Scientific group. CHD Exome+ Consortium. ExomeBP Consortium. T2D-GENES Consortium. GoT2DGenes Consortium. Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium. International Genomics of Blood Pressure (iGEN-BP) Consortium. UK Biobank CardioMetabolic Consortium BP working group Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017;49:403–415. [Google Scholar]
- 28.Wenzel P., Knorr M., Kossmann S., Stratmann J., Hausding M., Schuhmacher S., Karbach S.H., Schwenk M., Yogev N., Schulz E., et al. Lysozyme M-positive monocytes mediate angiotensin II-induced arterial hypertension and vascular dysfunction. Circulation. 2011;124:1370–1381. doi: 10.1161/CIRCULATIONAHA.111.034470. [DOI] [PubMed] [Google Scholar]
- 29.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zekavat S.M., Aragam K., Emdin C., Khera A.V., Klarin D., Zhao H., Natarajan P. Genetic association of finger photoplethysmography-derived arterial stiffness index with blood pressure and coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2019;39:1253–1261. doi: 10.1161/ATVBAHA.119.312626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q.W.,J., Hemani G., Bowden J., Small S.D. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. arXiv. 2018 https://arxiv.org/abs/1801.09652 arXiv:1801.09652. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ACEi = angiotensin converting enzyme inhibitor, ARB = angiotensin receptor blocker
ARDS = Acute respiratory distress syndrome
Data Availability Statement
UK Biobank individual-level data are available for request through the UK Biobank with application (https://www.ukbiobank.ac.uk). The code used toward genome wide association of pneumonia and subsequent two-sample Mendelian randomization is available at https://github.com/mzekavat/Pneumonia_BP_CellMed. Additionally, the full pneumonia genome wide association summary statistics have been uploaded onto the LocusZoom website (https://my.locuszoom.org/gwas/842685/). The published article includes all other data generated or analyzed during this study.


