Significance
The mechanotransduction (MT) complex in auditory hair cells converts the mechanical stimulation of sound waves into neural signals. Our findings reveal previously unrecognized physical and functional interactions of TMC1 and LHFPL5, two critical components of the MT complex, and provide insights into the molecular mechanism by which the D572N mutation in TMC1 causes deafness. Our findings also identify a missing link in the currently known physical organization of the MT micromolecular complex. Furthermore, by demonstrating the power of the microbead-based SiMPull assay for the biochemical examination of rare cells such as hair cells, this study opens up an avenue for the biochemical investigation of MT-complex proteins and other critical nonabundant proteins in hair cells.
Keywords: TMC1, LHFPL5, hair cells, D572N mutation, single-molecule pulldown (SiMPull)
Abstract
Transmembrane channel-like protein 1 (TMC1) and lipoma HMGIC fusion partner-like 5 (LHFPL5) are recognized as two critical components of the mechanotransduction complex in inner-ear hair cells. However, the physical and functional interactions of TMC1 and LHFPL5 remain largely unexplored. We examined the interaction between TMC1 and LHFPL5 by using multiple approaches, including our recently developed ultrasensitive microbead-based single-molecule pulldown (SiMPull) assay. We demonstrate that LHFPL5 physically interacts with and stabilizes TMC1 in both heterologous expression systems and in the soma and hair bundle of hair cells. Moreover, the semidominant deafness mutation D572N in human TMC1 (D569N in mouse TMC1) severely disrupted LHFPL5 binding and destabilized TMC1 expression. Thus, our findings reveal previously unrecognized physical and functional interactions of TMC1 and LHFPL5 and provide insights into the molecular mechanism by which the D572N mutation causes deafness. Notably, these findings identify a missing link in the currently known physical organization of the mechanotransduction macromolecular complex. Furthermore, this study has demonstrated the power of the microbead-based SiMPull assay for biochemical investigation of rare cells such as hair cells.
The mechanotransduction (MT) channel in sensory hair cells in the ear has been intensively investigated for >40 y, but the precise molecular composition of the channel remains enigmatic. Recently, transmembrane channel-like protein 1 (TMC1) and transmembrane inner ear expressed protein (TMIE) were suggested to be components of the MT channel (1–4). Emerging evidence indicates that TMC1 and TMIE are assembled into a macromolecular complex comprising several proteins, including protocadherin-15 (PCDH15), lipoma high-mobility group protein isoform C fusion partner-like 5 (LHFPL5), and calcium and integrin-binding family member 2 (CIB2) (5, 6), which are all indispensable for normal auditory MT.
The tetraspan membrane protein LHFPL5 was proposed to functionally couple the tip link to the MT channel in hair cells (7). Moreover, LHFPL5 expression was suggested to stabilize TMC1 expression in the MT complex in hair cells, but no physical interaction between the two proteins was detected in coimmunoprecipitation (co-IP) experiments in heterologous expression systems (9). Conversely, TMC1 and LHFPL5 were experimentally localized at the tip of shorter stereocilia in neonatal and adult mice (7–11). Given the close localization and functional interaction of TMC1 and LHFPL5, the reason for the observed lack of physical association between these two proteins is unclear.
Here, we investigate the physical and functional interactions between TMC1 and LHFPL5 by using multiple approaches, including our recently developed ultrasensitive microbead-based single-molecule pulldown (SiMPull) assay (12). We demonstrate that TMC1 physically interacts with LHFPL5 and that TMC1 expression is stabilized by LHFPL5 binding both in heterologous expression systems and in hair cells. Notably, we found that the deafness-causing mutation D572N in TMC1 disrupted TMC1-LHFPL5 interaction and destabilized TMC1 expression.
Results
Ectopic TMC1 and LHFPL5 Physically Interact and Mutually Stabilize Each Other.
To search for TMC1-binding partners, we performed yeast two-hybrid assays by using four human TMC1 (hTMC1) fragments as baits: TMC1-F1 (amino acids [aa] 1 to 199), TMC1-F2 (aa 200 to 461), TMC1-F3 (aa 462 to 634), and TMC1-F4 (aa 635 to 760); these four fragments divided the entire hTMC1 sequence based on a topology model featuring 6 transmembrane helices (13) that had been reported before the publication of more recent homology models featuring 10 transmembrane helices (4, 14). In the yeast two-hybrid assays, TMC1-F3 interacted with LHFPL5 (Fig. 1A). The interaction of TMC1-F3 with LHFPL5 was further confirmed in COS7 cells (SI Appendix, Fig. S1); by contrast, TMC1-F1, TMC1-F2, and TMC1-F4 did not interact with LHFPL5 (SI Appendix, Fig. S1). Moreover, ectopic FLAG-TMC1 and LHFPL5-HA coprecipitated from COS7 cells (Fig. 1B) and HEK293T cells (SI Appendix, Fig. S2), which further supported physical interaction between TMC1 and LHFPL5. Notably, the protein levels of both TMC1 and LHFPL5 increased markedly when the proteins were coexpressed, which indicated mutual stabilization of TMC1 and LHFPL5 (Figs. 1 B and C and 2). Our results also show that the LHFPL5 C-terminal half (aa 122 to 219) but not the N-terminal half (aa 1 to 120) bound to full-length TMC1 (Fig. 1 D and E). Although the LHFPL5 C-terminal protein was expressed at a lower level than full-length LHFPL5, presumably because of its high hydrophobicity, both proteins pulled down similar fractions of their input (Fig. 1E); these results indicate that the LHFPL5 C-terminal protein binds to TMC1 as well as full-length LHFPL5.
Fig. 1.
Ectopic TMC1 and LHFPL5 physically interact and mutually stabilize each other. (A) Human TMC1-F3 (aa 462 to 634) interacts with human LHFPL5 in yeast two-hybrid assays. Krev1/RalGDSwt, positive-control pair; Krev1/RalGDSm, negative-control pair; SD2, deficient in Leu and Trp; SD4, deficient in Leu, Trp, His, and Ura. (B) Ectopic FLAG-TMC1 and LHFPL5-HA in COS7 cells coprecipitate and mutually stabilize each other. (C) Summary of protein levels of TMC1 and LHFPL5 expressed alone (with mock vector, first two lanes in both halves of the blots) or together (third lane) in the experiment shown in B and in two other similar experiments. All values are normalized to the protein level of TMC1 or LHFPL5 expressed alone. **P = 0.0046, ***P = 0.0002; n = 3 independent biological replicates. (D and E) Full-length LHFPL5 (LHFPL5-HA) and the C-terminal half (122–219-HA) of the protein, but not the N-terminal half (1–121-HA) of LHFPL5, coprecipitate with FLAG-TMC1 expressed in HEK293T cells. Shown are representative results of three similar experiments. (D) Schematic topology of full-length and truncated LHFPL5, based on hydropathy plots. Terms/symbols used in this and other figures: IP, immunoprecipitation; IB, immunoblotting; actin, loading control; an asterisk (*) in Western blots, IgG band.
Fig. 2.
Deafness-causing mutations of TMC1 alter its physical and functional interactions with LHFPL5. (A) Schematic topology of hTMC1, based on a published homology model (4, 14). Numbers, amino acid positions; orange lines, TMC1-F3; small dots, recessive deafness mutations (M486T, P514L, C515R, W588X, and R604X) in TMC1-F3; large dot, dominant deafness mutations D572N/H in TMC1-F3. (B–E) Effect of TMC1 mutations on interaction and mutual stabilization of TMC1 and LHFPL5. FLAG-tagged WT or mutant hTMC1 was expressed alone or together with HA-tagged LHFPL5 in COS7 cells: M486T (B), P514L and C515R (C), D572N/H (D), and W588X and R604X (E). LHFPL5 was immunoprecipitated using rabbit anti-LHFPL5 antiserum. An asterisk (*) in Western blots indicates an IgG band resulting from cross-reactivity of second antibody; arrowhead in C and D, IgG band, which is visible in the following three lanes but is not separated from the TMC1 band in the three lanes on the right. (F) FLAG-tagged WT or mutant TMC1 and LHFPL5-HA were expressed separately in COS7 cells, and the lysates of these cells were mixed and immunoprecipitated with anti-FLAG antibody. (G) Summary of binding efficiency exhibited by WT and mutant TMC1 toward LHFPL5 in assays shown in B–E and in two other similar experiments. All values are normalized to WT TMC1. Different from WT TMC1: *P = 0.0222, **P ≤ 0.0033, ***P < 0.0001; n = 3 independent biological replicates. (H) Summary of relative increase of TMC1 and LHFPL5 protein levels after coexpression (Relative Increase After Coexpression) in assays shown in B–E and in two other similar experiments. All values are normalized to WT TMC1. Different from WT TMC1: D572N (P = 0.0029 for TMC1 and 0.0111 for LHFPL5); D572H (P = 0.0058 for TMC1 and 0.0043 for LHFPL5); W588X (P = 0.0339 for TMC1 and 0.0011 for LHFPL5); R604X (P = 0.0156 for TMC1 and 0.0004 for LHFPL5); n = 3.
The yeast two-hybrid assay results (Fig. 1) suggested that TMC1 potentially interacts directly with LHFPL5 because yeast cells are highly unlikely to express a protein that bridges two mammalian proteins. This notion of direct binding was further tested using purified TMC1 and LHFPL5. For such assays, both TMC1 and LHFPL5 should preferably be generated in and purified from bacteria for a “pairwise” pulldown (15, 16); however, TMC1 generated in bacteria was severely degraded. Thus, as an alternative, we used TMC1 purified from HEK293T cells; the protein was of high purity (SI Appendix, Fig. S3A), and, importantly, the purified TMC1 interacted with LHFPL5-GST but not with GST alone (SI Appendix, Fig. S3C). These data further support the direct interaction of TMC1 and LHFPL5.
Deafness Mutations D572N/H of TMC1 Impaired TMC1-LHFPL5 Interaction and Mutual Stabilization In Vitro.
Interestingly, hTMC1-F3 harbors at least seven deafness mutations (Fig. 2A) (13, 17–21), but how these mutations affect TMC1 normal function and impair hearing remain largely unexplored (22). Considering our aforementioned results (Fig. 1), we hypothesized that one of the deafness mutations might undermine LHFPL5 binding and thereby diminish TMC1-LHFPL5 mutual stabilization. Thus, we examined the impact of all seven mutations on LHFPL5 binding and TMC1-LHFPL5 mutual stabilization (Fig. 2A). Wild-type (WT) TMC1 and LHFPL5 are widely recognized as trapped in the endoplasmic reticulum (ER) in heterologous expression systems (7, 23, 24); similar to WT TMC1, all ectopically expressed TMC1 mutants were also retained in the ER (SI Appendix, Fig. S4 A and C). Moreover, coexpression of WT and D572N TMC1 with LHFPL5 did not alter the predominant ER localization of the TMC1 proteins (SI Appendix, Fig. S4B). Notably, whereas the mutations M486T, P514L, and C515R exerted little effect, D572N or D572H potently weakened LHFPL5 binding and the mutual stabilization (Fig. 2 B–D and G and H); conversely, both W588X and R604X markedly enhanced LHFPL5 binding and the mutual stabilization (Fig. 2 E and G and H). These results bolstered the notion that TMC1-LHFPL5 mutual stabilization depends on the physical interaction between the proteins. Why W588X and R604X produced the enhancing effect cannot be readily explained because this effect appears contrary to their deafness-causing phenotype; however, one possibility is that the large truncations caused by these nonsense mutations drastically alter TMC1 tertiary structure and thus impair TMC1 channel (or other) function while concurrently increasing TMC1 accessibility for LHFPL5 binding. Collectively, our results suggested that the D572N/H mutations cause deafness potentially by disrupting LHFPL5 binding and impairing TMC1-LHFPL5 mutual stabilization.
To determine whether D572N/H mutations adversely affect LHFPL5 binding because of the loss of the negative charge or the gain of residue bulkiness, we tested D572E and D572A mutations in our experiments (Fig. 2F); here, LHFPL5 and WT/mutant TMC1 were expressed in separate cells, and then the lysates of the cells were mixed and used in co-IP assays. We used this strategy because, in the absence of coexpression in the same cells of LHFPL5, which modulates the expression of WT and mutant TMC1 differentially, WT and mutant TMC1 were expressed at equal levels; this allowed us to not only compare the LHFPL5 binding of the WT and mutant proteins in a direct and unbiased manner, but also to avoid the technically challenging equalization of TMC1 protein amounts for Western blotting after co-IP. We found that both D572E and D572A mutations disrupted LHFPL5 binding (Fig. 2F). Like aspartate (D), glutamate (E) carries a negative charge but is slightly bulkier, whereas alanine (A) is less bulky but carries no charge. According to these data, both the charge and the size of D572 appear to be critical for LHFPL5 binding.
Technical Challenges and Our Solution for Examining TMC1-LHFPL5 Interaction In Vivo.
Protein-protein interactions (PPIs) observed in heterologous expression systems (Figs. 1 and 2) are prone to artifacts and require substantiation through studies on endogenous proteins. Conventional Western blotting and co-IP (pulldown) assays, two powerful and widely used techniques for detecting the expression of specific proteins and PPIs, typically require ∼104 to 105 cells (25) and ∼105 to 106 cells in a single assay, respectively. However, these conventional methods cannot be used for studying rare cells such as hair cells except when the target proteins are highly abundant (e.g., actin and prestin); this is because hair cells are extremely scarce: cochlear hair cells number ∼3,300 per mouse cochlea (26) and ∼15,000 per human cochlea (27). To study PPIs in hair cells and other rare cells, we developed the ultrasensitive microbead-based SiMPull technique. The method, which is described in detail in the study by Zhao et al. (12), was next used to assess TMC1-LHFPL5 interaction in hair cells.
To ensure that PPIs are tested rigorously in the microbead-based SiMPull assay, both the bait protein and the prey protein must be targeted by two antibodies that bind to distinct, nonoverlapping epitopes: one of the antibodies is for capturing the targeted protein onto the beads, and the other is for identifying the protein on the beads. We achieved this as follows: For TMC1, we used a mouse line carrying FLAG-tagged TMC1; in this line, which we previously generated in our laboratory, TMC1 can be recognized by both anti-FLAG antibody and a homemade anti-TMC1 antibody (11). For LHFPL5, we engineered an HA tag at the C terminus of mouse LHFPL5 by employing clustered regularly interspaced short palindromic repeat/CRISPR-associated protein 9 (CRISPR/Cas9) techniques (Fig. 3 A–C). First, the HA tag did not appear to affect LHFPL5 function in hearing, as per the results of auditory brainstem response (ABR) and distortion product otoacoustic emission (DPOAE) assays (Fig. 3 D–G). Second, LHFPL5-HA localization was examined in Lhfpl5HA/HA mice by using an anti-HA antibody; as in previous studies (7, 11), LHFPL5 puncta were detected in both outer hair cells (OHCs) and inner hair cells (IHCs) on postnatal day (P) 5 mice (Fig. 3 H and I). These results indicate that LHFPL5-HA functions similarly to WT LHFPL5. Third, a rabbit antibody against LHFPL5 was generated, and the avidity and specificity of the antibody were assessed using Western blotting and immunocytochemistry; in both assays, the antibody recognized LHFPL5 in COS7 cells and hair cells and exhibited little nonspecific binding (SI Appendix, Fig. S5). Thus, the anti-LHFPL5 antibody and anti-HA were used for microbead-based SiMPull of LHFPL5-HA.
Fig. 3.
Generation and characterization of Lhfpl5HA/HA mice. (A–C) Schematic of CRISPR/Cas9-based generation of a mouse line carrying an HA tag before the stop codon of Lhfpl5. Insertion of the HA tag was verified through DNA sequencing of homozygous Lhfpl5HA/HA mice (B). Lhfpl5HA mice were routinely PCR genotyped, wherein the HA-tag insertion resulted in a 27-nt shift in PCR products from the Lhfpl5HA mouse (C). Oligo donor, DNA oligo for precisely inserting the HA tag before the stop codon by using homology-directed repair; ORF, open reading frame; UTR, untranslated region. (D and E) Representative ABR traces elicited by click stimuli (D) and ABR thresholds in response to pure tones (E) in 1-mo-old Lhfpl5+/+ and Lhfpl5HA/HA mice (n = 3 each). (F and G) Representative DPOAE traces at L1 = L2 = 60 dB (E) and DPOAE thresholds across the 8- to 32-kHz range (F) in 1-mo-old Lhfpl5+/+ and Lhfpl5HA/HA mice (n = 6 each). (H and I) LHFPL5-HA immunostaining (green, anti-HA antibody) and actin labeling (red, phalloidin) in hair bundles of outer (H) and inner (I) hair cells in P5 Lhfpl5+/+ and Lhfpl5HA/HA mice. (Scale bar, 5 μm.)
TMC1 Physically Interacted with LHFPL5 in Cochlear Hair Cells.
TMC1-LHFPL5 interaction was examined in Tmc1FLAG/FLAG; Lhfpl5+/+ mice by using the microbead-based SiMPull assay (Fig. 4A). When LHFPL5 was immunoprecipitated, TMC1-FLAG was also captured (Fig. 4 B–E, I and II); here, when anti-FLAG was not included in the immunostaining, almost all of the signal was eliminated, which indicated that the signal was from anti-FLAG binding to the microbeads rather than from nonspecific binding of the second antibody. Furthermore, anti-FLAG specifically recognized TMC1 rather than some other nonspecific protein in the microbead-based SiMPull assay (Fig. 4 F–H) (SI Appendix, Fig. S6). Taken together, these results confirmed that the signal detected in SiMPull (shown in Fig. 4 B, II) is the genuine TMC1 signal.
Fig. 4.
TMC1 physically interacts with LHFPL5 in cochlear hair cells. (A) Experimental setup (schematic) and selection of imaging area for analysis (A′–C′) of microbead-based SiMPull assay. NeutrAvidin-coated agarose microbeads were modified by immobilizing biotinylated second antibody and anti-LHFPL5 antibody on their surface for capturing LHFPL5-HA (or LHFPL5) and TMC1-FLAG. TMC1-FLAG was immunostained using anti-FLAG and a fluorescently labeled second antibody. An imaging area of 25 × 25 μm (C′, region of interest [ROI] indicated in B′) was selected from a microbead (A′, bright field; B′, fluorescence image) for the analysis shown in B; imaging areas were similarly selected for analysis in other SiMPull assays. (Scale bars, 20 μm [A′ and B′] and 5 μm [C′].) (B and C) Representative results of microbead-based SiMPull assay on cochlear lysates of Tmc1FLAG/FLAG; Lhfpl5+/+ mice (groups I–II) and Tmc1FLAG/FLAG; Lhfpl5−/− mice (groups III–IV); B, G, and I show 25- × 25-μm imaging areas selected from microbeads (diameter: 40 to 70 μm) (see additional details in ref. 12). Cochlear lysates were immunoprecipitated using anti-LHFPL5 antiserum and immunostained with (II and IV) or without (I and III) anti-FLAG (TMC1), plus the second antibody (Alexa Fluor 488-conjugated goat anti-mouse IgG). Part of the cochlear lysates was subject to conventional Western blotting for prestin, an OHC marker used here as a loading control (C): Tmc1FLAG/FLAG; Lhfpl5+/+ (left lane) and Tmc1FLAG/FLAG; Lhfpl5−/− (right lane). IP, immunoprecipitation; IS, immunostaining. (Scale bar, 5 μm.) (D and E) Average signal intensity of an imaging area similar to that in B in seven randomly selected beads in the same experiment (D). A.U., arbitrary unit. (E) Summary of difference between the average signal intensity of groups II and I and groups IV and III from three independent biological replicates similar to B. Groups II and IV are normalized to groups I and III, respectively. ***P < 0.001, N.S., no significant difference. (F–H) Representative (G) and summary (H) results of the microbead-based SiMPull assay on cochlear lysates of Tmc1+/+ and Tmc1FLAG/FLAG mice. Cochlear lysates were immunoprecipitated using anti-TMC1 antiserum (SI Appendix, Fig. S6) and immunostained with anti-FLAG plus second antibody (Alexa Fluor 488-conjugated goat anti-mouse IgG). (Scale bar, 5 μm.) Part of the cochlear lysates was subject to conventional Western blotting for the loading control prestin (F) Tmc1WT/WT (left) and Tmc1FLAG/FLAG (right). G shows a 25- × 25-μm imaging area selected from a microbead. (H) Summary of signal intensity from three independent biological replicates similar to G. *P = 0.0196. The signal intensity of one biological replicate is the average signal intensity of an imaging area similar to that shown in F in seven randomly selected beads in the same experiment (Inset). ***P < 0.001. (I and J) Representative (I) and summary (J) results of a microbead-based SiMPull assay on cochlear lysates from Tmc1FLAG/FLAG; Lhfpl5HA/HA double-knockin mice. Cochlear lysates were immunoprecipitated using anti-LHFPL5 antiserum and subsequently immunostained without first antibody (I) or with anti-HA for LHFPL5 (II) or anti-FLAG for TMC1 (III), plus second antibody (Alexa Fluor 488-conjugated anti-mouse IgG). (Scale bar, 5 μm.) (J) Summary of signal intensity from three independent biological replicates. Different from group I: **P = 0.0045, *P = 0.0136; all values are normalized to group I. The signal intensity of one biological replicate is the average signal intensity of an imaging area similar to that shown in I in seven randomly selected beads in the same experiment (Inset). Different from group I: ***P < 0.001; n = 7. Mouse ages are indicated in the top-right corner of images in B, G, and I.
Importantly, knocking out LHFPL5 eliminated the TMC1 signal (Fig. 4 B–E, III and IV), which verified that the TMC1 signal was due to the capture of TMC1 by LHFPL5 and not because of nonspecific TMC1 binding to the beads. Accordingly, the results of another set of experiments performed on Tmc1FLAG/FLAG; Lhfpl5HA/HA double-knockin mice (generated by mating Tmc1FLAG/FLAG and Lhfpl5HA/HA mice) showed that LHFPL5 was pulled down by anti-LHFPL5 antiserum (used in Fig. 4B) together with TMC1 (Fig. 4 I and J). In a set of reciprocal pulldown experiments, anti-TMC1 antiserum pulled down TMC1 together with LHFPL5, further supporting the physical interaction of the two proteins (SI Appendix, Fig. S7).
Finally, the TMC1-LHFPL5 interaction was also examined using the proximity ligation assay (PLA), which is a commonly used sensitive approach for revealing in situ PPIs. Because the assay requires a pair of primary antibodies raised in different species, we used mice carrying both TMC1-FLAG and LHFPL5-HA for the PLA; this enabled us to use rabbit anti-LHFPL5 (or anti-TMC1) paired with mouse anti-FLAG (or anti-HA). We found that both the hair bundles and the soma of hair cells showed the PLA signal (SI Appendix, Fig. S8), which supported the notion that TMC1 and LHFPL5 interact at these locations.
TMC1 Was Stabilized by LHFPL5 in Cochlear Hair Cells.
To investigate whether LHFPL5 stabilizes TMC1 in vivo, we quantified TMC1 protein levels in Lhfpl5−/− mice by using microbead-based SiMPull; P7 mice were used to avoid the hair-cell degeneration that starts after P10 in Lhfpl5−/− mice (28). We pulled down TMC1-FLAG from Tmc1FLAG/FLAG; Lhfpl5+/+ and Tmc1FLAG/FLAG; Lhfpl5−/− littermates by using the anti-TMC1 antiserum and then detected TMC1-FLAG by using anti-FLAG. TMC1 was pulled down by anti-TMC1 from Tmc1FLAG/FLAG; Lhfpl5+/+ mice (Fig. 5 A, II), and anti-FLAG exclusion in immunostaining eliminated the signal (Fig. 5 A, I), which indicated that the signal was due to anti-FLAG specifically binding to TMC1 (Fig. 4 G and H). Notably, the TMC1 protein level markedly decreased in Tmc1FLAG/FLAG; Lhfpl5−/− mice (Fig. 5 A–D). These results indicate that LHFPL5 stabilizes TMC1 in cochlear hair cells.
Fig. 5.
TMC1 is stabilized by LHFPL5 in cochlear hair cells. (A and B) Representative results of a microbead-based SiMPull assay on P7 Tmc1FLAG/FLAG; Lhfpl5+/+ mice (groups I and II) and Tmc1FLAG/FLAG; Lhfpl5−/− mice (groups III and IV). Mouse cochlear lysates were immunoprecipitated using anti-TMC1 antiserum and subsequently immunostained (IS) with anti-FLAG (TMC1) antibody plus second antibody (Alexa Fluor 488-conjugated anti-mouse IgG); groups I and III: controls, IS without first antibody. (A) A 25- × 25-μm imaging area selected from a microbead. (B) Part of the cochlear lysates was subject to conventional Western blotting for prestin (OHC marker used as loading control): Tmc1FLAG/FLAG; Lhfpl5+/+ (left lane) and Tmc1FLAG/FLAG; Lhfpl5−/− (right lane). (Scale bar, 5 μm.) (C) Average signal intensity of an imaging area similar to that shown in A in seven randomly selected beads in the same experiment. ***P < 0.001. (D) Summary of difference between average signal intensity of groups II and I and groups IV and III from three independent biological replicates similar to A. All values are normalized to “group II – group I.” ***P < 0.001.
TMC1 expression in the hair bundles of hair cells in Lhfpl5−/− mice was also examined using immunocytochemistry. In P22 Lhfpl5−/− mice, hair bundles in OHCs and IHCs were deformed as reported previously (7), and the TMC1 signal was detected only weakly in stereocilia as compared with what was observed in Lhfpl5+/+ mice (SI Appendix, Fig. S9); this is consistent with published findings (9). Moreover, we observed a substantial reduction of the TMC1 signal in the soma in OHCs and IHCs in P7 Lhfpl5−/− mice (SI Appendix, Fig. S9). These results agree with the notion that LHFPL5 interacts with and stabilizes TMC1 not only in the hair bundle but also in the soma, which is in accord with the PLA results (SI Appendix, Fig. S8).
Deafness Mutation D569N in Mouse TMC1 Disrupted TMC1-LHFPL5 Interaction in Cochlear Hair Cells.
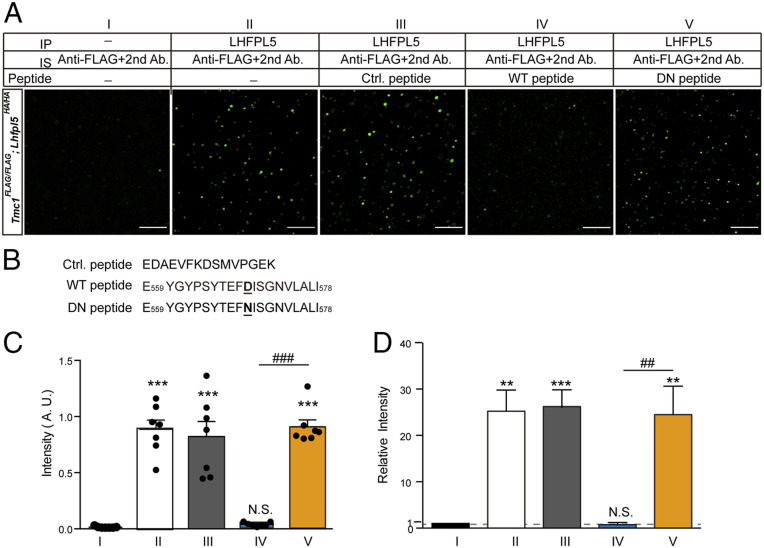
Deafness mutation D572N in hTMC1 disrupted LHFPL5 binding in vitro (Fig. 2); thus, we sought to determine whether this disruption also occurs in vivo. We tested how the D569N mutation in mouse TMC1 (mTMC1), which is homologous to D572N in hTMC1, affects LHFPL5 binding in cochlear hair cells. We reasoned that a short peptide containing D569 might compete with full-length TMC1 and disrupt TMC1-LHFPL5 interaction. Accordingly, addition of a 20-aa peptide containing D569 (WT peptide) almost eliminated TMC1-LHFPL5 interaction in the microbead-based SiMPull assay (Fig. 6), and this effect was specific because a peptide derived from TRPV1 failed to impair TMC1-LHFPL5 interaction (Fig. 6). Notably, mutating D569 to asparagine (N) eliminated the disrupting effect of the WT peptide (Fig. 6), which clearly suggests that D569 in TMC1 is essential for binding to LHFPL5 in cochlear hair cells.
Fig. 6.
D569 in mTMC1 (D572 in hTMC1) is critical for TMC1-LHFPL5 interaction in mouse cochlear hair cells. (A) Representative results of microbead-based SiMPull assay on cochlear lysates of Tmc1FLAG/FLAG; Lhfpl5HA/HA double-knockin mice in the absence of any peptide (I and II) or in the presence of control (III), WT (IV), or DN (V) peptide. Cochlear lysates were immunoprecipitated using anti-LHFPL5 antiserum and immunostained using anti-FLAG (TMC1) and second antibody (Alexa Fluor 488-conjugated anti-mouse IgG). Group I: control, anti-LHFPL5 antiserum omitted in IP. (Scale bar, 5 μm.) (B) Amino acid sequences of peptides used in the assay shown in A. Ctrl. peptide, a fragment of rTRPV1 used as a control; WT peptide, aa 559 to 578 of WT mTMC1, including D569 (underlined); DN peptide, aa 559 to 578 of D569N mTMC1. (C and D) Average signal intensity of a 25- × 25-μm imaging area similar to that shown in A in seven randomly selected beads in the same experiment (C). Different from group I: ***P < 0.001. (D) Summary of average signal intensity of three independent biological replicates similar to A; all values are normalized to group I. Different from group I: **P < 0.0017, ***P < 0.001, N.S., no significant difference. C and D, different from group IV: ###P < 0.001, ##P = 0.0017. One-way ANOVA with Tukey’s post hoc test was used for statistical analysis.
Discussion
Beurg et al. observed that LHFPL5 knockout led to drastic TMC1 reduction in the hair bundle of mouse auditory hair cells (9); this implied a functional interaction between TMC1 and LHFPL5, but no physical interaction between the two proteins was detected in the study (9). By contrast, we found in the present study that TMC1 physically interacted with LHFPL5 in both heterologous expression systems and hair cells by using multiple approaches: the yeast two-hybrid assay, conventional co-IP assays, pulldown assays performed using purified proteins, PLA, and, most importantly, our microbead-based SiMPull assay. We further demonstrated that the hTMC1-F3 fragment (aa 462 to 634) bound to LHFPL5 (Fig. 1) and, more specifically, that D572 in hTMC1-F3 (homologous to D569 in mTMC1) was critical for LHFPL5 binding: the naturally occurring deafness mutation D572N disrupted LHFPL5 binding both in heterologous expression systems and in hair cells (Figs. 2 and 6). Our results also indicate that the C-terminal half of LHFPL5 interacted with TMC1 (Fig. 1); conversely, the N-terminal half has been reported to engage in PCDH15 binding and LHFPL5 dimerization (29). The C-terminal part of LHFPL5 harbors several deafness mutations (28, 30, 31), but these are all autosomal-recessive mutations, which implies that the mutations likely damage the synthesis and trafficking of LHFPL5 rather than its TMC1 binding (see the 3rd and 4th sections in Discussion). In heterologous expression systems, the TMC1-LHFPL5 interaction appeared to occur in the ER because TMC1 was retained in the ER (SI Appendix, Fig. S4). Our PLA results also suggest that TMC1 probably interacts with LHFPL5 in both the MT complex and the ER in hair cells (SI Appendix, Fig. S8). Although interactions between various components of the MT complex have been reported, how all of the components are organized in the entire complex remains poorly understood. Our finding of TMC1-LHFPL5 interaction identifies a missing link in the currently known physical organization of the MT macromolecular complex.
Functionally, LHFPL5 binding was found here to stabilize global TMC1 expression in heterologous expression systems and in hair cells. In heterologous expression systems, TMC1 expression dropped by 77% in the absence of LHFPL5 (Fig. 1). Similarly, in microbead-based SiMPull assays, global TMC1 expression was measured to be decreased by 56% in LHFPL5 knockout hair cells (Fig. 5), and TMC1 staining was markedly diminished in the hair bundle in LHFPL5 knockout hair cells (SI Appendix, Fig. S9), which agrees with a previous report (9). Most notably, the D572N deafness mutation that disrupted LHFPL5 binding led to an ∼50% reduction in TMC1 expression in heterologous expression systems (Fig. 1). Intriguingly, Beurg et al. recently reported that the TMC1 immunostaining signal decreased by ∼66% in the hair bundle in homozygous D569N TMC1 mice and, accordingly, that the mechanoelectrical transduction (MET) current was also diminished by two-thirds in the mutant mice (22). These findings agree well with our hypothesis that D572 in hTMC1 is critical for LHFPL5 binding and TMC1 stabilization. We found that, in LHFPL5 knockout mice, 44% of TMC1 remained relative to the level in WT mice (Fig. 5); this finding indicates that LHFPL5 only modulates TMC1 stability and does not act as the sole determinant of TMC1 stability and further that TMC1 also interacts with other binding partners (and presumably performs other functions). This is consistent with our previous observation that TMC1 and LHFPL5 do not unfailingly colocalize in hair bundles (11).
Among >30 pathogenic mutations in hTMC1, M418K (equivalent to mouse Bth mutation or to G417R that probably causes an effect similar to M418K) and D572N/H are the only three dominant-negative mutations (32, 33). However, the mechanism by which the dominant and recessive mutations lead to abnormal TMC1 function and regulation remains mostly unexamined. In principle, a dominant-negative deafness mutation such as D572N could adversely affect the hearing function of WT TMC1 if the D572N mutant can still interact with the same components as WT TMC1 but block certain aspects of WT TMC1 function. The adverse effect could occur at the MT complex because D569N TMC1 was found to support MET channel function and was apparently transported to the cell surface of mouse hair cells and assembled into the MT complex (9, 22). The impaired interaction of D569N TMC1 with LHFPL5 resulted in diminished TMC1 stability (Figs. 5 and 6), which could potentially account for the MET current reduction in D569N TMC1 mice observed by Beurg et al. (22). Because the impaired LHFPL5 interaction appeared to also cause a loss of the tonotopic gradient of the MET current of OHCs in D569N TMC1 mice (22), it will be intriguing to examine whether a tonotopic gradient of LHFPL5 expression dictates the tonotopic gradient of TMC1 and the MET current (34). Thus, the tonotopic gradient of TMC1 and LHFPL5 expression could be assessed in WT and LHFPL5 knockout mice by using microbead-based SiMPull (Fig. 4). Interestingly, the D569N mutation also lowered the Ca2+ permeability of the MET channel but exerted no effect on the unitary conductance of the channel (22). On the basis of homology modeling, D569 was suggested to reside in the channel pore region of TMC1 (4, 14). Presumably, both diminished TMC1 stability and decreased Ca2+ permeability of the MET channel contribute to progressive hearing loss and, ultimately, to the destruction of D569N TMC1-expressing hair cells (22). Alternatively, D569N TMC1 might adversely affect WT TMC1 at an early point in the trafficking pathway by impairing TMC1 cell-surface trafficking or stability in the cytoplasm.
In contrast to dominant mutations, recessive mutations would be expected to nullify TMC1 function by completely impairing TMC1 folding, trafficking, or assembly into the MT complex, and thus the recessive mutations should exert no effect on WT TMC1 function in the MT complex. We also examined five recessive TMC1 mutants and found that they all maintained the ability to interact with LHFPL5, at least in heterologous expression systems (Fig. 2). Why these mutations display recessive phenotypes and do not interfere with WT TMC1 function is unclear. It appears unlikely that the mutant proteins are not synthesized or are completely degraded due to severe misfolding in hair cells because the mutants were expressed at least in heterologous systems. Another possibility is that the effect of the TMC1 mutation manifests itself only at the MT complex, but the cell-surface trafficking of the mutants or their assembly into the MT complex could be completely disrupted, similar to what occurs with the ∆F508 mutant of the CFTR channel: that is, deletion of a single amino acid (phenylalanine 508) leads to misfolding of the CFTR channel, and the mutant channel is trapped in the ER and does not reach the cell surface (35). It would be of interest to examine these possibilities in the future.
Our microbead-based SiMPull technique (12) was instrumental in the in vivo analysis of the expression and interaction of TMC1 and LHFPL5 in this study. Several components of the MT complex in hair cells have been identified in the last 10 y, and we can reasonably expect additional components to be identified in future studies. However, biochemical investigation of MT-complex components is in its infancy when compared with the considerably more sophisticated physiological, genetic, and cell biology studies on this complex that have been conducted to date. Because of the scarceness of the raw material (hair cells), previous biochemical studies of MT-complex components have relied mainly on heterologous overexpression systems; however, these systems are prone to artifacts, and the obtained results carry little weight (in most research fields) in the absence of validation through studies of endogenous proteins. The microbead-based SiMPull method (12) substantially simplifies the original SiMPull assay invented by Jain et al. (36) and makes the application of the powerful SiMPull technique in regular laboratories considerably more feasible. Notably, SiMPull potentially can also be used for studying the stoichiometry and the interaction kinetics of MT-complex proteins (12). Our hope is that this technique will markedly facilitate the biochemical investigation of MT-complex proteins and other critical nonabundant proteins in hair cells.
Materials and Methods
Antibodies and Phalloidin.
The following primary antibodies were used in this study: rabbit anti-TMC1 serum (homemade, against N-terminal 39-aa residues of hTMC1) (11), rabbit anti-LHFPL5 serum (homemade, against C-terminal aa 200 to 219 of human LHFPL5), mouse anti-FLAG (F1804; clone M2; Sigma-Aldrich), mouse anti-HA (MMS-101p; Covance), and goat anti-prestin (sc-22692; Santa Cruz Biotechnology). The second antibodies used were Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (ab150113; Abcam), Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody (ab150077; Abcam), and biotinylated goat anti-rabbit IgG antibody (65-6140; Thermo Fisher Scientific). Alexa Fluor 647-conjugated phalloidin (A22287; Thermo Fisher Scientific) was used for labeling F-actin. The anti-LHFPL5 serum was produced by immunizing rabbits housed in the animal care facility at the Hong Kong University of Science and Technology (ethics protocol number 2016040).
Complementary DNAs and Plasmids.
Human LHFPL5 and hTMC1 complementary DNAs (cDNAs) were purchased from the DNASU Plasmid Repository. For generating mammalian expression constructs, the cDNAs were amplified and subcloned into the pcDNA3 expression vector. The TMC1 fragments TMC1-F1 (aa 1 to 199), TMC1-F2 (aa 200 to 461), TMC1-F3 (aa 462 to 634), and TMC1-F4 (aa 635 to 760) were generated from full-length hTMC1 and inserted into the pcDNA3 expression vector. TMC1 point mutations—M486T, P514L, C515R, D572N/H/A/E, W588X, and R604X—were generated through site-directed mutagenesis from pcDNA3-FLAG-TMC1. The LHFPL5 fragments LHFPL5-1–121 and LHFPL5-122–219 were generated by using PCR from pcDNA3-LHFPL5-HA.
Yeast Two-Hybrid Screening.
A GAL4-based yeast two-hybrid system (Invitrogen) was used to screen for and analyze PPIs in yeast, following previously published procedures (37) with some modifications. The TMC1-F3 fragment was cloned into pDEST32 to generate the bait plasmid pDEST32-TMC1-F3, which contained an in-frame fusion of the GAL4 DNA-binding domain; the prey vector pDEST22 contained human cDNA collections in-frame fused to the GAL4-activating domain (Invitrogen). With the empty pDEST22 plasmid being used as a negative prey control, yeast two-hybrid screening was performed by transforming Mav203, the yeast strain harboring the bait vector pDEST32-TMC1-F3, with the prey vectors encoding a human lung cDNA expression library. Similar procedures were used for the TMC1-F1, TMC1-F2, and TMC1-F4 fragments. Yeast transformants were first grown on SD2 (Leu- and Trp-deficient) agar plates for selection of yeast cells containing both bait and prey vectors and then transferred to SD4 (Leu-, Trp-, His-, and Ura-deficient) plates to screen for proteins that potentially interact with the TMC1 fragment. Colonies that grew on the SD4 plates were picked and streaked onto another SD4 plate. The colonies that grew in SD4 were scored as “positive” colonies, and the prey vectors were recovered from these colonies and sequenced after amplification in Escherichia coli. Typically, each interaction was confirmed by transforming yeast Mav203 cells with the indicated bait and prey vectors and then allowing the transformants to grow on SD2 or SD4 agar plates for ∼3 d at 30 °C. Images of the colonies on both plates were recorded.
Cell Culture and Transfection.
HEK293T and COS7 cell lines were purchased from American Type Culture Collection; the cells were presumably authenticated by ATCC and were not further authenticated in this study. The cell lines, which routinely tested negative for mycoplasma contamination, were maintained in Dulbecco’s Modified Eagle Media (DMEM) supplemented with 10% fetal bovine serum and 100 U/mL penicillin and streptomycin in an atmosphere of 95% air/5% CO2 at 37 °C. All transfections were performed using polyethylenimine (23966-2; Polysciences), and the culture medium was changed to Opti-MEM during transfection. After 8 h of transfection, cells were cultured in normal DMEM and cultured for another 48 h before harvest.
Conventional Western Blotting and co-IP.
Western blotting and co-IPs were performed using procedures described previously (15) with minor modifications. For co-IPs, cells were plated onto 60-mm dishes and transfected 48 h before the assays. Cells were first lysed using a co-IP buffer (15) supplemented with proteinase inhibitors on ice for 30 min. Subsequently, the lysates were incubated with 2 μL of anti-TMC1 antiserum, anti-LHFPL5 antiserum, or anti-FLAG antibody at 4 °C for 2 h, and the mixtures were incubated with 30 μL of protein G agarose beads (17-0618-01; GE Healthcare) at 4 °C overnight with moderate shaking. Finally, the beads were washed thrice with the co-IP buffer, and the captured proteins were eluted by incubating the beads with 1× sodium dodecyl sulfate/polyacrylamide gel electrophoresis loading buffer for 2 h at room temperature and then immunoblotted. Protein bands in Western blots were quantified using Fiji software (https://fiji.sc/).
Purification of TMC1.
For purifying TMC1, we exploited the extremely high-affinity interaction of NeutrAvidin with Avi-tagged TMC1 that could be biotinylated in cells; this interaction can withstand high-stringency washing during purification and thus is desirable for obtaining high-purity and high-yield TMC1. We tagged hTMC1 with tdTomato-HA-HRV3C cleavage site-Avi at the C terminus and cloned it into an internal ribosomal entry site-containing bicistronic pcDNA3 vector that expresses codon-optimized hBirA biotin ligase separately. After transfection with the TMC1 expression vector for 48 h, HEK293T cells (8× 150-mm plates, cells at ∼80% confluence) were washed with phosphate-buffered saline (PBS) and suspended by gentle pipetting in 4 mL of PBS containing 1 tablet/10 mL protease-inhibitor mixture (11836153001; cOmplete Mini, Sigma-Aldrich) and 1 mM dithiothreitol. The cell suspension was mixed with 1% (vol/vol) Triton X-100, sonicated for 5 min in an ice bath, and gently stirred at 4 °C for 1 h for adequate cell lysis. After centrifugation at 15,700 × g at 4 °C for 15 min to remove insoluble cell debris, the cell lysate was mixed with NeutrAvidin agarose beads (29201, Thermo Scientific); the beads (80 μL) were prewashed four times with 1 mL of PBS and blocked with 1 mL of PBS containing 1% (wt/vol) BSA for 45 min at room temperature. The mixture of the cell lysate and beads was gently stirred at 4 °C for 2 h, and additional BSA was added to maintain a 1% (wt/vol) final concentration of BSA.
Subsequently, the beads were harvested through brief centrifugation and washed six times with 1.5 mL of PBS containing 0.2% (vol/vol) Nonidet P-40 and then two times with 1 mL of the co-IP buffer. The washed beads were resuspended in 200 μL of the co-IP buffer, mixed with 1 μL (2 U) of GST-tagged HRV3C protease (88946, Thermo Scientific), and gently stirred at 4 °C overnight. After removing the NeutrAvidin beads by centrifugation, the supernatant containing cleaved and biotinylated TMC1-tdTomato-HA was mixed with glutathione-Sepharose beads (17-0756-01; Sigma-Aldrich) and stirred gently at 4 °C for 1 h to remove GST-tagged HRV3C protease; the glutathione beads (50 μL) were washed four times with 1 mL of PBS and once with 1 mL of the co-IP buffer at room temperature. After removing the glutathione beads by centrifugation, the supernatant containing purified TMC1-tdTomato-HA was stored at –20 °C for up to 1 wk until use. All procedures were performed on ice unless indicated otherwise.
Purification of LHFPL5-GST and GST Pulldown Assays.
GST and LHFPL5-GST fusion proteins were produced in E. coli and purified as previously described (16). GST and LHFPL5-GST proteins (5 μg) were incubated with 30 μL of glutathione-Sepharose beads for 2 h at 4 °C in the co-IP buffer and then centrifuged at 500 × g for 1 min to pellet the beads. After washing once with 500 μL of the co-IP buffer, the beads were incubated with purified TMC1 at 4 °C overnight. Finally, the beads were washed three times (500 μL each time) with the co-IP buffer, and the proteins bound on the beads were eluted using 60 μL of 1× SDS gel loading buffer and analyzed using Western blotting.
Mice.
Tmc1FLAG mice and Lhfpl5− mice were generated using CRISPR/Cas9 genome-editing techniques (11). All animal procedures were approved by the University Committee on Research Practices at the Hong Kong University of Science and Technology (ethics protocol numbers 2015056 and 2018037).
Generation of LHFPL5-HA Knockin Mice by Using CRISPR/Cas9 Techniques.
The CRISPR/Cas9 technique was used to generate LHFPL5-HA knockin mice (Lhfpl5HA), which were maintained on the CBA mouse line.
Briefly, one pair of oligonucleotides for LHFPL5-HA single-guide RNA (sgRNA) (5′-caccggctgctggtcttagctcaag-3′ and 5′-aaaccttgagctaagaccagcagc-c-3′) was annealed and ligated into pX330 plasmid (Addgene) digested with BbsI (R0539S; New England Biolabs). Cas9 and the selected sgRNA were PCR amplified using the following in vitro transcription (IVT) primers: for LHFPL5-HA sgRNA—forward, 5′-ttaatacgactcactatagggctgctggtcttagctcaag-3′, and reverse, 5′-aaaagcaccgactcggtgcc-3′; for Cas9—forward, 5′-taatacgactcacta-tagggagaatggactataaggaccacgac-3′, and reverse, 5′-gcgagctctaggaattcttac-3′. Following published experimental procedures (38), the sgRNA and Cas9 were transcribed in vitro by using a Mega Short Script T7 Kit and a mMESSAGE mMACHINE T7 ULTRA Kit (AM1354 and AM1345; Thermo Fisher Scientific), respectively. All IVT products were purified using a Megaclear Kit (AM1908; Thermo Fisher Scientific). The purified sgRNA and Cas9 messenger RNA (mRNA) together with an oligo donor (synthesized by Integrated DNA Technologies) were mixed and diluted with RNase-free water to these final concentrations: 200 ng/μL single-stranded DNA, 100 ng/μL Cas9 mRNA, and 50 ng/μL sgRNA. The sequence of the oligo donor was as follows: 5′-ggctgtgggtggagaagtggctgcggtggtggctctctgaagccaagtgtgtctgggcctcttacccatacgatgttccagattacgcttgagctaagaccagcagccaaacctgtcctccctgagaaaggccatcagcggagtgttcaaagc-3′. The mixture was injected into mouse embryos by trained personnel at the Animal and Plant Care Facility of the Hong Kong University of Science and Technology.
Genotyping of Lhfpl5HA Mice.
Genomic DNA was extracted from mouse ears by incubating samples at 65 °C for 3 h in a buffer containing 10 mM Tris, pH 8.0, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.2% (wt/vol) Triton X-100, and 200 μg/mL Proteinase K; the samples were then heated for 10 min at 95 °C to deactivate Proteinase K. The DNA extract was subject to PCR by using the primers 5′-gcggtggtggctctctgaagccaag-3′ (sense) and 5′-catgacttaggccagcatcagtgccac-3′ (antisense), Pfu DNA polymerase, and the following reaction conditions: 5 min at 98 °C, followed by 32 cycles of 20 s each at 98 °C, 60 °C, and 68 °C and then a final extension for 5 min at 68 °C. This generated PCR products that were 241-nucleotides (nt) long (for Lhfpl5+) and 268-nt long (for Lhfpl5HA) and were clearly separated in DNA electrophoresis performed using 2% (wt/vol) agarose gels.
ABR and DPOAE Assays.
ABR and DPOAE measurements were performed on 4- to 5-wk-old mice, following previously published procedures (11).
Immunohistochemical Assays.
Cultured cells.
At 48 h after transfection with protein-expression vectors, COS7 cells were fixed with 4% paraformaldehyde (PFA) for 10 min, washed once with PBS, permeabilized, and blocked with PBS containing 0.2% Triton X-100 and 3% (wt/vol) BSA for 30 min at room temperature. Subsequently, the cells were incubated with mouse anti-FLAG/anti-HA antibodies or anti-LHFPL5 antiserum (all diluted 1:100) for 2 h at room temperature, washed three times with PBS, and then incubated with Alexa Fluor 488- or 647-conjugated goat anti-mouse or anti-rabbit IgG (1:500) and DAPI (1:2,000) for 1 h at room temperature. Finally, the cells were washed three times with PBS and mounted using Prolong Gold Antifade Mountant (P36930; Thermo Fisher Scientific).
Cochlear hair cells.
P7 and P60 mice were used for experiments. P60 mice were transcardially perfused with ice-cold PBS after anesthetization. Mice were euthanized by decapitation, and all cochleae were dissected in Hank’s Balanced Salt Solution (HBSS) containing 0.1 mM CaCl2. After fixation in 4% PFA for 24 h at 4 °C, the organ of Corti was further dissected, and then the tectorial membrane was removed. The organ of Corti was permeabilized with 0.5% Triton X-100 in PBS (30 min), blocked in 4% BSA in PBS (2 h), and then incubated (2 h, room temperature) with primary antibodies (1:50 anti-TMC1, 1:100 anti-LHFPL5, or 1:100 anti-HA) diluted in PBS containing 3% BSA. Finally, the samples were washed three times for 10 min with PBS, incubated with Alexa Fluor 488-conjugated second antibodies (1:500) and Alexa Fluor 647-phalloidin (1:40) for 1 h, washed three times with PBS, and mounted using Prolong Gold Antifade Mountant.
Microscopy for both cultured cells and mouse hair cells was performed using a Leica SP8 confocal microscope (Leica Microsystems). Leica LAS-AF imaging software and Fiji software were used for image acquisition and analysis, respectively.
Microbead-Based SiMPull.
Detailed experimental principles and procedures for SiMPull are described in the study by Zhao et al. (12).
Briefly, in one set of experiments, six mouse cochleae were dissected in HBSS containing 0.1 mM Ca2+ and then homogenized and lysed using 100 μL of SiMPull lysis buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% [vol/vol] Nonidet P-40) supplemented with proteinase inhibitors. Separately, 2 μL of NeutrAvidin agarose microbeads (50% slurry; mostly 40 to 70 μm in diameter) (29200, Thermo Fisher Scientific) were first incubated with 100 μL of washing buffer (SiMPull lysis buffer without Nonidet P-40) and 5 μL of biotinylated goat anti-rabbit IgG antibody for 10 min, spun down, and then resuspended with 100 μL of washing buffer, after which the microbeads were incubated with 3 μL of the first antibody against the bait protein for 30 min. After washing two times with 100 μL of washing buffer, the microbeads were resuspended in 100 μL of washing buffer, divided into several sample sets that were mixed with 30 μL of cochlear cell lysate premixed with or without 2 μg of peptides, and incubated for 30 min at room temperature. The sequences of the peptides used were the following: WT peptide of mTMC1, EYGYPSYTEFDISGNVLALI; D569N (DN) peptide of mTMC1, EYGYPSYTEFNISGNVLALI; control peptide (aa 824 to 838 of rat TRPV1), EDAEVFKDSMVPGEK.
Next, 2 μL of the first antibody against the prey protein was added into the microbead samples and incubated for 30 min. Here, the antibody used was from a species different from the species of the first antibody against the bait protein. After a single gentle wash with the washing buffer, 0.01 μL of Alexa Fluor 488-conjugated second antibody was added to the microbeads and incubated for 10 min. The samples were then washed three times with the washing buffer, and the microbeads were resuspended with 10 μL of washing buffer and transferred to a glass slide. After mounting with a clean coverslip and sealing with nail polish, the microbeads were examined under a Leica SP8 confocal microscope equipped with a 60× objective. All images were analyzed using Fiji software.
PLA.
A Duolink In Situ Orange Starter Kit Mouse/Rabbit (DUO92102; Sigma-Aldrich) was used according to the manufacturer’s instructions. Briefly, cochleae isolated from Tmc1FLAG/FLAG; Lhfpl5HA/HA double-knockin mice were fixed in 4% PFA for 24 h at 4 °C, and the organ of Corti was further dissected out in HBSS containing 0.1 mM CaCl2. The organ of Corti was blocked with Duolink Blocking Solution for 60 min at 37 °C and then incubated with primary antibodies (1:100 rabbit anti-LHFPL5/1:100 mouse anti-FLAG, in Duolink Antibody Diluent) at 4 °C overnight. After washing two times with Wash Buffer A for 5 min, the samples were incubated with a pair of PLA probes (1:5 in Duolink Antibody Diluent) for 60 min at 37 °C and then washed two times with Wash Buffer A for 5 min. Next, 40 μL of the ligation mixture (1 μL ligase with 5× Ligation Buffer) was applied to the samples and incubated for 60 min at 37 °C, and after washing two times with Wash Buffer A, the samples were incubated with 40 μL of the amplification mixture (0.5 μL of polymerase with 5× Amplification Buffer) for 100 min at 37 °C. Finally, the samples were washed two times with Wash Buffer B for 10 min and once with 0.01× Wash Buffer B for 1 min and then mounted using Duolink In Situ Mounting Medium.
Statistics.
All data are expressed as means ± SEM. Student’s two-tailed t test was used for statistical analysis of two groups, and one-way ANOVA with Tukey’s post hoc test was used for more than two groups in Fig. 4J and Fig. 6. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
The work was supported by Hong Kong Research Grants Council Grants GRF16111616, GRF16102417, and GRF16100218; National Natural Science Foundation of China-Hong Kong Research Grants Council Joint Research Scheme N_HKUST614/18; Shenzhen Basic Research Scheme Grants JCYJ20170818114328332 and SMSEGL20SC01-K (all six grants to P.H.); and in part by the Innovation and Technology Commission (Grant ITCPD/17-9) and Grants NFSC81873703 and NSFC31861163003 (to W.X.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011147117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Cunningham C. L., et al. , TMIE defines pore and gating properties of the mechanotransduction channel of mammalian cochlear hair cells. Neuron 107, 126–143.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim K. X., Fettiplace R., Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel–like proteins. J. Gen. Physiol. 141, 141–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan B., et al. , TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan B., et al. , TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99, 736–753.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giese A. P. J., et al. , CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat. Commun. 8, 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., et al. , Loss of CIB2 causes profound hearing loss and abolishes mechanoelectrical transduction in mice. Front. Mol. Neurosci. 10, 401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong W., et al. , TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell 151, 1283–1295 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahendrasingam S., Fettiplace R., Alagramam K. N., Cross E., Furness D. N., Spatiotemporal changes in the distribution of LHFPL5 in mice cochlear hair bundles during development and in the absence of PCDH15. PLoS One 12, e0185285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beurg M., Xiong W., Zhao B., Müller U., Fettiplace R., Subunit determination of the conductance of hair-cell mechanotransducer channels. Proc. Natl. Acad. Sci. U.S.A. 112, 1589–1594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurima K., et al. , TMC1 and TMC2 localize at the site of mechanotransduction in mammalian inner ear hair cell stereocilia. Cell Rep. 12, 1606–1617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., et al. , Localization of TMC1 and LHFPL5 in auditory hair cells in neonatal and adult mice. FASEB J. 33, 6838–6851 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Zhao Q., et al. , Fast and easy single-molecule pulldown assay based on agarose microbeads. bioRxiv, 2020.09.20.305177 (20 September 2020).
- 13.Kurima K., et al. , Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 30, 277–284 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Ballesteros A., Fenollar-Ferrer C., Swartz K. J., Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. eLife 7, e38433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y., et al. , A novel mechanism of control of NFκB activation and inflammation involving A2B adenosine receptors. J. Cell Sci. 125, 4507–4517 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W., et al. , The complex of TRIP-Br1 and XIAP ubiquitinates and degrades multiple adenylyl cyclase isoforms. eLife 6, e28021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brownstein Z., et al. , Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 12, R89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilgert N., et al. , Amino acid 572 in TMC1: Hot spot or critical functional residue for dominant mutations causing hearing impairment. J. Hum. Genet. 54, 188–190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitajiri S., Makishima T., Friedman T. B., Griffith A. J., A novel mutation at the DFNA36 hearing loss locus reveals a critical function and potential genotype-phenotype correlation for amino acid-572 of TMC1. Clin. Genet. 71, 148–152 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Santos R. L. P., et al. , Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum. Mutat. 26, 396 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tlili A., et al. , TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol. Neurotol. 13, 213–218 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Beurg M., Barlow A., Furness D. N., Fettiplace R., A Tmc1 mutation reduces calcium permeability and expression of mechanoelectrical transduction channels in cochlear hair cells. Proc. Natl. Acad. Sci. U.S.A. 116, 20743–20749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labay V., Weichert R. M., Makishima T., Griffith A. J., Topology of transmembrane channel-like gene 1 protein. Biochemistry 49, 8592–8598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B., et al. , TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron 84, 954–967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte R., Talamas J., Doucet C., Hetzer M. W., Single bead affinity detection (SINBAD) for the analysis of protein-protein interactions. PLoS One 3, e2061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willott J. F., Handbook of Mouse Auditory Research: From Behavior to Molecular Biology (CRC Press, 2001). [Google Scholar]
- 27.Pujol R., Nouvian R., Lenoir M., Hair cells: Overview. (2016). http://www.cochlea.eu/en/hair-cells. Accessed 22 September 2020.
- 28.Longo-Guess C. M., et al. , A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc. Natl. Acad. Sci. U.S.A. 102, 7894–7899 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge J., et al. , Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5. eLife 7, e38770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalay E., et al. , Mutations in the lipoma HMGIC fusion partner-like 5 (LHFPL5) gene cause autosomal recessive nonsyndromic hearing loss. Hum. Mutat. 27, 633–639 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Shabbir M. I., et al. , Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J. Med. Genet. 43, 634–640 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holt J. R., Pan B., Koussa M. A., Asai Y., TMC function in hair cell transduction. Hear. Res. 311, 17–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawashima Y., Kurima K., Pan B., Griffith A. J., Holt J. R., Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 467, 85–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beurg M., et al. , Variable number of TMC1-dependent mechanotransducer channels underlie tonotopic conductance gradients in the cochlea. Nat. Commun. 9, 2185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng S. H., et al. , Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63, 827–834 (1990). [DOI] [PubMed] [Google Scholar]
- 36.Jain A., Liu R., Xiang Y. K., Ha T., Single-molecule pull-down for studying protein interactions. Nat. Protoc. 7, 445–452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z., et al. , Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 26, 106–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.