Significance
We assembled one of the most contiguous genomes for the common goldfish and unveiled the genetic architecture of many well-known and anatomically interesting traits, owing to the sequencing of a large collection of goldfish varieties. The datasets that we have generated for candidate genes or genomic regions based on population genomics may help to elucidate the evolution of goldfish and goldfish varieties and may provide a resource for a wide range of studies with important implications for the use of goldfish as a model for vertebrate and fish genetics.
Keywords: Carassius auratus, goldfish, domestication, GWAS, genome evolution
Abstract
Goldfish have been subjected to over 1,000 y of intensive domestication and selective breeding. In this report, we describe a high-quality goldfish genome (2n = 100), anchoring 95.75% of contigs into 50 pseudochromosomes. Comparative genomics enabled us to disentangle the two subgenomes that resulted from an ancient hybridization event. Resequencing 185 representative goldfish variants and 16 wild crucian carp revealed the origin of goldfish and identified genomic regions that have been shaped by selective sweeps linked to its domestication. Our comprehensive collection of goldfish varieties enabled us to associate genetic variations with a number of well-known anatomical features, including features that distinguish traditional goldfish clades. Additionally, we identified a tyrosine-protein kinase receptor as a candidate causal gene for the first well-known case of Mendelian inheritance in goldfish—the transparent mutant. The goldfish genome and diversity data offer unique resources to make goldfish a promising model for functional genomics, as well as domestication.
Goldfish (Carassius auratus) were domesticated in ancient China from crucian carp (both are still considered the same species) (1–3), which is one of the most important farmed fish, with global aquaculture production of 3.096 million tons of crucian carp in 2018 (4). The appearance of red scales on normally gray or silver crucian carp was first recorded during the Chinese Jin Dynasty (AD 265 to 420) (3). During the Tang Dynasty (AD 618 to 907), goldfish with preferred phenotypes were selected to be raised in ornamental ponds and water gardens (5). In the Song Dynasty (AD 960 to 1279), the gold (yellow) variety of goldfish was the symbol of the imperial family, and goldfish became known as the “royal fish” while commoners were forbidden to raise these yellow goldfish (5). The goldfish was introduced into Japan (6) and Europe at the beginning of the 17th century (7) and introduced to North America ∼1850 where it quickly became popular (8).
Because goldfish can produce thousands of eggs and dozens of these small fish can be raised in the same pond, 1,000 y of domestication have been characterized by strong artificial selection. When describing goldfish, Charles Darwin once wrote, “Passing over an almost infinite diversity of color, we meet with the most extraordinary modifications of structure” (9), highlighting that goldfish provide a rich resource for investigating the genetics of diverse morphological features. The study of variation under domestication in goldfish supplied Darwin with the idea of selection, whether natural or artificial, as the driving force behind evolution (9). Over a century ago, William Bateson also considered goldfish promising study material for biological variation (10). Using genetic stocks of goldfish populations, Shisan C. Chen pioneered the study of modern genetics in China (11) and presented the first examples of traits exhibiting Mendelian inheritance in goldfish, including traits of transparency and mottling (12).
After over 1,000 y of domestication and breeding, hundreds of variants in body shape, fin configuration, eye style, and coloration exist, making goldfish an excellent genetic model system for fish physiology and evolution. Goldfish had also been used as a model for Mendelian genetics and biological variations before the development of the zebrafish system. In this study, to systematically understand these and other phenomena in goldfish, we have generated a high-quality reference genome for the common goldfish and have resequenced a large collection of 185 representative goldfish varieties covering major classification groups and famous ornamental lines, as well as 16 wild crucian carp individuals, to provide genomic insights into the evolution, domestication, and genetic basis of artificial selection in goldfish.
Results
Genome Sequencing, Assembly, and Annotation.
Through flow cytometry, we estimated the genome size of a 12-generation inbred line of a female common goldfish (G-12) at 1.8 gigabases (Gb). Approximately 140 Gb of sequence data generated using the PacBio RS II platform (SI Appendix, Table S1) were assembled using Canu (13) and polished with gigabases of Illumina paired-end sequences, yielding an initial 1.657-Gb draft assembly with contig N50 of 474 kilobases (kb) (SI Appendix, Table S2). We generated another 1,050,985 BioNano DNA molecules >150 kb to further correct the above genome assembly (SI Appendix, Table S3). The final assembly using the BioNano approach spanned 1.73 Gb with scaffold N50 of 606 kb (Table 1 and SI Appendix, Table S2 and Fig. S1).
Table 1.
Statistics of C. auratus genome assembly
| Assembly feature | C. auratus |
| Estimated genome size, Gb | 1.80 |
| No. of contigs | 5,888 |
| Contig N50, bp | 606,731 |
| Contig N90, bp | 137,868 |
| Longest contig, bp | 7,117,495 |
| No. of scaffolds | 1,770 |
| Scaffold N50, bp | 31,841,898 |
| Scaffold N90, bp | 24,975,486 |
| Longest scaffold, bp | 60,771,278 |
| Assembly length, bp | 1,739,655,870 |
| Assembled portion of genome, % | 96.11 |
| Repeat portion of assembly, % | 33.04 |
| Predicted gene models | 56,251 |
| Average coding sequence length, bp | 1,394 |
| Average exons per gene | 8.59 |
| Chromosomal assembled portion, % | 99.30 |
In total, 480 million 150-base pair (bp) reads were generated from three high-throughput chromatin conformation capture (Hi-C) libraries and uniquely mapped onto the draft assembly contigs using ALLHiC (14, 15), followed by manual correction (SI Appendix, Table S4). Approximately 99.30% (1,727.09 megabases [Mb]) of the assembled sequences were anchored onto the 50 pseudochromosomes (scaffold N50 31.84 Mb), and 95.75% (1,653.62 Mb) were oriented and ordered (SI Appendix, Table S5 and Fig. S2). To validate the Hi-C assembly, a genetic map of crucian carp with 8,487 single nucleotide polymorphism (SNP) markers and 50 linkage groups (16) was aligned to the assembled goldfish genome, which showed that the ordered scaffolds and the genetic map are highly concordant with one another, with average Pearson’s correlation coefficients of 0.923 (Fig. 1 and SI Appendix, Fig. S3). This goldfish genome assembly, based on an advanced generation inbred line, is more complete and of higher quality than the recently published draft goldfish genome “Wakin” (1) (SI Appendix, Fig. S4), but comparable to another goldfish genome published recently (2). CEGMA (17) and BUSCO (18) were used to recall 219 (88.7%) complete gene models from among 248 ultraconserved core eukaryotic genes and 4,344 (94.7%) complete gene models from among 4,584 conserved actinopterygian genes in our assembly (SI Appendix, Tables S6 and S7).
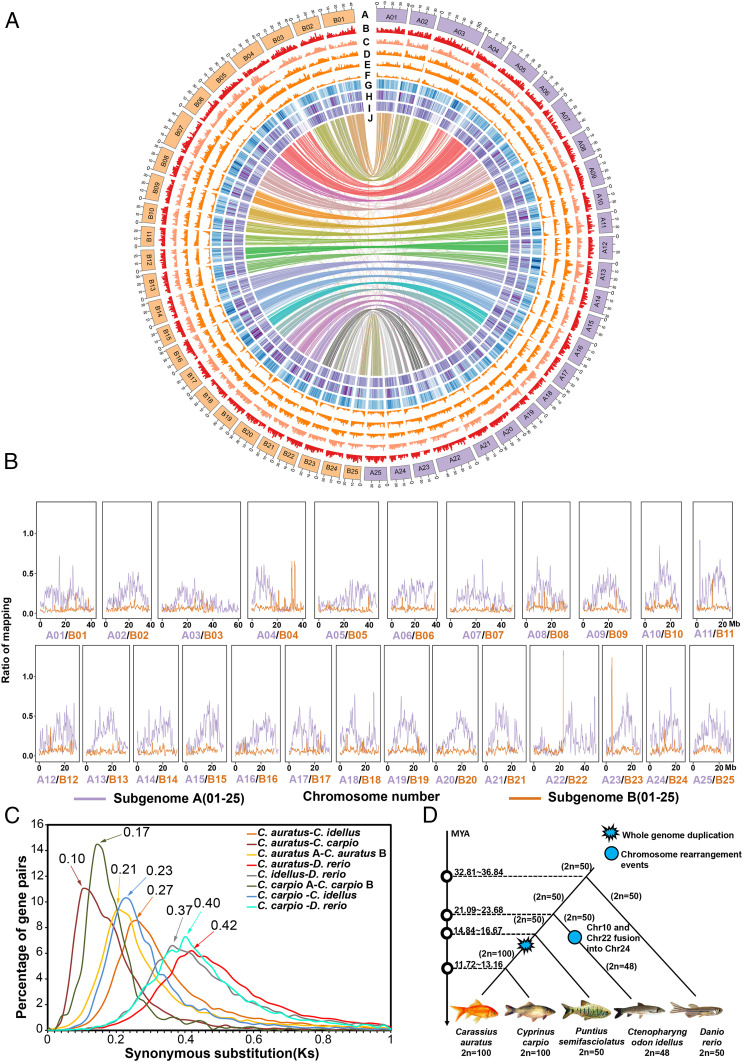
Fig. 1.
Goldfish genome features and their evolution. (A) The rings from outermost to innermost represent complete genomes in Mbp. Subgenome A is colored with purple, and subgenome B is colored with orange (ring A). Shown are gene density in goldfish (ring B), SNP density in the population of 201 goldfish (ring C), InDel density in the population of 201 goldfish (ring D), GC (guanine-cytosine) content of the whole goldfish genome (ring E), TEs in the whole goldfish genome (ring F), gene expression (ring G), chromosome collinearity between the subgenomes (ring H), gene expression (ring I), and chromosome collinearity between the subgenomes (ring J). Each signal was calculated in 500-kb sliding windows with 100-kb steps. (B) The proportion of P. semifasciolatus reads against mapped reads of goldfish. For patterns of the reads mapping for the other five Barbinae species, see SI Appendix, Figs. S9 and S10. (C) The distribution of nonsynonymous substitutions between selected pairs among taxa C. auratus (goldfish), C. carpio, C. idellus, D. rerio, illustrating the genetic distance between orthologous gene pairs (between two different taxa) or paralogous gene pairs (within the same taxon). (D) Genome duplications and chromosome rearrangements in C. auratus, C. carpio, C. idellus, P. semifasciolatus, and D. rerio.
The goldfish genome comprises 56,251 coding genes and 10,098 long noncoding transcripts (Table 1 and SI Appendix, Tables S5 and S8). Comparison with Cyprinus carpio, Ctenopharyngodon idellus, Danio rerio, and humans revealed 937 (4%) gene families unique to goldfish (SI Appendix, Fig. S5). Transposable elements (TEs) spanned 33.04% of the goldfish genome assembly, which was lower than that of common carp (39.20%) (19), “Wakin” (39.60%) (1), and zebrafish (54.30%) (20), but higher than the 33.00% for cave fish (21) and 30.68% for Oryzias latipes (22) (SI Appendix, Table S9). Potential centromeric regions were predicted for 38 of the 50 chromosomes, again highlighting the near completeness of the assembled goldfish genome (SI Appendix, Figs. S6 and S7 and Table S10).
Disentangling the Subgenomes in Goldfish.
For more than 50 y, it has been speculated that goldfish is a tetraploid species (23, 24). A recent study of cyprinids in the Barbinae (2n = 50) subfamily suggested that species in this subfamily might be the closest diploid ancestors of goldfish (25) and led to the hypothesis that Barbinae may be the progenitors of the diploid lineage leading to goldfish. We performed whole-genome shotgun sequencing of six representative diploid Barbinae species, including Puntius semifasciolatus, Hypsibarbus vernayi, Mystacoleucus marginatus, Balantiochelos melanopterus, Barbonymus schwanenfdi, and Hampala macrolepidota (SI Appendix, Fig. S8), and aligned an average of ∼14 million reads from these six Barbinae species to the goldfish genome assembly.
The 50 goldfish chromosomes can be clearly separated into two sets (subgenomes), as evidenced by the alignment of a disproportionate number of reads (average of 80.4%) to one of the homeologous genomes, despite overall similar sizes between pairs of homeologous chromosomes (Fig. 1 A and B and SI Appendix, Figs. S9 and S10 and Table S5). The biased proportion of mapped reads toward one homeologous chromosome (between 70.11% and 84.52%) is highly consistent across different chromosomes, suggesting that C. auratus originated from two ancestral lineages, one of which was common to Barbinae. Thus, we defined the set of chromosomes with the highest proportion of reads aligned between goldfish and Barbinae as subgenome A (ChrA01∼A25) and designated the remaining set as subgenome B (ChrB01∼B25). For comparison, SI Appendix, Table S11 shows how these different subgenomes relate to other (sub)genomes discussed in other studies. The repeat family was considered enriched in a subgenome by the criterion A/(A + B) − B/(A + B) ≥ 0.8 or B/(A + B) − A/(A + B) ≥ 0.8, resulting in one A-specific and six B-specific repeat families, which are the hAT-Ac and TcMar-Tc1 elements, respectively (26) (SI Appendix, Figs. S11–S13). Moreover, a phylogenetic tree was constructed based on a nuclear gene: i.e., the connective tissue growth factor-like gene, which has only one copy in diploid cyprinids but two copies in tetraploid cyprinids (SI Appendix, Fig. S14). The results showed that the inferred homolog from Barbinae species (P. semifasciolatus) was clustered with the gene from subgenome A in goldfish, with zebrafish and glass carp as outgroups. The results indicated that subgenome A may have originated from a progenitor species within the Barbinae subfamily whereas the diploid progenitor of subgenome B probably went extinct or derived from a yet unknown Cyprininae lineage (25, 27). We speculate that the origin of goldfish is due to an allotetraploidy event although other evolutionary events, such as autopolyploidy, cannot be ruled out, although much less likely. Chromosome numbers were designated in accordance with their collinearity to those of zebrafish (20) (SI Appendix, Fig. S15). The goldfish A and B subgenomes contain 28,133 and 26,141 genes, respectively (SI Appendix, Fig. S16 and Supplementary Text).
To explore any divergence in the transcriptional patterns between the two subgenomes, by means of RNA-seq analysis, we analyzed the expression patterns of 16,362 homeologous gene pairs in 10 tissues, including spleen, scale, muscle, midkidney, head kidney, gut, gill, dorsal fin, atrium, and testis (but see also ref. 2). In all of the examined tissues, 4,123 and 2,941 homeologs were inferred to be dominantly expressed in subgenomes A and B, respectively (Fig. 1B and SI Appendix, Figs. S16–S22). Apparently, the homeologous expression exhibited asymmetric expression patterns between the two subgenomes, and the genome-wide expression level dominance was overall more biased toward subgenome A in the goldfish genome (2).
Genome Evolution of Goldfish.
Large segmental inversions were observed for the homeologous chromosome pairs ChrA01/ChrB01, ChrA04/ChrB04, ChrA05/ChrB05, ChrA09/ChrB09, ChrA11/ChrB11, and ChrA15/ChrB15, despite a strong overall collinearity (Fig. 1B). These large segmental inversions might be one of the reasons for the allotetraploidy formation (28). According to the syntenic blocks detected in C. auratus, C. carpio, C. idellus, P. semifasciolatus, and D. rerio, the two subgenomes of goldfish (Ks [substitutions per synonymous site] = ∼0.17) and common carp (Ks = ∼0.21) diverged 13.28 to 16.67 million years ago (MYA) (Fig. 1 C and D), which was earlier than the divergence between goldfish and common carp that took place 7.81∼8.77 MYA (Ks = ∼0.10). We hypothesize that the whole-genome duplication (WGD) event occurred in the shared lineage of goldfish and common carp and, thus, that they may have the same number of chromosomes (2n = 100), which is twice the basic chromosome number (2n = 50) of diploid members of the Cyprininae subfamily, indicating these two species are tetraploid (Fig. 1D). Moreover, zebrafish, which is distantly and equally related to all carps, diverged from the last common ancestor (LCA) of C. carpio, C. auratus, and C. idellus ∼32.8 to 36.8 MYA (Fig. 1 C and D). Segmental inversions in ChrA04/ChrA14/ChrA19 occurred after the divergence of zebrafish and the lineage leading to the Cyprininae subfamily because the orientation of this chromosomal fragment was conserved among the extant Cyprininae genomes examined to date (SI Appendix, Figs. S15 and S23–S25).
Domestication and Population Structure of Goldfish.
To examine the evolutionary history and population structure of goldfish, we generated 4.3 terabases of sequence data with an average sequencing depth of ∼12.5× for a total of 201 samples, including 16 wild crucian carps and 185 representative goldfish variants (Datasets S1 and S2 and SI Appendix, Fig. S26, Table S1, and Supplementary Text). The wild crucian carps used were collected from 16 different locations in eastern China, including Zhejiang and Jiangsu (5) (SI Appendix, Fig. S26). We aligned 33 previously reported sequences of common carp (19) to our goldfish genome and constructed a distance-based phylogenetic tree by using ∼6.5 million biallelic SNPs genotyped in 234 samples, including common carp, crucian carp, and goldfish. The group containing common carp was clearly separated from those containing crucian carp and goldfish (SI Appendix, Fig. S27A), which was in accordance with the principal component analysis (PCA) based on the same SNP dataset (SI Appendix, Fig. S27B).
Phylogenetic reconstruction of the 201 crucian carp and goldfish samples showed that common goldfish was more closely related to crucian carp than to the other goldfish and divided the latter into two lineages (Fig. 2 A–C). Both principal component 1 (PC1) (11.29%) and PC2 (7.87%) generally distinguished common goldfish from Egg goldfish (dorsal fin absence) and Wen goldfish (dorsal fin presence) (SI Appendix, Figs. S28–S30), respectively. The phylogenetic tree also indicated that Wen goldfish could be further classified into three distinct subgroups, including Wenyu–Oranda, telescope, and Wen–Pompon, whereas the Egg goldfish could be classified into four subgroups, namely Eggfish–Lionhead, Egg–Pompon, and Chinese Ranchu, consistent with the PCA based on total SNPs (Fig. 2B and SI Appendix, Figs. S31–S37).
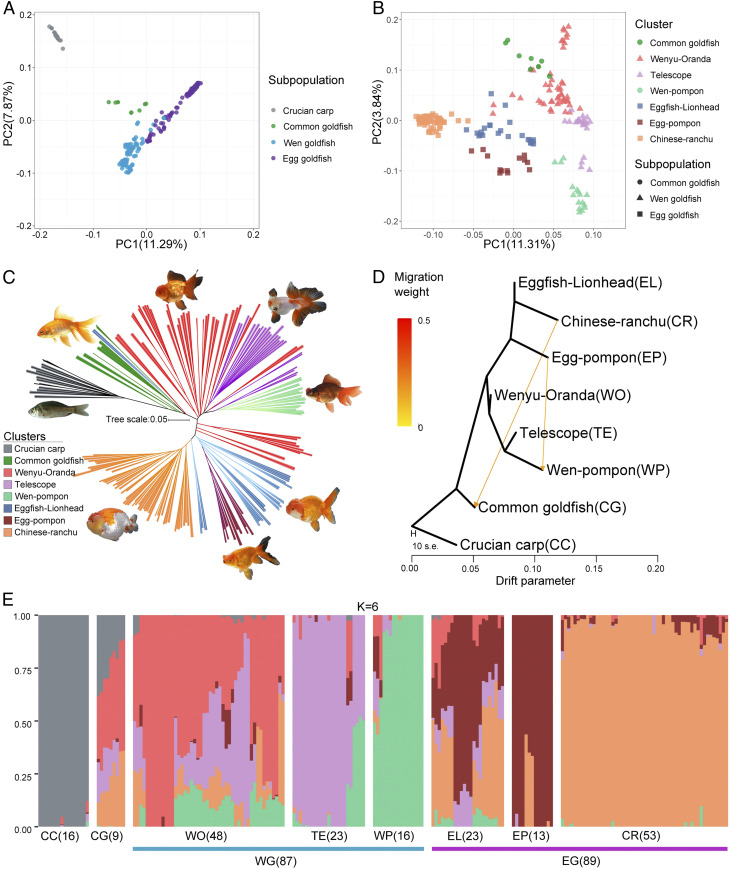
Fig. 2.
Population structure in goldfish. (A) Principal components of SNP variation. Samples from subpopulations of crucian carp (CC), common goldfish (CG), Wen goldfish (WG), and Egg goldfish (EG) and (B) Clusters of CG, Wenyu–Oranda (WO), Telescope eye (TE), Wen–Pompon (WP), Eggfish–Lionhead (EL), Egg–Pompon (EP), and Chinese–Ranchu (CR) are shown, with all of the samples in A, but excluding CC. The plots show the first two principal components. (C) Neighbor-joining clustering of CC, CG, WO, TE, WP, EL, EP, and CR based on genetic distance calculated from SNPs. Branch color indicates membership in one of the eight classified goldfish populations. The scale bar shows number of substitutions per site. (D) TreeMix analysis of 185 goldfish divided into seven clusters, with crucian carp samples serving as the outgroup. The arrows correspond to the direction of gene flow. (E) STRUCTURE plot for CC and goldfish. The distribution of the K = 6 genetic clusters is shown. The eight different populations and the sample number for each population are indicated after the abbreviated population names.
We further calculated drift parameters in light of the allelic variants across the 201 sampled genomes and constructed trees showing the phylogenetic relationships between goldfish and crucian carp (Fig. 2C). The direction of the gene flow for Wen–Pompon from Egg–Pompon (weight = 0.142) (Fig. 2D) indicated that it, including the Japanese Hanafusa, is the counterpart of the Chinese Pompon (Egg–Pompon) (27).
Domestication and Selective Sweeps.
To elucidate the genomics of domestication in goldfish, we analyzed the rate of decay of linkage disequilibrium (LD) (indicated by r2), Tajima’s D, and genetic diversity (π) for the four subpopulations (crucian carp [CC], common goldfish [CG], Wen, and Egg) (Fig. 3 A and B and SI Appendix, Figs. S38–S41). The decay of LD was dramatically faster in the crucian carp than in the goldfish (Fig. 3A), suggesting a considerably higher frequency of genetic recombination in the crucian carp while inbreeding is more common in goldfish. Moreover, LD decay in the Egg group was faster than in the Wen group, indicating stronger artificial selection in the Egg goldfish group, consistent with the estimates of the fixation index Fst, which quantifies the population differentiation (Fig. 3A and SI Appendix, Fig. S41). Compared with crucian carp (1.133), Tajima’s D values for the Egg group (1.770) and the Wen group (2.315) were considerably higher (Fig. 3B), which supports the hypothesized existence of a population genetic bottleneck during the domestication and strong artificial selection in goldfish. The different subgroups also displayed variations in genetic diversity (π) (SI Appendix, Fig. S38). The value of π increased from crucian carp (0.00059) to common goldfish (0.00124) and increased from common goldfish (0.00134) to both Wen goldfish and Egg goldfish (0.00297). These observations suggest that the process of domestication from crucian carp and common goldfish increased genetic diversity as a result of artificial selection (Fig. 3B and SI Appendix, Fig. S32), indicating the accumulation of significant genetic variation in goldfish after their domestication from crucian carp.
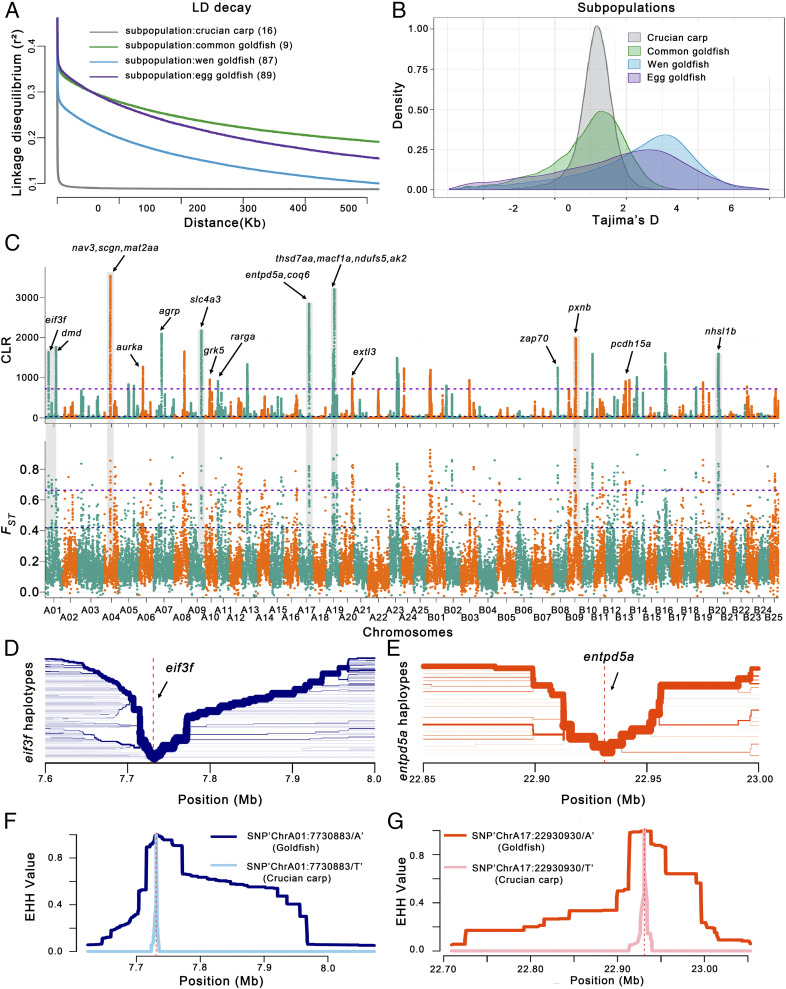
Fig. 3.
Genome-wide screening for domestication-associated selective sweeps in goldfish. (A) Decay of LD, (B) Tajima’s D value, and (C) whole-genome analysis of the domestication with selective sweeps inferred from the comparisons among common goldfish, Wen goldfish, Egg goldfish, and crucian carp. The genome-wide threshold of 2.5 was defined by the top 1% of Fst values. We calculated CLR scores to confirm selective sweeps based on domestication features; the highest CLR score was 5%. The arrows indicate the sweeps that occurred in goldfish during the domestication process, and the chromosome number of subgenome A and B is colored with purple and orange, respectively. Representative candidate genes in this region include: eif3f (Cau.A01G0002820), dmd (Cau.A01G0002960), nav3 (Cau.A04G0007000), scgn (Cau.A04G0010160), mat2aa (Cau.A04G0010480), aurka (Cau.A06G0010390), agrp (Cau.A07G0008060), slc4a3 (Cau.A09G0011550), grk5 (Cau.A10G0011480), rarga (Cau.A11G0000470), thsd7aa (Cau.A19G0007320), macf1a (Cau.A19G0007840), ndufs5 (Cau.A19G0007860), ak2 (Cau.A19G0007880), extl3 (Cau.A20G0012960), zap70 (Cau.B08G0005260), pxnb (Cau.B09G0001240), pcdh15a (Cau.B13G0010680), and nhsl1b (Cau.B20G0006630). (D and E) Haplotype bifurcation diagram in the goldfish population, with two illustrative examples shown with the extended haplotypes at the eif3f-allele of ChrA01 (D) and entpd5a-allele of ChrA17 (E). The haplotype bifurcation diagram visualizes the breakdown of LD at progressively longer distances from the core allele from the focal SNP, which is identified by a vertical dashed line. The thickness of the lines corresponds to the frequency of the haplotype. (F and G) EHH (extended haplotype homozygosity) for SNP “ChrA01:7730883/A/T” (diagnostic for eif3f) in the goldfish and crucian carp populations is shown with a dark and light blue line, respectively (F), and for SNP “ChrA17:22930930/A/T” (diagnostic for entpd5a) in the goldfish and crucian carp populations is marked with a deep and light red line, respectively (G).
To identify potential selective signals during goldfish domestication, we scanned genomic regions based on genome-wide calculations for selective sweeps by estimating Fst and the composite likelihood ratio (CLR) (28), and performed entropy analysis (Materials and Methods). To avoid the overrepresentation of goldfish data, we selected 33 individual goldfish and 16 individual wild crucian carp for this analysis. The top 1% of the –log (P) value covered 50 genomic regions with a total of 25.2 Mb and harboring 946 genes (Fig. 3C and Dataset S3). The overall polymorphism level (minor allele frequency) within the selected regions is 0.065 across all goldfish varieties, compared to 0.236 across the entire genome, representing a fourfold reduction of diversity, possibly due to selective breeding and domestication. The ∼25.2-Mb sequences displayed nonlinearity to each other (between the two subgenomes), which would indicate potential functional complementation of homeologous chromosomes in goldfish. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the 940 genes indicated that a significant portion of the candidate genes were related to morphogenesis, pigmentation, behavior, immune response/infectious diseases, energy metabolism, and response to hormones (Dataset S3). We also examined genes associated with phenotypes that have been observed in zebrafish mutants (https://zfin.org/) and found that 173 of the genes corresponding to 132 orthologs displayed mutant phenotypes in gene knockout lines in zebrafish, including phenotypes related to pigmentation, morphogenesis, and behavior that have undergone positive selection for over 1,000 y in goldfish (Dataset S3).
These selective sweeps were further scrutinized in the above 201 samples, and 393 genes indicated regions of completed selective sweep (Fig. 3 C and D and Dataset S4). Of the 393 genes, 21 representative candidate genes with a high degree of population differentiation (Fst), and highly negative Tajima’s D are indicated (Fig. 3C). Among these 21 genes, 13 are orthologous to genes for which there are knockout lines in zebrafish that display mutant phenotypes related to behavior (pcdh15a and agrp), decreased eye size (ndufs5), cell migration (nav3 and zap70), or decreased brain size (aurka) (Dataset S3). Haplotype bifurcation diagrams are shown for two examples, the eif3f-allele (Fig. 3 D and F) and the entpd5a-allele (Fig. 3 E and G). In the whole goldfish population, the average level of genetic diversity (π) of these 393 genes is 1.14E−4, compared to 1.16E−3 for all genes in the entire genome. These low-diversity genes likely contributed to the phenotypes associated with major domestication traits in goldfish.
Genome-Wide Association Study of Dorsal Fin in Domesticated Goldfish.
Darwin noted that goldfish exhibited “the most extraordinary modifications of structure” and recorded goldfish variants lacking a dorsal fin, with double anal fins, and with triple tails (29). The dorsal fin is a remarkable feature that distinguishes Wen goldfish from Egg goldfish, and the diminution of the dorsal fin is the key step in the evolution of Egg goldfish. To study the genetic segregation of the dorsal fin form, we crossed Chinese Ranchu (Egg goldfish) with Lionhead (Wen goldfish). The F1 population from the female Chinese Ranchu and male Lionhead contained 31% (62) normal, 4.5% (9) absent, and 64.5% (129) abnormal dorsal fins. In contrast, the reciprocal cross F1 population contained 25.5% (51) normal and 74.5% (149) abnormal dorsal fins (SI Appendix, Fig. S42), suggesting that this dorsal fin trait is controlled by multiple gene loci probably with maternal genetic effects.
Analysis of the homeolog retention of the associated genes for dorsal fin revealed that, of 222 genes located in the subgenome A, only 72 homeologs were present in subgenome B. These genes were retained less often (retention rates are ∼53% and ∼46% for genome-wide subgenomes A and B, respectively) than the average retained as two copies (retention rates are ∼89% and ∼83% for genome-wide subgenomes A and B, respectively) although the differences are not statistically significant based on a t test (P value = 0.1702). We further performed a genome-wide association study (GWAS) of the dorsal fin trait based on 96 controls (Wen goldfish) and 87 cases (Egg goldfish, excluding 2 fishes with ambiguous dorsal fins) (Fig. 4A). A total of 378 genes associated with the dorsal fin phenotype were detected in 8.96 Mb of genomic regions spread across 13 chromosomes. A total of 85.2% (322) of these genes were found on subgenome A, with significant portions being located on five chromosomes, including ChrA09 (84), ChrA07 (64), ChrA22 (56), ChrA16 (45), and ChrA05 (41) (Fig. 4B, SI Appendix, Fig. S43, and Dataset S5), a considerably higher proportion than the number of dorsal fin-associated genes (56) located on subgenome B (Fisher’s exact test, P value < 2.2e−16), suggesting an uneven subgenomic distribution of the genes associated with the dorsal fin. GO and KEGG analyses indicated that the candidate genes for this trait are putatively involved in the “cell surface receptor signaling” pathway, “signal transduction,” “transmembrane transport,” “skeletal system development,” and “primary metabolic process and organ nitrogen compound metabolic process” (Dataset S5).
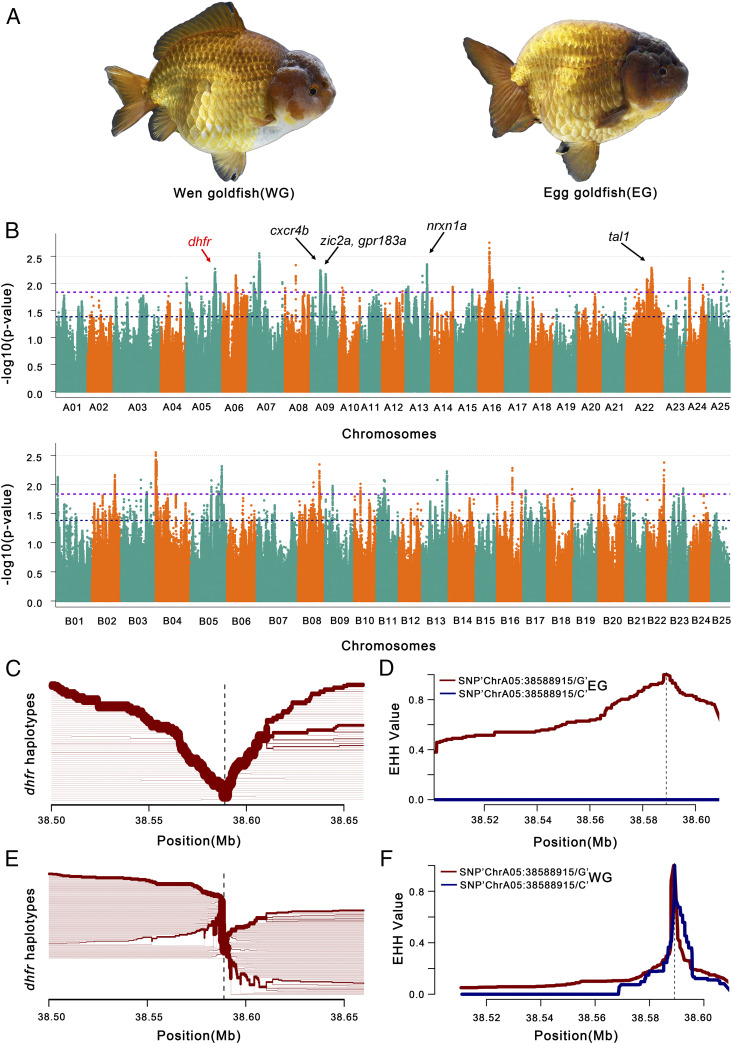
Fig. 4.
GWAS for dorsal fin-related traits in goldfish. (A) Pictures of Wen goldfish (WG, with dorsal fin) and Egg goldfish (EG, without dorsal fin). (B) GWAS for genes associated with dorsal fin in a Wen/Egg-goldfish population. Genes surrounding or within association peaks are indicated. Gene names are highlighted in bold black/red in candidate regions potentially related to dorsal fin development in goldfish. Representative candidate genes for dorsal fin-related traits include: dhfr (Cau.A05G0015640), cxcr4b (Cau.A09G0004970), zic2a (Cau.A09G0008030), gpr183a (Cau.A09G0007940), nrxn1a (Cau.A13G0000420), tal1 (Cau.A22G0009870), kif5ba (Cau.B02G0011950), and atp1a2a (Cau.B02G0012070). The chromosome number of subgenome A and B is colored with cyan and orange. The dashed lines show different genome-wide significance thresholds, respectively. (C and E) Haplotype bifurcation diagram in EG and WG subpopulations, starting from the two alleles at one of the representative significant GWAS SNP sites. The haplotype bifurcation diagram visualizes the breakdown of LD at progressively longer distances from the core allele from the focal SNP, which is identified by a vertical dashed line. The thickness of the lines corresponds to the frequency of the haplotype. We show the extended haplotype at the dhfr allele of EG subpopulation in C, relative to the shorter haplotypes at dhfr allele of WG subpopulation in E, which is in accordance with a selective sweep around the dhfr allele in the EG subpopulation. (D and F) EHH for SNPs “ChrA05:38588915” (diagnostic for dhfr) in EG subpopulation (D) and WG subpopulation (F) is shown with maroon line (for G) and blue line (for C), respectively.
It is noteworthy that 57 genes of these 378 genes correspond to 50 zebrafish orthologs for which gene knockout lines display mutant phenotypes (Dataset S5). The mutant phenotypes for 13 of these zebrafish genes (26%) are related to the development of the dorsal fin, including decreased occurrence of the dorsal longitudinal anastomotic vessel (itgb1a, rab13, and e2f8), dorsal abnormal (kif5ba), dorsal aorta abnormal (fev, cxcr4b, gpr183a, tal1, glrx2, and uchl5), ventralized (zic2a), and curved dorsal fin (atp1a2a and dhfr) (Dataset S5). A haplotype bifurcation diagram is shown for one example, dhfr (Fig. 4 C–F). We further scrutinized the genotypes of our 183 goldfish samples (excluding 2 samples with ambiguous dorsal fin) (Fig. 4E and Dataset S6) and found that 24 of the corresponding regions in goldfish do not contain annotated protein-coding exons while 246 genes in goldfish display genotypes that generally coincide with the presence/absence of the dorsal fin (Fig. 4E and Dataset S6). Eight representative genes are shown (Fig. 4E), including 7 genes related to dorsal development in zebrafish (dhfr, cxcr4b, zic2a, gpr183a, tal1, kif5ba, and atp1a2a), while nrxn1a has been associated with vertebral abnormalities in humans (30). Dihydrofolate reductase (dhfr) is a key enzyme in folate-mediated metabolism and is involved in the de novo mitochondrial thymidylate biosynthesis pathway. Recently, the enzymes regulating folate metabolism have been reported to be up-regulated in zebrafish fins 4 d postamputation (31). We speculate that dhfr may contribute, to some extent, to the presence/absence of a fin. These genes probably played key roles in the artificial selection of the Egg goldfish from the Wen goldfish, as well as in dorsal fin development (31).
Candidate Genes for a Classic Case of Mendelian Inheritance in Goldfish.
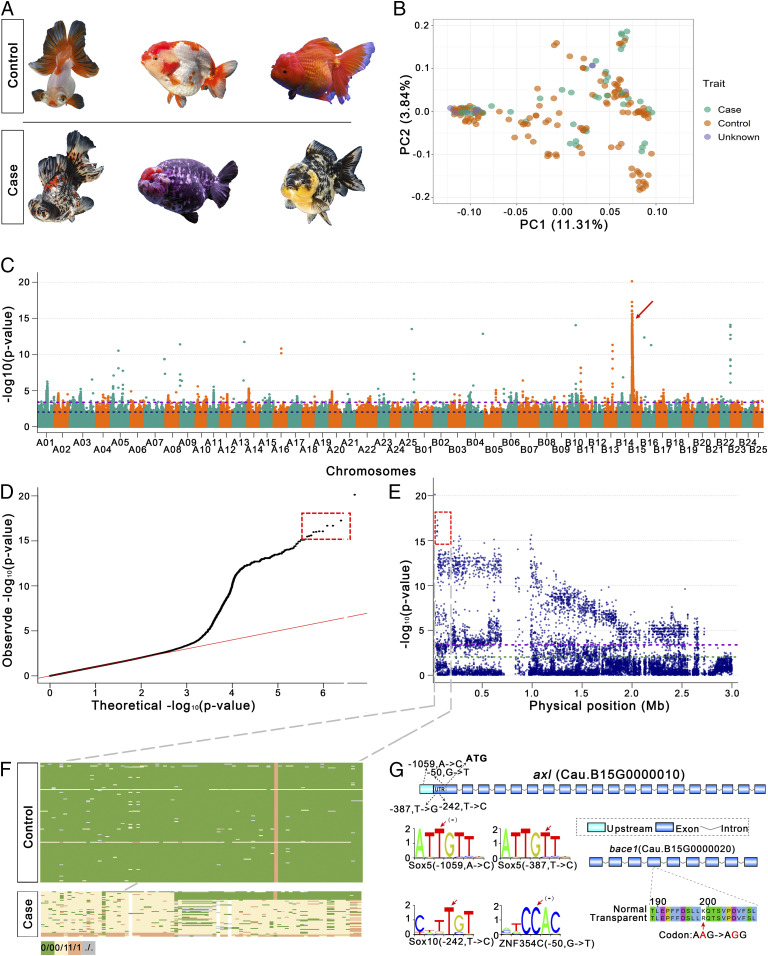
The transparent goldfish mutant was first recorded in 1579 (5) and is a classic case of Mendelian genetics in fish and even vertebrates with incomplete dominance at the T/t locus (12). However, the gene located at the T/t locus had not been identified after its first reporting in 1928. Goldfish with transparent scales are distributed across each of the subgroups in the panel of 185 fish phenotypes because the single gene controlling this transparent mutant has become widespread during goldfish breeding over the past 400 y. Using 48 cases and 127 controls, we detected a single strong association peak (−log10 (P) = 17.3) with five SNPs on one arm of ChrB15 (Fig. 5 A–E) that coincides with the incomplete dominant genetic pattern of the T/t locus (Fig. 5F). A genomic inversion was observed in this region in comparison to homeologous chromosome 15. Furthermore, we scrutinized the regions based on a larger variant dataset of the SNPs and insertions/deletions (InDels) of the population and found that segregation of 14 SNPs and two InDels coincides with the T/t locus, as suggested (12). In the candidate region ChrB15:28,558−153,105, we identified a gene, Cau.B15G0000010 (ChrB15:48,888−76,549), encoding a tyrosine-protein kinase receptor UFO-like protein containing four SNPs in predicted transcription factor-binding sites in its 5′ untranslated region (UTR) (Fig. 5G). A leukocyte tyrosine kinase has been related to pigment cell development in zebrafish (31), and members of the receptor tyrosine kinase family play roles in melanoma development in humans (32) although the roles of this gene family are not currently known in goldfish. In goldfish, a tyrosine-protein kinase receptor might regulate tyrosine kinase and thus result in pigmentation variation. In this region, under strong association, the neighboring gene Cau.B15G0000020 encoding a beta-secretase 1-like protein (bace1) with a nonsynonymous single-nucleotide mutation has been related to melanocyte migration in zebrafish (33) and in humans (34), which might explain the frequent development of black smudges or dots on goldfish with transparent scales (Fig. 5A). Although functional validation is still pending, we believe that the tyrosine-protein kinase receptor is a strong candidate for the T/t locus in goldfish.
Fig. 5.
GWAS for transparent scale-related traits in goldfish population. (A) Goldfish with transparent scales or translucent scales served as case and control, respectively. (B) PCA for the samples from case (transparent scale), control (translucent scale), and unknown populations based on whole-genome sequence data. PC1 and PC2 indicate scores of principal components 1 and 2, respectively. (C) Manhattan plot for transparent scale GWAS. The two dashed horizontal lines represent the thresholds for very high significance [top 0.1%, –log10 (P) = 3.38] and significance [top 1%, –log10 (P) = 2.03]. The arrow indicates the highly significant peaks. (D) Quantile–quantile plot for GWAS under a general linear model. (E) Local Manhattan plot surrounding the association peak on ChrB15. The double dashed horizontal lines represent the thresholds for high significance [top 0.1%, –log10 (P) = 3.38] and significance [top 1%, –log10 (P) = 2.03]. The boxed areas in D and E indicate the high-confidence SNPs. (F) Genotype analysis for the candidate region (Top) and gene order (Bottom) in the candidate region; green, yellow, orange, and gray blocks represent reference allele homozygous (0/0), heterozygous (0/1), variant allele homozygous (1/1), and missing genotype (./.), respectively. The vertical orange bar consists of orange blocks that represent variant allele homozygous (1/1). (G) Candidate genes and 5′ UTR regulatory elements are shown. Exons, introns, and 5′ UTR are represented with blue boxes, broken lines, and cyan boxes, respectively. The variant sites are marked with red arrows.
Discussion
Teleosts have undergone at least three rounds of WGD during their evolutionary past, of which the most recent is teleost-specific (Ts3R) and has been dated at 320 to 350 MYA (35, 36). Goldfish comprise one of the few teleost species that have undergone an additional, considerably more recent, WGD. Comparative genomics revealed that goldfish originated from a merger of two progenitor species, one of which is closely related to Barbinae species. The WGD in goldfish is assumed to originate from a hybridization event (allotetraploidy) between its two progenitor species before the divergence of common carp and goldfish, which dates to ∼13 to 16 MYA (Fig. 1C). The WGD in goldfish might have coincided with the end of the rapid elevation of the Tibetan Plateau (37) and accompanying global climate change and could have given the allotetraploid goldfish a selective advantage to survive in a quickly changed environment (38, 39). As is generally assumed for allopolyploid plants (40), goldfish also express genes predominantly residing on different subgenomes (SI Appendix, Fig. S20), suggesting that the two subgenomes probably diverged functionally. A recent study on common carp (41) also revealed subgenome dominance and the different expression of genes on homeologous chromosomes, generally supporting asymmetrical genome evolution in Cyprinidae. In addition, the WGD in goldfish seems primarily to have resulted in gene redundancy, increasing mutational robustness and likely shielding goldfish from the deleterious effects of mutations (42), thereby enabling artificial selection for mutations in key functional genes in goldfish. With the availability of this high-quality genome assembly, goldfish could become an excellent model system for genetic studies of dominant deleterious mutations in vertebrates.
Artificial selection for the dorsal fin was shown to have preferred variants from goldfish subgenome A, which was derived from the progenitor genome that is closely related to P. semifasciolatus. Both subgenomes may have differentially contributed to artificial selection for particular traits during the domestication of goldfish. A similar phenomenon has been observed in allotetraploid cotton, which displayed an asymmetric subgenome contribution to the long fiber trait (43). Although our GWAS analysis identified some genes previously known to be associated with the development of related traits in teleosts, we also identified many loci of unknown function. Very recently, the potentially associated genes relative to the dorsal fin loss phenotype were also identified in goldfish, and lrp6S was found to be located in subgenome S (corresponding to subgenome B in our notation) (SI Appendix, Table S11) (44). Based on classical genetics (crossing experiments), GWAS, and population history analyses in this study, we consider that the dorsal fin trait is likely controlled by multiple loci, as supported by our much wider sampling of goldfish; thus, the discrepancy between the current study and Kon et al. (44) is possibly caused by the differences of the two GWAS pupations. Further work will be necessary to functionally validate the specific genes governing these artificially selected traits.
The population genomic datasets that we have gathered related to candidate genes and genomic regions may help to elucidate the genetic basis of various traits in goldfish, may provide a resource for a wide range of studies, and may have important implications for the use of goldfish as a model for vertebrate genetic studies. A fully comprehensive understanding of goldfish genetics may also support the use of goldfish as promising material for the study of natural mutations and artificial selection and thus may provide connections with genome duplication, morphological evolution, and domestication in animals and plants in the future (45).
Materials and Methods
Additional materials and methods are described at length in SI Appendix, Supplementary Materials and Methods.
PacBio Sequencing.
Genomic DNA was extracted from goldfish blood. Concentrated and sheared DNA (6 µg) was size-selected using the BluePippin Automated DNA Size Selection System (Sage Science Inc.). SMRTbell libraries (∼20 kb) were prepared according to the protocol described by PacBio. The SMRT data were generated using the PacBio RSII system with P6-C4 chemistry (SI Appendix, Table S12).
Irys Optical Genome Maps.
Genome mapping was performed using the Bionano Genomics Saphyr System with NanoChannel array technology. Optical maps were assembled by scaffolding using Irys with default parameters.
Hi-C Library Construction and Sequencing.
Fresh blood was collected from goldfish to prepare genomic DNA for construction of Hi-C libraries at the BioMarker Technologies Company as follows (46). The paired-end 150-bp reads were generated by sequencing the libraries on the Illumina HiSeq X Ten platform.
Raw Data Preprocessing.
Reads with quality <0.75 or length <500 bp were then filtered out. Next, SMRT reads were corrected using the parameter corOutCoverage = 100, which enables correction of all of the input PacBio reads. The BioNano raw data were processed using the IrysView package (Bionano Genomics, San Diego, CA). Molecules with label signal-to-noise ratio (SNR) ≥3.0, average molecule intensity <0.6, and length >100 kb were retained for use in genome assembly.
Genome Assembly and Hi-C–Based Pseudochromosome Construction.
The initial de novo genome assembly of goldfish was performed with PacBio reads (80× sequence coverage) using Canu (13) (v1.6). Subsequently, the draft assembly was polished with 50× Illumina short reads using Pilon (47). ALLHiC (15) was used as previously described to scaffold the allotetraploid goldfish genome on chromosome-level scales.
Genetic Maps and Validation of Assembly.
To validate the accuracy of the chromosome-level assembly, a high-resolution genetic linkage map for crucian carp (16) with 8,487 SNP markers assigned to 50 linkage groups was applied to the 50 pseudochromosomes anchored by Hi-C using ALLMAPs (48).
Genome Annotation.
Consensus TE sequences for the goldfish genome were generated using RepeatModeler with a combination of de novo and homology strategies including two de novo repeat-finding programs, RECON (49) and RepeatScout (50), which we imported into RepeatMasker (www.repeatmasker.org/) to identify and cluster repetitive elements. Ab initio gene models were evaluated using transcript and protein evidence to select the most consistent model for each gene based on an Annotation Edit Distance (AED) value by performing first-pass gene annotation using MAKER (51).
Estimation of Species Divergence Time.
We used MCScanX (52) to identify syntenic blocks (regions with at least five colinear genes) between species and calculate Ks rates for syntenic genes using the Nei–Gojobori method, and the median Ks value was used to estimate the divergence time based on the C. idella evolution coefficient (53).
Population Structure Analysis.
We used PLINK (54) (v1.9) to calculate the proportions of heterozygotes, missing genotypes, and inbreeding coefficients for each sample. A genetic relatedness matrix (GRM) of all pairwise comparisons of samples was calculated using Genome-wide Complex Trait Analysis (GCTA) (v1.26.0) software (55) and visualized using the R package pheatmap (56). Population structure inference was performed using ADMIXTURE (57) (v1.23).
Population Parameter Estimation.
The LD decay analysis for genotypes in the variant call format (VCF) file was carried out using popLDdecay (58). We estimated the population parameters genetic diversity (π) and genetic distance (Tajima’s D) for each group and calculated the statistic (Fst) to measure population differentiation by comparing all samples to the crucian carp subpopulation using VCFtools (59) for each nonoverlapping 50-kb genomic block across the whole genome.
Selective Sweep Detection.
A set of SNP marker-specific crucian carp (C16) and 33 representative goldfish (G33) were filtered using PLINK (54) with the parameters –mac 10 –geno 0.2. We used this marker set to perform population structure analysis (PCA and TreeMix for four groups), and the results were confirmed by the previous results in this study.
Case-Control GWAS of Morphologic Traits.
PLINK (54) was used to perform case-control GWAS. λGC is defined as the ratio between the 1-degree-of-freedom χ2 value of the median P value and 0.455—the 1-degree-of-freedom χ2 value for a P value of 0.5. The top 0.1% of genetic corrected P value (PGC) was used as a threshold.
Supplementary Material
Acknowledgments
We thank Tianshan Xue for collecting some goldfish samples. This study was supported by the 13th Five-Year Plan for the Marine Innovation and Economic Development Demonstration Projects (FZHJ14) and Science and Technology Program of Fuzhou (2018-N-27). Y.V.d.P. acknowledges funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (Grant Agreement 833522).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005545117/-/DCSupplemental.
Data Availability.
We have submitted the goldfish genome and annotation to National Center for Biotechnology Information (NCBI). The goldfish genome is available from SAMN12618612. Goldfish RNA-Seq data and quality-filtered Illumina reads for the 185 resequenced goldfish genomes, 16 crucian carp, and 6 Barbinae species were deposited into the NCBI BioProject database under accession number PRJNA561458.
References
- 1.Chen Z. et al., De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole genome duplication. Sci. Adv. 5, eaav0547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo J. et al., From asymmetrical to balanced genomic diversification during rediploidization: Subgenomic evolution in allotetraploid fish. Sci. Adv. 6, eaaz7677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hervey G. F., Billardon-Sauvigny E.; Muséum National d’Histoire , “Introduction” in The Goldfish of China in the XVIII Century, Hervey G. F., Ed. (China Society, 1950), pp. 2–3. [Google Scholar]
- 4.FAO , FAO Yearbook of Fishery and Aquaculture Statistics. http://www.fao.org/fishery/static/Yearbook/YB2018_USBcard/navigation/index_intro_e.htm. Accessed 13 October 2020.
- 5.Chen S. C., A history of the domestication and the factors of the varietal formation of the common goldfish, Carassius auratus. Sci. Sin. 5, 287–321 (1956). [Google Scholar]
- 6.Matsui Y., Genetical studies on gold-fish of Japan. 2. On the Mendelian inheritance of the telescope eyes of gold-fish. J. Imp. Fish. Inst. 30, 37–46 (1934). [Google Scholar]
- 7.Boulenger E. G., “The fresh-water aquarium” in The Aquarium Book, Boulenger E. G., Ed. (Duckworth, London, 1925), pp. 150–178. [Google Scholar]
- 8.Mullertt H., “The history of the goldfish” in The Goldfish and Its Systematic Culture, Mullertt H., Ed. (Clarke, Cincinnati, OH, 1883), pp. 7–8. [Google Scholar]
- 9.Darwin C. R., “Duck-goose-peacock-turkey-guinea fowl-canary-bird-goldfish hive bees-silk moths” in The Variation of Animals and Plants under Domestication, Darwin C., Ed. (John Murray, 1868), p. 313. [Google Scholar]
- 10.Bateson W., “Introduction” in Materials for the Study of Variation: Treated with Especial Regard to Discontinuity in the Origin of Species, Bateson W., Ed. (Macmillan, London, 1894), pp. 1–75. [Google Scholar]
- 11.Chen S. C., Variation in external characters of Goldfish, Carassius auratus. Contrib. Biol. Lab. Sci. Soc. China Nanking 1, 1–64 (1925). [Google Scholar]
- 12.Chen S. C., Transparency and mottling, a case of mendelian inheritance in the goldfish Carassius auratus. Genetics 13, 434–452 (1928). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koren S. et al., Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu W., Ma X., Zhang Y., Li W., Wang Y., A novel conformable fractional non-homogeneous grey model for forecasting carbon dioxide emissions of BRICS countries. Sci. Total Environ. 707, 135447 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., et al. , Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 50, 1565–1573 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Liu H. et al., A high-density genetic linkage map and QTL fine mapping for body weight in crucian carp (Carassius auratus) using 2b-RAD sequencing. G3. G3 (Bethesda) 7, 2473–2487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parra G., Bradnam K., Korf I., CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Xu P. et al., Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46, 1212–1219 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Howe K. et al., The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGaugh S. E. et al., The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5, 5307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasahara M. et al., The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ohno S., Muramoto J., Christian L., Atkin N. B., Diploid-tetraploid relationship among old-world members of the fish family Cyprinidae. Chromosoma 23, 1–9 (1967). [Google Scholar]
- 24.Liu S. et al., Genomic incompatibilities in the diploid and tetraploid offspring of the goldfish × common carp cross. Proc. Natl. Acad. Sci. U.S.A. 113, 1327–1332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Gan X., Li J., Chen Y., He S., Cyprininae phylogeny revealed independent origins of the Tibetan Plateau endemic polyploid cyprinids and their diversifications related to the Neogene uplift of the plateau. Sci. China Life Sci. 59, 1149–1165 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Session A. M. et al., Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smartt J., “Introduction” in Goldfish Varieties and Genetics: A Handbook for Breeders, Smartt J., Ed. (Blackwell Science, 2008), pp. 1–10. [Google Scholar]
- 28.DeGiorgio M., Huber C. D., Hubisz M. J., Hellmann I., Nielsen R., SweepFinder2: Increased sensitivity, robustness and flexibility. Bioinformatics 32, 1895–1897 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Blenski P., Darwin: Voyage of the Beagle. Booklist 18, 40 (2019). [Google Scholar]
- 30.Zahir F. R. et al., A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1α. J. Med. Genet. 45, 239–243 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Kang J. et al., Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Easty D. J., Gray S. G., O’Byrne K. J., O’Donnell D., Bennett D. C., Receptor tyrosine kinases and their activation in melanoma. Pigment Cell Melanoma Res. 24, 446–461 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y. M. et al., Distant insulin signaling regulates vertebrate pigmentation through the Sheddase Bace2. Dev. Cell 45, 580–594.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochin L. et al., BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10658–10663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaillon O. et al., Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Nakatani Y., Takeda H., Kohara Y., Morishita S., Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17, 1254–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spicer R. A. et al., Constant elevation of southern Tibet over the past 15 million years. Nature 421, 622–624 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Fawcett J. A., Maere S., Van de Peer Y., Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. U.S.A. 106, 5737–5742 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van de Peer Y., Mizrachi E., Marchal K., The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Bird K. A., VanBuren R., Puzey J. R., Edger P. P., The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 220, 87–93 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Xu P. et al., The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat. Commun. 10, 4625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comai L., The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Wang M. et al., Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Kon T. et al., The genetic basis of morphological diversity in domesticated goldfish. Curr. Biol. 30, 2260–2274.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Braasch I., Genome evolution: Domestication of the allopolyploid goldfish. Curr. Biol. 30, R812–R815 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Burton J. N. et al., Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker B. J. et al., Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang H. et al., ALLMAPS: Robust scaffold ordering based on multiple maps. Genome Biol. 16, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao Z., Eddy S. R., Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12, 1269–1276 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price A. L., Jones N. C., Pevzner P. A., De novo identification of repeat families in large genomes. Bioinformatics 21 (suppl. 1), i351–i358 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Cantarel B. L. et al., MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Li J., Paterson A. H., MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29, 1458–1460 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Wang Y. et al., The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 47, 625–631 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Chang C. C. et al., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J., Lee S. H., Goddard M. E., Visscher P. M., GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolde R., pheatmap: Pretty Heatmaps. R Version 1.0.12. https://CRAN.R-project.org/package=pheatmap. Accessed 13 October 2020.
- 57.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C., Dong S. S., Xu J. Y., He W. M., Yang T. L., PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Danecek P. et al.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have submitted the goldfish genome and annotation to National Center for Biotechnology Information (NCBI). The goldfish genome is available from SAMN12618612. Goldfish RNA-Seq data and quality-filtered Illumina reads for the 185 resequenced goldfish genomes, 16 crucian carp, and 6 Barbinae species were deposited into the NCBI BioProject database under accession number PRJNA561458.