Digitized natural history records, now numbering in the billions (1), span widely across the tree of life and provide the foundation for numerous recent advances in biodiversity research (2, 3). Mechanistic insights are emerging for old questions, including how diversity has expanded and contracted through Earth’s history (4), how species have come to occupy the wide range of ecological roles observed on land and sea alike (5, 6), and how the millions of species on Earth will respond to a rapidly changing climate in the future (7). Fundamentally, such studies require an understanding of both how individual organisms are classified to species and how species are related in their evolutionary history. In deep time, where fossils provide scattered snapshots of historical diversity, taxonomic resolution is particularly elusive. Molecular data are entirely absent, the fossil record contains numerous gaps in time and space, and fossil preservation presents a host of challenges for evaluating the shape and structure of diagnostic anatomical traits. Numerous fossil specimens remain poorly resolved, particularly in their evolutionary relationships to modern taxa, clouding the temporal and geographic resolution of biodiversity in deep time. For widespread ecologically important clades like plants, this limits our ability to reconstruct the dynamics of ancient ecosystems (8).
In PNAS, Romero et al. (9) present a deep-learning–based approach for classifying some of the fossil record’s most widely documented yet vexing historical material—fossil pollen (8, 10). Paired with an ecological and climatic understanding of the distributions of plant groups today, taxonomically resolved pollen studies provide an important lens for paleobotanical diversity and data for paleoclimatic inference (8, 10). Romero et al. (9) show how this record can be further refined with deep learning. The authors examine a locally rare but geographically widespread fossil morphospecies historically distributed through Africa and South America between 59.2 and 7.2 Ma. They classify individual fossil specimens according to their affiliation with modern plant genera (Fig. 1) and, in turn, suggest that the nanoscale diversity of pollen shape and texture in this widespread taxon represents far greater evolutionary diversity than appreciated so far. When classified into modern genera, the ages of these fossils imply evolutionary splits that predate those established with molecular techniques, impacting estimates of diversity through time and our understanding of the rise of species. Other identifications suggest modern African genera were previously established in South America and have since gone locally extinct, with implications for biogeography and historical distribution of ecological diversity. The authors provide an in-depth comparison between the deep-learning–based classification and a traditional morphometric approach that relies on measurements obtained by hand from images. The analysis reveals how deep-learning–based taxonomic identifications for pollen fossils can reshape our understanding of when and where plants were historically distributed and, in turn, how they evolved.
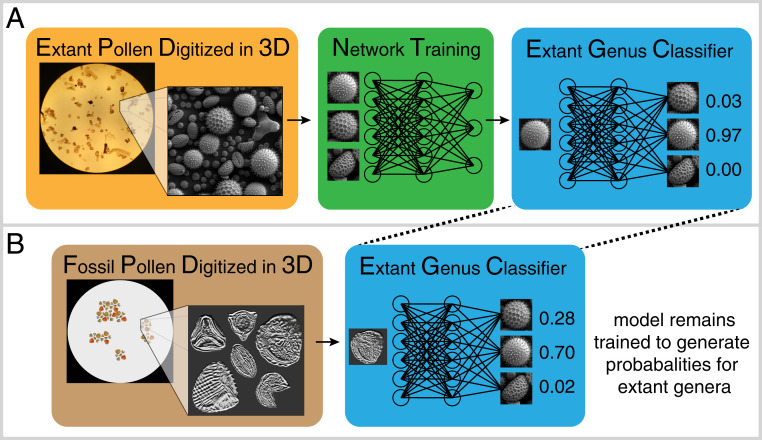
Fig. 1.
A workflow for classifying fossil pollen with deep learning. (A) Extant pollen specimens are digitized using a nondestructive microscopy technique. Images are then used to train a neural network (middle block). Once trained, the model processes images and generates probabilities for membership in modern genera. (B) Fossil pollen is processed with the same microscopy technique, and images are passed through the trained classifier from A. The model quantifies the membership of fossil specimens in modern taxonomic groups. Images shown here are merely illustrations.
The crux of the authors’ approach is the combination of recently developed high-resolution microscope technology (11) with deep convolutional neural networks (CNNs), powerful machine-learning models developed for image pattern recognition and classification. Neural networks are not new—early concepts were developed in the mid-20th century and CNNs emerged in the 1980s—but computational advances have rapidly improved their accuracy over the last decade (12). The methodological framework is relatively straightforward—a dataset of images with known labels is used to train a model to generate accurate classifications for data with unknown labels. Training is accomplished by feeding the known data (e.g., red, green, and blue color values contained in the pixels of a digitized image) through the model and iteratively adjusting model parameters to generate accurate labels (e.g., taxon known to be represented in the image). Protocols and algorithms for training accurate CNNs are rapidly advancing, and transfer learning (13), a process by which pretrained CNNs are modified and adjusted for new applications, has lowered the threshold for the number of training images required, allowing relatively data depauperate implementations. Where thousands of examples per class were once needed, Romero et al. (9) train a relatively accurate model with 16 extant plant genera and a mere 459 specimens.
The power of CNNs is that they are “deep nets”—multilayered statistical models that interpret information hierarchically to generate accurate predictions. For pollen, this means early model layers likely represent generalized shapes (oval, oblong, spherical) and deeper layers represent fine-scale differences in surface texture. This is why transfer learning works—a CNN trained to identify household items has already learned quite a bit about recognizing shapes. This hierarchical interpretation of visual cues is also not unlike the way a traditional taxonomist might work to classify an organism, and deep learning is now being applied widely for automated species detection and ecosystem monitoring (reviewed in ref. 14) and was recently developed in a study of extant pollen (15). Romero et al. (9) show how CNNs trained to identify modern taxa can be paired with imaging techniques to classify organisms deep in the fossil record.
The models developed by Romero et al. require a specific type of microscope image as input, so in this way restrict the widespread generality of their model to other forms of data. This disadvantage is far outweighed, however, by the fact that deep learning applied in this context is “trait agnostic”—because the input is an entire image or set of three-dimensional (3D) image slices, the relevant features that discriminate between classes are not predefined but rather learned innately. Traditional approaches, and indeed many other machine-learning algorithms, require a predefined set of traits (“features” in machine-learning parlance) on which to learn (16); assessments of morphological diversity through time and space thus rely on which traits are chosen. The automated learning of discriminative features through deep learning instead allows an objective quantification of highly complex traits like 3D shapes, as shown by Romero et al. (9), or colors, as was recently examined in moths (17). There are certainly drawbacks—deep-learning models are often constrained by the set of classes used for training, so called “supervised” learning, but new methods are being developed to cope with taxonomic error in species identification (18). Perhaps more fundamentally, the biological significance and origin of complex learned features are not obvious, and methods are needed to understand the biological basis of deep-learning–based classifications. Advances in the science of deep learning are coming rapidly and require time to be integrated in a biodiversity context. For example, the geometry (19) and the intrinsic dimensionality (20) of the learned feature space may yet hold promise for understanding how deep-learning models quantify the physical differences between taxonomic groups, potentially allowing metric assessments of learned representations across disparate taxonomic groups.
Sophisticated digitized images of biological specimens are now a critical piece of the biodiversity information pipeline (21). The utility of these specimen-based artifacts has yet to be fully explored, however, and many challenges remain. Foremost, mass digitization (imaging) of specimens, not to mention specimen preservation itself, was in many cases initiated before the potential of deep learning in biodiversity research was well understood. Making use of biological material that was otherwise not intended to be photographed raises numerous questions about how these massive samples of convenience can and should be used to generate deep-learning–based insights. In some cases, as in Romero et al.’s contributions here, purpose-built deep-learning models will be needed to process the products of specific imaging techniques. In all cases, however, biologists will need to carefully consider how the hypothesis testing framework and likelihood-based methods of today can be integrated with the complex and relatively understudied science of deep learning. With over 800,000 unidentified fossil records currently in the world’s primary biodiversity data aggregator (1, 22), there are countless insights on the biodiversity of deep time waiting to be uncovered.
Footnotes
The author declares no competing interest.
See companion article, “Improving the taxonomy of fossil pollen using convolutional neural networks and superresolution microscopy,” 10.1073/pnas.2007324117.
References
- 1.The Global Biodiversity Information Facility , What is GBIF? https://www.gbif.org/what-is-gbif. Accessed 6 October 2020.
- 2.Funk V. A., Collections‐based science in the 21st century. J. Syst. Evol. 56, 175–193 (2018). [Google Scholar]
- 3.Nelson G., Ellis S., The history and impact of digitization and digital data mobilization on biodiversity research. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20170391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knope M. L., Bush A. M., Frishkoff L. O., Heim N. A., Payne J. L., Ecologically diverse clades dominate the oceans via extinction resistance. Science 367, 1035–1038 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Pigot A. L., et al. , Macroevolutionary convergence connects morphological form to ecological function in birds. Nat. Ecol. Evol. 4, 230–239 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Sallan L., Friedman M., Sansom R. S., Bird C. M., Sansom I. J., The nearshore cradle of early vertebrate diversification. Science 362, 460–464 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Trisos C. H., Merow C., Pigot A. L., The projected timing of abrupt ecological disruption from climate change. Nature 580, 496–501 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Mander L., Punyasena S. W., On the taxonomic resolution of pollen and spore records of Earth’s vegetation. Int. J. Plant Sci. 175, 931–945 (2014). [Google Scholar]
- 9.Romero I. C., et al. , Improving the taxonomy of fossil pollen using convolutional neural networks and superresolution microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 28496–28505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards K., Fyfe R., Jackson S., The first 100 years of pollen analysis. Nat. Plants 3, 17001 (2017). [Google Scholar]
- 11.Romero I. C., Urban M. A., Punyasena S. W., Airyscan superresolution microscopy: A high-throughput alternative to electron microscopy for the visualization and analysis of fossil pollen. Rev. Palaeobot. Palynol. 276, 104192 (2020). [Google Scholar]
- 12.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Yosinski J., Clune J., Bengio Y., Lipson H., How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 27, 3320–3328 (2014). [Google Scholar]
- 14.Christin S., Hervet É., Lecomte N., Applications for deep learning in ecology. Methods Ecol. Evol. 10, 1632–1644 (2019). [Google Scholar]
- 15.Dunker S., et al. , Pollen analysis using multispectral imaging flow cytometry and deep learning. New Phytol., 10.1111/nph.16882 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Bengio Y., Courville A., Vincent P., Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Wu S., et al. , Artificial intelligence reveals environmental constraints on colour diversity in insects. Nat. Commun. 10, 4554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villon S., et al. , A new method to control error rates in automated species identification with deep learning algorithms. Sci. Rep. 10, 10972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen U., Chung S., Lee D. D., Sompolinsky H., Separability and geometry of object manifolds in deep neural networks. Nat. Commun. 11, 746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansuini A., Laio A., Macke J. H., Zoccolan D., Intrinsic dimension of data representations in deep neural networks. Adv. Neural Inf. Process. Syst. 32, 6111–6122 (2019). [Google Scholar]
- 21.Hedrick B. P., et al. , Digitization and the future of natural history collections. Bioscience 70, 243–251 (2020). [Google Scholar]
- 22.The Global Biodiversity Information Facility , GBIF occurrence download. GBIF.org. 10.15468/dl.u73tcg. Accessed 6 October 2020. [DOI]