Significance
Kinetoplastids is a group of flagellated unicellular eukaryotic parasites including human pathogens, such as Trypanosoma cruzi and Leishmania spp., etiologic agents of Chagas disease and leishmaniasis. They are potentially lethal, affecting more than 20 million people worldwide. Therapeutic strategies are extremely limited and highly toxic. The finding of new molecular targets represents one venue for the development of new therapeutic strategies. In this work, we present the structure of the full mitoribosomes from two kinetoplastids, Leishmania tarentolae and T. cruzi, thus far uncharacterized. We also reveal the structure of an large subunit assembly intermediate harboring 16 different factors. In addition to defining a subset of novel kinetoplastid-specific factors, our results shed light on the mitochondrial ribosomal RNA maturation process.
Keywords: kinetoplastids, mitochondrial ribosome, cryo-electron microscopy, ribosome biogenesis, mRNA translation
Abstract
Kinetoplastids are unicellular eukaryotic parasites responsible for such human pathologies as Chagas disease, sleeping sickness, and leishmaniasis. They have a single large mitochondrion, essential for the parasite survival. In kinetoplastid mitochondria, most of the molecular machineries and gene expression processes have significantly diverged and specialized, with an extreme example being their mitochondrial ribosomes. These large complexes are in charge of translating the few essential mRNAs encoded by mitochondrial genomes. Structural studies performed in Trypanosoma brucei already highlighted the numerous peculiarities of these mitoribosomes and the maturation of their small subunit. However, several important aspects mainly related to the large subunit (LSU) remain elusive, such as the structure and maturation of its ribosomal RNA. Here we present a cryo-electron microscopy study of the protozoans Leishmania tarentolae and Trypanosoma cruzi mitoribosomes. For both species, we obtained the structure of their mature mitoribosomes, complete rRNA of the LSU, as well as previously unidentified ribosomal proteins. In addition, we introduce the structure of an LSU assembly intermediate in the presence of 16 identified maturation factors. These maturation factors act on both the intersubunit and the solvent sides of the LSU, where they refold and chemically modify the rRNA and prevent early translation before full maturation of the LSU.
Kinetoplastids are unicellular eukaryotic parasites, causative agents of several human and livestock pathologies (1). They are potentially lethal, affecting more than 20 million people worldwide (1). Owing in part to their parasitic nature, they strongly diverged from other eukaryotic model species. Kinetoplastids evolved to live in and infect a large variety of eukaryotic organisms in very different molecular environments. Consequently, beyond the general similarities, kinetoplastid species have diverged evolutionarily from one another, and their protein sequence identity can be relatively low. They have a single large mitochondrion, a crucial component of their cellular architecture (2), where gene expression machineries have also largely diverged, notably their mitochondrial ribosomes (mitoribosomes) (3–10). These sophisticated RNA-protein complexes translate the few mRNAs still encoded by mitochondrial genomes. The mitoribosomes’ composition and structure diverged greatly from their bacterial ancestor, with the kinetoplastid mitoribosomes the most extreme case described to date, with highly reduced rRNAs and more than 80 supernumerary ribosomal proteins (r-proteins) compared with bacteria, completely reshaping the overall ribosome structure.
Recent structural studies performed in Trypanosoma brucei have highlighted the particularities of this mitoribosome structure and composition, as well as the assembly processes of the small subunit (SSU) (3, 6). However, despite the very comprehensive structural characterization of the full T. brucei SSU and its maturation, several pivotal aspects related to the large subunit (LSU) have remained uncharacterized. For instance, a large portion of the LSU at the intersubunit side, including the whole rRNA peptidyl-transfer center (PTC) along with several r-proteins, were unresolved (3). Moreover, in contrast to the SSU, nearly nothing is known about LSU maturation and assembly. More generally, the maturation of the mitoribosomes in all eukaryotic species remains largely underexplored. Here we present a cryo-electron microscopy (EM) investigation of the full mature mitoribosomes from two different kinetoplastids, Leishmania tarentolae and Trypanosoma cruzi. In addition, we reveal the structure of an assembly intermediate of the LSU displaying unprecedented details on rRNA maturation in these very singular mitoribosomes, some of which likely can be generalized to the maturation of all rRNA.
To obtain high-resolution reconstructions of the full kinetoplastid mitoribosomes, we purified mitochondrial vesicles from both L. tarentolae and T. cruzi and directly purified the mitoribosomes from the sucrose gradient (Methods) (SI Appendix, Fig. S1). All of our collected fractions were analyzed by nano-liquid chromatography tandem mass spectrometry (LC-MS/MS) (SI Appendix, Tables S1 and S2) to determine their proteomic composition. We collected micrographs from multiple vitrified samples corresponding to different sucrose gradient density peaks for both species, and following image processing, we obtained cryo-EM reconstructions of the full mitoribosomes, as well as of the dissociated SSU. Moreover, we also derived reconstructions of what appeared to be an assembly intermediate of L. tarentolae LSU. After extensive rounds of two-dimensional (2D) and three-dimensional (3D) classification and refinement we obtained the structure of L. tarentolae and T. cruzi complete and mature mitoribosomes at 3.9 Å and 6 Å, respectively (SI Appendix, Figs. S2–S4). Other notable features include the intersubunit contacts and two distinct rotational states in T. cruzi. Similarly to T. brucei (3), our cryo-EM analysis revealed a reconstruction of an early initiation complex from T. cruzi at 3.1 Å for the body and 3.2 Å for the head of the SSU (SI Appendix, Fig. S5). Further focused refinement on the LSU, SSU head, and SSU body generated reconstructions at 3.6, 3.8, and 4 Å, respectively, for L. tarentolae (SI Appendix, Figs. S2 and S3), and 3.7 and 4.5 Å for T. cruzi LSU and SSU, respectively (SI Appendix, Fig. S4). Combined, these reconstructions, along with the MS data allowed us to build nearly complete atomic models, with only few protein densities remaining unidentified.
General Description of Mature Kinetoplastid Mitoribosomes
Even though T. cruzi and L. tarentolae proteins are of relative modest sequence identity (∼70%) for such closely related species (7), the tertiary structures of the r-proteins and rRNAs, as well as the overall structure of the mitoribosomes, were nearly identical (SI Appendix, Fig. S4). Both ribosomes are large complexes formed mainly by proteins (Fig. 1 and SI Appendix, Fig. S4), 68 in the LSU and 54 in the SSU (SI Appendix, Table S3), with rRNA constituting only 15% of the total mass. The SSU was reconstructed to high resolution and appears similar to what was previously observed (3, 6). We obtained reconstructions of the latter in the context of the mature ribosomes from L. tarentolae and T. cruzi, but also dissociated in the context of what appears to be a partial initiation complex from T. cruzi, bound to mt-IF3, which accumulated naturally in our samples (Fig. 1 and SI Appendix, Fig. S5). In the presence of mt-IF3, the 9S rRNA (in particular the highly reduced helix 44) and uS12m are clearly stabilized (SI Appendix, Fig. S5E); however, in the context of both mature ribosomes, mt-IF3 is no longer present, and the 9S rRNA along with uS12m at the subunit interface are significantly more flexible, resulting in a lower resolution. In the T. cruzi mitoribosome reconstructions, two distinct rotational states were observed, strongly suggesting a fully assembled mitoribosome structure capable of undergoing different conformational states, even considering the flexible h44 and uS12m region (SI Appendix, Fig. S4). In contrast to what is observed in prokaryotes and other mitoribosomes, very few of the intersubunit bridges are strictly conserved, and most of them rely on protein–protein interactions. However, more of the observed intersubunit bridges are spatially conserved compared with other known ribosomes but involve kinetoplastid-specific r-proteins (SI Appendix, Fig. S8).
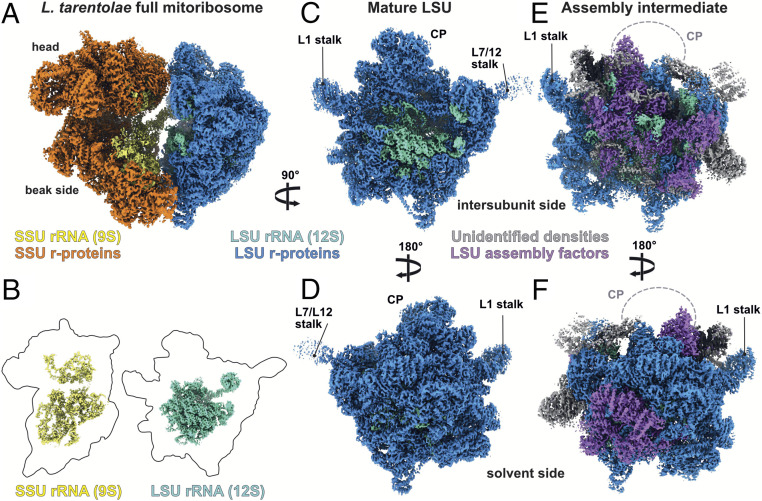
Fig. 1.
Cryo-EM reconstructions of the mature and assembly intermediate L. tarentolae mitoribosome. (A) Composite cryo-EM map of the full and mature mitoribosome of L. tarentolae. SSU is depicted in orange shades, with proteins in orange and rRNA in yellow, and LSU is depicted in blue, with proteins in dark blue and rRNA in aquamarine. (B) Relative sizes of the rRNA (colored segmented densities) compared with the r-proteins (thick black contours). (C–F) Cryo-EM reconstructions of the mature LSU viewed from the intersubunit (C) and solvent (D) sides, compared with the LSU assembly intermediate subunit (E and F). Densities corresponding to assembly factors are shown in purple; gray represents unidentified factors.
The reconstruction of the LSU derived from the full mature ribosome (Fig. 1) allowed us to build the entire 12S rRNA, including domains IV and V at the interface subunit (Figs. 1 B and C and 2), revealing the catalytic PTC as well as its intertwined interactions with r-proteins. The structure of domains IV and V is globally conserved even when compared with classical prokaryotic ribosomes (Fig. 2). Among the 68 LSU characterized proteins, uL2m, uL14m, and extensions of bL19m and mL68, previously unaccounted for (3), were visualized and detected by MS (Fig. 3). These proteins and extensions squeeze through the rRNA and stabilize it in a very intricate manner compared with the classical prokaryotic and cytosolic ribosomes. Surprisingly, compared to what was previously described on the solvent side of the LSU in T. brucei (3), proteins mL67, mL71, mL77, mL78 and mL81, localized close to the peptide channel exit, are absent (Figs. 1 and 3). However, these proteins are present in the assembly intermediate, as described and discussed below.
Fig. 2.
Comparison of the mature and assembly intermediate 12 rRNA. Comparison between mature (A–C) and assembly intermediate (D–F) 12S rRNA. Each rRNA domains is colored differently. The rRNA is presented from the intersubunit view (B–E) and a side view (C–F). (A) Superimposition of the 12S mature rRNA with E. coli 23S rRNA, shown in gray, demonstrating that the structure of domain IV (orange) and the PTC are highly conserved. The main differences between the mature and assembly intermediate rRNA are observed in domain II, where the L7/12 stalk is folded differently; in domain IV, which is entirely unfolded (D); and in domain V, which is unfolded from H90 to H92 (E) and where H75 is entirely lifted up (F). Most of the maturation factors act on domain V, which is accessible because of the unfolded domain IV.
Fig. 3.
Atomic models of L. tarentolae mitoribosome and LSU assembly intermediate. (A) Overall model of L. tarentolae mitoribosome. 12S rRNA is colored in cyan (LSU) and the 9S rRNA in brick (SSU). Ribosomal proteins are shown in shades of blue (LSU) and shades of yellow (SSU), according to their conservation. (B and C) Solvent side (B) and intersubunit side (C) of the mature LSU with each individual r-proteins annotated and displayed with different colors. (D and E) Solvent side (D) and intersubunit side (E) of the LSU assembly intermediate. Conserved proteins in the mature LSU (B and C) are shown in gray, and assembly factors are colored in different shades and individually annotated.
General Description of the LSU Assembly Intermediate
During L. tarentolae 3D classification, a class of particles presenting LSU-like features was found to naturally accumulate in our sample (SI Appendix, Fig. S2). This class is rather distinct from classical LSU classes observed previously. After refinement and postprocessing, this class resulted in a 3.4-Å reconstruction of the complex (Fig. 1). Comparison of this reconstruction with the mature LSU allowed us to visualize significant differences. Indeed, a large portion of the 12S rRNA appeared to be unfolded, or at least highly flexible at the intersubunit face (Fig. 2). In addition, large well-resolved protein densities were observed occupying the position of the rRNA at this region. Moreover, the whole central protuberance (CP) was missing, and several additional protein densities were observed on the solvent side (Fig. 1 E and F). Therefore, this complex clearly appears to correspond to an LSU assembly intermediate. Our cryo-EM reconstructions, along with the LC-MS/MS analysis, allowed us to build an atomic model of this LSU assembly intermediate (Fig. 3 D and E). Compared with the mature LSU, this assembly intermediate lacks 18 core r-proteins but includes 16 maturation factors (SI Appendix, Fig. S6), including mL67, mL71, mL77, mL78, and mL81, which were previously described as r-proteins (3). On the interface, domain IV of the 12S rRNA appears to be unfolded, and several portions of the rRNA undergo refolding or modifications. Since this assembly intermediate is similar in part to the state B recently described for T. brucei, the factor names presented here use the same nomenclature (11).
Solvent Side
On the solvent side, close to the peptide channel exit, a crown of proteins composed of mL67, mL71, mL77, mL78, and mL81 was observed in the assembly intermediate (Fig. 4), but not on the full mature mitoribosome. The analysis of their structure and interaction network suggests two main roles. Parts of them hold the 3′ and 5′ extremities of the 12S rRNA. Indeed, the 5′ end of the 12S rRNA is surrounded by several positively charged residues of mL67 that coordinate and stabilize this extremity (Fig. 4A and SI Appendix, Fig. S7). On the 3′ end, mL67 again, with the help of mL81 as well as r-proteins bL17m and mL85, stabilize this extremity, which is more flexible in the mature LSU (Fig. 4B), as suggested by its lower density in the latter. Thus, in the context of the assembly intermediate, these proteins (mainly mL67) hold both 12S rRNA extremities, most likely stabilizing the rRNA in its r-protein shell during its maturation.
Fig. 4.
Specific factors hold the 12S rRNA termini and probe the peptide channel exit on the solvent side of the immature LSU. On the solvent side, five maturation factors act together to hold the extremities of the rRNA (A and B) and probe the peptide exit channel (C and D). The 12S 5′ termini (A) is held by several positively charged residues of mL67, and the 3′ extremity (B) is stabilized by mL67 and mL81 as well as by r-proteins mL85 and bL17m. U34 and U1176 indicate the terminal rRNA residues. The very start and end of the 12S rRNA are not visible, as they are disordered in the solvent (SI Appendix, Fig. S7 A and B). (C and D) The peptide channel of the assembly intermediate (Top) and mature (Bottom) LSU from top (C) and cut (D) views. Proteins mL71, mL77, and mL78 reshape the peptide exit channel by slightly extending and splitting it in two in the assembly intermediate compared with the mature LSU (C). Dashed white contours delimit the peptide channel exit orifices in the mature and immature LSUs. Maturation factor mL71 completely blocks the exit channel through its N-terminal domain (D). Dashed yellow lines delimit the peptide channel in the longitudinal axis. In C and D, proteins and rRNAs are depicted in surface representation with cyan-colored rRNA in D.
Moreover, next to the rRNA extremities, the peptide channel exit is entirely different than the mature LSU, as proteins mL71, mL77, and mL78 reshape the peptide exit. These proteins slightly extend and split the channel exit into two parts (Fig. 4C); however, the channel itself is completely blocked by the N-terminal part of mL71, preventing the subunit from engaging in active translation (Fig. 4D), similar to mL45, which prevents translation until the mitoribosome is bound to the mitochondrial inner membrane in mammals (12). On maturation, these factors are disengaged, and a classical peptide channel is restored, similar in structure to bacterial ribosomes (Fig. 4 C and D and SI Appendix, Fig. S7). Thus, we hypothesize that these proteins, previously observed in T. brucei (3), are actually maturation factors, and that the LSU reconstruction of T. brucei is more likely to be a late assembly intermediate (Fig. 5B). On their dissociation from the mature LSU, a novel protein that we term mL109 replaces mL67 and could mark the maturation of this region (Fig. 4).
Fig. 5.
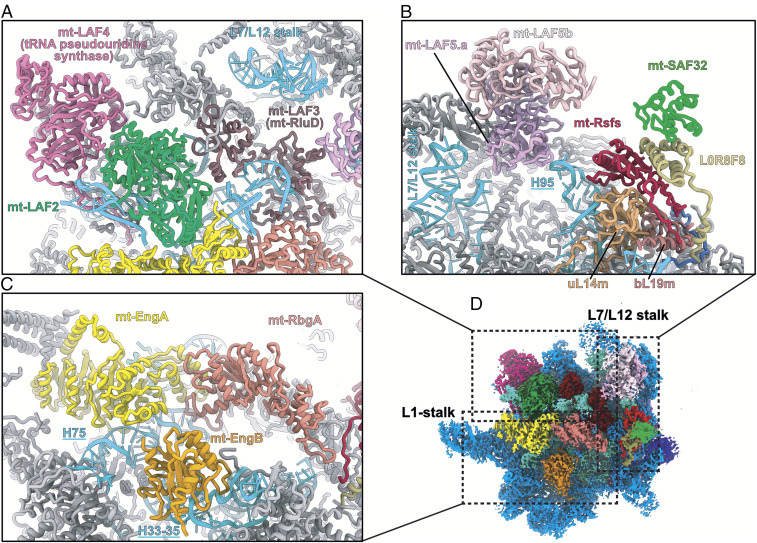
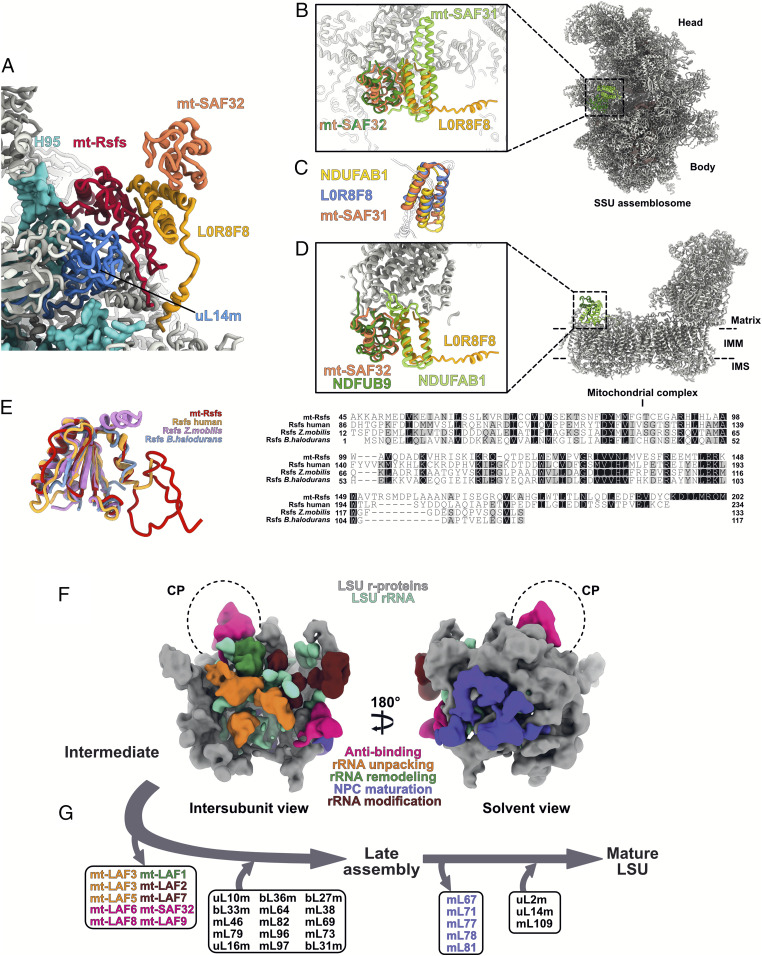
Key factors and schematic view of the stepwise assembly. (A) Zoom-in view of the mt-Rsfs:L0R8F8:mt-SAF32 factors involved in anti-subunit association. mt-Rsfs is the homolog of the bacterial protein Rsfs (F), and mt-SAF32 and L0R8F8 are also found in mitochondrial respiratory complex I as supernumerary proteins. (B) mt-SAF32 and L0R8F8 superimposed with mt-SAF32 and mt-SAF31 found in the SSU assemblosome of T. brucei (6) (PDB ID code 6SGB). (C) L0R8F8, NDUFAB1, and mt-SAF31 are LYR proteins characterized by the LYR-protein fold (three antiparallel helices). IMM, inner mitochondrial membrane; IMS, for intermembrane space. (D) mt-SAF32 and L0R8F8 superimposed with NDUFB9 and NDUFAB1, respectively, here from the yeast mitochondrial complex I (34) (PDB ID code 6RFR). Homolog proteins are annotated using the human nomenclature of complex I proteins. (E) The high structural conservation of Leishmania mt-Rsfs is illustrated by structural superimposition and sequence alignment with the human mitochondrial Rsfs (PDB ID code 5OOL) as well as two bacterial Rsfs (PDB ID codes 3UPS and 2O5A). (F) Schematic view of the LSU assembly intermediate of L. tarentolae presented from the intersubunit and solvent sides. Maturation factors are colored by function. (G) Starting from the LSU assembly intermediate in F, the dissociation of maturation factors and the association of r-proteins are shown, considering the T. brucei LSU (3) as a late assembly intermediate. Maturation factors are colored as in F, and r-proteins are shown in black.
Intersubunit Side
On the intersubunit side, a large portion of the 12S rRNA, corresponding to domain IV, appears to be unfolded, and several proteins occupy its position, acting on the immature domain V. Based on our cryo-EM reconstructions coupled with LC-MS/MS analysis and structural and sequence homologies, 11 factors were identified bound to the intersubunit side (Fig. 6). We term these proteins “mt-LAFs” for mitochondrial LSU assembly factors, consistent with the proposed nomenclature (6, 11). Some of these factors belong to the family of GTPases and RNA helicases/isomerases, classic actors in ribosome biogenesis, and were observed with their respective ligands and rRNA targets (Fig. 7E). Others are RNA modification enzymes. Some of these factors exhibit homology with previously identified bacterial and eukaryotic maturation factors; however, most of them have never been structurally captured in an assembly intermediate context.
Fig. 6.
Overall view of the maturation factors on the intersubunit side. On the intersubunit side of the immature LSU, 11 maturation factors act together to remodel and modify the rRNA (D). (A) mt-LAF4 and mt-LAF2 occupy the position of the CP. Mt-LAF2 and mt-LAF3 interact with remodeled rRNA. (B) Mt-Rsfs together with L0R8F8 and mt-SAF32 interact with uL14m and H95 to block the SSU association. (C) Arrangement of the GTPase triptych. The GTPases mt-EngA and mt-EngB bind the rRNA H75 and H37, respectively, while mt-RbgA occupies a central place in the complex and creates a bridge between the GTPases and mt-LAF3 and mt-Rsfs.
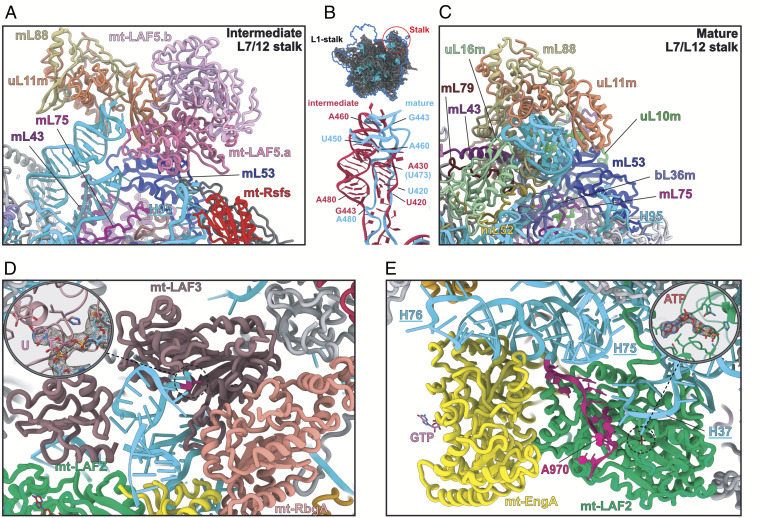
Fig. 7.
Maturation of the Leishmania mitochondrial LSU rRNA. (A) Details of how maturation factors interact with and affect the 12S RNA L7/12 stalk region of the intermediate LSU. The mt-LAF5 homodimer contacts uL11m and mL53. The two helices of the immature L7/12 stalk, as well as the helix H95, are adjacent to the dimer. (B) Overlap of the two forms of rRNA L7/12 stalk structure, with immature shown in red and mature in blue. (C) L7/12 stalk region of the mature LSU. (D) Insight into the rRNA modification by the pseudouridine synthase mt-LAF3. For catalyzation, the rRNA is refolded into a helix structure, probably to be recognized by mt-LAF3, and the uridine substrate is flipped out of the helix and engaged in the catalytic core of the enzyme (SI Appendix, Fig. S7C). (E) The two factors mt-EngA and mt-LAF2 bind jointly to the rRNA. The DEAD-box helicase mt-LAF2 is in an active state with a single-strand RNA of the immature H88, shown in burgundy, in its catalytic core and loaded with an ATP molecule. The dotted circle corresponds to magnified region.
The first noticeable difference with the mature LSU is the absence of a CP. Indeed, eight proteins composing the CP are missing, and some of the surrounding proteins adopt different conformations (SI Appendix, Fig. S9). In the assembly intermediate, the binding of the CP block is prevented by mt-LAF2 and mt-LAF4 (Fig. 6A and SI Appendix, Figs. S9 and S7). The factor mt-LAF2 is a DEAD-box RNA helicase in which both the catalytic and ATP-binding sites are conserved (13) and the ATP ligand is clearly observed (Fig. 7E). It occupies the spatial positions of bL27m, bL31m, bL33m, and mL82. The role of this family of helicase proteins is to unwind RNA structures and/or RNA-protein complexes; thus, they are highly involved in ribosome maturation. Mt-LAF2 shows similarity with Has1, previously described in yeast pre-60S maturation, and with Mss116, involved in yeast mitoribosome maturation (14, 15) (SI Appendix, Fig. S10). Here it contacts two different segments of the 12S rRNA, the tip of H37 loop (residues 341 to 346), shifting the whole helix compared with the mature LSU (Fig. 7E), and a single strand (residues 968 to 975) that becomes part of H88 from domain V after its maturation. Thus, mt-LAF2 appears to be involved in the maturation of domain V rRNA. The second factor, mt-LAF4, is in direct contact with mt-LAF2 but does not contact rRNA. Compared with the mature LSU, it occupies the place of mL38, mL69, and mL82. It shows high homology with tRNA pseudouridine synthases; however, several residues of the active site are not conserved (SI Appendix, Fig. S11), hinting that the protein has lost its original function here. Its role seems to be as a placeholder for CP r-proteins, acting together with mt-LAF2 to prevent the premature association of these proteins. When mt-LAF2 and 4 are disengaged, the CP r-proteins (composed of eight proteins) could dock onto the LSU (SI Appendix, Fig. S9). Moreover, the mL59/64 C-terminal part that is flexible in the assembly intermediate could contact and stabilize the CP in the mature LSU context.
A large portion of the 12S rRNA is unfolded, which corresponds to the entire domain IV (Fig. 2), and several maturation factors are present in the region that will later be occupied by the rRNA after its maturation, including a triptych of GTPases: mt-RbgA, mt-EngB, and mt-EngA (Fig. 6C and SI Appendix, Fig. S9). While some of these GTPases have already been identified in bacteria (16, 17) (SI Appendix, Table S3), here they were observed acting together on a maturating ribosomal complex at the reported resolution. GTPases have widespread roles in the cell (18). These enzymes usually exist in two states, active and inactive, with the transition between the states triggered by GTP hydrolysis. This results in conformational changes of the enzyme that allow its binding to or release from specific targets. Mt-EngA, similar to its bacterial homolog, also known as GTPase Der, has two characteristic G-domains (16). Its role is crucial for domain IV PTC folding in bacteria, a role that is likely conserved here for mt-EngA, given their similar binding sites. mt-EngB is bound to H33-35 and appears here to prevent uL2m association. Mt-RbgA (homolog to the bacterial GTPase RbgA) (19) is in contact with mt-mt-EngA and mt-EngB; however, it is the only GTPase that does not make significant contacts with the rRNA (Fig. 6C), suggesting that it either has already performed its function or is still awaiting its rRNA target (SI Appendix, Fig. S12B).
Three RNA modification enzymes were found in the assembly intermediate structure: one pseudouridine synthase, termed mt-LAF3, and two methyltransferases forming a homodimer, termed mt-LAF5a and b. The protein mt-LAF3 is a pseudouridine synthase D family enzyme that shows high homology with RluD, a bacterial enzyme responsible for the modification of several residues on H69 in domain IV of 23S rRNA (20, 21) (SI Appendix, Fig. S11). In our reconstruction, we clearly observe a uridine residue flipped out of an RNA regular helix to engage the catalytic core of the enzyme (Fig. 7D and SI Appendix, Fig. S7). Mt-LAF5a and b are RNA 2′O-methyltransferases belonging to the SpoU family, enzymes widely involved in methylation of the 23S rRNA in prokaryotes (22, 23) (SI Appendix, Fig. S13). They are located at the L7/12 stalk region (Fig. 7A). Surprisingly, the L7/12 stalk structure in the immature rRNA is radically different than its mature form. Indeed, the L7/12 stalk rRNA undergoes drastic structural rearrangements between the immature state, where it is shaped into two adjacent large regular helices (Fig. 7A), and the mature L7/12 stalk (Fig. 7C), where it does not form any RNA helices. Interestingly, even though the fold is radically different, the immature L7/12 stalk rRNA still spans the entire length and interacts with the distal uL11m (Fig. 7B) that retains its position in the mature L7/12 stalk. While the nearby homodimeric factors mt-LAF5.a/mt-LAF5.b are in close proximity to the rRNA, they cannot be unambiguously interpreted to bind it directly. mt-LAF5.a/mt-LAF5.b contact uL11m and mL53, inducing the shift of the L7/12 stalk r-protein block compared with the mature structure. Moreover, several surrounding r-proteins, such as uL10m and uL16m, are absent. Mt-LAF5a and b are most likely the homologs of the NHR SpoU described in Streptomyces actuosus (22), which are involved in an adenine methylation in H43 of the L7/12 stalk. Owing to their position in our assembly intermediate, it is possible that they modify the L7/12 stalk rRNA. The latter will be reshaped into its mature structure afterward, probably under the action of a helicase analog to mt-LAF2 (SI Appendix, Fig. S10C).
Finally, a complex formed by three assembly factors is found bound to uL14m, close to the L7/12 stalk. This complex is formed by mt-Rsfs, directly contacting uL14m and the tip of H95 (residues 1,133 to 1,136), mt-SAF32, and L0R8F8 (Fig. 5 A and 6 B and SI Appendix, Fig. S7). This complex is nearly identical to the MALSU1:L0R8F8:mt-ACP complex observed in the human mitoribosome LSU assembly intermediate (24). Indeed, each factor shows high homology to their mammalian homologs, with mt-SAF32 corresponding to mt-ACP and mt-Rsfs corresponding to MALSU1. The latter shows high homology to the conserved and well-characterized bacterial ribosome silencing factor (Rsfs) (Fig. 5E). Rsfs is known to prevent premature subunit association during diminished nutrient availability, a molecular mechanism mimicked by eIF6 in eukaryotes (25, 26). However, in mitoribosomes, as has been shown in humans, the Rsfs homolog alone is not able to prevent subunit association, and the anti-subunit association activity can be achieved only in the presence of mt-SAF32 and L0R8F8.
Discussion
Here we describe the structures of the full kinetoplastid mitoribosomes from L. tarentolae and T. cruzi. These reconstructions allowed us to characterize the complete 12S rRNA of the large subunit, revealing the as-yet uncharacterized domains IV and V including the PTC. While kinetoplastid mitoribosomes represent one of the most extreme cases of rRNA reduction (4, 6), these regions are structurally conserved compared with bacterial and other mitochondrial ribosomes (Fig. 2). These rRNA domains represent part of the conserved functional core of the ribosome in which the peptidyl transfer is catalyzed, in addition to serving as a conserved binding surface to the small subunit (27).
On the small ribosomal subunit, the 9S rRNA appears to be more flexible than its counterpart LSU 12S rRNA, as indicated by its lower densities at the intersubunit face (Fig. 1). In our study, its structure was derived from the SSU/mt-IF3 complex, in which the 9S rRNA densities are solid and interpretable, as corroborated by previous studies (3, 6). The reason for the accumulation of these SSU/mt-IF3 complexes is unclear, but it could reflect a relatively low level of translation initiation in T. cruzi, at least at the growth stage at which the parasites were harvested. This complex is also somewhat different from that observed in humans, where mt-IF3 does not contact uS12m and a N-terminal extension contacts uS7m and mS37 (28, 29), which was not observed here (SI Appendix, Fig. S5E). In kinetoplastids, mt-IF3 is required not only for proper translation initiation, but also for stabilization of the SSU subunit interface by locking uS12 and the reduced h44 helix. This requirement could be related to the fairly high hydrophobicity index of uS12m. Indeed, recruiting such a hydrophobic r-protein to the negatively charged rRNA surface may require the presence of mt-IF3 to stabilize it on the SSU intersubunit face rRNA (SI Appendix, Fig. S5). The biological rationale for the presence in kinetoplastids mitoribosomes of such a hydrophobic r-protein (uS12m) embedded in the rRNA at this strategic region remains to be unveiled but could be related to the regulation of the nuclear-mitochondrial crosstalk.
We also describe an assembly intermediate of the large subunit of the L. tarentolae mitoribosome. Assembly intermediates in mitoribosomes have only been structurally characterized in mammals for the LSU (24) and in T. brucei for the SSU (6). Here our reconstruction enabled the identification of 16 protein factors involved in various ways in the LSU maturation. Some of these factors appear to be conserved in bacteria, while the rest are specific to the kinetoplastid mitoribosome. The kinetoplastid-specific factors are found on a region of the LSU that has evolutionally diverged the most in terms of structure, i.e., on the solvent side of the LSU. The positions and interactions of these factors suggest two main roles. First, they maintain the partially unfolded and maturing rRNA 3′ and 5′ extremities in the LSU protein shell until the maturation of the 12S rRNA is complete (Fig. 4). Second, they prevent any premature translation by blocking the peptide channel exit. This molecular mechanism is a known feature of immature LSU/ribosomes. For instance, during cytosolic 60S maturation, the Rei1 protein probes and clots the nascent peptide tunnel (30). Another example is found in the human mitoribosome, where mL45 similarly blocks the channel until the binding of the ribosome to the inner mitochondrial membrane is complete (12).
The other maturation factors have homologs found in bacterial, cytosolic and mitochondrial ribosome biogenesis processes. They act on the intersubunit side of the LSU, where the 12S rRNA is mostly conserved and to some extent similar to bacteria. Among these conserved factors are three modification enzymes, two 2′O-methyltransferase (mt-LAF5a and b), and one pseudouridine synthase (mt-LAF3). The global structures and active sites of mt-LAF5 and 3 are conserved (SI Appendix, Figs. S11 and S13). Mt-LAF5a and b are RNA 2′O-methyltransferases that belong to the SpoU family. Compared with the bacterial enzymes, their structure as well as their dimerization are conserved. Their catalytic core, involved in SAM binding, is conserved, as is their N-terminal domain, involved in dsRNA binding (SI Appendix, Fig. S13). In the assembly intermediate, mt-LAF5a and b are located near three possible targets: H95 and the two helices of the immature L7/12 stalk. Several of these enzymes are involved in methylation of the 23S rRNA in prokaryotes (22, 23).The NHR SpoU described in S. actuosus (22) is involved in an adenine methylation in H43 of the L7/12 stalk. Given that mt-LAF5a and b are located close to the L7/12 stalk, it is possible that their target is similar to NHR SpoU. Therefore, the folding of the immature L7/12 stalk in two helices would be crucial for mt-LAF5a and b to recognize their target and perform methylation. The protein mt-LAF3 is a pseudouridine synthase D family enzyme that shows high homology to RluD, a conserved bacterial enzyme responsible for the pseudouridylation of several residues on H69 in domain IV of 23S rRNA (20, 21). This enzyme is highly conserved in all Rlu bacterial enzymes (SI Appendix, Fig. S11) and is essential for ribosome biogenesis and cell growth in bacteria (31). In our reconstruction, we clearly observe a uridine residue flipped out of an RNA regular helix to engage the catalytic core of the enzyme, as previously characterized for these enzymes (Fig. 7D and SI Appendix, Fig. S11), strongly suggesting that this factor is catalytically functional.
To date, only a few examples of rRNA modifications have been characterized in mitoribosomes, with the best ones described in humans (32). Only three types of modification have been identified in the human mitoribosomal rRNA: nucleobase methylation, 2′-O-methylation, and pseudouridylation. Given the conservation level of mt-LAF5 and 3 (SI Appendix, Figs. S11 and S13), as well as the rRNA regions that they target, it is expected that other modifications found in bacteria (33) and mitoribosomes (32) in domains IV and V might also be conserved in kinetoplastid mitoribosomes, such as Um2552 in Escherichia coli, equivalent to mitoribosome human Um1369 (32). In our reconstruction, we also observed the presence of an inactive pseudouridine synthase, mt-LAF4, as deduced by the lack of conservation of critical catalytic amino acids compared with bacterial, human, and yeast homologs (SI Appendix, Fig. S10A). Its location on the LSU assembly intermediate suggests its repurposing as an antibinding factor, as it interferes with the binding of the CP (SI Appendix, Fig. S9).
Three GTPases, all homologs of bacterial proteins, are also found in our reconstruction (SI Appendix, Fig. S12). The bacterial homologs of two of them, mt-EngA and mt-RbgA, were previously described in the context of the 50S (16, 17). Mt-EngA, in the GTP state (SI Appendix, Fig. S11A), interacts with H74-75 of domain V and mt-RbgA that contacts mt-EngA through its N-terminal domain. Similar to the described roles of these proteins in bacteria, they bind the assembly intermediate rRNA to facilitate the folding of domain IV and V helices (16, 17). The third GTPase, mt-EngB, is homologous to EngB/YihA in bacteria that was structurally characterized alone, outside the context of the maturing LSU. Here we show that in the L. tarentolae LSU assembly intermediate, mt-EngB contacts helices H33–35 of domain II and occupies the place of uL2m (SI Appendix, Fig. S11C). Finally, the DEAD-box RNA helicase mt-LAF2 is found on the immature H88 (SI Appendix, Fig. S10). Comparison with other structures of DEAD-box enzymes revealed that, aside from the presence of a flexible insertion domain between the DEAD and helicase domains, the overall organization of the domains is conserved. Ligand and RNA binding are also performed in a conserved fashion. mt-LAF2 is in the closed state of its cycle, acting on a single-strand portion of H88, confirming its activity. Although the role of the flexible insertion domain between the DEAD and helicase domains is unknown, it does not appear to hamper function. This factor has no obvious bacterial homologs but shows similarity to Has1 (15) and Mss116 (14). While Has1 participates in the maturation of H16 in the pre-60S cytosolic ribosome, Mss116 appears to be an mt-LAF2 yeast mitochondrial homolog. The structure of Mss116 was solved alone, out of the context of the maturing yeast mitochondrial ribosome (14). Therefore, our structure provides invaluable insight into the molecular basis of the maturation function of this protein.
Finally, the mt-Rsfs:L0R8F8:mt-SAF32 complex is composed of the bacteria-conserved Rsfs factor (mt-Rsfs) and two repurposed respiratory complex I components (mt-SAF32 and L0R8F8) (34–36). The latter are supernumerary subunits of the complex I in eukaryotes; that is, they are not part of the core proteins found in bacterial complex I (37) but were acquired during respiratory complex I evolution in mitochondria (38, 39). Interestingly, they interact together in the same fashion as in the mitochondrial complex I (Fig. 5 A–E). Moreover, in the T. brucei SSU assemblosome (6), mt-SAF32 binds mt-SAF31, which has an LYR (the conserved tripeptide motif leucine, tyrosine, and arginine) domain. LYR domains are characterized by a specific fold composed of three antiparallel helices. L0R8F8, found in our structure, also contains a LYR domain; thus, mt-SAF32 most likely recognizes the characteristic LYR fold to bind to mt-SAF31 and mt-LAF9 in the context of SSU and LSU assembly intermediates. Even though respiratory chain components in kinetoplastids appear to be strongly divergent from the rest of the eukaryotes (40), the presence of L0R8F8:mt-SAF32 in both human LSU assembly intermediate and complex I, in addition to the presence of mt-SAF32 in kinetoplastid SSU assembly intermediates, suggests a multifaceted role of these proteins and hint at possible crosstalk between mitoribosome maturation and respiratory complex assembly. The natural accumulation of the LSU assembly intermediate in our sample could be promoted by the presence of the mt-Rsfs:L0R8F8:mt-SAF32 complex, which might be stress-induced or caused by diminished nutrient availability, likewise in bacteria. Interestingly, as its name already indicates, mt-SAF32 is also found in the SSU assembly intermediates, likewise in a peripheral position of the intersubunit side (6).
In conclusion, our cryo-EM structures of kinetoplastid mitoribosomes, especially the as-yet uncharacterized LSU assembly intermediate, shed new light on ribosome maturation. Even though several proteins are specific to kinetoplastids, most of the maturation factors present in our structure have homologs in other eukaryotes, mitochondria, or bacteria (SI Appendix, Table S3). Thus, our structure unveils the mode of actions of these factors, for which no high-resolution structures in native conditions exist. It also highlights the fact that despite being highly divergent, kinetoplastid mitoribosomes retained several canonical maturation pathways for their rRNAs and likely follow an analogous mechanistic pattern (Fig. 5 F and G). Indeed, in concert, a set of factors intercalate with the premature rRNA (GTPases) to mechanically unpack different premature rRNA domains, allowing RNA helicases to disentangle and unwind the rRNA and several families of modification enzymes to access their targets. The remodeling of the rRNA is also aided by recruitment of the r-proteins early during the maturation process. This is especially true in the kinetoplastid mitochondria, where most of the r-proteins are already recruited at the observed stage and form a thick protein shell holding the rRNA together. At the same time, various antiassociation factors prevent premature binding of r-proteins and SSU joining.
Despite large environmental and host diversities in which these parasites evolve, their mitoribosomes are conserved structurally among kinetoplastids. Therefore, our structures could serve as a basis for future experiments to develop more effective and safer kinetoplastid-specific therapeutic strategies. Finally, as kinetoplastids undergo extensive mitochondrial reorganization (41) during their life cycle, in both vectors and hosts, it would also be interesting to study mitoribosomes from different stages of development from the parasite’s life cycle.
Methods
Mitochondria were purified from T. cruzi and L. tarentolae on Histodenz step gradients. The mitoribosomes were consequently purified on sucrose gradients, then analyzed using the single-particle cryo-EM technique. The cryo-EM movies were acquired on a Talos Arctica (200 kv) transmission and scanning transmission electron microscope equipped with a Falcon III direct detector camera (Thermo Fisher Scientific). Movie stacks were processed mainly using Relion. In parallel, the mitoribosome samples were analyzed by MS to deduce their compositions.
Extended Materials and Methods.
Purification of T. cruzi and L. tarentolae mitochondria.
Mitochondria purification was done following protocols adapted from Hauser et al. (42) and Schneider et al. (43). In brief, cells were resuspended in SoTE buffer (600 mM sorbitol, 20 mM Tris⋅HCl pH 7.5, and 1 mM EDTA) to a final concentration of 2.5 × 109 cells/mL and lysed by nitrogen cavitation at 80 bar for 1 h. After pressure release the lysate was centrifuged and resuspended in SoTM buffer (600 mM sorbitol, 20 mM Tris⋅HCl pH 7.5, and 5 mM MgCl2) for DNase I treatment over 15 min. EDTA was added to stop DNase treatment, and the sample was centrifuged and then resuspended in two volumes of SoTE containing 50% Histodenz (Sigma Aldrich D2158). For L. tarentolae, the fraction was loaded onto a 21.7%/25%/28.3%/31.6% Histodenz step gradient. Gradients were centrifuged for 45 min at 100,000 × g, and the mitochondrial fraction was collected at the 25%/28.3% interface. For T. brucei, the fraction was loaded onto a 18%/21%/25%/28% Histodenz step gradient, and the mitochondrial fraction was collected at the 21%/25% interface.
Purification of mitochondrial ribosomes.
For mitoribosome purification, the protocol adapted from Waltz et al. (10) was applied. In brief, mitochondria were resuspended in lysis buffer (20 mM Hepes-KOH pH 7.6, 100 mM KCl, 20 mM MgCl2, 1 mM DTT, 1% Triton X-100, and 2% DDM, supplemented with protease inhibitors [Complete EDTA-free]) to a concentration of 1 mg/mL and incubated for 15 min in 4 °C. Lysate was clarified by centrifugation at 30,000 × g for 20 min at 4 °C. The supernatant was loaded on a 40% sucrose cushion in Monosome buffer (same as lysis buffer without Triton X-100 and with 0.02% DDM) and centrifuged at 235,000 × g for 3 h at 4 °C. The crude ribosome pellet was resuspended in Monosome buffer and loaded on a 10 to 30% sucrose gradient in the same buffer and run for 16 h at 65,000 × g. Fractions corresponding to mitoribosomes were collected, pelleted, and resuspended in Monosome buffer (SI Appendix, Fig. S1).
Grid preparation.
Here 4 µL of sample at a concentration of 2 µg/µL was applied onto Quantifoil R2/2 300-mesh holey carbon grid, which had been coated with a thin home-made continuous carbon film and glow-discharged. The sample was incubated on the grid for 30 s and then blotted with filter paper for 2 s in a temperature- and humidity-controlled Vitrobot Mark IV (4 °C; 100% humidity; blot force 5), followed by vitrification in liquid ethane precooled by liquid nitrogen.
Single-particle cryo-EM data collection.
For the two datasets (full and dissociated complexes), data collection was performed on a Talos Arctica instrument (Thermo Fisher Scientific) at 200 kV using the EPU software (Thermo Fisher Scientific) for automated data acquisition. Data were collected at a nominal underfocus of −0.5 to −2.7 µm at a magnification of 120,000×, yielding a pixel size of 1.24 Å. Micrographs were recorded as movie stack on a Falcon III direct electron detector (Thermo Fisher Scientific). Each movie stack wase fractionated into 20 frames for a total exposure of 1 s, corresponding to an electron dose of 60 ē/Å2.
EM image processing.
Drift and gain correction and dose weighting were performed using MotionCor2 (44). A dose-weighted average image of the whole stack was used to determine the contrast transfer function with the software Gctf (45). The following process has been achieved using RELION 3.0 (46). Particles were picked using a Laplacian of Gaussian function (minimum diameter 280 Å; maximum diameter 480 Å). For the L. tarentolae full mitoribosome, after 2D classification, a total of 303,140 particles were extracted with a box size of 400 pixels and binned fourfold for 3D classification into six classes. Three classes depicting high-resolution features, consisting of 82,060 particles, were selected for refinement, yielding 3.9 Å resolution. After focused refinement with a mask on the LSU and the body and head of the SSU, resolutions of 3.6, 4, and 3.8 Å, respectively, were obtained. For the assembly intermediate of the LSU, after 2D classification, 215,120 particles were extracted with a box size of 400 pixels and binned threefold for 3D classification into eight classes. Two classes, consisting of 59,200 particles, depicting high-resolution features were selected for refinement, yielding 3.4 Å resolution.
For the T. cruzi full mitoribosome reconstruction, 126,340 particles were selected after 3D classification for refinement, resulting in a 6 Å resolution reconstruction, which was further refined to 3.7 Å for the LSU and 4.5 Å for the SSU. For the T. cruzi mt-IF3 SSU complex, 148,180 particles were selected after 3D classification for refinement, resulting in a 3.5 Å resolution reconstruction, which was further refined to 3.1 Å for the SSU body and 3.2 Å for the SSU head. Determination of the local resolution of the final density map was performed using ResMap (47).
Structure building and model refinement.
The atomic models of the kinetoplastid mitoribosomes were built into the high-resolution maps using Coot, PHENIX, and Chimera. Atomic models from the E. coli ribosome (Protein Data Bank [PDB] ID code 5KCR) (48) and Trypanosoma mitoribosome (PDB ID code 6HIV) (3) were used as starting points for protein identification and modelization. For the Leishmania mitoribosome, atomic models were build using Leishmania major protein sequences owing to the poor quality of the available data for L. tarentolae. The online SWISS-MODEL service was used to generate initial models for bacterial and mitochondria conserved r-proteins. Models were then rigid body fitted to the density in Chimera (49), and all subsequent modeling was done in Coot (50). For the LSU ribosomal RNA, the 23S rRNA from E. coli was docked into the maps and used as a template for positioning and reconstruction. Extensions were built as stretches of polyalanine and mutated to the adequate sequences. A combination of regularization and real-space refinement was performed in Coot for each protein. The global atomic model was then subjected to real space refinement cycles using the phenix.real_space_refine PHENIX function (51), during which protein secondary structure, Ramachandran and side chain rotamer restraints were applied. Several rounds of refinement (manual in Coot and automated using phenix.real_space_refine) were performed to obtain the final models, which were validated using the built-in validation tool of PHENIX, based on MolProbity. For the assembly intermediate, the model was built with the mature complex as a template. Refinement and validation statistics are summarized in SI Appendix, Table S4.
Proteomic and statistical analyses of mitochondrial ribosome composition.
MS analyses of the ribosome fractions were performed at the Strasbourg-Esplanade proteomic platform and performed as described previously (10, 52). In brief, proteins were trypsin-digested, and MS analyses were performed by nano LC-ESI-MS/MS analysis on a QExactive+ mass spectrometer (Thermo Fisher Scientific). Quantitative label-free proteomics analysis was performed through in-house bioinformatics pipelines.
Figure preparation.
Figures featuring cryo-EM densities as well as atomic models were visualized with UCSF ChimeraX (53) and Chimera (49).
Supplementary Material
Acknowledgments
This work has benefitted from the facilities and expertise of the Biophysical and Structural Chemistry platform at IECB, CNRS UMS3033, Inserm US001, University of Bordeaux. We thank A. Bezault for assistance with the Talos Arctica electron microscope and J. Chicher and P. Hamman of the Strasbourg-Esplanade proteomic analysis for the proteomic analysis. This work was funded by a European Research Council Starting Grant (TransTryp ID: 759120) and the IdEx junior excellence chair program of the Université de Bordeaux (to Y.H.) and the French National Program “Investissement d’Avenir” (Labex MitoCross), administered by the Agence National de la Recherche (ANR-11-LABX-0057_MITOCROSS, to M.S.). This project was supported by Inserm, CNRS, and the University of Bordeaux.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011301117/-/DCSupplemental.
Data Availability.
The cryo-EM maps of the mitoribosome have been deposited at the Electron Microscopy Data Bank (accession codes EMD-11821, EMD-11796, EMD-11829, and EMD-11846), and corresponding atomic models have been deposited in the Protein Data Bank (PDB ID codes 7AM2, 7AIH, 7ANE, and 7AOR). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD018981). All other study data are provided in the main text and SI Appendix.
References
- 1.Stuart K., et al. , Kinetoplastids: Related protozoan pathogens, different diseases. J. Clin. Invest. 118, 1301–1310 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson L., Thiemann O. H., Savill N. J., Alfonzo J. D., Maslov D. A., Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. U.S.A. 97, 6986–6993 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramrath D. J. F., et al. , Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 362, eaau7735 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Waltz F., Giegé P., Striking diversity of mitochondria-specific translation processes across eukaryotes. Trends Biochem. Sci. 45, 149–162 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Bieri P., Greber B. J., Ban N., High-resolution structures of mitochondrial ribosomes and their functional implications. Curr. Opin. Struct. Biol. 49, 44–53 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Saurer M., et al. , Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery. Science 365, 1144–1149 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Jackson A. P., et al. , Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr. Biol. 26, 161–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai N., Brown A., Amunts A., Ramakrishnan V., The structure of the yeast mitochondrial ribosome. Science 355, 528–531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amunts A., Brown A., Toots J., Scheres S. H. W., Ramakrishnan V., The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waltz F., Soufari H., Bochler A., Giegé P., Hashem Y., Cryo-EM structure of the RNA-rich plant mitochondrial ribosome. Nat. Plants 6, 377–383 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Jaskolowski M., et al. , Structural insights into the mechanism of mitoribosomal large subunit biogenesis. Mol. Cell 79, 629–644.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Kummer E., et al. , Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 560, 263–267 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Mallam A. L., Sidote D. J., Lambowitz A. M., Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. eLife 3, e04630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng R., Smith E., Barrientos A., Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab. 27, 645–656.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou D., et al. , Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 10, 120–130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., et al. , Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Res. 42, 13430–13439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seffouh A., et al. , Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate. Nucleic Acids Res. 47, 10414–10425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittinghofer A., Vetter I. R., Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Pausch P., et al. , Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA. J. Biol. Chem. 293, 19699–19709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivaraman J., Iannuzzi P., Cygler M., Matte A., Crystal structure of the RluD pseudouridine synthase catalytic module, an enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli. J. Mol. Biol. 335, 87–101 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Vaidyanathan P. P., Deutscher M. P., Malhotra A., RluD, a highly conserved pseudouridine synthase, modifies 50S subunits more specifically and efficiently than free 23S rRNA. RNA 13, 1868–1876 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H., et al. , Crystal structure of the nosiheptide-resistance methyltransferase of Streptomyces actuosus. Biochemistry 49, 6440–6450 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Mosbacher T. G., Bechthold A., Schulz G. E., Structure and function of the antibiotic resistance-mediating methyltransferase AviRb from Streptomyces viridochromogenes. J. Mol. Biol. 345, 535–545 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Brown A., et al. , Structures of the human mitochondrial ribosome in native states of assembly. Nat. Struct. Mol. Biol. 24, 866–869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., et al. , Structure of ribosomal silencing factor bound to Mycobacterium tuberculosis ribosome. Structure 23, 1858–1865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khusainov I., et al. , Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach. Nat. Commun. 11, 1656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melnikov S., et al. , One core, two shells: Bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Khawaja A., et al. , Distinct pre-initiation steps in human mitochondrial translation. Nat. Commun. 11, 2932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koripella R. K., et al. , Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit. iScience 12, 76–86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greber B. J., et al. , Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164, 91–102 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Del Campo M., Ofengand J., Malhotra A., Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA 10, 231–239 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rebelo-Guiomar P., Powell C. A., Van Haute L., Minczuk M., The mammalian mitochondrial epitranscriptome. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 429–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polikanov Y. S., Melnikov S. V., Söll D., Steitz T. A., Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parey K., et al. , High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease. Sci. Adv. 5, eaax9484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agip A. A., et al. , Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 25, 548–556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soufari H., Parrot C., Kuhn L., Waltz F., Hashem Y., Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM. Nat. Commun. 11, 5195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baradaran R., Berrisford J. M., Minhas G. S., Sazanov L. A., Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray M. W., Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4, a011403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sazanov L. A., A giant molecular proton pump: Structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16, 375–388 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Valach M., Léveillé-Kunst A., Gray M. W., Burger G., Respiratory chain complex I of unparalleled divergence in diplonemids. J. Biol. Chem. 293, 16043–16056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakob M., et al. , Mitochondrial growth during the cell cycle of Trypanosoma brucei bloodstream forms. Sci. Rep. 6, 36565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauser R., Pypaert M., Häusler T., Horn E. K., Schneider A., In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J. Cell Sci. 109, 517–523 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Schneider A., Bouzaidi-Tiali N., Chanez A.-L., Bulliard L., ATP production in isolated mitochondria of procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 111, 87–94 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arenz S., et al. , Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome. Proc. Natl. Acad. Sci. U.S.A. 113, 7527–7532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waltz F., et al. , Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 5, 106–117 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps of the mitoribosome have been deposited at the Electron Microscopy Data Bank (accession codes EMD-11821, EMD-11796, EMD-11829, and EMD-11846), and corresponding atomic models have been deposited in the Protein Data Bank (PDB ID codes 7AM2, 7AIH, 7ANE, and 7AOR). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD018981). All other study data are provided in the main text and SI Appendix.