Significance
The role of viruses in shaping microbial communities in the marine photic zone has been studied extensively in recent years. However, temporal patterns of abundance and activity of highly abundant viral groups and its relation to host abundance are still mostly unconstrained. By tracking changes in rank abundance patterns, we report evidence for seasonal succession in key viral groups alongside a persistence and low transcriptional activity of a large bank population. In addition, we show that high virion abundance does not necessarily translate to high host infection levels, suggesting the coexistence of a diverse repertoire of diel activity and life history traits, even within viral taxonomic groups. Our results highlight the diversity of viral life strategies in the ocean.
Keywords: marine viruses, seasonal succession, virus–host interaction, diel oscillation
Abstract
Virus–microbe interactions have been studied in great molecular details for many years in cultured model systems, yielding a plethora of knowledge on how viruses use and manipulate host machinery. Since the advent of molecular techniques and high-throughput sequencing, methods such as cooccurrence, nucleotide composition, and other statistical frameworks have been widely used to infer virus–microbe interactions, overcoming the limitations of culturing methods. However, their accuracy and relevance is still debatable as cooccurrence does not necessarily mean interaction. Here we introduce an ecological perspective of marine viral communities and potential interaction with their hosts, using analyses that make no prior assumptions on specific virus–host pairs. By size fractionating water samples into free viruses and microbes (i.e., also viruses inside or attached to their hosts) and looking at how viral group abundance changes over time along both fractions, we show that the viral community is undergoing a change in rank abundance across seasons, suggesting a seasonal succession of viruses in the Red Sea. We use abundance patterns in the different size fractions to classify viral clusters, indicating potential diverse interactions with their hosts and potential differences in life history traits between major viral groups. Finally, we show hourly resolved variations of intracellular abundance of similar viral groups, which might indicate differences in their infection cycles or metabolic capacities.
Viruses of marine microorganisms outnumber their hosts and are considered the most abundant biological entities in the ocean (1). These viruses compose the largest reservoir of genetic diversity in the oceans, and they are major participants in oceanic biotic and abiotic processes (2–5). It is estimated that 50% of marine microbial production is mediated by virus-induced release of dissolved organic matter or viral shunt (6, 7). Research suggests that viruses alter microbial primary and secondary production (8, 9), impact the population dynamics and diversity of microbial communities (10), and play an indispensable role in marine biogeochemical fluxes (11).
Viruses rely on their host for propagation, and their abundance is predicted to follow that of their hosts (kill-the-winner model (12), and the bank model (13)). The bank model suggests that in any given environment, a small fraction of the viral community is highly abundant while the rest are rarer, waiting for the right conditions (i.e., host) to propagate. These models have been useful for describing, for the most part, the local and global distribution of free viruses in marine samples (2, 10, 13); however, our understanding of the relationship between abundance variation of marine viruses and host interaction remains limited. Furthermore, viruses depend on their host metabolism (14), often by manipulating the metabolic and transcriptional machinery using auxiliary metabolic genes (AMGs) (10, 15). Recent environmental studies have reported several viral AMGs expressed in diurnal patterns, coupled with their host metabolism and reproduction cycle (16–20). To date, only a few environmental studies of diel patterns in marine viruses (16, 17, 20) and seasonality effects on viral communities (19, 21–25) have been reported, and none of these incorporate both diel and seasonal time scales.
Here we describe seasonal and diel abundance patterns of viral communities in the Red Sea (specifically, double-strand DNA viruses of prokaryotes) and show how these patterns can be used to gain insights into viral life history traits and the potential interactions of viruses with their hosts. To resolve seasonal and diel patterns we collected diel samples of marine coastal water during two seasons. We separated the cellular DNA fraction (gDNA or metagenome), free virus fraction (vDNA or virome), and cellular RNA fraction (RNA or metatranscriptome; SI Appendix, Fig. S1B), allowing us to look at these fractions in both seasonal and diel time scales. We used the bank model (13) as a conceptual framework to classify viral active or inactive state based on their seasonal abundance across sample types (i.e., RNA, gDNA, or vDNA), that is, classifying the abundant viruses as the active group and the rest as the bank group (inactive) (Fig. 1A). Our results suggest that many of the seasonally abundant viruses go through a seasonal succession and that the high similarity in viral richness across seasons is largely derived from low-abundance viruses (bank). Furthermore, we assigned a taxonomic and ecological classification to hundreds of viral contigs with different patterns of abundance in the viral and cellular fractions. This classification indicates that similar seasonal abundance patterns of free viruses do not directly translate to similar abundance of these viruses associated with their hosts (cellular samples), illustrating limitations of studies entirely based on viral metagenomes (i.e., viromes). Finally, we observed many potential virus–host interactions (predicted by virus abundance in the cellular and RNA samples) having a differential signature between light and dark hours of the day. Such light-dependent interactions are prevalent in cyanophages (viruses infecting Cyanobacteria) but also include several viruses predicted to infect a heterotrophic host.
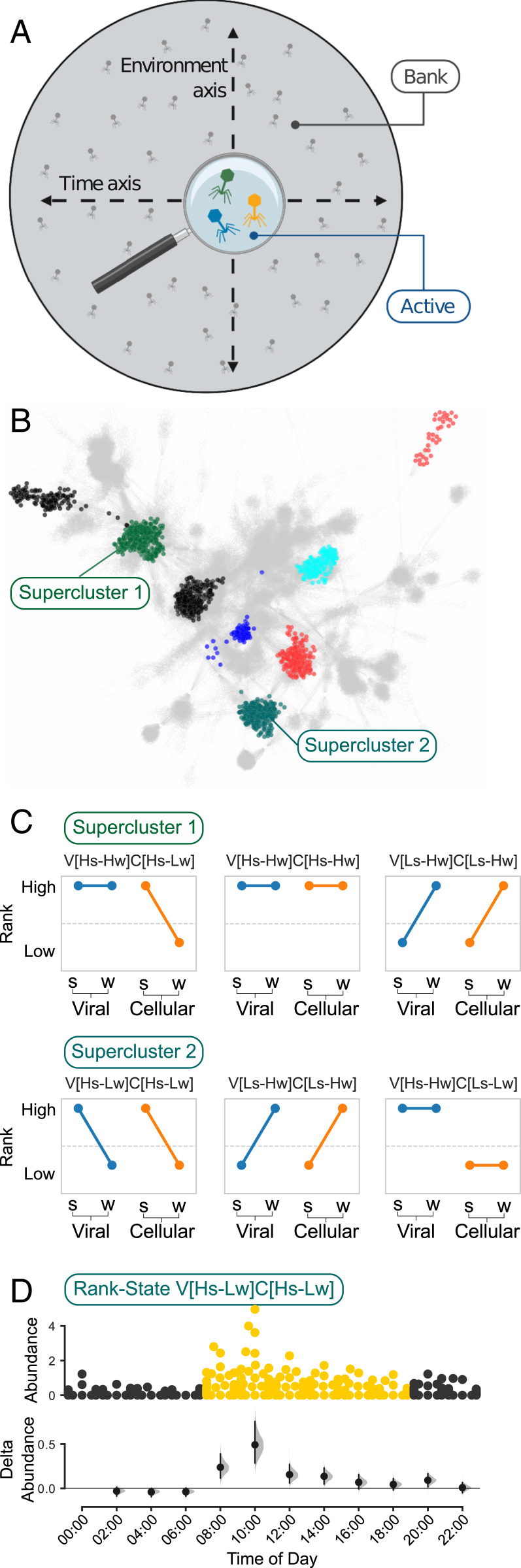
Fig. 1.
Classifying viruses by seasonal abundance patterns. (A) Using the bank model (13) as a conceptual framework, the viral contigs were divided into a high-abundance (active) group and a low-abundance (bank) group. (B) Contigs from the active group were annotated and clustered into VCs. Colored clusters represent examples of distinct superclusters containing several VCs. (C) The rank abundance of each viral contig in the viral and cellular DNA samples was used to classify its seasonal abundance patterns (rank state; see text for annotation). Left connected line (blue) represents the rank of a virus in the free-viral fraction in the summer and winter samples. Similarly, the connected line on the right (orange) represents the virus abundance in the cellular DNA fraction. (D) Diel expression patterns of VCs can be linked to a rank state. x axis indicates sampling time of day. (Upper) Abundance distribution of viral contigs by time point. Point color represents samples collected in the dark (black) and light (yellow). y axis indicates contig abundance (RPKM). (Lower) Estimation plot (31) displaying the effect size as a 95% confidence interval (1,000 bootstraps) of the mean differences between each time point compared against midnight as a reference group.
Results and Discussion
Two sets of 24-h time series samples, with 2-h intervals between samplings, were collected for this study. The first set was collected during the summertime (11 to 12 August 2015) when the water column was stratified, and the second was collected during late winter (7 to 8 February 2016) when the upper water column was mixed. In addition, we used metagenomic data from a diel sample of Red Sea pelagic water collected during the autumn (October 2012) (26) (SI Appendix, Fig. S1A). Assembled contigs (length 5 kb) from all 48 DNA samples (cellular and viral fractions) were filtered for predicted viral contigs using two virus classifiers (Materials and Methods). A total of 32,496 contigs were assigned as viral by both algorithms (minimum probability cutoff 0.75) and were mostly enriched in the viromes compared to the metagenomes. These contigs were considered of viral origin for downstream analysis.
The rank abundance curve of free viruses in the summer and winter samples was heavy-tailed (SI Appendix, Fig. S2A), and the measured abundance evenness ranged between 0.004 and 0.326 with different indices (Simpson evenness, EQ, and Evar; SI Appendix, Fig. S2B, and Materials and Methods). Evenness indices are useful for describing the distribution of species in a community (27, 28). For example, in a community where all species have the same abundances (perfectly even), evenness is equal to 1. In contrast, in a very uneven community, where a few species have high abundance and most species have low abundances, the evenness will be closer to 0 (29). The observed values across evenness indices seem to suggest that the viral community in our samples is composed of a relatively small number of viruses with high abundance, while the majority have a very low abundance, in agreement with the bank model as well as other reports (13, 16).
The rank reordering of free viruses in the diel samples per season or between seasons showed that on average the mean rank change between seasons is 2.4-fold higher compared to diel changes (SI Appendix, Fig. S2C, and Materials and Methods). This could indicate a seasonal change in dominance of the different viral groups. Interestingly, the turnover of free viruses (vDNA) between summer and winter [measured as the proportion of viruses either gained or lost relative to the total number of viruses observed across both time periods (29)] was significantly lower compared to the turnover of intracellular active viruses (i.e., RNA and gDNA; SI Appendix, Fig. S2D). These observations not only provide further support to the existence of a persistent bank population of free viruses as previously reported (13, 16, 25) but also highlight the large seasonal change in the intracellular active population.
To further examine the group of abundant viruses, we pooled the most abundant viral contigs, contributing to 80% of the viral abundance in either season or sample type (SI Appendix, Figs. S2E and S3). The presence–absence of these abundant viruses across seasons shows that most of them are present in the high-abundance group only in one of the seasons (SI Appendix, Fig. S2F). These results suggest that low-abundance viruses (bank) compose most of the viruses that are present across seasons (25), while the top-ranking members of the viral community undergo a seasonal succession.
Together, the seasonal change of ranks and the persistent low-abundance viral population support the bank model where low-abundance viruses in one environment (stratified summer water in our case) increase in abundance as the environment changes (e.g., winter water column mixing) and their host abundance increases (13, 16). Furthermore, in the winter samples, our alpha diversity measures were generally higher, indicating a larger active population fraction than in the summer samples where the alpha diversity measures were generally lower. In turn, this could indicate an increase in both the diversity and abundance of potential hosts (14% unique contigs in the viral fraction in each of the summer and autumn samples vs. 52% in the winter samples; SI Appendix, Fig. S2F). A similar mechanism where high seasonal host diversity generates high viral diversity has been recently hypothesized to be in effect in the Arctic marine environment (30). Our observations suggest that seasonal variations in host diversity could drive an increase in viral diversity in a coastal and more temperate marine environment and are not confined to the Arctic.
We classified the high-abundance viral group into viral clusters (VCs) based on a gene-sharing network (32) (Fig. 1B and Materials and Methods), and a taxonomic annotation was assigned based on the identity of Refseq viral genomes found in that cluster or according to the closest Refseq genomes that represented a distinct hub. We kept the annotation at the viral genus level (33) (e.g., cyanopodovirus, SAR11 myovirus, etc.) and considered highly connected VCs with the same taxonomic annotation as viral superclusters. Most of the VCs did not cluster with a Refseq representative and were classified as uncharacterized Red Sea virus in our data (SI Appendix, Fig. S4), accentuating the high proportion of yet uncharacterized viruses in the environment (2, 16, 30).
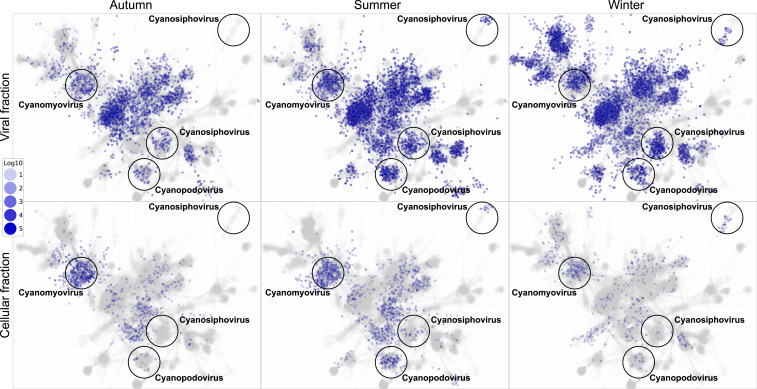
A closer look at the abundance of different viral superclusters across seasons and sample types reveals different patterns for distinct clusters. For example, cyanophages, one of the most abundant viral groups in our data, were grouped into four superclusters according to their viral families, Cyanomyoviridae, Cyanopodoviridae, and two Cyanosiphoviridae clusters (Fig. 2). The cyanomyovirus cluster (300 contigs) was abundant in both viral and cellular samples in all seasons. The cyanopodovirus cluster (220 contigs) was abundant in the viral samples across seasons, while in the cellular samples it was highly abundant in the summer but mostly absent from the autumn and winter samples. One cyanosiphovirus cluster (Fig. 2, Lower Right, 175 contigs) was abundant in the viral samples across all seasons but almost absent from the cellular samples. In contrast, another cyanosiphovirus cluster (Fig. 2, Upper Right, 34 contigs) showed a distinct difference in abundance of VCs within the same supercluster between summer and winter. VCs 251_0 and 550_0 were abundant in the summer samples and VCs 281_0 and 419_0 in the winter samples (in both viral and cellular samples). The observed variability in the seasonal abundance within these closely related viral superclusters highlights the different life history traits of viral populations and subpopulations in terms of possible host range and lifestyle, suggesting potential adaptations to changing host landscapes. Indeed, a comparison of the most abundant microbial taxonomic groups shows considerable differences between summer and winter (SI Appendix, Fig. S5).
Fig. 2.
Seasonal virus abundance overlaid on a gene sharing network. Each node is a viral genome/contig. Color intensity represents log abundance (RPKM). (Top) Viral fraction. (Bottom) Cellular DNA fraction. Labeled are cyanophage superclusters discussed in the text.
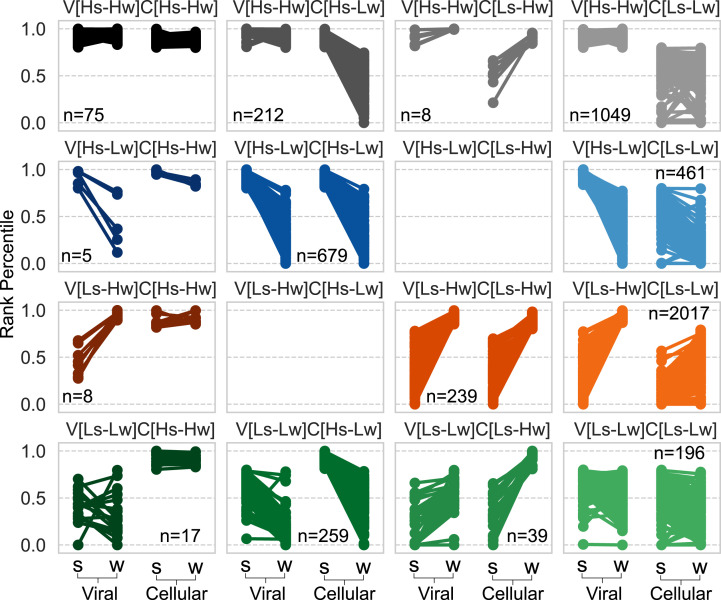
To systematically assign an ecologically relevant classification to viruses based on abundance patterns we focused on the most abundant viral contigs and looked for seasonal changes in their rank percentile (0 to 1) in the viral and cellular DNA fractions (Fig. 1C). Specifically, we grouped these contigs based on their intracellular and extracellular seasonal rank changes, where contigs ranked in the top 20% were considered as active and lower-ranking contigs as bank (Materials and Methods). The combination of two seasons (summer and winter) and two sample types (viral fraction and cellular DNA fraction) yields 16 possible combinations, representing the relationship between a virus’s abundance in the free virus fraction and its abundance when interacting with its host in the cellular fraction, in a changing environment (Fig. 3). Furthermore, as relative abundance measurements arising from metagenomic studies have been shown to contain a systematic bias (34, 35), we validated the rank-based results using the centered log-ratio transform (CLR) which is invariant to such bias (35) (SI Appendix, Fig. S6, and Materials and Methods). We termed these abundance–sample combinations rank states and annotated them as follows: S, summer; W, winter; H, high ranks (active group); L, low ranks (bank group); V, virome samples; and C, cellular metagenome samples. For example, high abundance of a group in the viral sample in both summer and winter is annotated as Viral[Highsummer-Highwinter] and abbreviated as V[Hs-Hw].
Fig. 3.
Representation of viral rank states of the most abundant viral contigs group, including 16 possible rank state combinations, ranging from viruses that are highly abundant in both seasons and samples (V[Hs-Hw]C[Hs-Hw], n = 75; Top Left) to those with lower abundance across seasons and samples (V[Ls-Lw]C[Ls-Lw], n = 196; Bottom Right).
Furthermore, the extent of rank change can also be deduced, and for example, contigs that undergo no change in rank across seasons are easily identified (Fig. 3). Classifying the Red Sea viral community using this framework reinforces the observation of various abundance patterns for different viral superclusters, while assigning each viral contig an ecological context. The summer-abundant free viruses (V[Hs-Lw]) represented 21.75% from the total number of classified viral contigs, while their winter counterparts (V[Ls-Hw]) represented 43%. In contrast, the abundance of viral contigs in the cellular DNA fraction was inverted, where the summer-abundant intracellular contigs (C[Hs-Lw]) represented 21.85% from the total number of classified viral contigs, while the winter-abundant contigs (C[Ls-Hw]) represented only 5.53% (Fig. 3 and SI Appendix, Table S1).
These results further suggest that the abundance of free viruses alone (as measured in the viral fraction) does not directly correlate to their abundance in the cellular fraction and vice versa and thus cannot reliably serve as a predictor for infection levels. For example, this approach highlights viral contigs with a V[Ls-Lw]C[Hs-Hw] rank state (Fig. 3, Bottom Left) that in the free viruses fraction show seasonal variability in ranks within the bank group, while steadily ranked among the active group in the cellular DNA fraction in both seasons. As the majority of contigs in this rank state appear to be of uncharacterized SAR11 viruses (based on major capsid protein gene homology), the traits that enable this pattern are still mostly unknown. However, a recent discovery of prophages in cultured and metagenomic sequences of SAR11 phages (36) suggests that lysogeny is a very plausible explanation for these seasonal patterns as these temperate phages appear to be produced continuously during the infection cycle with higher production during nutrient limitations.
Temperate phages were found to dominate summer blooms in the Southern Ocean close to Antarctica, and the induction of lysis was correlated with bacterial production and increase in chl a (37). However, none of the abundant contigs in our samples were assigned to rank states V[Hs-Lw]C[Ls-Hw] or V[Ls-Hw]C[Hs-Lw]. Our interpretation of this result is that no phages appearing in the cellular DNA fraction in one season bloomed in the free virus fraction in the next season, as would be expected from seasonally induced lysogenic phages.
While the methods used in this study are not sufficient to assess the prevalence of temperate phages in the Red Sea, our observations along with a recent report (38) suggest that lysogeny might not be as dominant in the Red Sea as it is in the Southern Ocean (37). However, further, targeted research into the prevalence and identity of temperate phages in the Red Sea is needed.
Diel cycles lie at the basis of ocean productivity as sunlight is the most readily available source of energy in the photic zone (39), driving microbial community-wide gene expression patterns (40). Many of the diel patterns of highly expressed genes in the microbial community in our samples, including several viral transcripts, presented a strong correlation with a daylight cycle and no seasonal hierarchical order. This correlation was robust across seasons and functions of the microbial community, indicative of potential community-level trait–diel relationship and functional redundancy among these seasonally varying communities (41, 42) (SI Appendix, Fig. S7 and Table S3).
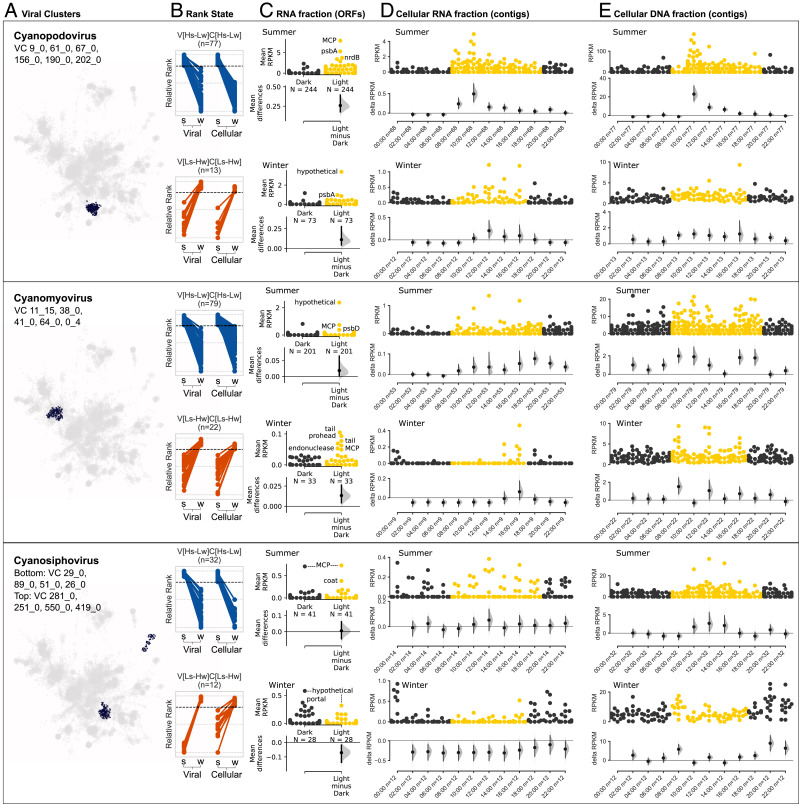
Since virus proliferation strongly depends on its host metabolism (14), we followed the patterns observed in the microbial community to examine seasonality and daylight effect on the intracellular activity of selected viral groups (16). Generally, in the RNA fraction we observed an increase in the transcript abundance of the cyanopodovirus supercluster at 08:00 (first sample collected in light) and a peak at 10:00 in the summer sample with a slight shift in the winter samples (Fig. 4D, Top). This peak included significantly higher expression in the light (Fig. 4C, Top) of phage structural genes and key cyanophage AMGs such as psbA (subunit of Photosystem II) and nrdB (nucleotide metabolism), indicating active infection. Abundance of cyanopodoviruses in the cellular DNA fraction peaked at 10:00 in the summer and was again shifted in the winter sample (Fig. 4E, Top), consistent with the shift in transcription of viral genes observed in the cellular RNA fraction. In addition, a clear difference in the diel pattern can be observed between contigs originating from different rank states, despite the fact that they are part of the same viral supercluster (Fig. 4, Top). The cyanomyovirus (Fig. 4D, Middle) supercluster’s transcript abundance also increased with the onset of light and fluctuated during the day, with most genes at their highest around 18:00. Expressed viral genes included structural genes as well as AMGs such as psbD (Fig. 4C, Middle). The abundance patterns of cyanomyoviruses in the cellular DNA fraction displayed multiple peaks, most pronounced at 10:00 to 12:00 and at 16:00 to 18:00 (Fig. 4E, Middle). The cyanosiphovirus supercluster’s transcript abundance peaked at midnight and noon, while their cellular abundance increased during daytime, peaking at noon (Fig. 4, Bottom).
Fig. 4.
Intracellular diel abundance patterns of (Top) cyanopodovirus VCs, (Middle) cyanomyovirus VCs, and (Bottom) cyanosiphovirus VCs. (A) Superclusters of cyanophage VCs by family. (B) Cyanophage VCs contigs that were classified into two rank states with strong seasonal abundance pattern (V[Hs-Lw]C[Hs-Lw] and V[Ls-Hw]C[Ls-Hw]). (C) Light–dark expression in the cellular RNA fraction of the viral ORFs from the contigs in B. (D and E) Diel distribution in the cellular RNA (D) and DNA (E) fractions of viral contigs from B. x axis indicates hours of the day when a sample was collected and the number of viral contigs expressed. (Upper) Abundance distribution of viral contigs by time point (each point represents a contig). Point color represent samples collected in the dark (black) and light (yellow). y axis indicates contig abundance (RPKM). (Lower) Estimation plot (31) displaying the effect size as a 95% confidence interval (1,000 bootstraps) of the mean differences between each time point compared against midnight as a reference group.
To understand the finer details of these patterns and their seasonality, we examined the cellular RNA and cellular DNA samples for light–dark abundance patterns of viral contigs by VC (441 VCs with counts 1 reads per kilo base per million mapped reads (RPKM) in the light or dark samples, where the light–dark label was determined by the photosynthetic active radiation measurements at the time of sampling; SI Appendix, Fig. S1C). This analysis revealed 97 VCs (22%) with at least one rank state that displayed differential light–dark abundance (Mann–Whitney U test, P < 0.05; SI Appendix, Fig. S8A and Table S2). Within these 97 VCs, most of the light–dark differential signal in either the cellular or RNA fractions originated from contigs in only four rank states (24.63% V[Ls-Hw]C[Ls-Lw], 23.18% V[Hs-Lw]C[Hs-Lw], 11.59% V[Ls-Lw]C[Hs-Lw], and 10.14% V[Ls-Hw]C[Ls-Hw]). Moreover, most of the viral RNA signal in the light–dark differential VCs came from only two rank states of viruses with distinct summer (27.27% V[Hs-Lw]C[Hs-Lw]) and winter (31.81% V[Ls-Hw]C[Ls-Hw]) abundance patterns. This can be explained by most of the VCs composing these two rank states being cyanophages (Table S2). This signal was very low or undetectable in other rank states, indicating that most of the detected viral RNA transcripts are from viruses that show a strong seasonal pattern, both extracellularly and intracellularly (Table S2). When examining the seasonal patterns of the microbial groups in the cellular and RNA fractions, it appears that the seasonal microbial communities are largely different from one another, sharing only 19.5% of the contigs (23,219 shared contigs). Additionally, while the Synechococcus population is highly abundant and active across our seasonal samples, changes in the dominance of unicellular picoeukarya algae in the winter and Prochlorococcus in the summer (SI Appendix, Fig. S5) are consistent with previous reports in this region (43, 44).
Indeed, as would be expected from these patterns of potential microbial hosts, a large proportion of the VCs with significant light–dark differential abundance was part of the cyanophage groups (488 contigs in 16 VCs and 9 rank states; SI Appendix, Fig. S8B and Table S2). Some cyanophages have been shown to have a light-dependent lytic infection cycle, where the infection is initiated in the early morning and progresses throughout the day when energy fluxes are at their peak (through photosynthetic activity), promoting viral propagation (18, 45–47). Recent work has also shown that some Prochlorococcus viruses are capable of adsorption and even replication in the dark (47).
The cellular and RNA diel abundance of some of the cyanophage groups in our data (cyanopodoviruses and cyanomyoviruses) show a pattern that peaks during light hours, suggesting that the infection of these cyanophages indeed follows diel energy fluxes as previously suggested (16, 18, 47, 48). Summer-abundant cyanophages (in rank state V[Hs-Lw]C[Hs-Lw]) show an increase in intracellular abundance during light hours in all three families. In winter-abundant cyanophages (V[Ls-Hw]C[Ls-Hw]), similar daytime increase was observed for cyanopodoviruses and cyanomyoviruses but not for cyanosiphoviruses (Fig. 4). The variation in intracellular abundance patterns by viruses infecting similar hosts might indicate differences in the infection cycle or the metabolic capabilities of these viruses as has been recently described for three different cultured Prochlorococcus viruses (47, 49). That is to say, based on our results we hypothesize that a high diversity exists in the timing and rate of various key aspects of infection such as absorption, virion production, and lysis for these viruses in their natural environment.
The contrast between the detection of viral transcripts in the different cyanophage families, where the podoviruses and myoviruses show a clear transcriptional increase at the onset of light, as opposed to the less clear pattern in the siphoviruses, could result from lack of AMGs in the latter (based on currently described genomes) (50). Further manual inspection of several complete cyanosiphovirus genomes in our samples (length 40 kb) confirmed the absence of known AMGs in these genomes (51, 52). However, this hypothesis cannot be validated using our data and requires additional experimental validation. These results signify that while many cyanophages display a clear intracellular increase in abundance during daytime, the hourly resolved variations might indicate differences in their infection cycles or capacities to tinker host metabolism with AMGs.
Interestingly, our approach also detected a large group of uncharacterized Red Sea viruses (947 contigs in 61 VCs and 9 rank states) with light–dark differential abundance signal (SI Appendix, Fig. S8B and Table S2). Many of these putatively infect heterotrophic microbial hosts (based on proximity to a RefSeq viral genome in the sequence similarity network; SI Appendix, Fig. S4). This large group of viruses exhibits diverse abundance patterns and potentially different diel-dependent life history traits. For example, contigs in this group with a high summer presence in both the viral and cellular fractions (V[Hs-Lw]C[Hs-Lw]) showed an increase in intracellular abundance during light hours (a handful in the RNA and the majority in the DNA fraction), while with their winter counterparts (V[Ls-Hw]C[Ls-Hw]), such an increase was only observed in the cellular DNA and not in the RNA (SI Appendix, Fig. S9, Top). This result extends the previously described diel and seasonal activity repertoire of viruses of heterotrophic microbial hosts in the marine environment (16, 17).
Differential light–dark virus–host interaction is thus prevalent among viruses in the active population in the photic marine environment and is not restricted to cyanophages. However, while the uncharacterized Red Sea virus group is larger than the cyanophages group, the prevalence of contigs with detectable differential light–dark RNA signal was more pronounced in the latter (SI Appendix, Table S2). In contrast, we could not detect any diel abundance patterns in the vDNA fraction of the bank population, and the abundance levels in the gDNA and RNA fractions were too low for analysis.
While highly abundant in the free virus fraction, we observed low abundance of transcripts from viruses of heterotrophic bacteria (SAR116 as a prime example; SI Appendix, Fig. S9, Bottom) despite the strong transcriptional diel patterns of many of their potential heterotrophic bacteria hosts in this study (see photoautotroph to heterotroph ratio in SI Appendix, Fig. S7 and Table S3). Our results are consistent with previous reports which mostly detected cyanophage transcripts in environmental metatranscriptomic samples (16), indicating their high levels of intracellular transcription in comparison to highly abundant heterotroph viruses (e.g., SAR11 and SAR116 viruses). In addition, viruses infecting heterotrophic microbes such as SAR11, SAR116, and MGII Euryarchaea (26, 53, 54) have been previously reported to have lower intracellular signal (26, 55). Notwithstanding the high abundance of free viruses, these results indicate that the fraction of actively infected heterotrophic hosts is low in our samples, especially in comparison to cyanobacteria and their viruses. It is possible that the low signal in the RNA and cellular DNA fractions is due to an experimental bias or to the fact that they operate on longer and slower life cycles (53, 54). However, our observations were consistent across multiple seasons, samples, and technical replicates.
The discrepancy between the infection levels of cyanophages and that of viruses of heterotrophs suggests a fundamental difference in lifestyle and infection dynamics between these groups. It is most intriguing, then, that viruses of heterotrophs remain in high proportions in the bank, and future research is needed for understanding their underlying infection dynamics in the marine environment. In light of recent reports of temperate phages in SAR11 and constant production of viral particles (36, 56), our results hint that such mechanisms likely govern the abundance patterns of this as well as other groups of phages of heterotrophs in the marine environment.
Conclusions
Here we have elucidated previously unknown complex seasonal and diel patterns of abundance and activity of marine viruses of the photic zone. We used the bank model (13) as a conceptual framework where the most abundant viruses are considered active and the remaining majority as bank. In addition, we classified viruses based on their seasonal changes in ranks. The bank group persisted across seasons, while many contigs in the active group showed seasonal change in rank abundance in either the viral or cellular fractions. We speculate that the seasonal viral response is facilitated by a stable bank population that is readily activated upon increase in abundance of a potential host, in accordance with kill-the-winner and bank models (10, 12, 13).
A negative correlation between virus production (burst size) and virus survival has been previously demonstrated even between viruses of Escherichia coli (57). Thus, beyond the straightforward implications regarding the infection cycle of these viral groups (bank and active) in the marine environment, questions arise regarding their life history traits and challenge the prevailing view of virus decay rates (7). Specifically, the mechanisms that allow for viral groups to persist in the bank population despite apparent low infection levels are currently not being considered in most models.
In taxonomically similar viral superclusters we detected a variety of rank states. This could suggest that at the general supercluster-level abundance, some viral groups might appear to be stable despite changes in host composition. However, when examining the rank states of these superclusters, seasonal patterns emerge. These patterns, we hypothesize, are a result of a considerable number of members of the viral population having a broad host range and their being able to operate in a changing host landscape (58–60). Our observation that the taxonomical clustering does not conform with the rank state classification supports the theoretical finding that virus–host interactions cannot be easily deduced from linear relationship in their abundance in surface water samples (1).
Our observed diversity in diel patterns of viral transcripts and intracellular abundance, within both VCs and rank states, provides environmental support to recent culture-based evidence (47, 49) and suggests that it is more widespread. Our observations in the Red Sea of seasonal changes in the ranks of the most abundant virus groups are supported by previous long-term studies in marine and freshwater environments (21, 22, 24, 37, 61, 62), and our observations are somewhat different from a recent report of mostly stable viral populations in a 5-y time series study at the San Pedro Ocean Time Series (25).
Additionally, our data suggest that high viral abundance in the viral fraction might not necessarily translate into high infection levels, as is evident in the case of many heterotrophic viruses with almost no detectable intracellular presence, despite being highly abundant in the viral fraction. This could suggest a consistently low production of virions (36) (below detection levels), allochthonous inputs, extremely slow decay rates, or another as yet unknown mechanism supporting this intracellular and extracellular abundance discrepancy.
The approach used in this study facilitated the discovery of several coexisting viral populations that exhibit various seasonal and diel patterns. The prevalence of differential diel light–dark abundance patterns of viral contigs across seasons might indicate the convergence of infection and propagation strategies employed by viruses as a function of the recurrent diel metabolic patterns of their hosts. Furthermore, the repertoire of possible viral lifestyles and infection dynamics, manifested in rank states in this study, suggests underlying differences within major virus groups in the marine environment.
Additional fundamental differences between viruses of marine photosynthetic microbes and viruses of heterotrophs appear to exist, and further investigation is needed to understand how life history traits differ between these groups and whether they can explain the patterns we observed. The VCs and rank states reported here can serve as potential targets for such future research, which could be fundamental for understanding the contribution of viruses to the structure of microbial assemblages and the resulting impact on the dynamics of viral shunt, nutrient cycles, and the marine food web.
Materials and Methods
Sampling Site.
Evident seasonal succession patterns of microbial communities have been previously described in the Gulf of Aqaba (44, 63), largely driven by its oligotrophic nature and annual stratification-mixing cycles (63, 64). Additionally, changes in viral communities have also been observed between different seasons and between stratified and mixed water column (23, 65, 66).
Sample Collection and DNA/RNA Extraction.
Water samples were collected every 2 h during a span of 24 h, from the Interuniversity Institute for Marine Sciences (IUI) pier in Eilat from a depth of 2 to 3 m (surface water). At each time point, two 20-L containers of seawater were initially filtered through a GF/D glass microfiber filter (Whatman) with a nominal pore size of 2.7 m to remove large eukaryotic cells. One 20-L container was used to collect two fractions: cellular genomic DNA (cellular DNA fraction or metagenome) and, after filtration, viral DNA (viral fraction, virome, or free viruses). The second container was used to collect RNA (cellular RNA fraction or metatranscriptome). A similar set of samples was collected on 11 to 12 August 2015 (summer samples) and on 7 to 8 February 2016 (winter samples). The cellular DNA fraction was collected onto a 142-mm 0.22-m Durapore filter (Millipore) using a peristaltic pump. DNA was extracted using the alkaline-lysis protocol. For the viral DNA, the flow through of the 0.22-m filters was treated with iron chloride (FeCl3) for the precipitation of viral particles, as described in ref. 67. The precipitate was collected on a 0.22-m Durapore filter (Millipore) and washed with 10 mL of calcium oxalate solution. The washed precipitate was further concentrated using a 100-kDa Centricon filter (Millipore) and purified on a CsCl gradient followed by DNase treatment as described in ref. 68. DNA extraction was performed using Wizard minicolumns (Promega) as described in ref. 69. The metatranscriptome fraction was collected onto a 142-mm 0.22-m Durapore filter (Millipore) using a peristaltic pump. After collection, the filters were transferred immediately to a screw cap containing 1 mL of RNAlater (Ambion) and frozen in liquid nitrogen. Total handling time was less than 15 min. Total RNA extraction was done using the mirVana RNA isolation kit (Ambion), followed by DNA removal with Turbo DNase (Ambion) and cleanup using the RNeasy MinElute Kit (QIAGEN).
Sample Processing and Sequencing.
The DNA samples were sheared using Covaris E220 with the following parameters: 10% duty factor, 45 s duration time, 200 cycles per burst, 175 W peak incident power, and a temperature of 6 ○C. The RNA samples were fragmented using a library preparation kit (NEBNext) with 5 min in 75 ○C. The mean fragment lengths (without adapters) of the DNA and RNA samples were 404 and 350 bp, respectively. None of the samples were amplified prior to sequencing-library preparation, nor were rRNA depletion protocols applied in order to avoid possible bias resulting from these steps (70) and loss of considerable amount of RNA during the rRNA depletion process. Libraries were constructed with 10 ng of DNA per sample using NEBNext Ultra II DNA Library Prep Kit with 12 PCR cycles for the DNA samples and 100 ng with NEBNext Ultra RNA Library Prep Kit with 15 PCR cycles for the RNA samples. All samples were paired-end (PE) sequenced at the Technion Sequencing center on Illumina Hiseq 2500, where the DNA samples sequenced with 2 ×125 bp and the RNA samples with 2 ×100 bp.
Quality Control and Assembly of Short Reads.
Reads were trimmed using Trim Galore version 0.4.4 (https://github.com/FelixKrueger/TrimGalore) with default parameters. Data for the 48 different DNA samples (24 vDNA and 24 gDNA) were assembled separately using IDBA-UD (71) with default parameters.
Annotation.
The assembled contigs from all of the different samples were concatenated in a single FASTA file and dereplicated to keep those 5,000 bp using vsearch version 2.6.2 (72) with the options “–derep_fulllength –minseqlength 5000 –maxseqlength 5000000.” The dereplicated contigs were then screened using Mash version 2.0 (73) for similarities to PhiX (GenBank accession no. HM775309.1), a common control used during Illumina sequencing runs which can result in spurious assemblies (74). Using the Mash results, those contigs with similarities to PhiX were discarded from further analysis. Open reading frames (ORFs) of the remaining contigs were predicted by Prodigal version 2.6.3 (75) using the parameter -p meta. ORFs longer than 300 nt were dereplicated using vsearch with the options “–derep_fulllength –minseqlength 300 –maxseqlength 5000000.” ORFs were taxonomically annotated using either BLASTn best-hit against NCBI nr, Diamond version 0.8.38 (76), or blastp best-hit to the protein sequences from the Prokaryotic Virus Orthologous Groups database (77). ORFs appearing in the RNA data were assigned a functional category using eggNOG mapper version 1 (78), with mapping mode Diamond and default parameters.
Relative Abundance Estimations.
The relative abundance of ORFs was calculated using Salmon version 0.8.2 (79). A total of 16,047,552 ORFs predicted from 3,878,543 assembled contigs and 3,224 Refseq genomes (bacteria, archaea, plastid, protozoa, and viruses; accessed 2 February 2017) were dereplicated using vsearch “–derep_fulllength” (10,893,032 unique ORFs) and used to create a Salmon index. The Refseq genomes used were selected by classifying the unassembled reads with Kraken version 0.10.4-beta (80) and a custom database composed by all Refseq available genomes (bacterial, archaeal, and viral; accessed 2 February 2017). The list of taxa assigned to the reads was retrieved and processed to keep only those genomes not considered as sample-processing contamination sources (81). The ORF abundance in the 72 datasets (metagenomes, metatranscriptomes, and viromes) was quantified with the index using Salmon in the quasi-mapping mode with the following parameters: “–meta –incompatPrior 0.0 – seqBias –gcBias –numAuxModelSamples 2500000 –numBootstraps 100 –validateMappings.” Quantification results were processed by tximport (version 1.10.0) (82), followed by normalization with edgeR (v3.24.2) (83). Reads per kilobase per million were calculated from the normalization results by the edgeR function edgeR::rpkm.
Clustering Diel Gene Expression Patterns.
Predicted ORFs were quantified as mentioned above (Salmon and tximport), followed by normalization with Bioconductor R package DESeq2 (84) by applying a regularized log transformation (rlog) to the Salmon counts data (baseMean 2). The normalized counts were clustered using Gaussian mixture models coupled with Dirichlet process (DP-GP) (85) with default parameters. The ORFs clustered into 18 and 23 clusters in the summer and winter datasets, respectively, based on the similarity of their diel expression patterns.
Mapping, Binning, and Prediction of Viral Contigs.
Short reads were mapped to the assembled contigs using Bowtie2 version 2.3.4.1 (86) and Samtools 1.3.1 (87). The mapping files were used to generate a depth coverage matrix using Metabat2 script jgi_summarize_bam_contig_depths and binned with Metabat2 version 2.12.1 (88) with parameters -m 1500 -s 10000. Prediction of viral contigs was performed using two independent classifiers: 1) VirFinder (89), R package version 1.1 with default parameters and modEPV_k8.rda model for predicting both prokaryotic and eukaryotic viruses, and 2) MARVEL (90) version 0.1 with default parameters.
Evenness and Rank Change Estimation.
Evenness was measured for the abundance of viral contigs per diel time points in the summer and winter samples using the codyn R package (91) community_structure function for Simpsons evenness, EQ, and Evar (27) indices, as described in ref. 29. Rank change was measured based on the concepts presented in ref. 29. Briefly, mean rank shift was measured by taking the sum of absolute rank differences between consecutive time points, divided by the total number of unique viral contigs in both time points. Rank change was measured by dividing the mean rank shift by the total number of unique viral contigs in all time points, making the measure independent of species richness (29). For the seasonal rank change we used the geometric mean of viral contigs across diel time points per season as the input for the process described above.
Taxonomic Classification of Prokaryote Viruses and Network Visualization.
Taxonomic assignment of viral contigs from the active group was performed with vConTACT (Viral CONTigs Automatic Clustering and Taxonomy) 2.0 (32) on the CyVerse cyberinfrastructure platform (92). Viral contigs were clustered into VCs based on a gene-sharing network, where each VC was assigned a taxonomic annotation based on the identity of Refseq viral genomes found in the same VC or according to the closest Refseq genomes that represented a distinct hub (node degree average node degree. The vConTACT analysis was performed with the following parameters: protein clusters generation method, Markov Cluster Algorithm (MCL); protein–protein similarity method, Diamond; VC generation method, Clustering with Overlapping Neighborhood Expansion (ClusterONE); and reference database, National Center for Biotechnology Information (NCBI) Bacterial and Archaeal Viral RefSeq V85 with International Committee on Taxonomy of Viruses (ICTV) + NCBI taxonomy. Network visualization was performed using Cytoscape version 3.7.1.
Rank State Assignment and CLR Transformation.
Grouping contigs into active and bank groups relies on setting a cutoff to differentiate between the two. In the absence of a formal definition for the bank model (13), we used the highest proportion of the most abundant contigs group in a single season as a baseline (15%; Fig. 2E). Next, we aimed to set the cutoff such that the widest array of rank states are represented (by at least five contigs), while keeping it strict enough to avoid introducing noisy patterns in some contigs. (This means that very strict cutoffs eliminate the assignment of some rank states, while very permissive ones allow the assignment of seasonal contigs as nonseasonal and vice versa.) Having tested a range of cutoffs for the top 10 to 30% (5% increments), setting the top 20% as the active group represented the maximum realized rank state diversity with minimum noise. Furthermore, to avoid misrepresenting borderline cases as seasonal contigs (e.g., a move from the top 19% to the top 21%), we defined a seasonal change between the active and bank as a minimum of 20% change in rank percentile and at least one order of magnitude change in abundance (RPKM). To further validate that the rank state patterns persist when analyzed with bias-insensitive measures (35), we used the ALDEx2 (ANOVA-like differential expression) R package to visualize the rank states based on the CLR of the viral abundances (that is, the centered log ratio of each virus to the geometric mean of all viruses). Specifically, we used the aldex.clr function to calculate the transformation with the denominator parameter set to iqlr and 128 Monte Carlo samples (93) (SI Appendix, Fig. S6).
Supplementary Material
Acknowledgments
We thank IUI in Eilat, Israel, for providing access to their pier as well as work space and the Genomics Centers at the Biomedical Core Facility and Lorry I. Lokey Interdisciplinary Center for Life Sciences and Engineering at the Technion for library preparation and sequencing. We also thank Dan Fishman for sample preparation, Itai Sharon for help with metagenome assemblies, Sheila Roitman and Faris Salama for their help with sampling, David Cohen of the Physics Department at the Technion for his help with the HPC ATLAS cluster used for the analyses, and Michael McLaren for invaluable comments on our manuscript. This work was funded by a European Commission European Research Council Advanced grant 321647, Israel Science Foundation grant 143/18, the Technion’s Lorry I. Lokey Interdisciplinary Center for Life Sciences and Engineering and the Russell Berrie Nanotechnology Institute, and the Louis and Lyra Richmond Memorial Chair in Life Sciences (to O.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010783117/-/DCSupplemental.
Data Availability.
The assembled viral contigs, along with raw sequence data, are available from the European Nucleotide Archive under accession no. PRJEB35627. SI Appendix, Tables S1–S5, can be downloaded from https://doi.org/10.17605/osf.io/b74mt.
Code Availability
Scripts and notebooks for reproducing the rank state analysis are available at https://doi.org/10.17605/osf.io/b74mt.
References
- 1.Wigington C. H., et al. , Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 1, 15024 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Brum J. R., et al. , Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Paez-Espino D., et al. , Uncovering Earth’s virome. Nature 536, 425–430 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Puxty R. J., Millard A. D., Evans D. J., Scanlan D. J., Viruses inhibit CO2 fixation in the most abundant phototrophs on Earth. Curr. Biol. 26, 1585–1589 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Guidi L., et al. , Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelm W. S., Suttle A. C., Viruses and Nutrient Cycles in the Sea (American Institute of Biological Sciences, Washington, DC, 1999), vol. 49. [Google Scholar]
- 7.Fuhrman J. A., Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Brussaard C. P. D., et al. , Global-scale processes with a nanoscale drive: The role of marine viruses. ISME J. 2, 575–578 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Jiao N., et al. , Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Breitbart M., Bonnain C., Malki K., Sawaya A. N., Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Jover L. F., Chad Effler T., Buchan A., Wilhelm S. W., Weitz J. S., The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 12, 519–528 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Thingstad T. F., Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328 (2000). [Google Scholar]
- 13.Breitbart M., Rohwer F., Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Weinbauer M. G., Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Warwick-Dugdale J., Buchholz H. H., Allen M. J., Temperton B., Host-hijacking and planktonic piracy: How phages command the microbial high seas. Virol. J. 16, 15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aylward F. O., et al. , Diel cycling and long-term persistence of viruses in the ocean’s euphotic zone. Proc. Natl. Acad. Sci. U.S.A. 114, 11446–11451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida T., et al. , Locality and diel cycling of viral production revealed by a 24 h time course cross-omics analysis in a coastal region of Japan. ISME J. 12, 1287–1295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puxty R. J., Evans D. J., Millard A. D., Scanlan D. J., Energy limitation of cyanophage development: Implications for marine carbon cycling. ISME J. 12, 1273–1286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieradzki E. T., Ignacio-Espinoza J. C., Needham D. M., Fichot E. B., Fuhrman J. A., Dynamic marine viral infections and major contribution to photosynthetic processes shown by spatiotemporal picoplankton metatranscriptomes. Nat. Commun. 10, 1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolody B. C., et al. , Diel transcriptional response of a California current plankton microbiome to light, low iron, and enduring viral infection. ISME J. 13, 2817–2833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Needham D. M., et al. , Short-term observations of marine bacterial and viral communities: Patterns, connections and resilience. ISME J. 7, 1274–1285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagarete A., Chow C.-E. T., Johannessen T., Fuhrman J. A., Thingstad T. F., Sandaa R. A., Strong seasonality and interannual recurrence in marine myovirus communities. Appl. Environ. Microbiol. 79, 6253–6259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dekel-Bird N. P., et al. , Diversity and evolutionary relationships of T7-like podoviruses infecting marine cyanobacteria: Evolutionary relationships of T7-like cyanopodoviruses. Environ. Microbiol. 15, 1476–1491 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Arkhipova K., et al. , Temporal dynamics of uncultured viruses: A new dimension in viral diversity. ISME J. 12, 199–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ignacio-Espinoza J. C., Ahlgren N. A., Fuhrman J. A., Long-term stability and red queen-like strain dynamics in marine viruses. Nat. Microbiol. 5, 265–271 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Alon P., Yutin N., Flores-Uribe J., Sharon I., Koonin V. E., Béjà O., Novel abundant oceanic viruses of uncultured marine group II Euryarchaeota. Curr. Biol. 27, 1362– 1368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith B., Wilson J. B., A consumer’s guide to evenness indices. Oikos 76, 70–82 (1996). [Google Scholar]
- 28.Haegeman B., Hamelin J., Moriarty J., Neal P., Dushoff J., Weitz J. S., Robust estimation of microbial diversity in theory and in practice. ISME J. 7, 1092–1101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avolio M. L., et al. , A comprehensive approach to analyzing community dynamics using rank abundance curves. Ecosphere 10, e02881 (2019). [Google Scholar]
- 30.Gregory A. C., et al. , Marine DNA viral macro-and microdiversity from pole to pole. Cell 177, 1109–1123.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho J., Tumkaya T., Aryal S., Choi H., Adam C.-C., Moving beyond p values: Data analysis with estimation graphics. Nat. Methods 16, 565–566 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Jang H. B., et al. , Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Simmonds P., Aiewsakun P., Virus classification—Where do you draw the line? Arch. Virol. 163, 2037–2046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooks J. P., Challenges for case-control studies with microbiome data. Ann. Epidemiol. 26, 336–341.e1 (2016). [DOI] [PubMed] [Google Scholar]
- 35.McLaren M. R., Willis D. A, Callahan B. J., Consistent and correctable bias in metagenomic sequencing experiments. Elife 8, e46923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris R. M., Kelsy R. C., Hvorecny K. L., Kollman J. M., Lysogenic host–virus interactions in sar11 marine bacteria. Nat. Microbiol. 5, 1011–1015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brum J. R., Hurwitz B. L., Schofield O., Ducklow H. W., Sullivan M. B., Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 10, 437–449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashy R. A., Agustí S., Low host abundance and high temperature determine switching from lytic to lysogenic cycles in planktonic microbial communities in a tropical sea (Red Sea). Viruses 12, 761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Field C. B., Behrenfeld M. J., Randerson J. T., Paul F., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Aylward F. O., et al. , Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc. Natl. Acad. Sci. U.S.A. 112, 5443–5448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vellend M., The Theory of Ecological Communities (MPB-57) (Princeton University Press, 2016), vol. 75. [Google Scholar]
- 42.Louca S., et al. , Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Lindell D., Anton F., Post. Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol. Oceanogr. 40, 1130–1141 (1995). [Google Scholar]
- 44.Al-Najjar T., Badran M. I., Richter C., Meyerhoefer M., Sommer U., Seasonal dynamics of phytoplankton in the Gulf of Aqaba, Red Sea. Hydrobiologia 579, 69–83 (2007). [Google Scholar]
- 45.Lindell D., et al. , Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Fridman S., et al. , A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected prochlorococcus cells. Nat. Microbiol. 2, 1350–1357 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Liu R., Liu Y., Chen Y., Zhan Y., Zeng Q., Cyanobacterial viruses exhibit diurnal rhythms during infection. Proc. Natl. Acad. Sci. U.S.A. 116, 14077–14082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldbauer J. R., et al. , Nitrogen sourcing during viral infection of marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A., 116, 15590–15595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demory D., et al. , Linking light-dependent life history traits with population dynamics for prochlorococcus and cyanophage. mSystems 5, e00586-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan M. B., et al. , Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4, e234 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A., et al. , CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic. Acids Res., 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puxty R. J., Millard A. D., Evans D. J., Scanlan D. J., Shedding new light on viral photosynthesis. Photosynth. Res. 126, 71–97 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y., et al. , Abundant SAR11 viruses in the ocean. Nature 494, 357–360 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Kang I., Oh H.-M., Kang D., Cho J.-C., Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc. Natl. Acad. Sci. U.S.A. 110, 12343–12348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alonso-Sáez L., Morán X. A. G., Clokie M. R. J., Low activity of lytic pelagiphages in coastal marine waters. ISME J. 12, 2100–2102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y., et al. , Pelagiphages in the podoviridae family integrate into host genomes. Environ. Microbiol. 21, 1989–2001 (2019). [DOI] [PubMed] [Google Scholar]
- 57.De Paepe M., Taddei F., Viruses’ life history: Toward a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4, e193 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Avrani S., Schwartz D. A., Lindell D., Virus-host swinging party in the oceans: Incorporating biological complexity into paradigms of antagonistic coexistence. Mob. Genet. Elements 2, 88–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz D. A., Lindell D., Genetic hurdles limit the arms race between prochlorococcus and the t7-like podoviruses infecting them. ISME. J. 11, 1836–1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Jonge P. A., Nobrega F. L., Brouns S. J. J., Dutilh B. E.. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 27, 51–63 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Parsons R. J., Breitbart M., Lomas M. W., Carlson C. A., Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 6, 273–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheryl-EmilianeChow T., Fuhrman J. A., Seasonality and monthly dynamics of marine myovirus communities. Environ. Microbiol. 14, 2171–2183 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Labiosa R. G., Arrigo K. R., Genin A., Monismith S. G., van Dijken G., The interplay between upwelling and deep convective mixing in determining the seasonal phytoplankton dynamics in the Gulf of Aqaba: Evidence from SeaWiFS and MODIS. Limnol. Oceanogr. 48, 2355–2368 (2003). [Google Scholar]
- 64.Wolf-Vecht A., Paldor N., Brenner S.. Hydrographic indications of advection/convection effects in the gulf of elat. Deep Sea Res. Part A. Oceanogr. Res. Pap. 39, 1393–1401 (1992). [Google Scholar]
- 65.Parsons R. J., Breitbart M., Lomas M. W., Carlson C. A., Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern sargasso sea. ISME J. 6, 273–284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldsmith D. B., Brum J. R., Hopkins M., Carlson C. A., Breitbart M., Water column stratification structures viral community composition in the Sargasso Sea. Aquat. Microb. Ecol. 76, 85–94 (2015). [Google Scholar]
- 67.Poulos B. T., John S. G., Sullivan M. B.. Iron chloride flocculation of bacteriophages from seawater. Methods Mol. Biol. 1681, 49–57 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Thurber R. V., Haynes M., Breitbart M., Wegley L., Rohwer F., Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4, 470–483 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Vik D. R., et al. , Putative archaeal viruses from the mesopelagic ocean. PeerJ 15, e3428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lahens N. F., et al. , IVT-seq reveals extreme bias in RNA sequencing. Genome Biol. 15, R86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng Yu., Henry C., Leung M., Yiu S. M., Chin. F. Y. L., IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Rognes T., Flouri T., Nichols B., Quince C., Mahé F., Vsearch: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ondov B. D., et al. , Mash: Fast genome and metagenome distance estimation using MinHash. Genome. Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukherjee S., Huntemann M., Ivanova N., Kyrpides N. C., Pati A., Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand. Genomic Sci. 10, 18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using diamond. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Grazziotin A. L., Koonin E. V., Kristensen D. M., Prokaryotic virus orthologous groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 45, D491–D498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huerta-Cepas J., et al. , Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wood D. E., Salzberg S. L., Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salter S. J., et al. , Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soneson C., Love M. I., Robinson M. D., Differential analyses for rna-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McDowell I. C., et al. , Clustering gene expression time series data using an infinite Gaussian process mixture model. PLoS Comput. Biol. 14, e1005896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ben L., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H., et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang D., et al. , Metabat 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren J., Ahlgren N. A., Lu Y. Y., Fuhrman J. A., Sun. F.. Virfinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amgarten D., Lucas P., Braga P., da Silva A. M., Setubal J. C., Marvel, a tool for prediction of bacteriophage sequences in metagenomic bins. Front. Genet. 9, 304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hallett L. M., et al. , codyn: An R package of community dynamics metrics. Methods Ecol. Evol. 7, 1146–1151 (2016). [Google Scholar]
- 92.Merchant N., et al. , The iplant collaborative: Cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 14, e1002342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gloor G., Aldex2: ANOVA-like differential expression tool for compositional data (ALDEX Manual Modular 20, Bioconductor, 2015), pp. 1–11.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled viral contigs, along with raw sequence data, are available from the European Nucleotide Archive under accession no. PRJEB35627. SI Appendix, Tables S1–S5, can be downloaded from https://doi.org/10.17605/osf.io/b74mt.