Significance
Ribosomal proteins (RPs) are essential structural components of ribosomes. In many instances, RPs are encoded in small families of genes, suggesting possible neofunctionalization for some family members. This is the case of the uL18 family of proteins that developed during plant evolution and which is composed of eight members in most Angiosperms. Our study shows that two members associate with organellar ribosomes while two other uL18-Like proteins have become intron-specific splicing factors acting in mitochondria and plastids, respectively. Our study shows that the RNA binding capacity of RPs can be diverted to create factors acting in intron splicing.
Keywords: ribosomal protein, intron splicing, plants, mitrochondria, chloroplasts
Abstract
Production and expression of RNA requires the action of multiple RNA-binding proteins (RBPs). New RBPs are most often created by novel combinations of dedicated RNA-binding modules. However, recruiting existing genes to create new RBPs is also an important evolutionary strategy. In this report, we analyzed the eight-member uL18 ribosomal protein family in Arabidopsis. uL18 proteins share a short structurally conserved domain that binds the 5S ribosomal RNA (rRNA) and allows its incorporation into ribosomes. Our results indicate that Arabidopsis uL18-Like proteins are targeted to either mitochondria or chloroplasts. While two members of the family are found in organelle ribosomes, we show here that two uL18-type proteins function as factors necessary for the splicing of certain mitochondrial and plastid group II introns. These two proteins do not cosediment with mitochondrial or plastid ribosomes but instead associate with the introns whose splicing they promote. Our study thus reveals that the RNA-binding capacity of uL18 ribosomal proteins has been repurposed to create factors that facilitate the splicing of organellar introns.
The evolution of eukaryotic cells has involved the acquisition of highly specialized energy-producing organelles such as mitochondria and plastids, both of which are of bacterial origin (1, 2). The transformation of the original endosymbionts into specialized organelles involved a massive reduction of their genomes and a progressive loss of their autonomy. A large number of host-derived factors, either acquired or recruited during evolution, became necessary for proper expression of the essential genes present in modern mitochondrial and plastid genomes. The interplay between ancient bacterial-derived processes and eukaryotic-derived functions resulted in organellar gene expression mechanisms that are more complex than those of modern bacteria (3, 4). Consequently, the production and the expression of mitochondrial and plastid transcripts is highly complex, especially in plants. Angiosperm mitochondrial and plastid genomes produce an array of monocistronic and polycistronic transcripts that undergo nucleolytic processing, extensive sequence modification through C-to-U RNA editing, and removal of multiple group II introns prior to translation. Each of these RNA processing steps requires the action of dedicated ribonucleoprotein complexes that are still poorly characterized. Several classes of nuclear-encoded RNA-binding proteins were found to play roles in mitochondrial and/or plastid RNA expression including the pentatricopeptide repeat (PPR) proteins (5, 6) and other kinds of helical repeat protein families (7), which all adopt similar solenoid-like structures exposing key amino acids for RNA binding (7–9). Other protein families like RNA recognition motif (RRM) factors, multiple organellar RNA editing factors (MORF), chloroplast RNA splicing and ribosome maturation (CRM), or plant organellar RNA recognition (PORR) families as well as maturases also play roles in plant organellar RNA expression (10–14). Some of these factors were created out of generic RNA-binding domains (such as the RRM domain) while others evolved by hijacking proteins with different functions such as the CRM proteins that are likely derived from a bacterial protein involved in ribosome maturation (11). Ribosomal proteins (RPs) represent another large class of conserved and abundant RNA-binding proteins. Although they are primarily involved in protein synthesis, extraribosomal functions have been described in a few cases (15–17). Plant RPs are often encoded by small multigene families comprising up to eight active genes, suggesting potential specialization or extraribosomal activities for some family members (18, 19). In this study, we investigated the function of a small protein family in Arabidopsis whose members were annotated to be homologs of the uL18 RP. The detailed analysis of two of these uL18 homologs revealed that they have lost the ability to associate with ribosomes and have become organellar group II intron splicing factors.
Results
The uL18 Protein Family Comprises Eight Members in Arabidopsis.
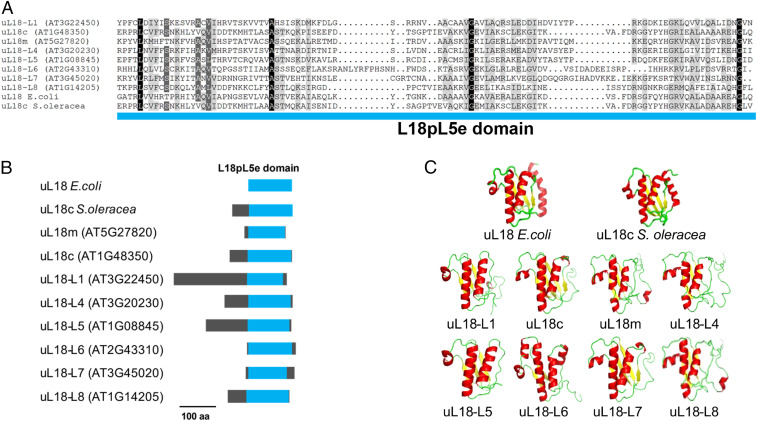
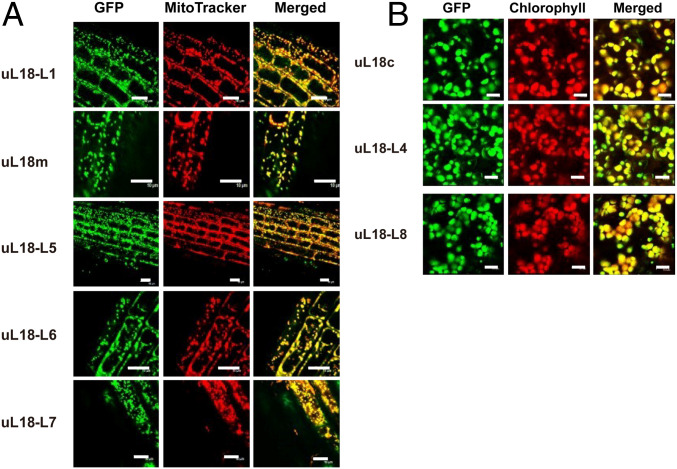
While analyzing sequence variability among RPs in Arabidopsis, we noticed that eight proteins were classified in the L18pL5e superfamily. These proteins share a rather degenerate but structurally conserved C-terminal RNA-binding domain forming three α-helices and three and a half β-sheets that was shown to bind the 5S ribosomal RNA (rRNA) (20) (Fig. 1). C-terminal GFP translational fusions with each of these uL18-Like proteins revealed that five members are targeted to mitochondria and three to plastids (Fig. 2). Multiple sequence alignments indicated that uL18-Like members are more homologous to the Escherichia coli uL18 protein than to the cytosolic uL18, strongly suggesting a bacterial origin for the uL18-Like family (SI Appendix, Fig. S1). Moreover, two members of the uL18 family, AT1G48350 and AT5G27820, have sequences and sizes very close to the E. coli uL18 protein. A protein 81% identical to the mature form of AT1G48350 was found in the structure of the spinach (Spinacia oleracea) chloroplast ribosome (21), indicating that it most likely corresponds to the plastid uL18 protein in Arabidopsis. We thus named this protein uL18c in agreement with the new RP nomenclature (22). Similarly, the AT5G27820 protein was recently found to be a component of the mitochondrial ribosome (23) and was thus termed uL18m (Fig. 1). Our GFP fusion analysis confirmed the plastid and mitochondrial targeting of Arabidopsis uL18c and uL18m, respectively (Fig. 2). The function of the six other proteins could not be recognized from their sequence and were named uL18-Like (or uL18-L) proteins. Potential orthologs could be easily identified for most uL18-Like members in both monocots and dicots, supporting their essential character for organelle functions (SI Appendix, Fig. S1). The only exception is the uL18-L6 protein, which appears to be absent in monocotyledons.
Fig. 1.
The Arabidopsis nuclear genome encodes eight different uL18-Like proteins. (A) Sequence alignment of the C-terminal L18pL5e domain of the eight uL18-Like Arabidopsis proteins, the uL18 protein from E. coli and the uL18c from S. oleracea. The residues are colored according to the percentage of conservation from dark gray (100% identical) to light gray (50% identical). (B) Diagrams showing in blue the position of the L18pL5e domain in Arabidopsis uL18-Like, E. coli uL18, and S. oleracea uL18c proteins. (C) Three-dimensional structures depicting the three α-helices and three and a half β-sheets of the L18pL5e RNA binding domain as present in the E. coli uL18 (Protein Data Bank [PDB] ID code: 4YBB) and the S. oleracea plastid uL18c (PDB ID code: 5MMM) and predicted for the eight uL18-Like Arabidopsis proteins. β-sheets are shown in yellow and α-helices in red. The predicted structural models for the eight uL18-Like Arabidopsis proteins were generated with Phyre2.
Fig. 2.
The Arabidopsis uL18 and uL18-Like proteins are transported into mitochondria or plastids. Confocal microscope images showing the subcellular distribution of GFP translational fusions involving the indicated uL18 and uL18-Like proteins. (A) Pictures taken from root cells of young Arabidopsis transgenic plants. (Left) GFP fluorescence. (Center) Mitotracker Red marker. (Right) Merged signals. (B) Pictures taken from leaf cells of Arabidopsis transgenic plants. (Left) GFP fluorescence. (Center) Chlorophyll fluorescence. (Right) Merged signals. (Scale bars, 10 µm.)
ul18-l1 Mutant Plants Have a Strongly Retarded Growth Phenotype.
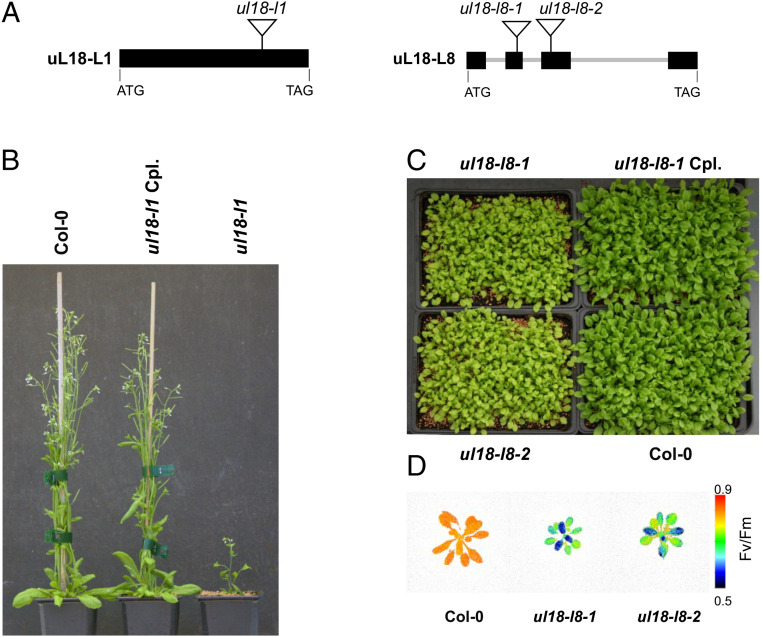
To gain insight into the function of uL18-Like proteins, we first identified a mutant in the uL18-L1 gene (Fig. 3A). Under conventional greenhouse conditions, homozygous ul18-l1 plants displayed a marked slow growth phenotype and harbored small twisted leaves (Fig. 3B). The mutant plants were late flowering and produced seeds with lower germination capacity compared to the wild type (Fig. 3B). All these phenotypes could be complemented by the expression of an HA-tagged copy of uL18-L1 in the mutant, supporting the view that the observed alterations were due to the inactivation of AT3G22450 (Fig. 3B). The mitochondrial localization of uL18-L1 was then confirmed by probing total, plastid, and mitochondrial protein fractions prepared from complemented plants (SI Appendix, Fig. S2). Collectively, the results demonstrated the strict mitochondrial localization of the uL18-L1 protein.
Fig. 3.
ul18-l1 and ul18-l8 mutants display altered developmental phenotypes. (A) Schematic representations of uL18-L1 and uL18-L8 gene structures with exons shown as black boxes and introns as thick gray lines. Triangles indicate the positions of T-DNA insertions in corresponding mutant alleles. (B) Photograph of 8-wk-old plants showing the reduced size of ul18-l1 homozygous mutants compared with WT and genetically complemented (Cpl) plants. (C) Photograph of 4-wk-old plants at the rosette stage showing the slightly reduced size and pale green color of ul18-l8 homozygous mutants compared with WT and genetically complemented (Cpl) plants. (D) False-color images representing Fv/Fm ratios of 4-wk-old WT (Col-0), ul18-l8-1, and ul18-l8-2 plants. The color scale indicates Fv/Fm signal intensities.
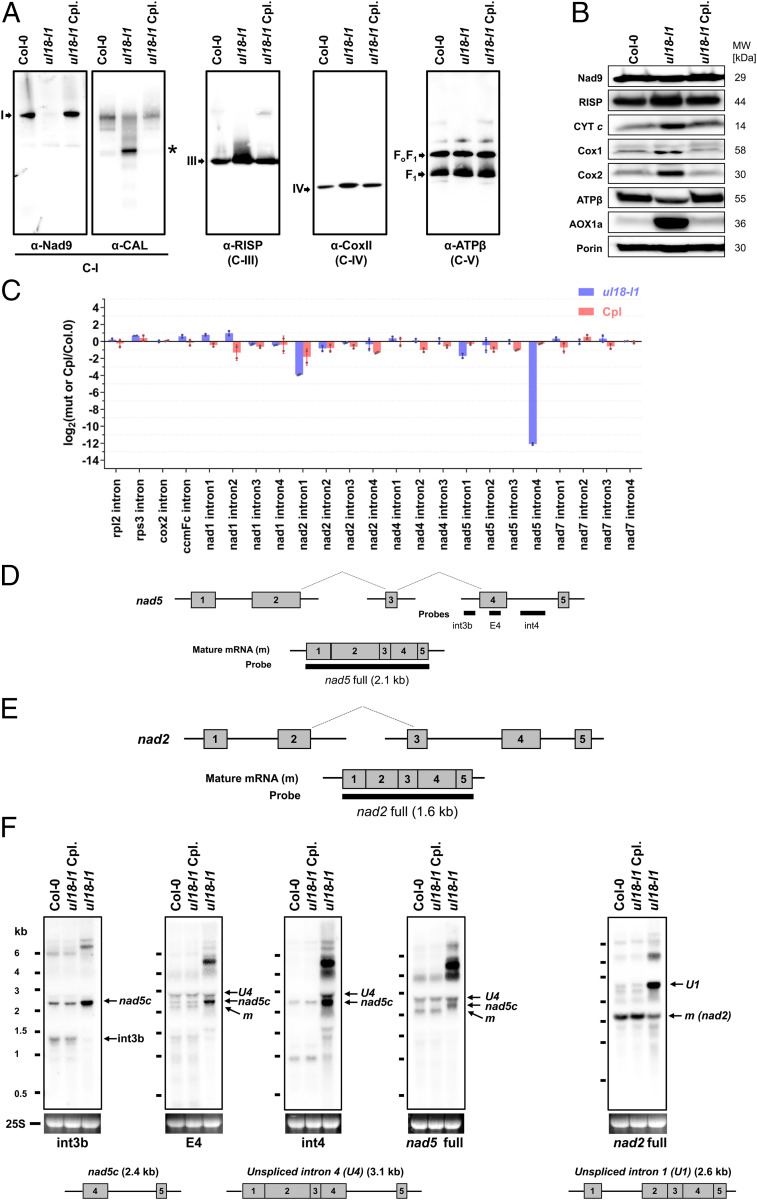
ul18-l1 Mutant Plants Show Strongly Reduced Levels of Respiratory Complex I.
The mitochondrial localization of the uL18-L1 protein strongly suggested that the developmental alterations of ul18-l1 plants could result from altered respiration. We thus monitored the levels of the different respiratory complexes in comparison with the wild type by blue-native gel analysis. Immunoblot analyses revealed only trace amounts of fully assembled complex I in ul18-l1 plants and an overaccumulation of complex IV as well as a 450-kDa complex I assembly intermediate in the mutant (Fig. 4A). In-gel activity staining confirmed the marked reduction of respiratory complex I in ul18-l1 plants and an overabundance of complex IV (SI Appendix, Fig. S3), which correlated with increased steady-state levels of Cox1, Cox2, and cytochrome c (Fig. 4B). The reduction in complex I activity in the ul18-l1 mutant was further confirmed by in vitro root growth assays, which revealed a much weaker sensitivity of ul18-l1 plants to rotenone compared to the wild type (SI Appendix, Fig. S3B). Although rotenone reduces root growth in both WT and mutant plants, at the highest dose the inhibition in the wild type is close to 50%, whereas it is limited to only 10% in the ul18-l1 mutant. Further, we could detect an overexpression of several components of the alternative respiratory pathway in ul18-l1 plants, including the alternative mitochondrial NADH dehydrogenase NDB4 and the alternative oxidase (Fig. 4B and SI Appendix, Fig. S4). These results indicated that uL18-L1 is crucial for the biogenesis and activity of mitochondrial complex I.
Fig. 4.
ul18-l1 plants are defective in complex I and are impaired in nad5 intron 4 and nad2 intron 1 splicing. (A) Immunodetection of respiratory complexes in blue native polyacrylamide gel electrophoresis blots made from mitochondrial extracts of WT (Col-0), ul18-l1 mutant, and complemented (Cpl) plants. Antibodies to Nad9 and CAL proteins were used to detect fully assembled and assembly intermediates of complex I (C-I), respectively. Complex III (C-III) was detected with antibodies to the RISP protein, complex IV (C-IV) with anti-Cox2 antibodies, and complex V (C-V) with anti-ATPβ antibodies (see SI Appendix, Table S2 for details). Respiratory complexes are designated by Roman numerals, and the asterisk designates a complex I assembly intermediate. (B) SDS/PAGE immunoblots performed on total mitochondrial protein extracts prepared from the indicated genotypes (see A for details) and probed with antibodies to subunits of respiratory complex I (Nad9), complex III (RISP), complex IV (Cox1 and Cox2), the complex V (ATPβ), cytochrome c (CYT c), and the alterative oxidase (AOX). Porin was used as protein loading control. (C) qRT-PCR analysis measuring the splicing efficiencies of mitochondrial introns in ul18-l1 (blue bars) and complemented (Cpl, red bars) plants. The bars depict log2 ratios of splicing efficiencies in ul18-l1 and complemented plants to the wild type (Col-0). Three technical replicates and two independent biological repeats were used for each genotype. SD are indicated. (D) Schematic representation of the three nad5 precursor transcripts, which are fused by two trans-splicing events to produce the mature (m) nad5 mRNA. Gray boxes indicate exons. Introns and 5′ and 3′ UTRs are shown as thin lines. The probes used to interpret RNA gel blot results are also indicated. (E) Schematic representation of nad2 precursors and mature mRNA, using the same iconography as in D. (F) RNA gel blots showing the accumulation profiles of nad5 and nad2 transcripts in the ul18-l1 mutant compared to WT (Col-0) and complemented plants (Cpl). Used probes are indicated below hybridization results. Ethidium bromide staining of the 25S ribosomal RNA serves as a loading control. Schematic representations and sizes of the nad5 transcript with unspliced intron 4 (nad5 U4), nad5c, and nad2 transcript with unspliced intron 1 (nad2 U1) are depicted below the blots to help interpreting the results. The apparent size of nad5 U4 is smaller than expected due to a congestion by near-sized 25S rRNA (24). RNA marker sizes are indicated (kilobases). E, exon; int, intron; m, mature mRNAs.
The uL18-L1 Protein Is Required for the Splicing of Two Mitochondrial Introns.
Because of their homology to uL18, we suggested that uL18-type proteins may be RNA-binding proteins as well and that the uL18-L1 protein could therefore play a key role in the production of one or several mitochondria-encoded RNAs. To test this hypothesis, we determined the steady-state levels of mature and precursor mitochondrial transcripts by qRT-PCR (SI Appendix, Fig. S5) and calculated the splicing efficiency of mitochondrial introns in both WT and ul18-l1 plants (Fig. 4C). A highly pronounced decrease in the splicing efficiency of nad5 intron 4 as well a weaker reduction for nad2 intron 1 splicing could be detected in ul18-l1 plants (Fig. 4C). A global overexpression of other premature and mature mitochondrial messenger RNAs (mRNAs) was also detected in the mutant (SI Appendix, Fig. S5 A and B). This response is often observed in respiratory mutants (24, 25) and likely plays a role in compensating for their decreased respiratory capacity. RNA gel blots confirmed these observations and indicated that no detectable amount of mature nad5 mRNA could be found in the ul18-l1 mutant (Fig. 4 D–F). The decrease in nad2 intron 1 splicing was also verified, but the production of mature nad2 mRNA was much less affected than that of nad5 and is therefore less likely to limit the production of complex I in ul18-l1 plants (Fig. 4 and SI Appendix, Fig. S5). Altogether, these results indicated that the uL18-L1 protein is essential for the cis-splicing reaction involving nad5 intron 4 and, to a lesser extent, also for the removal of nad2 intron 1.
The ul18-l8 Mutant Plants Display Altered Photosynthesis.
To further explore the functions of uL18-Like proteins, we isolated two transfer DNA (T-DNA) insertion mutants, ul18-l8-1 and ul18-l8-2, in the uL18-L8 gene, whose encoded protein is transported into plastids (Fig. 2B). Both mutant lines displayed a pale green phenotype associated with a slightly retarded growth (Fig. 3C). These alterations could be reversed by expression of an HA-tagged copy of uL18-L8 in one of the mutants, confirming that they were produced by the inactivation of uL18-L8. Pigment composition revealed that both mutants have a slight decrease in chlorophyll and carotenoid contents (SI Appendix, Table S4). Chlorophyll fluorescence further showed that the maximum quantum efficiency of PSII (Fv/Fm) was decreased by around 11% in both allelic mutants (SI Appendix, Fig. S4 and Fig. 3D) and that fraction of open PSII centers (qL) decreased by 40%. Moreover, immunoblots of subcellular fractions revealed that uL18-L8 is enriched in isolated chloroplasts and is detected solely in the stromal fraction (SI Appendix, Fig. S2). Overall, this analysis demonstrated that the loss of the uL18-L8 protein has a moderate but significant impact on the light energy efficiency of photosynthesis.
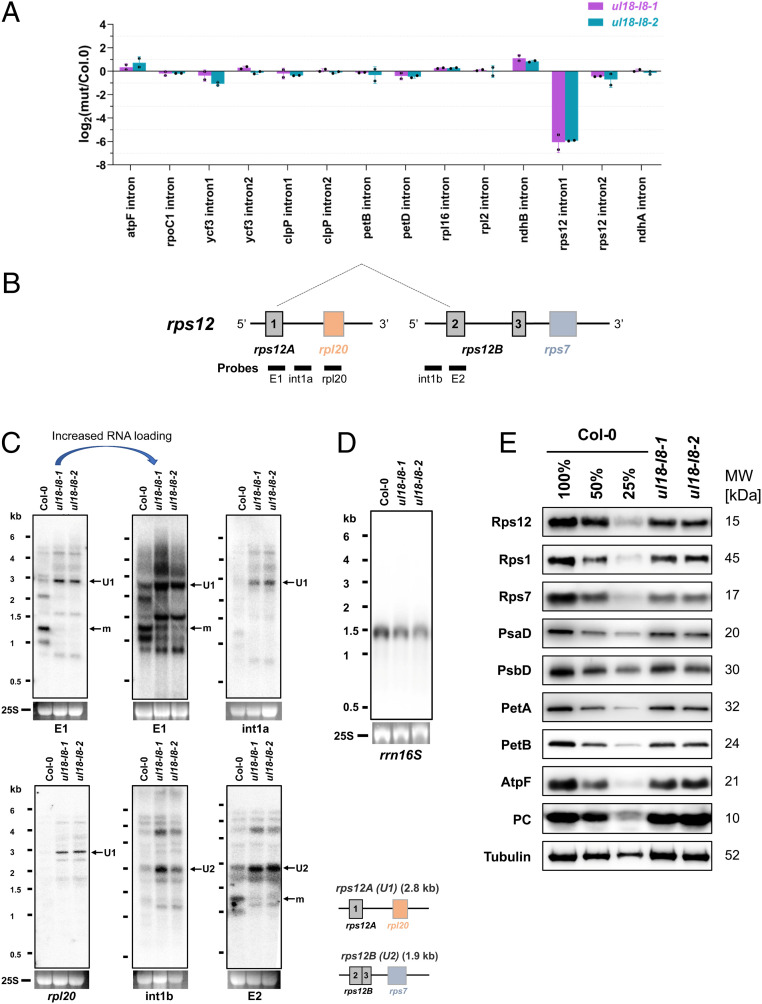
The uL18-L8 Protein Is Required for the Splicing of the First Intron of rps12 in Plastid.
To get insight into the function of uL18-L8, qRT-PCR analysis was performed to comparatively measure the steady-state level of plastid-encoded transcripts in ul18-l8 plants. We first observed that no plastid transcript suffered from global mRNA destabilization in either of the mutants (SI Appendix, Fig. S6). However, an ∼64-fold decrease in the splicing efficiency of the first intron of the rps12 transcript could be detected (Fig. 5A). This reduction is partially compensated by a general overaccumulation of plastid transcripts, resulting in mature rps12 mRNA being around 13-fold less abundant in ul18-l8 plants compared to the wild type (SI Appendix, Fig. S6A). The mature rps12 ORF is contained in a dicistronic transcript with rps7 and is produced via a trans-splicing reaction involving a first precursor comprising the first exon of rps12 plus rpl20 and a second precursor carrying the last two rps12 exons as well as the rps7 gene (Fig. 5B). RNA gel blot analysis showed that very low amounts of mature rps12 dicistronic mRNA are detected in ul18-l8 plants and that both rps12 precursor transcripts overaccumulate compared to the wild type (Fig. 5C). The splicing efficiency of introns contained in plastid tRNAs was then evaluated by RNA blot analysis and did not show any reduction (SI Appendix, Fig. S7A). It however revealed that most mature plastid tRNAs overaccumulated in both mutants (SI Appendix, Fig. S7A). Further RNA blots showed normal steady-state levels for the 23S, 4.5S, and 5S rRNAs in ul18-l8 plants (SI Appendix, Fig. S7B), whereas 16S rRNA levels show an ∼30% reduction in the mutants (Fig. 5D and SI Appendix, Fig. S7C). Immunoblots of leaf extracts revealed that reduced amounts of Rps12 could be detected in the ul18-l8 mutants (Fig. 5E), which is coherent with the reduction of rps12 intron splicing detected in these plants (Fig. 5A). Indeed, a reduction in the steady-state levels of all tested plastid-encoded proteins (Rps12, Rps1, Rps7, PsbD, PetA, PetB, and AtpF) was also observed in ul18-l8 plants. These proteins accumulated at about 50% of WT levels, indicating that a 13-fold decrease in mature rps12 mRNA production in ul18-l8 mutants results in a reduction of only half of Rps12 protein level and plastid translation. Conversely, the level of the nuclearly encoded plastocyanin was found unchanged in the mutants (Fig. 5E).
Fig. 5.
The ul18-l8 mutants have reduced levels of rps12 intron 1 splicing and underaccumulate plastid-encoded proteins. (A) qRT-PCR analysis measuring the splicing efficiencies of plastid introns in ul18-l8 mutants. The bars show log2 ratios of splicing efficiencies in ul18-l8 plants to the wild type (Col-0). Three technical replicates and two independent biological repeats were used for each genotype; SDs are indicated. (B) Schematic representation of the two rps12 precursor transcripts that are fused by trans-splicing to produce the mature (m) rps12 mRNA. Gray and colored boxes indicate exons. Introns and 5′ and 3′ UTRs are shown as thin lines. The probes used to interpret RNA gel blot results are shown. (C) RNA gel blot analyses showing the accumulation patterns of rps12 transcripts in ul18-l8 mutants in comparison with the wild type. Used probes are specified below each blot. A second blot with increased RNA loading (15 μg) was done for the rps12 E1 probe to better visualize the low amounts of mature rps12 transcript that accumulate in ul18-l8 mutants. Ethidium bromide staining of the 25S ribosomal RNA serves as a loading control. Schematic representations and sizes of unspliced rps12A (U1) and rps12B (U2) precursor transcripts are depicted below the blots. RNA marker sizes are indicated (kilobases). E, exon; int, intron; m, mature mRNA. (D) RNA gel blot showing the abundance of the 16S rRNA in the wild type (Col-0) and in ul18-l8 mutants. (E) Immunodetection of photosynthetic chain proteins on SDS/PAGE blots in ul18-l8 mutants compared to the wild type. Total proteins purified from the indicated genotypes were probed with antibodies to subunits of the chloroplast ATP synthase (AtpF), photosystem II (PsbD), photosystem I (PsaD), the cytochrome b6f complex (PetA and PetB), RPs (Rps12, RPS1, and Rps7) and the plastocyanin. Tubulin was used as protein loading control. Dilution series of proteins extracted from wild type (Col-0) were used for signal comparison. Protein molecular masses (MW) of detected proteins are indicated on the right.
Overall, these results demonstrated that the uL18-L8 protein is required for the trans-splicing reaction involving the first intron of rps12 in plastids and that ul18-l8 mutants consequently produce reduced levels of the Rps12 RP, resulting in a significant decrease in the production of several plastid-encoded proteins and 16S rRNA accumulation.
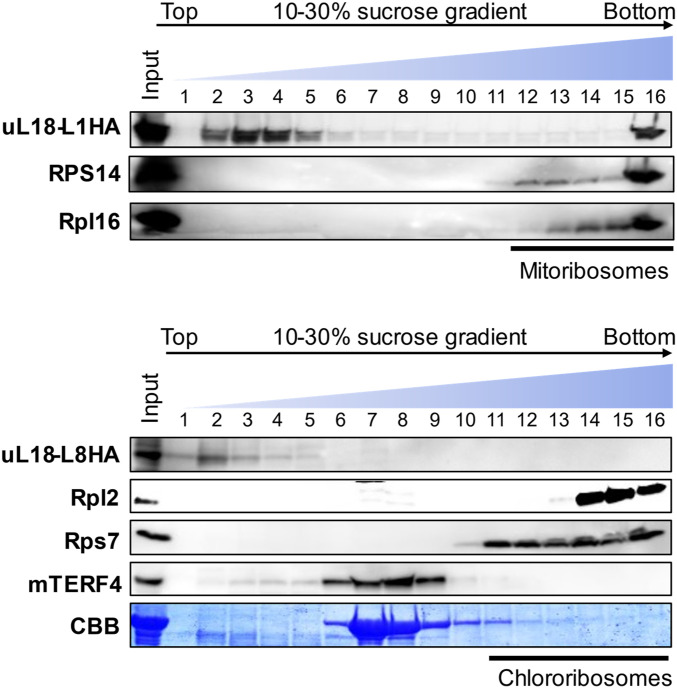
uL18-L1 and uL18-L8 Do Not Associate with Mitochondrial or Plastid Ribosomes.
To see whether the uL18-L1 and uL18-L8 proteins conserved a capacity to associate with mitochondrial or plastid ribosomes, the sizes of complexes containing these two proteins were measured by density gradient sedimentation. Most previously identified mitochondrial or plastid group II intron splicing factors reside in high molecular mass ribonucleoprotein complexes that are, however, lighter than ribosomes (12, 26). Mitochondrial or stromal extracts prepared from ul18-l1 and ul18-l8-1 complemented plants were fractionated on sucrose gradient and the recovered fractions were analyzed by immunoblot assays. Both uL18-Like proteins were found in particles that were much smaller than mitoribosomes or chlororibosomes (Fig. 6). Interestingly, uL18-L8 was found in lighter fractions compared with the plastid splicing factor mTERF4 (27). Identical results were obtained with overexpressing Arabidopsis cell lines in which the detection of uL18-L1 and uL18-L8 is greatly facilitated compared to organelles purified from complemented plants (SI Appendix, Fig. S8A). Both proteins could be immunoprecipitated from these transgenic lines but none of the tested anti-ribosome antibodies indicated coenrichment with mitoribosomes for uL18-L1 or chlororibosomes for uL18-L8 (SI Appendix, Fig. S8B).
Fig. 6.
uL18-L1 and uL18-L8 are not components of ribosomes. Mitochondrial (Upper) and stromal (Lower) extracts fractionated on sucrose gradients. An equal volume of each recovered fraction was analyzed by protein gel blot with the indicated antibodies. Mitochondrial Rpl16 and RPS14 as well as plastid Rpl2 and Rps7 were used to localize mitochondrial or plastid ribosomes along the gradients. mTERF4 is a protein required for the splicing of multiple plastid group II introns (27). The αHA antibody detects uL18-L1-3HA and uL18-L8-3HA fusion proteins. Coomassie brilliant blue (CBB) staining of the membrane detecting the position of the large Rubisco subunit (RbcL) in the gradient is shown below the hybridization results.
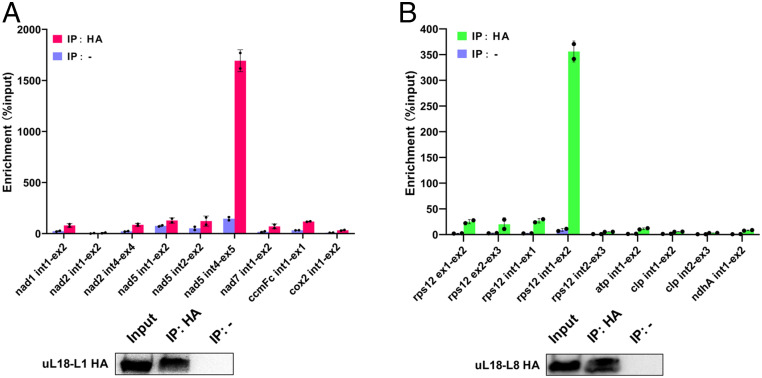
uL18-L1 and uL18-L8 Associate In Vivo with Introns Whose Splicing They Facilitate.
To identify intron RNAs that associate with uL18-L1 and uL18-L8 in vivo, both proteins were immunoprecipitated from overexpressing Arabidopsis cell lines with HA antibody and coenriched RNAs were purified from the coimmunoprecipitates. Obtained RNAs were then used for cDNA synthesis and analyzed by qPCR using primer pairs amplifying mitochondrial or plastid precursor mRNAs. Enrichment ratios were calculated in comparison to control immunoprecipitations performed with IgG-Protein A magnetic beads. We observed a very high and specific enrichment of uL18-L1 with nad5 intron 4 and uL18-L8 with rps12 intron 1 (Fig. 7). Interestingly, nad2 intron 1 was not coenriched in uL18-L1 immunoprecipitates, suggesting that nad2 intron 1 splicing defect detected in ul18-l1 plants could be a secondary effect of complex I deficiency as observed in several independent Arabidopsis complex I mutants (28–32). Both uL18-type proteins thus associate with their genetic target in vivo, supporting a direct role in the splicing of the concerned introns.
Fig. 7.
uL18-L1 and uL18-L8 specifically associate with nad5 intron 4 and rps12 intron 1, respectively. (A) Analysis of RNAs that coimmunoprecipitate with uL18-L1-3HA. Total extracts from transgenic Arabidopsis cell lines expressing uL18-L1-3HA were used for immunoprecipitation with anti-HA antibody (IP+) and IgG-Protein A magnetic beads (IP−) as negative control. Coimmunoprecipitated RNAs were analyzed by qRT-PCR using primer pairs positioned across intron/exon junctions of the indicated mitochondrial transcripts. Enrichment ratios calculated with the delta Ct method are shown. (B) qRT-PCR analysis of RNAs coimmunoprecipitating with uL18-L8-3HA. Similar experiment as in A except that cells expressing uL18-L8-3HA were used and that primers pairs amplifying plastid transcript precursors were employed to analyze coimmunoprecipitated RNAs. Immunoblot results of input and immunoprecipitated fractions using the HA antibody (IP+) or IgG-Protein A magnetic beads (IP−) are shown.
Discussion
The uL18 RNA Binding Domain Was Recruited Early in the Evolution of Terrestrial Plants to Produce Intron-Specific Splicing Factors for Organelles.
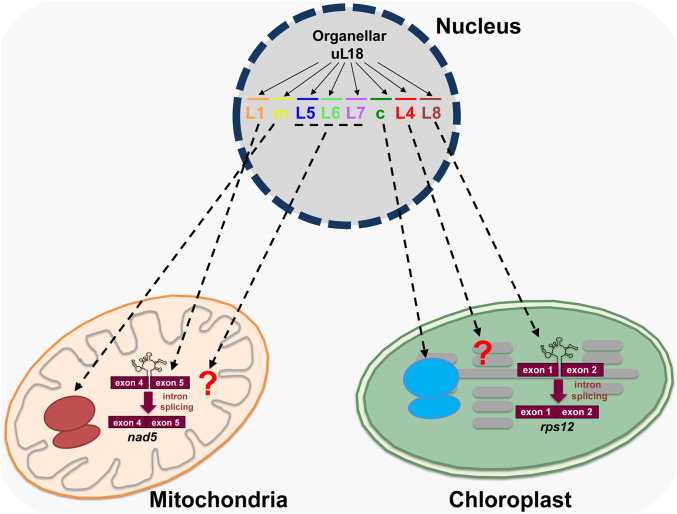
RPs are essential components of ribosomes that help structuring rRNAs to allow them to function optimally in protein synthesis. In recent years, several lines of evidence have documented that RPs can carry ribosome-independent functions (15, 33, 34). In this report, we reveal that two Arabidopsis proteins of the uL18 family play essential roles in group II intron splicing in chloroplasts or mitochondria. The uL18-L1 protein was found to be required for the splicing of last intron contained in nad5 pre-mRNAs in mitochondria, whereas the plastid uL18-L8 is required for the removal of rps12 intron 1 (Fig. 8). In both mutants, the observed splicing reductions are associated with an overaccumulation of various splicing intermediates whose analysis will be instructive to better understand the mechanism and orchestration of splicing of group II introns in organelles. Moreover, these two factors are not similarly required for the splicing reactions they promote. The loss of uL18-L1 results in a very strong reduction in nad5 intron 4 splicing while the decrease in rps12 intron 1 splicing in ul18-l8 plants is much less pronounced and leads to moderate pale-green photosynthetic mutants. Other nuclear-encoded transfactors such as PPR4 or EMB2654 have proven to be more critical for rps12 intron 1 splicing, and their loss leads to embryo lethality in Arabidopsis and seedling-lethal plants in maize, highlighting the essentiality of Rps12 and plastid translation for plant survival (35–38). Arabidopsis knockdown mutants for these two genes result in severely discolored plants that are not always associated with a greater decrease in rps12 intron 1 splicing compared to ul18-l8 mutants (36–38). However, side-by-side comparisons to notably measure residual Rps12 protein in these different mutants appears indispensable to explain their phenotypic differences. The uL18-L1 protein is found in ribonucleoprotein complexes (RNPs) whose sizes are close to those corresponding to splicing factors such as the nMAT2 protein (Fig. 6 and SI Appendix, Fig. S8). Conversely, the uL18-L8 protein was detected in smaller RNP complexes compared to the general plastid splicing factor mTERF4. Further analyses determining the composition of these different plastid splicing complexes need to be performed to determine the origin of this size difference. The functions for these two uL18-Like factors cannot be considered as extraribosomal functions sensu stricto, since both uL18-L1 and uL18-L8 proteins were not found to reside in mitochondrial or plastid ribosomes, respectively (Fig. 6). Our results nevertheless support the capacity of RP binding domains to accommodate new RNA targets, and reveal that this class of proteins can acquire functions in intron splicing. These observations add support to an important evolutionary paradigm in which existing genes are duplicated and become diversified to support new functions. In the present cases, duplicated and subsequently divergent copies of uL18m or uL18c were likely selected to play roles in organellar intron splicing at some point in the evolution of plants. The search for uL18 homologs from bacteria to seed plants indicates that the diversification of the uL18 family occurred early in land plant evolution. In bacteria or algae, a single uL18 gene can be generally recognized, which encodes a protein sharing strong homologies with the RP uL18c (SI Appendix, Fig. S1). Nonvascular plants encode a few more uL18-Like proteins whose closest homologs correspond to the uL18m, uL18c, and uL18-L4 proteins. Gymnosperms and angiosperms encode similar corteges of uL18 and uL18-Like proteins as in Arabidopsis, with the intriguing exception of uL18-L6 that is specific to dicotyledonous plants (SI Appendix, Fig. S1). It thus appears that the expansion of the uL18 family was initiated in nonvascular basal plants and that the first diverging uL18 member may have been the uL18-L4 gene. These observations nicely corroborate previous analyses indicating that divergence of organellar introns from the classic structure of group II introns has occurred early in the evolution of terrestrial plants (39–41).
Fig. 8.
Graphical summary depicting the functional diversification of the uL18 RP family in Angiosperms. The uL18 protein family comprises eight members that evolved from ancestral organellar uL18 genes. Five members are transported into mitochondria and three into plastids. Two uL18 proteins (uL18m and very likely uL18c) have retained the original uL18 function and are subunits of organellar ribosomes. Among the other six uL18-Like members, uL18-L1 and uL18-L8 have evolved toward splicing and specifically associate in vivo with introns whose splicing they promote.
Organellar introns derive from bacterial counterparts and can be classified as group I or group II based on specific structural characteristics. Introns contained in vascular plants organellar genomes are mostly of type II and fold into a conserved structure comprising six helical domains (DI–DVI) that radiate from a central hub (42). Autonomous group II introns encode a maturase that is essential for intron mobility and splicing. Most organellar introns in seed plants show, however, degenerate structures compared to canonical group II introns or lack structural features that are important for removal (40, 41). Unlike in bacteria, these deviant group II introns are maintained in plant organellar genomes as splicing-only elements. They may be mobilized via unusual splicing mechanisms, but a tolerance to structural changes permitted by the selection of a compensatory accessory machinery is an evolutionary strategy that is chosen to maintain efficient splicing of derivatized introns in organelles (41, 43). Various kinds of RNA-binding proteins have been recruited to play roles in organellar intron splicing like aminoacyl tRNA synthetases, a peptidyl-tRNA hydrolase, pseudouridine synthase homologs, or proteins derived from a bacterial ribosome assembly factors (11, 44–47). Minor evolutionary tinkering thus seems sufficient to confer novel activities in intron splicing to these RNA-binding proteins, as for the uL18 homologs. By associating with their target intron, these factors could form ribonucleoprotein complexes that help intron folding and stabilize their structure in a catalytically active form (48). Advanced biochemical and structural analyses are however necessary to understand how uL18-Like proteins along with other proteinaceous factors facilitate group II intron splicing in plant organelles. Effectively, the coaction of several splicing factors is generally required for intron splicing in plant organelles. In the case of nad5 intron 4, three nuclear-encoded PPR proteins [MISF68 (24), ZmSMK9 (49), and ZmSMR1 (50)] in addition to uL18-L1 are required for its removal from pre-mRNAs. Similarly, in addition to uL18-L8, splicing factors from various protein families, namely the PPR protein PPR4 and EMB2654 (35–38), the CRM protein CAF2 (51), the PTH protein CRS2 (47), and the DEAD box helicases protein RH3 (52) also participate in rps12 intron 1 splicing. It remains to be clarified how these factors collaborate or whether they act independently from each other in intron splicing.
The uL18-L1 and uL18-L8 Proteins Could Associate with 5S rRNA-like Structures in Their Target Intron.
In most cases, the evolutionary recruitment of RPs toward new sites is permitted because the structure of newly targeted mRNA regions resembles that of rRNA domains to which they associate in the ribosomes (53–60). The uL18 protein is the most important factor for incorporating the 5S rRNA into the 50S ribosomal subunit. The 5S rRNA is a small RNA of around 120 nucleotides and adopts a conserved tertiary structure comprising various helices as well as terminal and internal loops (61). The most important structural determinants for uL18 anchoring are contained in its β domain comprising two helices plus one internal and a terminal loop (20). Upon binding, the uL18 protein increases the stability of 5S rRNA, modifies its structure, and enhances its affinity of uL5 (62, 63). Our structural predictions showed that C-terminal regions of uL18-L1 and uL18-L8 adopt the same structure as ribosomal uL18s and that the three β-sheets forming the RNA binding surface of uL18 are also found in the predicted structures of these two uL18-Like proteins (Fig. 1). This strongly suggests that the recruitment of uL18-Like proteins in splicing was permitted because regions within nad5 intron 4 and rps12 intron 1 likely resemble the β domain of 5S rRNA, at least enough so that uL18-L1 and uL18-L8 proteins could associate with them in a productive way. Interestingly, the recent structures of mammalian mitoribosomes showed that the uL18-binding domain can accommodate different but structurally similar RNA targets since tRNAs serve as architectural replacements for the 5S rRNA in these ribosomes (64, 65). Therefore, the search for regions within nad5 intron 4 and rps12 intron 1 that form secondary structures similar to the β domain of 5S rRNA may reveal the binding sites of uL18-L1 and uL18-L8. As uL18 does with 5S rRNA, the binding of uL18-L1 and uL18-L8 could induce local structural changes to the RNA regions to which they bind and, thereby, help their target intron to fold in an active form. These structural modifications may also permit the recruitment of other protein factor(s) necessary for splicing, similarly to the impact that the bacterial uL18 has on increasing the uL5-binding affinity for 5S rRNA. Finding the exact binding sites of uL18-L1 and uL18-L8 within their target introns represents an important challenge to better understand the role of uL18-L1 and uL18-L8 in group II intron splicing. Deciphering the functions of other uL18-Like proteins will reveal whether they also play roles in organellar intron splicing or whether they are involved in other RNA expression steps in plant organelles. The case of the uL18 family is an as-yet undescribed example in which a set of six independent members still cohabit with the original factors (uL18c and uL18m) from which they are derived. This provides a unique opportunity to understand the functional transitions that led the uL18 ribosomal-Like proteins to acquire roles in organellar intron splicing and maybe other RNA expression processes. Together, these results will better document how RPs can evolve toward new functions in RNA metabolism and how they can adapt to associate and process new RNA targets.
Materials and Methods
Plant materials, preparation of organelles, immunodetection of proteins, subcellular distributions, RNA analysis, RNA immunoprecipitation assays, blue native gel, sucrose density gradient centrifugation, transformation of Arabidopsis cell suspension cultures, and analysis of photosynthesis and pigment composition are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Agence Nationale de la Recherche (ANR) MITRA Grant ANR-16-CE11-0024-01 (to H.M.) and the China Scholarship Council (to C.W.). The Institut Jean-Pierre Bourgin’s (IJPB's) benefits from the support of Saclay Plant Sciences Grant ANR-17-EUR-0007. This work has benefited from the support of IJPB’s Plant Observatory technological platforms. We thank Professor Gregory Brown (McGill University) for critical reading of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004075117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Martijn J., Vosseberg J., Guy L., Offre P., Ettema T. J. G., Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gould S. B., Waller R. F., McFadden G. I., Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Barkan A., Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155, 1520–1532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammani K., Giegé P., RNA metabolism in plant mitochondria. Trends Plant Sci. 19, 380–389 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Small I. D., Peeters N., The PPR motif–A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Lurin C., et al. , Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammani K., et al. , Helical repeats modular proteins are major players for organelle gene expression. Biochimie 100, 141–150 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Yin P., et al. , Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Shen C., et al. , Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7, 11285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruwe H., Kupsch C., Teubner M., Schmitz-Linneweber C., The RNA-recognition motif in chloroplasts. J. Plant Physiol. 168, 1361–1371 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Barkan A., et al. , The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA 13, 55–64 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroeger T. S., Watkins K. P., Friso G., van Wijk K. J., Barkan A., A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. U.S.A. 106, 4537–4542 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francs-Small C. C., et al. , A PORR domain protein required for rpl2 and ccmF(C) intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 69, 996–1005 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Schmitz-Linneweber C., Lampe M. K., Sultan L. D., Ostersetzer-Biran O., Organellar maturases: A window into the evolution of the spliceosome. Biochim. Biophys. Acta 1847, 798–808 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Warner J. R., McIntosh K. B., How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aseev L. V., Boni I. V., [Extraribosomal functions of bacterial ribosomal proteins]. Mol. Biol. (Mosk.) 45, 805–816 (2011). [PubMed] [Google Scholar]
- 17.Lu H., Zhu Y. F., Xiong J., Wang R., Jia Z., Potential extra-ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae. Microbiol. Res. 177, 28–33 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Savada R. P., Bonham-Smith P. C., Differential transcript accumulation and subcellular localization of Arabidopsis ribosomal proteins. Plant Sci. 223, 134–145 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Bonen L., Calixte S., Comparative analysis of bacterial-origin genes for plant mitochondrial ribosomal proteins. Mol. Biol. Evol. 23, 701–712 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Scripture J. B., Huber P. W., Analysis of the binding of Xenopus ribosomal protein L5 to oocyte 5 S rRNA. The major determinants of recognition are located in helix III-loop C. J. Biol. Chem. 270, 27358–27365 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N., The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 36, 475–486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ban N., et al. , A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24, 165–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waltz F., et al. , Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 5, 106–117 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Wang C., Aubé F., Quadrado M., Dargel-Graffin C., Mireau H., Three new pentatricopeptide repeat proteins facilitate the splicing of mitochondrial transcripts and complex I biogenesis in Arabidopsis. J. Exp. Bot. 69, 5131–5140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., et al. , The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3´ end of its 5´-half intron. Nucleic Acids Res. 45, 6119–6134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keren I., et al. , AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammani K., Barkan A., An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 42, 5033–5042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Longevialle A. F., et al. , The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19, 3256–3265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koprivova A., et al. , Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 285, 32192–32199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haïli N., et al. , The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiol. 170, 354–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haïli N., et al. , The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 41, 6650–6663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colas des Francs-Small C., et al. , The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 165, 1409–1416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhavsar R. B., Makley L. N., Tsonis P. A., The other lives of ribosomal proteins. Hum. Genomics 4, 327–344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Liao W.-J., Liao J.-M., Liao P., Lu H., Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz-Linneweber C., et al. , A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18, 2650–2663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aryamanesh N., et al. , The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 173, 1164–1176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadini L., et al. , Trans-splicing of plastid rps12 transcripts, mediated by AtPPR4, is essential for embryo patterning in Arabidopsis thaliana. Planta 248, 257–265 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Lee K., et al. , The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 100, 1193–1207 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Malek O., Knoop V., Trans-splicing group II introns in plant mitochondria: The complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA 4, 1599–1609 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoop V., The mitochondrial DNA of land plants: Peculiarities in phylogenetic perspective. Curr. Genet. 46, 123–139 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Bonen L., “Evolution of mitochondrial introns in plants and photosynthetic microbes” in Advances in Botanical Research, L. Maréchal-Drouard, Ed. (Elsevier, Amsterdam, 2012), pp. 155–186. [Google Scholar]
- 42.Bonen L., Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8, 26–34 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Maier U. G., et al. , Complex chloroplast RNA metabolism: Just debugging the genetic programme? BMC Biol. 6, 36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akins R. A., Lambowitz A. M., A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell 50, 331–345 (1987). [DOI] [PubMed] [Google Scholar]
- 45.Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P., The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 7, 473–483 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perron K., Goldschmidt-Clermont M., Rochaix J. D., A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18, 6481–6490 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenkins B. D., Barkan A., Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20, 872–879 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fedorova O., Solem A., Pyle A. M., Protein-facilitated folding of group II intron ribozymes. J. Mol. Biol. 397, 799–813 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan Z., et al. , ZmSMK9, a pentatricopeptide repeat protein, is involved in the cis-splicing of nad5, kernel development and plant architecture in maize. Plant Sci. 288, 110205 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Chen Z., et al. , PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize. J. Exp. Bot. 70, 5245–5258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostheimer G. J., et al. , Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 22, 3919–3929 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asakura Y., Galarneau E., Watkins K. P., Barkan A., van Wijk K. J., Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 159, 961–974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillier M., et al. , Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: A possible case of ribosomal RNA-messenger RNA molecular mimicry. RNA 8, 878–889 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serganov A., Polonskaia A., Ehresmann B., Ehresmann C., Patel D. J., Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 22, 1898–1908 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stelzl U., et al. , RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon. J. Biol. Chem. 278, 28237–28245 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merianos H. J., Wang J., Moore P. B., The structure of a ribosomal protein S8/spc operon mRNA complex. RNA 10, 954–964 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scott L. G., Williamson J. R., The binding interface between Bacillus stearothermophilus ribosomal protein S15 and its 5′-translational operator mRNA. J. Mol. Biol. 351, 280–290 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Nevskaya N., et al. , Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA. Nucleic Acids Res. 33, 478–485 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choonee N., Even S., Zig L., Putzer H., Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 35, 1578–1588 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iben J. R., Draper D. E., Specific interactions of the L10(L12)4 ribosomal protein complex with mRNA, rRNA, and L11. Biochemistry 47, 2721–2731 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Delihas N., Andersen J., Generalized structures of the 5S ribosomal RNAs. Nucleic Acids Res. 10, 7323–7344 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bear D. G., Schleich T., Noller H. F., Garrett R. A., Alteration of 5S RNA conformation by ribosomal proteins L18 and L25. Nucleic Acids Res. 4, 2511–2526 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A., The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Brown A., et al. , Structure of the large ribosomal subunit from human mitochondria. Science 346, 718–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greber B. J., et al. , The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515, 283–286 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.