Significance
The multidrug transporter P-glycoprotein protects tissues from xenobiotics and other toxic compounds by pumping them out of cells. This transporter has been implicated in altering the bioavailability of chemotherapeutic drugs and in the development of multidrug resistance in tumor cells. Despite decades of research, the modulation of P-glycoprotein to overcome drug resistance in the clinic has not been successful. Here, by substituting a group of 14 conserved residues in homologous transmembrane helices 6 and 12 with alanine, we generated a mutant that exhibits a change in the direction of transport from export to import for certain drug substrates including the taxol derivative flutax-1. The ability to convert P-glycoprotein into a drug importer provides a strategy to combat cancer drug resistance.
Keywords: ABC transporter, drug transport, multidrug resistance, P-glycoprotein, mechanism
Abstract
P-glycoprotein (P-gp), also known as ABCB1, is a cell membrane transporter that mediates the efflux of chemically dissimilar amphipathic drugs and confers resistance to chemotherapy in most cancers. Homologous transmembrane helices (TMHs) 6 and 12 of human P-gp connect the transmembrane domains with its nucleotide-binding domains, and several residues in these TMHs contribute to the drug-binding pocket. To investigate the role of these helices in the transport function of P-gp, we substituted a group of 14 conserved residues (seven in both TMHs 6 and 12) with alanine and generated a mutant termed 14A. Although the 14A mutant lost the ability to pump most of the substrates tested out of cancer cells, surprisingly, it acquired a new function. It was able to import four substrates, including rhodamine 123 (Rh123) and the taxol derivative flutax-1. Similar to the efflux function of wild-type P-gp, we found that uptake by the 14A mutant is ATP hydrolysis-, substrate concentration-, and time-dependent. Consistent with the uptake function, the mutant P-gp also hypersensitizes HeLa cells to Rh123 by 2- to 2.5-fold. Further mutagenesis identified residues from both TMHs 6 and 12 that synergistically form a switch in the central region of the two helices that governs whether a given substrate is pumped out of or into the cell. Transforming P-gp or an ABC drug exporter from an efflux transporter into a drug uptake pump would constitute a paradigm shift in efforts to overcome cancer drug resistance.
The emergence of drug resistance in cancer is a major clinical obstacle. The multidrug-resistance–linked plasma membrane transporter ABCB1 or P-glycoprotein (P-gp) effluxes various toxic amphipathic and hydrophobic compounds from cells, including anticancer drugs (1–4). Structurally, P-gp is comprised of two transmembrane domains (TMDs), each having six transmembrane helices (TMHs) and one cytosolic nucleotide-binding domain (NBD). Recent cryo-electron microscopy (cryo-EM) studies have shown that human P-gp exists in both inward open and inward closed conformations (5, 6). Substrates bind to P-gp in the inward open conformation and are then effluxed. The process is associated with conformational changes induced by ATP binding and hydrolysis by the NBDs (7).
Earlier studies revealed that the transport of substrates mediated by P-gp involves a number of specific residues in the transmembrane (TM) region. Some of these residues directly interact with the substrates, while others help to achieve the proper conformation of the protein for the translocation process. Previously, we generated P-gp mutants with single, double, or triple mutations that alter the binding, but not the transport of various substrates (8). We also generated a P-gp variant with 15 residues in the drug-binding pocket mutated to tyrosine that could transport most of the substrates despite increased hydrogen-bonding potential (9). These studies revealed the high degree of permissiveness of the drug-binding pocket of P-gp. To further investigate the mechanism of this transporter we decided to introduce multiple mutations in a pair of homologous helices known to be involved in the binding and translocation of substrates. Using this strategy, we generated a mutant named TMH 1,7 that has six mutations each in TMH 1 and TMH 7. This mutant has diminished polyspecificity, as it is able to transport only 3 of the 25 substrates tested (10).
In this study, we focus on another pair of homologous helices, TMH 6 and TMH 12, which form part of the drug-binding cavity through which substrates are translocated. To test the hypothesis that mutations in multiple residues in these homologous TMHs would significantly modify the drug-binding pocket resulting in altered or loss of binding and transport of substrates, the 14A mutant of human P-gp was generated in which seven conserved residues from TMH 6 and seven conserved residues from TMH 12 were mutated to alanine. Our results showed that the 14A mutant could not efflux most of the substrates tested. While P-gp is known to be able to pump many diverse toxic substances out of cells, we were surprised to find that the 14A mutant actually gained the ability to import certain substrates including rhodamine123 and flutax-1. So in effect, the mutations had caused a change in the direction of transport for those substrates. We found that this uptake function of the 14A mutant is ATP-binding– and hydrolysis-dependent, and similar to other ABC importers, the mutant transporter exhibits a narrow substrate specificity. Further mutagenesis of additional residues identified a possible switch in the central region of these two helices that determines the direction of the substrate transport. Our findings suggest that the ability to change the direction of transport of an ABC drug transporter from efflux to influx might provide a strategy to kill cancer cells specifically expressing such transporters.
Results and Discussion
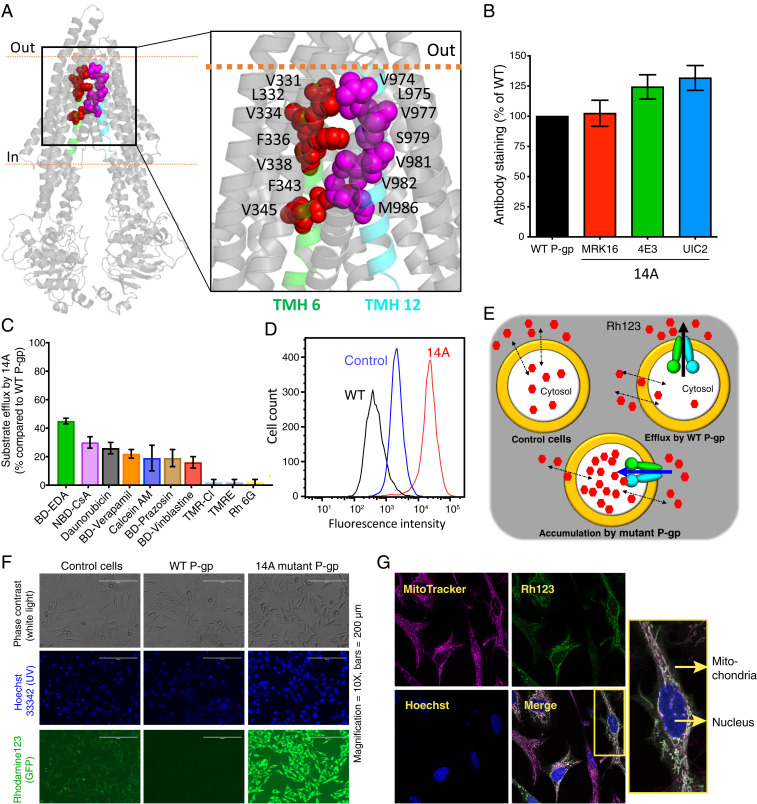
Previous mutagenesis, structural, and in silico studies revealed that at least 20 residues in TMH 6 and TMH 12 are involved in substrate binding and transport (11–15). However, their precise roles had not been fully explored. Consequently, to understand the role of these homologous helices in the polyspecificity of the transporter and in drug translocation across the membrane, we generated 14A mutant P-gp in which seven residues from TMH 6 (V331, L332, V334, F336, V338, F343, V345) and seven from TMH 12 (V974, L975, V977, S979, V981, V982, M986) were mutated to alanine. The locations of these residues are shown in Fig. 1A. Most of the mutated residues are phylogenetically conserved, hydrophobic, and/or aromatic; Gly and Pro residues were unaltered to conserve the structural integrity of the protein (SI Appendix, Fig. S1A).
Fig. 1.
Transport direction is reversed by mutations in TMHs 6 and 12 of P-gp. (A) Inward open human P-gp structure (PDB ID: 6QEX) highlighting the location of residues mutated in 14A. The residues selected for mutation in TMH 6 and TMH 12 are shown as red and magenta spheres, respectively. (B) Relative surface expression of 14A detected by human P-gp–specific antibodies MRK-16, 4E3, and UIC2, with binding to WT P-gp taken as 100%. (C) Relative efflux of various substrates by 14A, with efflux by WT P-gp taken as 100%. Although 25 substrates (list given in ref. 10) were tested, results for only the top 10 are shown (mean ± SD, n ≥ 3). (D) A typical histogram showing uptake of Rh123 by 14A. Untransduced cells were used as a control for equilibration (referred to as control cells here and in subsequent figures), and Rh123 efflux from WT P-gp–expressing cells is shown for comparison. (E) Cartoon representation of substrate uptake by 14A as compared to efflux by WT protein. (F) Accumulation of Rh123 was determined by fluorescence microscopy. In the top row are phase contrast images, in the middle row are nuclei stained with Hoechst dye, and the bottom row of images show Rh123 fluorescence signal. (G) Confocal microscopy of HeLa cells expressing 14A after uptake of Rh123 (green). MitoTracker (pink), and Hoechst (blue) are markers for mitochondria and nuclei, respectively. The merged image shows colocalized sections (white), and a single cell is enlarged in the inset. GFP, green fluorescent protein; TMR-CI, tetramethylrosamine chloride; TMRE, tetramethylrhodamine, ethyl ester.
To characterize the 14A mutant, we used the BacMam baculovirus-based expression system in HeLa cells. Flow cytometry with three human P-gp–specific antibodies [MRK16, UIC2, and 4E3 (5, 16–18)] and western blotting with C219 showed that the expression of the mutant was comparable to that of wild-type (WT) P-gp (Fig. 1B and SI Appendix, Fig. S1B). Thus, mutations of these 14 residues did not affect the mutant P-gp expression, trafficking to the cell surface, or overall conformation. Next, we checked the transport of several known fluorescently labeled substrates by using a flow cytometry-based steady-state transport assay (10). The 14A mutant P-gp–mediated efflux of most substrates was significantly decreased or abolished (Fig. 1C). Of the 25 tested substrates, only partial efflux of BODIPY FL ethylenediamine (BD-EDA) (40% compared to WT P-gp) by 14A was observed, confirming the importance of residues in TMH 6 and 12 for binding and efflux of substrates. [In previous studies, efflux below 30% of WT was considered to be insignificant transport activity (9, 10).]
Surprisingly, we found that rhodamine 123 (Rh123), instead of being effluxed, as occurs with WT P-gp, accumulated in HeLa cells expressing 14A at much higher levels than in the untransduced control cells (Fig. 1 D and E), suggesting the uptake of this substrate mediated by the mutant transporter. Fluorescence microscopy using live cells expressing 14A displays Rh123 accumulation in the cytosol and not on the cell surface (Fig. 1F). Confocal microscopy-based colocalization analysis with markers of mitochondria, the endoplasmic reticulum (ER), and nuclei (MitoTracker, ER-tracker, and Hoechst 33342, respectively) showed that despite increased concentrations, Rh123 localized in mitochondria, as occurs in control cells [Fig. 1G and SI Appendix, Fig. S2 (19)].
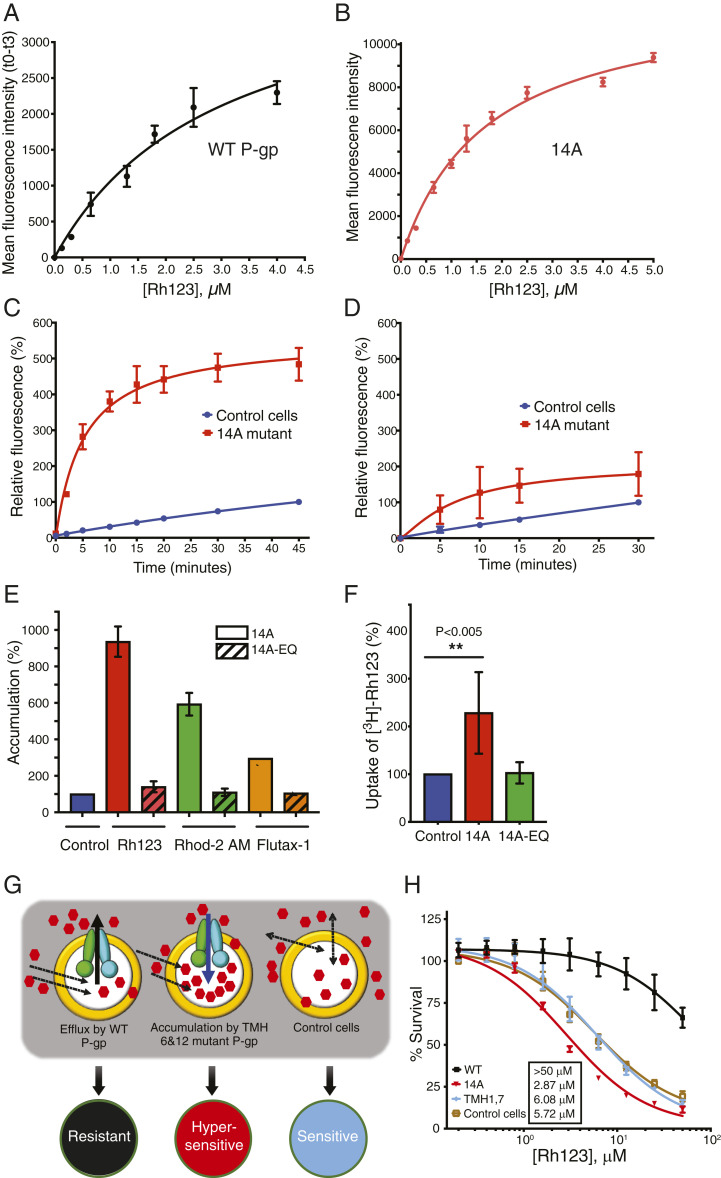
Next, we studied the kinetics of Rh123 accumulation in cells expressing the 14A mutant. The rates of Rh123 efflux by WT P-gp and uptake by 14A were compared. The Km for Rh123 uptake by 14A was 1.69 ± 0.12 µM, whereas the Km for efflux by WT P-gp was 2.98 ± 0.51 µM (Fig. 2 A and B). HeLa cells expressing 14A achieved maximum Rh123 accumulation in 15 min (t1/2 = 4.45 ± 0.34 min) (Fig. 2C). Uptake by the 14A mutant was also temperature-dependent, with optimal activity at 37 °C, which is the same for efflux by the WT protein (9). These results demonstrate that Rh123 uptake by 14A is a time- and concentration-dependent saturable process with kinetic properties very similar to those for its efflux by WT P-gp. The kinetics of Rh123 uptake in untransduced control cells (blue curves in Fig. 2 C and D) indicate slower, nonsaturable uptake, consistent with equilibration by passive diffusion.
Fig. 2.
Characterization of Rh123 uptake by the 14A mutant. (A) Efflux of Rh123 from ATP-depleted WT P-gp–expressing cells after 20 min equilibration at different concentrations of Rh123. Efflux was calculated as the difference in mean fluorescence intensity (MFI) of cells at 0 and 3 min. (B) Uptake was determined by measuring the MFI of 14A-expressing cells when incubated with various Rh123 concentrations for 3 min. (C) Cells (control or 14A-expressing) were incubated with Rh123 (1.3 µM) for the indicated times, and MFI was measured. MFI of control cells (after 45 min) was taken as 100%, and relative fluorescence was calculated. (D) Same as in C, except with ATP-depleted cells. (E) Accumulation of Rh123, Rhod-2 AM, and Flutax-1 by 14A (solid bars) and ATP hydrolysis-deficient 14A-EQ (E556Q/E1201Q, diagonal bars). (F) ATP-depleted cells (untransduced control and expressing 14A or 14A-EQ) were equilibrated with Rh123 (1.3 µM). After washing, cells were exposed to [3H]-Rh123 (0.25 µM) for 20 min, and intracellular radioactivity was measured. The significance was calculated using one-way ANOVA (**P < 0.005). (G) Schematic of cell hypersensitization to anticancer drug substrates. Efflux activity of WT P-gp confers resistance to drugs compared with cells that do not express P-gp (sensitive). However, uptake mediated by the 14A mutant increases the intracellular concentration of drugs, resulting in hypersensitization. (H) The 14A mutant P-gp hypersensitizes HeLa cells to Rh123. Cytotoxicity assay with HeLa cells (untransduced or expressing either WT, TMH 1,7, or 14A). After 48 h incubation with 0–50 µM Rh123, survival was estimated by calculating IC50 (mean ± SD of at least three replicates) values given in the box.
Because the efflux of substrates by WT P-gp depends on ATP binding and subsequent hydrolysis, we tested whether the uptake activity of the 14A mutant also requires ATP as the energy source. To address this question, we used two approaches. First, we depleted ATP from the cells expressing 14A by adding 2-deoxyglucose and sodium azide, followed by an uptake assay under ATP-depleted conditions. As shown in Fig. 2D, uptake was drastically decreased even after 30 min (compare to Fig. 2C). In the second approach, we generated the ATP hydrolysis-deficient 14A-E556Q/E1201Q (14A-EQ) mutant (6, 20) with a total of 16 mutations. Rh123 did not accumulate in cells expressing the 14A-EQ mutant (Fig. 2E), confirming the results obtained with ATP depletion assays (Fig. 2D).
In addition to Rh123, a few other substrates were also imported by 14A, most of which were rhodamine derivatives (10). They included dihydrorhodamine 123 (DHR123, with fourfold accumulation), which is oxidized to Rh123 in the cells (21), Rhod-2 acetoxymethyl (AM) (five- to sixfold accumulation), and X-Rhod-1 AM (twofold accumulation). An anticancer drug analog Flutax-1 (a paclitaxel [trade name Taxol] derivative) was also imported with accumulation ranging from two- to threefold more than in control cells (Fig. 2E). We also confirmed the lack of uptake of Rhod-2 AM and Flutax-1 by the 14A-EQ mutant (Fig. 2E). Thus, the import function of 14A is ATP hydrolysis-dependent. No other tested rhodamine analog was found to accumulate in the cells (SI Appendix, Fig. S3A), indicating that no specific physicochemical properties of substrates determine the 14A mutant-mediated uptake, as they vary in molecular weight, charge, structure, and polarity (SI Appendix, Fig. S3B).
ATP-dependent transporters carry out substrate transport against a concentration gradient (22). To determine if Rh123 uptake occurred against the gradient, we performed an uptake assay using [3H]-Rh123. After equilibrating ATP-depleted 14A-expressing cells with nonradioactive Rh123 (1.3 µM) (SI Appendix, Fig. S4), [3H]-Rh123 was added at a concentration five times lower (0.25 µM) in glucose-containing medium to replenish ATP, and [3H]-Rh123 accumulation in cells was quantified 20 min later. We found that the [3H]-Rh123 level was 2.3 ± 0.9-fold higher in 14A-expressing cells as compared to control cells or cells expressing 14A-EQ (Fig. 2F). Thus, [3H]-Rh123 accumulated inside 14A-expressing cells against a steep concentration gradient.
P-gp is known to confer resistance to several anticancer drug substrates. We tested whether increased uptake by the 14A mutant hypersensitized HeLa cells to Rh123 (see schematic diagram of the assay in Fig. 2G). Cells overexpressing 14A were exposed to increasing concentrations (0.25–50 µM) of Rh123 in a cytotoxicity assay. Cells expressing WT P-gp, which efflux Rh123, and cells not expressing exogenous P-gp (untransduced) or expressing TMH 1,7 P-gp, which do not transport Rh123 (10), were used as controls. Fig. 2H shows that 14A-expressing cells are hypersensitized to Rh123, with half-maximal inhibitory concentration (IC50) values decreasing from 6.08 ± 0.77 µM (TMH 1,7 control cells) to 2.87 ± 0.3 µM. As expected, cells expressing WT P-gp were resistant even at 50 µM. This result confirms that an engineered ABC efflux transporter can function as an importer, thereby increasing the accumulation of chemotherapy drugs in cancer cells, leading to cell death. Cytotoxicity tests with other drugs showed that cells expressing the 14A mutant are marginally sensitized to paclitaxel and doxorubicin, but not to colchicine or etoposide (SI Appendix, Table S1), confirming the limited specificity of the import function of the mutant.
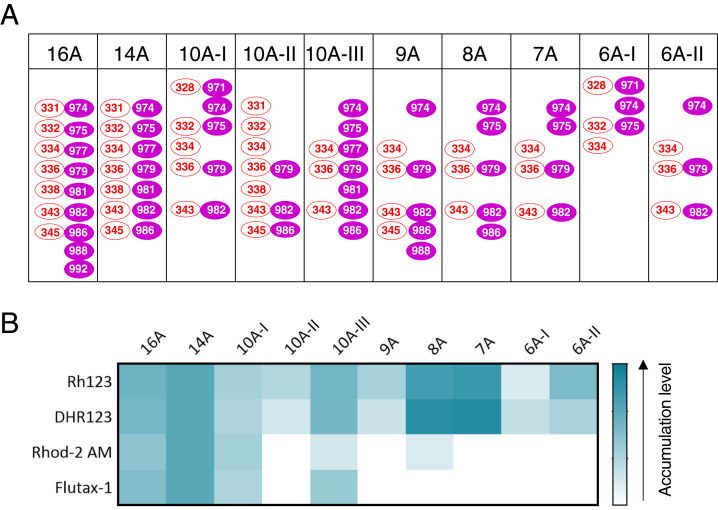
Because the binding and efflux of substrates by P-gp involves several residues in TMH 6 and 12, we assumed that multiple residues might also be involved in its uptake function. To identify the key residues required for binding and uptake of Rh123, we generated a series of mutant proteins in which different combinations of residues from TMH 6 and TMH 12 were substituted with alanine. A few additional residues adjacent to the residues mutated in the 14A mutant in the same TMHs within the drug-binding pocket were also mutated in some variants (Fig. 3A and SI Appendix, Table S2). We determined the uptake profiles of all of the mutants by testing the same four substrates (Rh123, DHR123, Rhod-2 AM, and Flutax-1) that were found to accumulate in 14A mutant-expressing cells (Fig. 3B and SI Appendix, Fig. S5). Only the 16A, 10A-I, and 10A-III mutant P-gps could import all four substrates, but to a lesser extent than 14A. The remaining mutants (10A-II, 9A, 8A, 7A, 6A-I, and 6A-II) could mediate the uptake of only Rh123 and DHR123. Thus, at least three specific residues from TMH 6 and seven from TMH 12 are required for uptake of the four substrates; on the other hand, just six residues (three from both TMH 6 and 12) are necessary for the uptake of Rh123. Although both 8A and 10A-III have the same three residues from TMH 6 (Fig. 3A), 10A-III has two extra mutations in TMH 12 (V977A and V981A), which appear to be critical for the uptake of all four substrates by the 10A-III mutant. Also, 8A shows marginal uptake of Rhod-2 AM, which does not occur with the 7A mutant that lacks the V986A mutation. This indicates that these three residues must contribute to the uptake of Rhod-2 AM and Flutax-1. Similarly, a comparison of mutated residues in 6A-II and 7A shows that the L975A mutation enhances the uptake of Rh123 and DHR123 (Fig. 3B and SI Appendix, Fig. S5; 11-fold accumulation by 7A versus sevenfold accumulation mediated by 6A-II mutant).
Fig. 3.
Identification of the residues of TMH 6 and TMH 12 that govern the import of substrates by the 14A mutant. (A) Schematic showing residues mutated in different P-gp variants in TMH 6 (red ovals) and TMH 12 (magenta-filled ovals). (B) Heatmap showing the accumulation of Rh123, DHR123, Rhod-2 AM, and Flutax-1 by 14A and other mutants. The heat map shows the relative levels of accumulation of each substrate, with the darkest being the highest level of uptake of a given substrate (white panels indicate no accumulation).
We also assessed the efflux profile of these mutants for other substrates. Mutants 16A, 14A, 10A-I, and 10A-III, which mediated uptake of four substrates, lost the ability to efflux most of the substrates (SI Appendix, Figs. S6 and S7). On the other hand, 10A-II, 9A, 8A, 7A, 6A-I, and 6A-II, which accumulate only Rh123, could efflux some of the substrates, thus mediating transport in either direction, depending on the substrate.
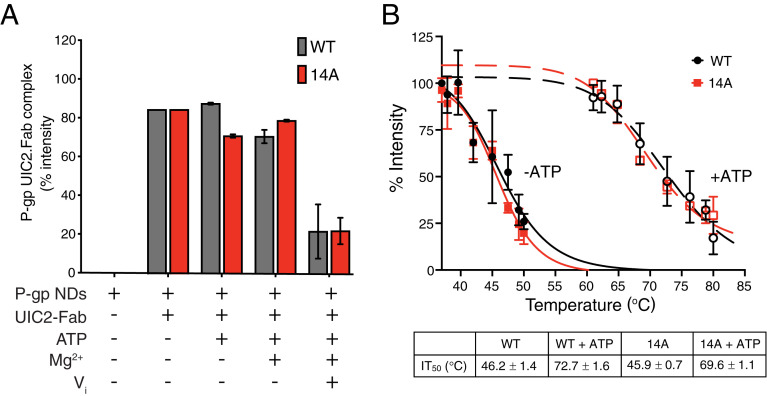
We next investigated whether the conformational changes that are associated with ATP or ligand binding are different in the14A mutant. The fact that ATP binding and hydrolysis was required for the uptake function (Fig. 2) suggested that the 14A mutant may follow the same transport cycle as the WT. To assess the overall conformation of the 14A mutant, we compared the binding of the conformational-sensitive antibody UIC2 (Fab region), the sensitivity to trypsinization, and the ATP-dependent changes in thermal stability. The ATPase activity of purified 14A in nanodiscs is inhibited by tariquidar, which also occurs with the WT, but Rh123 only marginally stimulated it (SI Appendix, Fig. S8A). The UIC2-Fab binding to purified WT and 14A mutant P-gp reconstituted in nanodiscs was assayed at different steps of the ATP hydrolysis cycle (Fig. 4A and SI Appendix, Fig. S8). We found that UIC2 only dissociates from WT P-gp and 14A in the vanadate (Vi)-trapped posthydrolysis conformation. The susceptibility of WT and 14A to trypsin digestion showed that both proteins required similar concentrations of trypsin for degradation (SI Appendix, Fig. S9), indicating that they most likely have similar conformations. Previously, we have shown that ATP binding, under nonhydrolyzing conditions, favors the thermal stabilization of WT P-gp; thus, the inward open conformation is more sensitive to heat treatment when compared to the inward closed conformation (23). We found that WT and 14A show similar thermal stability profiles both in the absence and presence of ATP (Fig. 4B).
Fig. 4.
Conformational changes in the 14A mutant are very similar to those occurring in WT P-gp. (A) WT and 14A show comparable binding to UIC2 Fab in different steps of the ATP hydrolysis cycle. The histogram shows the quantification of bands corresponding to WT or 14A P-gp: UIC2-Fab complex (native-gels presented in SI Appendix, Fig. S8B). (B) WT and 14A proteins show similar thermal stability in the presence or absence of ATP. The disappearance of monomeric WT or 14A P-gp bands as a function of incubation temperature (37–80 °C) was determined as described in Materials and Methods. Data presented as mean ± SD, n = 3. P-gp NDs, P-gp nanodiscs.
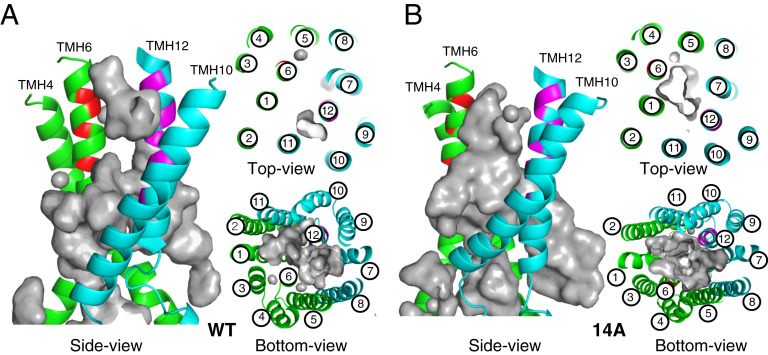
To further understand the effects of mutations in TMHs 6 and 12 on the structure of P-gp that change the direction of transport from efflux to uptake, we conducted a series of molecular dynamics (MD) simulations of the transporter embedded in a lipid bilayer (see Materials and Methods). All models were based on the recently determined structure of human P-gp with bound paclitaxel (Protein Database [PDB] ID: 6QEX) (5). In addition to the control, unaltered 6QEX.pdb structure (paclitaxel-WT), models of the apo protein with the WT sequence and the 14A mutations (Apo-WT and Apo-14A), and the paclitaxel-bound and 14A mutations (Taxol-14A) were generated. The TM region of the control system was found to be relatively unchanged after 1,000 ns: the rmsd from the starting structure of the Cα’s of the 12 TMHs from 200–1,000 ns was only 1.2 Å. While the rmsd for Apo-WT is ∼2.3 Å, the Apo-14A system remains similar to the control, with an approximate rmsd of only 0.8 Å. As the bound paclitaxel likely contributes to the stability of this region (5), we focused on the two trajectories of apo models to better understand the effects of the mutations. Examining only the areas where mutations were introduced in TMH 6 (residues 331–345) and TMH 12 (974–986), which form the narrowest part of the lumen, brings to light significant differences in the Cα rmsd among the Apo-WT and Apo-14A proteins (SI Appendix, Fig. S10). A larger fluctuation was seen in the Apo-WT system, with an approximated rmsd range of 1.4 vs. 0.5 Å, suggesting less stability. The distances among six pairs of Cα atoms at similar positions along TMH 6 and TMH 12 throughout the simulation are shown in SI Appendix, Fig. S11A. This shows that the greater deviation of the Apo-WT system is due to the helices moving closer together as the dynamics progress. Also, the helices of the Apo-WT system adopt curved conformations compared to the mutated, Apo-14A system (SI Appendix, Fig. S11B). This is consistent with alanine residues enhancing the rigidity and linearity of protein α-helices (24). As expected, the differential change in distance between TMH 6 and TMH 12 causes significant differences in the size and shape of the TMH lumen of the two systems. As seen in the side, top, and bottom views of WT and 14A (Fig. 5 A and B), the lumen of the Apo-WT system is occluded at the outer leaflet of the membrane at the position of the mutations, whereas the lumen of the Apo-14A system has an open “channel” in this region (top view in Fig. 5 A and B). This suggests how the mutations may allow for uptake, as the path is open for substrates to enter the lumen from outside the cell.
Fig. 5.
MD simulations of TMH 6 and 12 of the 14A mutant and WT P-gp. Cartoon representations of Apo-WT (A) and Apo-14A (B) using the paclitaxel-bound cryo-EM structure (PDB ID: 6QEX) of P-gp after 1,000-ns MD simulation: different views (side, top, and bottom) relative to the lipid bilayer. TMD1 helices are presented in green, and TMD2 are in cyan. Mutated residues from TMH 6 and TMH 12 are colored red and magenta, respectively. The gray surface depicts the binding cavity/empty space inside the transmembrane region. WT and 14A views from the extracellular region (top) and from the cytosolic end (bottom) highlight the depth and size of the binding cavity.
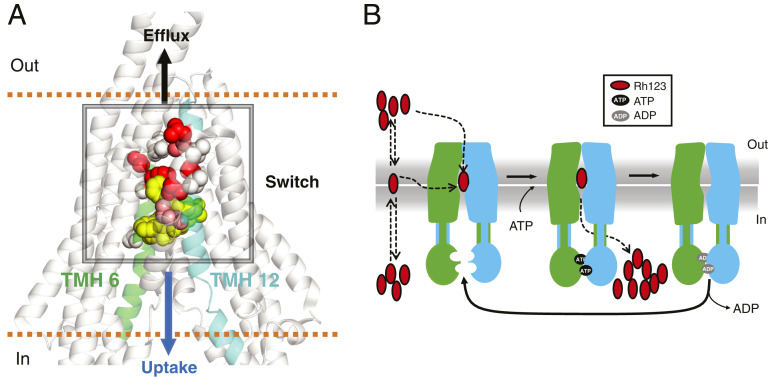
The analyses of transport function of several mutants with alanine substitution of 6 to 16 total residues of TMHs 6 and 12 (Figs. 2 and 3 and SI Appendix, Figs. S5–S7) and MD simulations (Fig. 5 A and B) suggest the presence of a switch in the central region of these two helices that governs whether a given substrate is pumped out of (efflux) or into (influx) the cell (Fig. 6A). Exactly how the substrate enters the binding cavity during the import process remains elusive. Comparison of the atomic structures of the 14A mutant (not yet available) with WT P-gp and other ABC importers will be critical for understanding the mechanism of the switch. It is possible that Rh123 enters the 14A transporter directly from the extracellular region, as suggested by the wider opening observed in the MD simulations (Fig. 5 A and B), or Rh123 can be partitioned in the membrane bilayer, with access to the drug-binding pocket occurring in a fashion similar to WT P-gp (7, 25). Subsequent conformational changes lead to intracellular translocation of Rh123 with concomitant ATP hydrolysis, while ADP release resets the transporter to the inward open conformation. Fig. 6B is a schematic of the proposed mechanism of uptake by the 14A mutant.
Fig. 6.
Identification of a switch region that controls the direction of transport mediated by P-gp and the proposed mechanism of substrate uptake by the 14A mutant. (A) Proposed switch in the central region of TMH 6 and 12. The transport profile of several mutants with mutations ranging from a total of 6 to 16 residues in TMH 6 and 12 (Figs. 2 and 3 and SI Appendix, Figs. S5–S7) suggests the presence of a switch that governs whether the substrate is transported out of or into the cell. Residues substituted with Ala, resulting in loss of efflux and gain of uptake function, are shown in spheres with different shades, with darker colors representing a greater contribution to the switch. The relative position of paclitaxel (yellow spheres) is based on the PDB ID: 6QEX structure. (B) Schematic of proposed Rh123 uptake mechanism. Rh123 has access to the binding site either directly from the extracellular region or through the membrane. ATP binding induces the dimerization of the NBDs. Subsequent conformational change leads to intracellular translocation of Rh123 with concomitant ATP hydrolysis. (How the efflux of Rh123 by the 14A mutant is blocked remains to be elucidated.) ADP release and separation of NBDs resets the protein to the inward-open conformation.
Thus, in this study, the approach of introducing multiple mutations in homologous TMHs 6 and 12 enabled us to observe a change in the direction of transport for certain substrates from efflux to uptake by P-gp. Among the 48 human ABC proteins, only two exhibit an uptake function, transporting substrates into the cytosol. They are ABCA4, which is present in retinal photoreceptor cells and imports N-retinylidene–phosphatidylethanolamine (26), and the lysosomal ABCD4 transporter, which mediates uptake of cobalamin (27). Some plant ABC transporters have also been suggested to import substrates, including auxin transport by Arabidopsis thaliana PGP4 (28) and its homolog in Coptis japonica MDR1 for alkaloid berberine (29). These transporters as well as bacterial importers including mycobacterial IrtAB (30) and the Escherichia coli vitamin B12 transporter BtuCD (31) and others (32) exhibit a very narrow substrate specificity. Similarly, concerning uptake by the 14A mutant, we observed a narrow substrate specificity, indicating that this may be a common characteristic of ABC importers compared to the polyspecificity observed in exporters.
The ability of the 14A mutant to hypersensitize cancer cells to drugs by mediating their uptake provides an approach to selectively kill P-gp–expressing cancer cells, thereby improving the efficiency of chemotherapy. Recent developments in nonintegrating DNA therapeutics could be a promising approach for such targeted therapies. A new class of DNA nanovectors could be used for sustained enhanced expression of transgenes such as the 14A mutant, without the risk of vector-associated cell toxicity (33). Altering the direction of transport of P-gp (and potentially other drug efflux pumps) could improve the delivery of chemotherapy in specific cancers. In addition, by screening libraries of small molecules or drugs, it may be possible to identify therapeutic agents that switch the direction of transport of an ABC drug transporter from efflux to influx and could be used to kill the cells expressing such transporters. Further studies on the switch mechanism of P-gp and other ABC drug efflux pumps will help to exploit the uptake function for therapeutic use.
Materials and Methods
Chemicals and Antibodies.
[3H]-rhodamine 123 (40 mCi/mmol) was custom-synthesized by Moravek, Inc. BD-verapamil was purchased from Setareh Biotech, and sources for other fluorescent compounds, reagents, and antibodies are described previously (10).
Cell Lines and Culture Conditions.
HeLa cells purchased from American Type Culture Collection were maintained in Dulbecco’s modified Eagle’s Medium (Gibco, Thermo Fisher Scientific) supplemented with 10% Fetal Bovine Serum, 5 mM l-glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin at 37 °C in 5% CO2 as described previously (9, 10).
Generation of P-gp Mutants in BacMam Baculovirus.
Certain residues in TMH 6 and TMH 12 were selected for mutation, with the selection rationale described in the main text. For generation of the 14A and 6A-II mutants, selected residues were substituted with alanine in the human P-gp sequence using the GeneArt gene synthesis method (Life Technologies, Thermo Fisher Scientific), and BacMam viruses were generated as described earlier (10, 34).
BacMam Baculovirus Transduction of HeLa Cells, Expression of WT and Mutant P-gp, and Transport of Fluorescent Substrates.
Previously described procedures (9, 10, 35) were used for the transduction of HeLa cells with the WT or mutant P-gp BacMam baculovirus, determination of cell surface expression using human P-gp specific antibodies MRK-16, UIC2, and 4E3 and assessment of transport of fluorescent substrates by flow cytometry.
Fluorescence Microscopy-Based Transport Assay.
The procedure followed was similar to one described earlier (35). Briefly, transduction was carried out as described in the earlier section using HeLa cells seeded in a 24-well plate format (50,000 cells/well). After 20–22 h, cells were washed twice with phosphate-buffered saline (PBS), and Rh123 was added at a 1.3-µM concentration to 1 mL Iscove’s Modified Dulbecco’s Medium (IMDM) containing 5% fetal bovine serum (FBS). Untransduced cells were used as a control. The transport assay was carried out for 45 min at 37 °C. Ten minutes before completion of the transport assay, Hoechst 33342 was added to the cells. The cells were washed with cold PBS three times, and 0.5 mL PBS was added to each well. Live cells were visualized in an Evos AMG microscope at 10× magnification in transmitted light (to visualize the cells) and in the green region (for fluorescence of Rh123) and ultraviolet (UV) region (to visualize Hoechst stained nuclei).
For confocal microscopy, glass-bottom culture dishes (35 mm Petri dish, 10 mm Microwell, MatTek Corp.) were used. Transduction of HeLa cells and transport was carried out as described above. Cells were stained with 0.5 µM MitoTracker Deep Red FM (far-red, 5 min, 37 °C), 0.5 µM ER tracker (red, 5 min, 37 °C), 5 µg/mL Hoechst 33342 (UV) (5 min, 37 °C), and 1.3 µM Rh123 (green, 45 min, 37 °C) dyes. Live cells were imaged immediately after staining at 60× magnification under the corresponding fluorescence filters. Colocalization analysis was done using a Zeiss LSM 780 microscope (NCI, CCR core facility) and ZEN Lite software (Zeiss Microscopy).
Kinetics of Rh123 Efflux and Uptake.
HeLa cells were transduced with the respective P-gp variant as described in the section above. Cells were trypsinized and used for kinetic analyses, as follows. Time-course of Rh123 accumulation: HeLa cells (untransduced control or expressing 14A) were incubated with Rh123 (1.3 µM) for the indicated time periods, and MFI was measured using flow cytometry. MFI after 45 min in control cells was taken as 100%, and relative fluorescence was calculated. Rh123 efflux by WT P-gp: HeLa cells (control and expressing WT P-gp) after trypsinization were washed with PBS (containing 1 mM MgCl2 and 0.1 mM CaCl2) and depleted of ATP by addition of 20 mM 2-deoxyglucose and 5 mM sodium azide for 10 min at 37 °C, as described earlier (35). In the same medium, Rh123 was added to the cells at the indicated concentrations for 20 min for equilibration. After incubation, cells were washed with ice-cold PBS/Ca2+/Mg2+ and resuspended in Rh123-free buffer; the level of Rh123 was taken as 100% (time = 0). Efflux was initiated by transferring the cells to PBS containing 1 mM MgCl2 and 0.1 mM CaCl2 supplemented with 55 mM glucose at 37 °C for 3 min (approximate t1/2 for efflux). Efflux was calculated by subtracting the fluorescence intensity after 3 min at a given Rh123 concentration from that at 0 min. Rh123 accumulation by the 14A mutant: HeLa cells (control and expressing 14A) were incubated with indicated concentrations of Rh123 for 3 min (approximate t1/2 for accumulation). Accumulation was calculated by directly measuring the MFI at 3 min at a given Rh123 concentration. ATP requirement for Rh123 accumulation: HeLa cells (untransduced control and cells expressing 14A) were depleted of ATP as described above and incubated with Rh123 (1.3 µM) at 37 °C for the indicated time points, followed by washing cells and measuring the level of cellular Rh123 by flow cytometry as described earlier (9, 10).
Transport Assay with [3H]-Rh123.
To analyze the accumulation of Rh123 against the concentration gradient, HeLa cells were transduced with 14A in a 6-well plate monolayer. Cells were depleted of ATP as described in the section above and exposed to cold Rh123 (1.3 µM) in IMDM + 5% FBS for 15 min. After washing, cells were exposed to IMDM + 5% FBS containing [3H]-Rh123 at lower concentrations (0.25 µM; ∼0.5 µCi/well) for 20 min. Cells were washed three times with cold PBS and lysed by PBS containing 2% Triton X-100 for 30 min. The lysate was transferred to scintillation vials containing 20 mL Bio Safe 2 scintillation fluid (Research Products International). Intracellular radioactivity was determined using the liquid scintillation counter LS6500 (Beckman).
Cytotoxicity Assays.
Cytotoxicity assays were performed with HeLa cells expressing either WT P-gp or 14A. After transduction with baculovirus (without adding butyrate), cells were plated in white 96-well plates at varying drug concentrations followed by incubation for 48 h at 37 °C. Percent survival was calculated using Cell Titer Glo reagent (Promega). Killing curves were plotted, and IC50 values were calculated using GraphPad Prism 8.1.2. All of the experiments were performed at least three times.
Preparation of Membrane Vesicles of High Five Insect Cells.
High Five insect cells (Invitrogen, Thermo Fisher Scientific) were infected with recombinant baculovirus carrying WT or mutant P-gp with a tobacco etch virus (TEV) protease-cleavage site and a His6-tag in the C-terminal end, as described previously (36, 37). Membrane vesicles were prepared by hypotonic lysis of cells expressing WT or mutant P-gp followed by differential centrifugation to collect the membranes. Total protein in membrane vesicles was quantified by a colorimetric assay using Amido black B as described earlier (38).
SDS/PAGE and Western Blotting.
HeLa cells (0.5–1.0 × 106) expressing WT or mutant P-gp were lysed by sonication and freeze–thaw cycles in a buffer containing 10 mM Tris-Cl pH 8.0, 0.1% Triton X-100, 10 mM MgSO4, 2 mM CaCl2, 1% aprotinin, 1 mM AEBSF, 2 mM DTT, and 20 µg/mL nuclease. Samples (lysates of 60,000 cells/lane) were resolved by 7% Tris-acetate SDS/PAGE gels (Invitrogen, Thermo Fisher Scientific), and western blotting was done using C219 (anti–P-gp, 1:2,000 dilution) and GAPDH-6C5 (1:20,000 dilution) monoclonal antibodies (primary) and HRP-conjugated mouse IgG secondary antibody (1:10,000 dilution). The blots were developed using an ECL western blotting detection kit (GE Healthcare). The quantification of P-gp band signals was done using ImageJ (NIH) (39). For quantification, each experiment was repeated at least three times, and values represent mean ± SD.
Purification of 14A Mutant P-gp and Reconstitution in Nanodiscs.
Membrane vesicles of High Five cells expressing WT or 14A mutant P-gp were solubilized with DDM/CHS and purified using Ni-NTA affinity chromatography and size-exclusion chromatography as described previously (40). Nanodiscs were prepared by combining sonicated E. coli lipids, MSP1D1 and WT or 14A P-gp (200:4:1 ratio) in the presence of 0.8 g/mL SM2 Biobeads (Bio Rad) and purified by size-exclusion chromatography using nanodisc (ND) buffer (40 mM Hepes pH 7.3, 150 mM NaCl, and 5 mM DTT).
ATPase Activity.
The ATPase activity of WT and 14A was measured as described earlier (9, 10), and the Vi-sensitive ATPase activities were analyzed and plotted using GraphPad Prism 8.1.2.
Determination of WT and 14A Mutant P-gp Trypsin Sensitivity.
Membrane vesicles (50 µg) of High Five cells containing WT or 14A were incubated at 37 °C for 5 min followed by treatment with increasing concentrations of L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-trypsin for another 5 min at 37 °C. The reaction was stopped by the addition of 25 µg of soybean trypsin inhibitor. Samples in the SDS/PAGE buffer were incubated for 20 min at 37 °C and then resolved by SDS/PAGE (10 µg/lane). Protein bands were stained with InstantBlue (Sigma). The loss of monomeric P-gp was quantified using the ImageJ program and plotted with GraphPad Prism 8.1.2 software.
Binding of UIC2-Fab to WT and 14A Mutant P-gp and Native Gel Electrophoresis.
Nanodiscs containing WT or 14A mutant P-gp (3 µg) were first incubated in the absence or presence of UIC2-Fab (10 µg) for 5 min in the ATPase assay buffer at 37 °C, and then ATP, MgCl2, and/or Vi (1, 2, and 0.5 mM, respectively) were added to the samples to a final volume of 20 µL. Samples were incubated for 5 min at 37 °C. Native PAGE sample buffer (Thermo Fisher Scientific) was added, and samples were loaded in a 4–12% Bis-Tris NuPAGE gel (Thermo Fisher Scientific) and run according to the manufacturer’s instructions. Gels were fixed and destained with a 50% ethanol, 10% acetic acid solution and restained with InstantBlue (Sigma-Aldrich). Gel band intensities were quantified using ImageJ (39).
Effect of Temperature on the Stability of WT and 14A Mutant P-gp.
Membrane vesicles containing WT or 14A mutant P-gp were incubated at a range of temperatures (37–80 °C) for 10 min in the presence or absence of ATP as described previously (23). Next, 5× gel sample buffer was added and incubated for 20 min at 37 °C. Samples were then run on a 7% Tris-Acetate NuPAGE gel and stained with InstantBlue (Sigma-Aldrich).
In Silico Analysis.
MD simulations were performed with the Nanoscale Molecular Dynamics (NAMD) 2.13b2 software package (41). For the WT and mutants, the transporter protein was embedded in a square bilayer patch of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids. The defining structural topologies and parameters were those of the CHARMM36 all-atom force-field (42). Taxol was modeled with CHARMM-GUI (43). Periodic boundary conditions were used for a rectangular cell of starting dimensions: 118.0 × 115.0 × 168.0 Å. This allowed for a minimum distance between the protein and the cell wall of 12.0 Å. The system was hydrated with TIP3P water molecules, and sodium and chloride ions were added to electrically neutralize and provide an ionic strength of 150 mM using Visual Molecular Dynamics (44). Regular electrostatic and van der Waals forces were calculated with a CUTOFF and SWITCHDIST of 12.0 and 8.0 Å, respectively. The temperature and pressure were maintained at 310.0 °K and 1 ATM with the Langevin and Langevin-Piston methods, respectively. The SETTLE mechanism was used to restrain covalent bond lengths to equilibrium values, allowing for an integration time-step of 2.0 fs. Trajectories were calculated for a minimum of 1,000.0 ns.
Supplementary Material
Acknowledgments
We thank George Leiman for editing this manuscript and Drs. John Golin and Tanaji Talele for constructive comments. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov/). The study was funded by the Intramural Program of the NIH, the National Cancer Institute, and the Center for Cancer Research; and M. M. was supported in part by the Fellowship from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016270117/-/DCSupplemental.
Data Availability.
All data are included in the main paper or in SI Appendix.
References
- 1.Ambudkar S. V., Kimchi-Sarfaty C., Sauna Z. E., Gottesman M. M., P-glycoprotein: From genomics to mechanism. Oncogene 22, 7468–7485 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Eckford P. D., Sharom F. J., ABC efflux pump-based resistance to chemotherapy drugs. Chem. Rev. 109, 2989–3011 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Gottesman M. M., Fojo T., Bates S. E., Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Juliano R. L., Ling V., A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta 455, 152–162 (1976). [DOI] [PubMed] [Google Scholar]
- 5.Alam A., Kowal J., Broude E., Roninson I., Locher K. P., Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y., Chen J., Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lusvarghi S., Robey R. W., Gottesman M. M., Ambudkar S. V., Multidrug transporters: Recent insights from cryo-electron microscopy-derived atomic structures and animal models. F1000 Res. 9, 17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chufan E. E., et al. , Multiple transport-active binding sites are available for a single substrate on human P-glycoprotein (ABCB1). PLoS One 8, e82463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vahedi S., Chufan E. E., Ambudkar S. V., Global alteration of the drug-binding pocket of human P-glycoprotein (ABCB1) by substitution of fifteen conserved residues reveals a negative correlation between substrate size and transport efficiency. Biochem. Pharmacol. 143, 53–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sajid A., Lusvarghi S., Chufan E. E., Ambudkar S. V., Evidence for the critical role of transmembrane helices 1 and 7 in substrate transport by human P-glycoprotein (ABCB1). PLoS One 13, e0204693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szewczyk P., et al. , Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr. D Biol. Crystallogr. 71, 732–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loo T. W., Clarke D. M., Cross-linking of human multidrug resistance P-glycoprotein by the substrate, tris-(2-maleimidoethyl)amine, is altered by ATP hydrolysis. Evidence for rotation of a transmembrane helix. J. Biol. Chem. 276, 31800–31805 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Loo T. W., Clarke D. M., Drug-stimulated ATPase activity of human P-glycoprotein requires movement between transmembrane segments 6 and 12. J. Biol. Chem. 272, 20986–20989 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Crowley E., et al. , Transmembrane helix 12 modulates progression of the ATP catalytic cycle in ABCB1. Biochemistry 48, 6249–6258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chufan E. E., Sim H. M., Ambudkar S. V., Molecular basis of the polyspecificity of P-glycoprotein (ABCB1): Recent biochemical and structural studies. Adv. Cancer Res. 125, 71–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vahedi S., et al. , Mapping discontinuous epitopes for MRK-16, UIC2 and 4E3 antibodies to extracellular loops 1 and 4 of human P-glycoprotein. Sci. Rep. 8, 12716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie T. K., Kwon H., Atkins W. M., Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J. Biol. Chem. 286, 39489–39496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okochi E., Iwahashi T., Tsuruo T., Monoclonal antibodies specific for P-glycoprotein. Leukemia 11, 1119–1123 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Darzynkiewicz Z., Traganos F., Staiano-Coico L., Kapuscinski J., Melamed M. R., Interaction of rhodamine 123 with living cells studied by flow cytometry. Cancer Res. 42, 799–806 (1982). [PubMed] [Google Scholar]
- 20.Sauna Z. E., Nandigama K., Ambudkar S. V., Exploiting reaction intermediates of the ATPase reaction to elucidate the mechanism of transport by P-glycoprotein (ABCB1). J. Biol. Chem. 281, 26501–26511 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Sakurada H., Koizumi H., Ohkawara A., Ueda T., Kamo N., Use of dihydrorhodamine 123 for detecting intracellular generation of peroxides upon UV irradiation in epidermal keratinocytes. Arch. Dermatol. Res. 284, 114–116 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Stein W. D., Litman T., Channels, Carriers, and Pumps: An Introduction to Membrane Transport (Elsevier, UK, ed. 2, 2015). [Google Scholar]
- 23.Lusvarghi S., Ambudkar S. V., ATP-dependent thermostabilization of human P-glycoprotein (ABCB1) is blocked by modulators. Biochem. J. 476, 3737–3750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Neil K. T., DeGrado W. F., A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250, 646–651 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Quazi F., Lenevich S., Molday R. S., ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 3, 925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu D., et al. , Cryo-EM structure of human lysosomal cobalamin exporter ABCD4. Cell Res. 29, 1039–1041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santelia D., et al. , MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 579, 5399–5406 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Shitan N., et al. , Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc. Natl. Acad. Sci. U.S.A. 100, 751–756 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold F. M., et al. , The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 580, 413–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locher K. P., Lee A. T., Rees D. C., The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science 296, 1091–1098 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Swier L. J. Y. M., Slotboom D.-J., Poolman B., “ABC importers” in ABC Transporters–40 Years on, George A. M., Ed. (Springer International Publishing, Cham, 2016), pp. 3–36. [Google Scholar]
- 33.Bozza M., et al. , Novel non-integrating DNA nano-S/MAR vectors restore gene function in isogenic patient-derived pancreatic tumor models. Mol. Ther. Methods Clin. Dev. 17, 957–968 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla S., Schwartz C., Kapoor K., Kouanda A., Ambudkar S. V., Use of baculovirus BacMam vectors for expression of ABC drug transporters in mammalian cells. Drug Metab. Dispos. 40, 304–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sajid A., et al. , Synthesis and characterization of bodipy-FL-cyclosporine A as a substrate for multidrug resistance-linked P-glycoprotein (ABCB1). Drug Metab. Dispos. 47, 1013–1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandra M., et al. , Human P-glycoprotein exhibits reduced affinity for substrates during a catalytic transition state. Biochemistry 37, 5010–5019 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Kerr K. M., Sauna Z. E., Ambudkar S. V., Correlation between steady-state ATP hydrolysis and vanadate-induced ADP trapping in human P-glycoprotein. Evidence for ADP release as the rate-limiting step in the catalytic cycle and its modulation by substrates. J. Biol. Chem. 276, 8657–8664 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Schaffner W., Weissmann C., A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56, 502–514 (1973). [DOI] [PubMed] [Google Scholar]
- 39.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandigama K., Lusvarghi S., Shukla S., Ambudkar S. V., Large-scale purification of functional human P-glycoprotein (ABCB1). Protein Expr. Purif. 159, 60–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33-38–27-38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the main paper or in SI Appendix.