Significance
Here, a genome-scale CRISPR/Cas9 screen in epithelial cells using Salmonella, a prototypical intracellular pathogen, led to the discovery of a new role for IFN-I signaling. We found that this canonical antiviral signaling pathway controls the subcellular localization, protein content, pH, and protease activity of lysosomes. IFN-I signaling-dependent lysosomal acidification was associated with heightened Salmonella virulence gene expression and escape into the cytosol, and in vivo studies confirmed a role for epithelial IFN-I signaling in promoting Salmonella systemic infection. We propose that IFN-I signaling-mediated control of lysosome function contributes to host responses to diverse intracellular pathogens and viruses. Investigating how cytokine signaling modifies organelle function will deepen understanding of the cell biology of innate defense.
Keywords: type I interferon, lysosome, Salmonella pathogenesis, intestinal epithelium, mucosal defense
Abstract
Organelle remodeling is critical for cellular homeostasis, but host factors that control organelle function during microbial infection remain largely uncharacterized. Here, a genome-scale CRISPR/Cas9 screen in intestinal epithelial cells with the prototypical intracellular bacterial pathogen Salmonella led us to discover that type I IFN (IFN-I) remodels lysosomes. Even in the absence of infection, IFN-I signaling modified the localization, acidification, protease activity, and proteomic profile of lysosomes. Proteomic and genetic analyses revealed that multiple IFN-I–stimulated genes including IFITM3, SLC15A3, and CNP contribute to lysosome acidification. IFN-I–dependent lysosome acidification was associated with elevated intracellular Salmonella virulence gene expression, rupture of the Salmonella-containing vacuole, and host cell death. Moreover, IFN-I signaling promoted in vivo Salmonella pathogenesis in the intestinal epithelium where Salmonella initiates infection, indicating that IFN-I signaling can modify innate defense in the epithelial compartment. We propose that IFN-I control of lysosome function broadly impacts host defense against diverse viral and microbial pathogens.
Microbial pathogens have evolved varied virulence strategies to modulate host cell function (1, 2). A common mechanism, employed by all viral and some bacterial pathogens, is to enter host cells, where they co-opt cellular functions while simultaneously evading extracellular threats, such as innate and adaptive immune mechanisms (3, 4). Inside cells, intracellular pathogens interact with and exploit host cell organelles to support their proliferation (5). As a result of their intimate relationships with and manipulation of varied host cell functions, intracellular pathogens have proven to be outstanding tools to probe basic eukaryotic cell biology (6).
Compared to knowledge of how microbial pathogens interact with phagocytic cells, less is known about the landscape of pathogen-epithelial cell interactions at barrier sites where most infections originate (7). The human foodborne pathogen Salmonella enterica serovar Typhimurium (Stm) is a model intracellular bacterium that initially invades and subsequently kills intestinal epithelial cells (IECs) before spreading systemically to distal sites via circulating phagocytes (8). Stm’s entry into and initial trafficking inside IECs is well characterized, and a hallmark of Stm infection is the formation of the Salmonella-containing vacuole (SCV), a dynamic lysosome-like compartment that is permissive for Stm replication (9, 10). Several canonical and noncanonical cell death pathways have been proposed to underlie Stm-induced IEC cytotoxicity (11–15), but a global approach to understand how this phenotype is controlled has not been undertaken.
One striking feature of the host response to Stm is the induction of IFN-Is, which include IFNα and IFNβ (16). IFN-Is are cytokines that, once secreted, bind the IFN-I receptor (IFNAR1/2) to activate JAK-STAT signaling, triggering the expression of an intracellular antimicrobial transcriptional program consisting of over 400 IFN-stimulated genes (ISGs) (17). Due to the large size of the “interferome” and the complex interactions of ISGs with thousands of additional cellular proteins (18), knowledge of the full spectrum of IFN-I-mediated changes in cellular function is incomplete. Although IFN-Is are known to play critical roles in antiviral responses, their functions in bacterial infection are less clear, and IFN-I signaling has been reported to be either protective or detrimental to the host depending on the specific bacterial pathogen (19).
Here, we carried out a genome-scale CRISPR/Cas9 screen to identify the host factors that contribute to Stm’s cytotoxicity to IECs. This screen revealed IFN-I signaling as a key susceptibility factor for cytotoxicity in IECs and led to our discovery of a role for IFN-I signaling in lysosomal localization and function, including the modification of this organelle’s pH, protease activity, and protein content. Organellar proteomics revealed that 11 ISGs were enriched in lysosomes following IFN-I stimulation, several of which were found to directly impact lysosomal pH. IFN-I signaling-dependent lysosomal acidification was associated with heightened Stm virulence gene expression, and in vivo studies confirmed a role for epithelial IFN-I signaling in promoting IEC cell death and systemic Stm infection. IFN-I signaling-mediated control of lysosome function likely contributes to host responses to diverse intracellular pathogens and viruses.
Results
A CRISPR/Cas9 Screen Identifies IEC Factors Required for Stm Cytotoxicity.
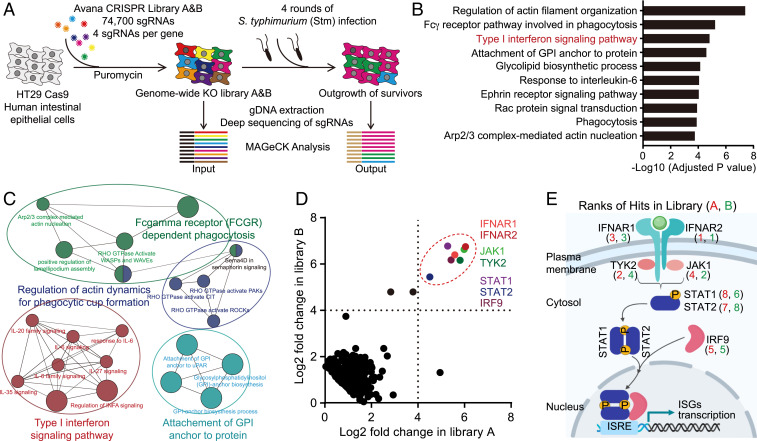
We performed a multiround genome-wide CRISPR/Cas9 loss-of-function screen in the human colonic epithelial cell line HT29-Cas9 (20) to identify IEC genes that confer resistance to Stm cytotoxicity (Fig. 1A). HT29 cells are efficiently invaded and subsequently killed by Stm, providing a strong selective force to enrich for guide RNAs targeting host factors that modulate cytotoxicity. The screen identified known pathways that sensitize cells to Stm infection, including those involved in regulation of actin dynamics (the Arp2/3 complex and Rac signaling proteins), which are important in pathogen invasion (Fig. 1 B and C and Dataset S1) (21, 22). Genes linked to pathways not previously directly linked to Stm virulence, including the Fc-γ receptor-dependent phagocytic and glycosylphosphatidylinositol (GPI) anchor modification pathways were also enriched (Fig. 1 B and C). Strikingly, the top "hits" of the screen were remarkably coherent—the seven most enriched guide RNAs from both libraries screened corresponded to genes in the IFN-I signaling pathway, including the receptor (IFNAR1/IFNAR2), adaptor (JAK1/TYK2), and transcription factor (STAT1/STAT2/IRF9) components of the system (Fig. 1 D and E), suggesting that IFN-I signaling is a major driver of Stm-mediated cytotoxicity in IECs.
Fig. 1.
A CRISPR/Cas9 screen identifies IEC factors required for Stm cytotoxicity. (A) Workflow for CRISPR/Cas9 Stm cytotoxicity screen in HT29-Cas9 cells. (B) Adjusted P values for selected enriched gene ontology (GO) terms from GO-analyzed hits in the Stm cytotoxicity screen (upper threshold set at P < 1 × 10−3). (C) Cytoscape visualization of enriched pathways. (D) Scatterplots showing normalized read enrichment of specific single guide RNAs (sgRNAs) in two libraries (A and B) after four rounds of Stm infection. Genes involved in IFN-I signaling are delineated by the dashed red circle. (E) Overview of IFN-I signaling pathway. Numbers correspond to hit ranks in each library. See also Dataset S1.
IFN-I Promotes Stm Cytotoxicity in IECs.
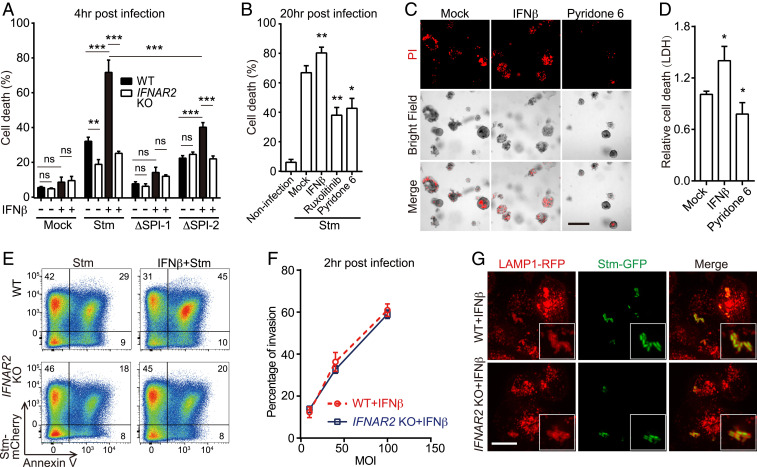
Stm induces IFN-I production during infection (16), but the function of IFN-I signaling in IECs is unknown. A clonal knockout (KO) of IFNAR2, the top enriched hit in both libraries, was constructed in the HT29-Cas9 background (SI Appendix, Fig. S1A) to validate the screen findings. At both early (4-h) and late (20-h) infection time points, IFNAR2 KO cells were more resistant to Stm-induced cell death than the wild-type (WT) parental line (Fig. 2A and SI Appendix, Fig. S1 B and C). Priming cells with IFNβ (a major IFN-I), conditions that mimic the multiple rounds of the original screen, further sensitized WT but not IFNAR2 KO cells to death (Fig. 2A). To complement these findings, we treated WT cells with chemical inhibitors of JAK-STAT signaling, the downstream target of activated IFNAR1/2. Similar to IFNAR2 KO cells, treatment with the JAK inhibitors ruxolitinib and pyridone-6 also diminished Stm-induced death in WT cells, indicating active IFN-I signaling is required for this phenotype (Fig. 2B and SI Appendix, Fig. S1D). These findings were recapitulated in primary human-derived small intestinal organoids. In this more physiologic culture system, IFNβ priming of organoids also increased cell death associated with Stm infection, and treatment of organoids with pyridone-6 had the opposite effect (Fig. 2 C and D). Together, these data support the idea that IFN-I signaling promotes Stm pathogenicity in IECs.
Fig. 2.
IFN-I promotes Stm cytotoxicity in IECs. (A) Survival of IFNβ-primed or IFNβ-unprimed WT or IFNAR2 KO HT29 cells 4-h post-WT or mutant Stm infection by lactate dehydrogenase (LDH) assay. (B) Survival of mock or drug-treated WT HT29 cells 20-h post-WT Stm infection. (C) Representative images of IFNβ or pyridone-6–primed or unprimed human small bowel enteroids 20-h post-WT Stm infection. Propidium iodide (PI) staining was used to detect cell death. (Scale bar, 100 μm.) (D) Enteroid survival 20-h post-WT Stm infection. (E) Flow cytometry of IFNβ-primed or IFNβ-unprimed WT and IFNAR2 KO HT29 cells 20-h post-mCherry-Stm infection and stained with Annexin V-fluorescein isothiocyanate (FITC). (F) Flow cytometric quantification of invasion of HT29 cells by mCherry-Stm. (G) Representative images of LAMP1-RFP-expressing HeLa cells 4-h post-green fluorescent protein (GFP)-Stm infection. Boxed insets depict higher magnification showing bacterial colocalization with LAMP1-RFP. (Scale bar, 10 μm.) Data shown are means ± SD from three independent experiments. See also SI Appendix, Figs. S1 and S2.
The observation that IFN-I promotes Stm-induced IEC death is consistent with prior data that this cytokine enhances necroptosis in Stm-infected macrophages (23). However, it is not clear whether macrophage and epithelial cell responses to Stm infection are similar; furthermore, it is known that Stm-induced cell death in macrophages is invasion independent (24). Thus, we next tested whether IFN-I-promoted epithelial cell death depended on SPI-1 or SPI-2, critical Salmonella pathogenicity islands that each encode type 3 secretion systems required for cellular invasion and intracellular survival, respectively (25). SPI-1–deficient (ΔprgH) Stm did not induce epithelial cell death in any condition, confirming that IEC cytotoxicity requires cell invasion (Fig. 2A). In contrast, SPI-2–deficient (ΔssaV) Stm led to reduced but still detectable levels of cytotoxicity that remained sensitive to IFNβ priming, suggestive of both SPI-2–dependent and SPI-2–independent mechanisms of intracellular Stm-induced cytotoxicity (Fig. 2A).
In support of the population-level LDH assays, flow cytometry of HT29 or HeLa cells infected with fluorescent Stm and stained with the cell death probe Annexin-V indicated that IFN-I only influenced cell death in the population of cells that contained intracellular Stm (Fig. 2E and SI Appendix, Fig. S1 E–G). In addition, we found that IFN-I signaling did not impact Stm invasion (Fig. 2F and SI Appendix, Fig. S2A), nor did it influence bacterial association with the early endosomal marker RAB5, late endosomal marker RAB7, or lysosomal marker lysosomal-associated membrane protein 1 (LAMP1) (26) (Fig. 2G and SI Appendix, Fig. S2 B–F). Together, these data suggest that IFN-I–mediated sensitization of epithelial cells to Stm occurs downstream of cell invasion and initial SCV formation.
IFN-I Regulates Lysosome Localization and Function.
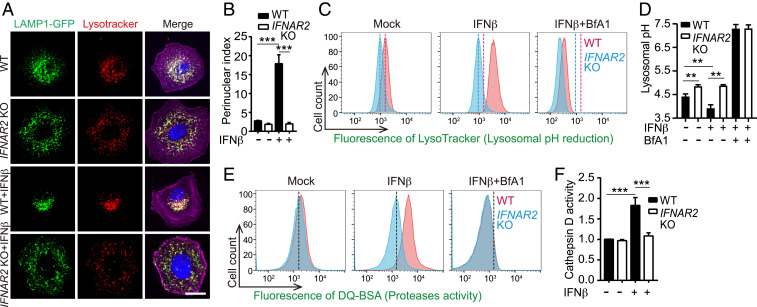
During our analyses of SCV formation, we unexpectedly observed that IFN-I signaling alters the subcellular localization of lysosomes in epithelial cells even in the absence of infection. In HeLa cells, lysosomes (identifiable as LAMP1+/Lysotracker+ costaining organelles) were scattered throughout the cytoplasm under basal conditions. Following IFNβ stimulation, lysosomes relocalized to the perinuclear region (Fig. 3 A and B and Movies S1 and S2); lysosome relocalization was not observed in IFNAR2 KO HeLa cells, confirming that this response was dependent on IFN-I signaling.
Fig. 3.
IFN-I signaling regulates lysosomal positioning, acidity, and protease activity. (A) Representative images of lysosome (LAMP1-GFP+/Lysotracker+ compartment) distribution in WT and IFNAR2 KO HeLa cells with or without 16 h of IFNβ stimulation. Nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI) and actin (purple) was stained with phalloidin. (Scale bar, 5 μm.) (B) Lysosomal perinuclear indices in cells described in A. (C) Lysotracker fluorescence of WT or IFNAR2 KO HeLa cells ± 16 h of treatment with IFNβ or the lysosomal acidification inhibitor bafilomycin A1 (Bfa1). Vertical dashed lines indicate the mean fluorescence value of the mock control in WT (red) or IFNAR2 KO (blue) cells. (D) Absolute lysosomal pH values from FITC-dextran labeling of WT or IFNAR2 KO HeLa cells ± 16 h of treatment with IFNβ or Bfa1. (E) DQ-Green BSA fluorescence in cells treated identically to C. (F) Relative cathepsin D activity in WT and IFNAR2 KO HeLa cells ± 16 h of IFNβ treatment. Statistical analysis was performed by two-tailed Student’s t test (**P < 0.01 and ***P < 0.001). Data shown are means ± SD from, at least three independent experiments. See also SI Appendix, Fig. S3.
Furthermore, IFNβ priming led to significantly higher intensities of two different fluorescent lysosomal pH reporters (Lysotracker and Lysosensor) in WT but not IFNAR2 KO cells, indicating that IFN-I signaling lowers lysosome pH (Fig. 3C and SI Appendix, Fig. S3 A–C). Ratiometric microscopy with a third dye, Lysosensor Yellow/Blue, corroborated this trend (SI Appendix, Fig. S3 D and E). We also measured FITC-dextran fluorescence coupled to a standard pH curve to quantify the absolute lysosomal pH (27) in cells from each condition. Consistent with the relative reporter measurements, treatment of WT cells with IFNβ significantly lowered average lysosomal pH from 4.4 to 3.9; IFNAR2 KO cells displayed a raised basal pH value of 4.9 that was unchanged by IFNβ treatment (Fig. 3D).
Additional experiments with fluorescent reporters of the activities of lysosomal proteases (DQ-BSA, ref. 28), or cathepsin D (a major lysosomal protease) revealed that IFNβ treatment stimulated their activities in an IFNAR2-dependent fashion (Fig. 3 E and F and SI Appendix, Fig. S3F). These findings are consistent with the understanding that the activity of most resident lysosomal enzymes, including cathepsins and other degradative enzymes, is positively regulated by acidic pH (29). In contrast, staining with fluorescent reporters of endocytic activity (dextran-568) suggested that IFN-I signaling does not impact general endocytic trafficking (SI Appendix, Fig. S3G). IFNβ-induced lysosomal acidification and protease activation were abolished by the addition of the v-ATPase inhibitor Bfa1 (30) (Fig. 3 C–E), demonstrating that IFN-I signaling primarily relies on the conventional lysosomal acidification machinery to modulate lysosome function.
Together, these observations reveal that IFN-I signaling promotes epithelial cell lysosomal relocalization, acidification, and degradative activities without broadly affecting intracellular trafficking. Besides epithelial cells, such as HeLa and HT29, IFNβ treatment also reduced lysosomal pH in monocyte/macrophage-like THP-1 cells (SI Appendix, Fig. S3H), suggesting that IFN-I signaling controls lysosomal acidification outside of epithelial cell lineages.
The ISG IFITM3 Regulates Lysosomal Function and Stm Cytotoxicity.
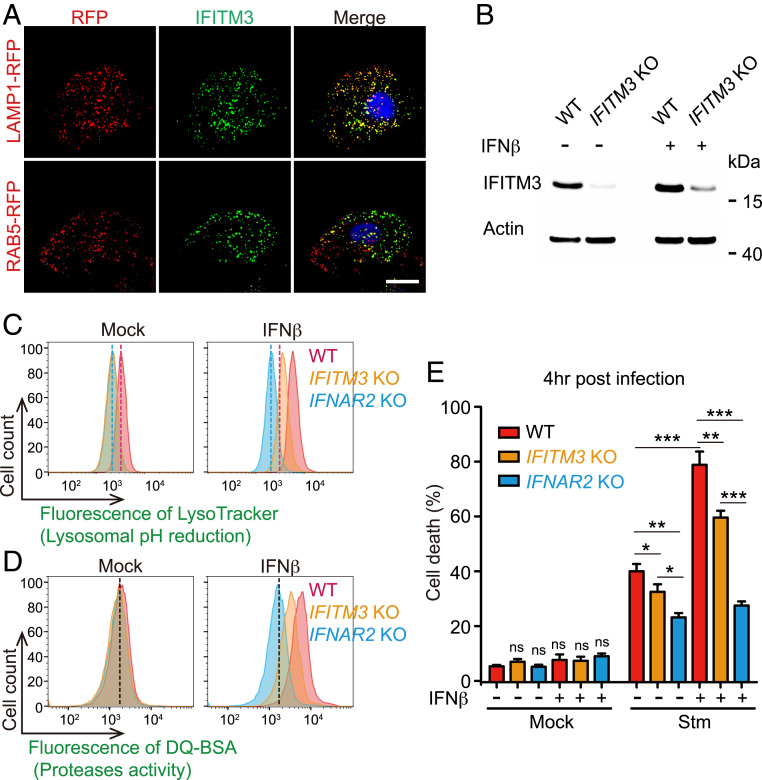
Although IFN-γ (IFN-II), which in contrast to IFN-I promotes resistance to Stm (31), can alter phagosomal activity (32), mechanisms of IFN-I, and more broadly, cytokine control of lysosome function have not been described. To investigate the mechanism by which IFN-I signaling regulates lysosome acidification and function, we first took a candidate-based approach and investigated IFITM3. This transmembrane ISG has antiviral activity and is thought to reside in the endosomal trafficking system and to interact with the lysosomal v-ATPase complex (33, 34), suggesting a potential role for this protein in lysosome function. Immunofluorescence analysis revealed that IFITM3 colocalized with LAMP1 but not RAB5, confirming that IFITM3 is a lysosomal protein (Fig. 4A). Remarkably, lysosomal pH in an IFITM3 KO cell line was elevated in both basal and IFNβ-primed conditions relative to WT cells, partially phenocopying the IFNAR2 KO, and suggesting that IFITM3 contributes to IFN-I–meditated lysosomal remodeling (Fig. 4 B–D). IFITM3’s contribution to basal pH levels is consistent with the tonic activities of IFNs observed in diverse mammalian cell types (35). The IFITM3 KO cell line was more resistant to Stm-induced cell death than the WT parental line both before and after IFNβ priming (Fig. 4E), suggesting that this ISG also contributes to IFN-I signaling enhancement of Stm cytotoxicity.
Fig. 4.
The ISG IFITM3 regulates lysosomal function and Stm cytotoxicity. (A) Representative images of LAMP1-RFP or RAB5-RFP-expressing HeLa cells stained with IFITM3 antibody (GFP). Nuclei (blue) were stained with DAPI. (Scale bar, 5 μm.) (B) Immunoblotting for IFITM3 in WT and IFITM3 KO HeLa cells ± 16 h of IFNβ treatment. (C and D) Flow cytometry of LysoTracker Red (C) and DQ-Green BSA fluorescence (D) in WT, IFITM3 KO, and IFNAR2 KO HeLa cells ± 16 h of IFNβ treatment. (E) Survival of IFNβ-primed or IFNβ-unprimed WT, IFITM3, or IFNAR2 KO HeLa cells 4 h post-Stm infection. Statistical analysis was performed by two-tailed Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001). Data shown are means ± SD from three independent experiments.
Discovery of ISGs with Roles in Lysosomal pH Regulation.
Given that both lysosomal pH and degradative activity in IFITM3 KO cells were still somewhat sensitive to IFNβ priming (Fig. 4 C and D), we hypothesized that additional ISGs regulate lysosome function. To identify these factors, we used a recently described lysosome immunopurification (LysoIP) system (36) to profile the proteomes of intact lysosomes from WT or IFNAR2 KO cells in basal or IFNβ-stimulated states. The purity and integrity of the lysosome samples was confirmed by verifying the presence of luminal cathepsin D and the absence of cytosolic and Golgi apparatus markers (Fig. 5A and SI Appendix, Fig. S4). Quantitative profiling revealed that the abundances of ∼15 proteins, most of them ISGs, were increased in purified lysosomes upon IFNβ stimulation (Fig. 5B and Dataset S2). Spectral counts for IFITM3 were enriched in lysosomes from IFNβ-treated cells, supporting the imaging above (Fig. 4A) and providing validation of the dataset. Immunoblots of purified lysosomal and cytoplasmic fractions from naive and IFNβ-treated cells with antibodies to IFITM3 further corroborated this observation, with the known cytosolic ISG IFIT3 serving as a negative control in this assay (Fig. 5A).
Fig. 5.
Identification of ISGs with roles in lysosomal pH regulation. (A) Immunoblotting for known (LAMP1, CTSD) and suspected (IFITM3) lysosomal proteins in whole-cell lysates (T), and purified lysosomes (IP). (B) Relative fold change scatterplot of protein abundance in lysosomes purified from WT or IFNAR2 KO HeLa cells ± 16 h of IFNβ treatment. Colored dots indicate proteins that are known ISGs. (C) Quantification of mean fluorescence intensity from flow cytometry of Lysotracker staining in WT or ISG KO cells ± 16 h of IFNβ treatment. Statistical analysis was performed by two-tailed Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001). Data shown are means ± SD from three independent experiments. See also SI Appendix, Fig. S4 and Dataset S2.
We constructed KO cell lines for most of the lysosomally enriched ISGs and assessed their contributions to the pH of this degradative organelle (Fig. 5C). Although most of the tested ISGs did not appear to influence lysosomal pH, two of them (SLC15A3 and CNP), like IFITM3, contributed to both basal and IFN-I–mediated lysosomal acidification (Fig. 5C). In contrast to IFITM3, SLC15A3, a lysosome-resident proton-coupled histidine and di-tripeptide transporter (37) and CNP, 2′,3′-cyclic nucleotide 3′ phosphodiesterase, are not known to be linked to v-ATPase activity.
IFN-I Stimulates Intracellular Stm Virulence Gene Expression and Promotes SCV Damage and Stm Cytosolic Exposure.
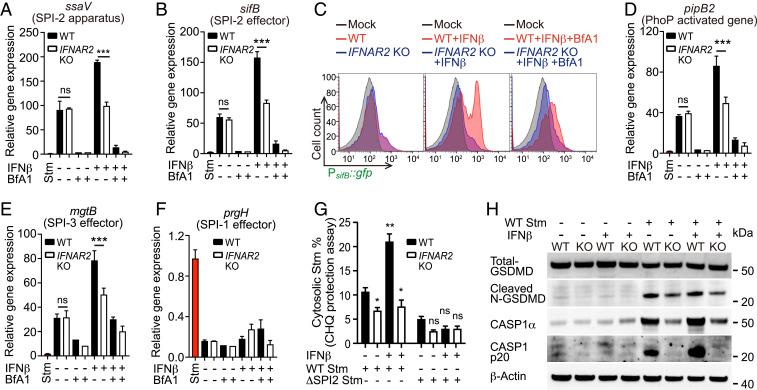
We hypothesized that IFN-I’s role in promoting lysosomal acidification might explain why IFN-I signaling enhances Stm cytotoxicity because acidic pH is thought to stimulate expression of SPI-2-encoded and other virulence genes (38, 39). Consistent with this idea, IFNβ priming of host cells was associated with IFNAR2-dependent heightened intracellular Stm SPI-2–encoded gene expression (Fig. 6 A and B). Furthermore, treatment with Bfa1 abolished IFNβ induction of SPI-2 expression (Fig. 6 A and B), suggesting that SCV acidification is the primary mechanism of IFN-I–enhanced SPI-2 induction. Analyses of SPI-2 gene expression using flow cytometry and a fluorescent PsifB::gfp Stm reporter strain (40), confirmed this phenotype at single bacterial cell resolution (Fig. 6C). Similar expression trends were also observed in known acid-induced virulence-associated genes that are not encoded within SPI-2, such as pagD (41) (Fig. 6 D and E and SI Appendix, Fig. S5A). This is consistent with the observation above (Fig. 2A) that SPI-2–deficient Stm retain some cytotoxicity. Importantly, the expression of SPI-1 genes, which encode invasion-specific functions, was not altered in infections with IFNβ priming or in IFNAR2 KO cells (Fig. 6F and SI Appendix, Fig. S5B). Together, these data suggest that IFN-I–mediated acidification of lysosomes promotes intracellular Stm virulence gene expression.
Fig. 6.
IFN-I signaling promotes Stm virulence gene expression and SCV rupture. Relative induction of SPI-2 (ssaV) (A) (sifB) (B), PhoP-induced virulence gene (pagD) (D), SPI-3 effector (mgtB) (E), and SPI-1 effector (prgH) (F) in intracellular Stm from WT and IFNAR2 KO HeLa cells ± 16 h of drug treatment at 4 hpi. Data are normalized to transcript levels in LB-cultured Stm (red). (C) Flow cytometry of intracellular PsifB::gfp Stm isolated from WT and IFNAR2 KO HeLa cells ± 16 h of drug treatment at 4 hpi. LB-cultured Stm were used as the mock control. (G) Proportions of CHQ-resistant (cytosolic) Stm from WT or IFNAR2 KO HeLa cells ± 16 h of IFNβ treatment at 1 hpi. (H) Immunoblot analysis of cell lysates from uninfected or 4-hpi-infected WT and IFNAR2 KO HeLa cells ± 16 h of IFNβ treatment. Statistical analysis was performed by two-tailed Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001). Data shown are means ± SD from three independent experiments. See also SI Appendix, Fig. S5.
The Stm virulence program can lead to the breakage of the SCV, exposing the pathogen to the host cytosol (42, 43). To assess whether the pathogen was cytosol exposed, infected cells were treated with high concentrations of gentamicin, an antibiotic that can penetrate into cells at high concentrations (44). Stm in IFNβ-treated WT cells were markedly more sensitive to gentamicin than bacteria in IFNβ-treated IFNAR2 KO cells (SI Appendix, Fig. S5C), suggesting that IFN-I activation of Stm virulence gene expression promotes SCV rupture and facilitates the pathogen’s access to the cytosol. Chloroquine (CHQ) protection assays to quantitate the proportion of cytosolic intracellular Stm complemented the gentamicin sensitivity data. In WT cells infected with WT Stm, IFNβ priming led to a doubling of cytosolic bacteria. Priming did not increase the fraction of cytosolic Stm in IFNAR2 KO cells (Fig. 6G), which had a lower cytosolic Stm proportion than naive WT cells. In contrast, when a SPI-2–deficient (ΔssaV) Stm was used in the CHQ protection assays, there was uniformly diminished Stm exposure to the cytosol in both WT and IFNAR2 KO cells, regardless of IFNβ priming, indicating virulence gene induction is required for cytosolic exposure even at early (1-h postinfection [hpi]) timepoints, when population-level SPI-2 induction was not detected (SI Appendix, Fig. S5D). Additionally, ∼60% of WT Stm stained positive for galectin-3 (GAL3), a marker of SCV damage (45), in infected IFNβ-primed WT HeLa cells, whereas <20% of Stm were GAL3+ in infected IFNAR2 KO cells (SI Appendix, Fig. S5 E–G). Less than 5% of ssaV Stm in infected WT or IFNAR2 KO cells were GAL3+, providing further evidence linking Stm intracellular virulence gene expression with both basal and IFN-I–enhanced SCV rupture and escape into the cytosol. The residual SPI-2–independent cytosolic Stm in infected cells is consistent with the observation above (Fig. 2A) of SPI-2–independent cytotoxicity in IECs.
Since cytosolic pathogen exposure is known to induce inflammasome activation and pyroptosis, we performed immunoblotting for the total and activated forms of caspase-1 (CASP1) and gasdermin-D (GSDMD). Consistent with prior reports, Stm infection triggered inflammasome activity (46). Strikingly, both CASP1 and GSDMD cleavages tracked with the level of cytosolic Stm with the highest activation levels in infected WT cells treated with IFNβ (Fig. 6H and SI Appendix, Fig. S5H). Together, these data suggest a model that explains why IFN-I signaling was a hit in the CRISPR/Cas9 screen: IFN-I signaling-dependent lysosome acidification stimulates intracellular Stm virulence gene expression, which promotes SCV rupture and subsequent cytosolic Stm-induced cytotoxicity.
IFN-I Promotes Epithelial Stm Pathogenesis In Vivo.
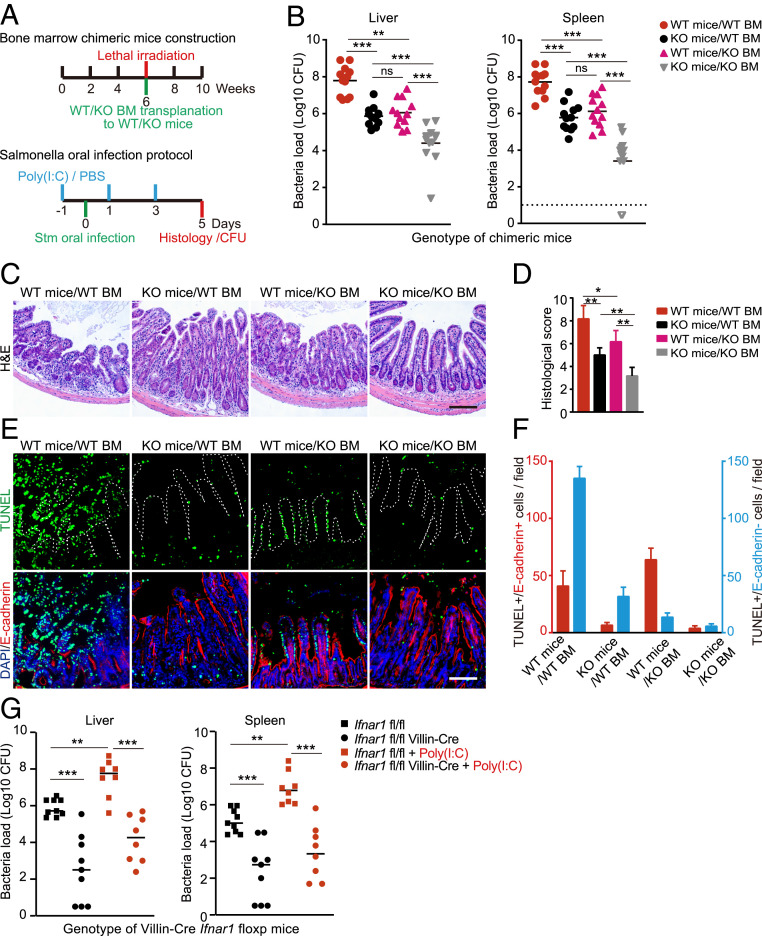
To dissect the importance of IFN-I signaling in the context of in vivo Stm infection, we used bone marrow transfers to generate chimeric C57BL/6 mice that had Ifnar1 deleted in only the hematopoietic compartment or in other bodily tissues, including epithelial surfaces (SI Appendix, Fig. S6A). Following intraperitoneal (i.p.) delivery of polyinosinic:polycytidylic acid (poly[I:C]) to induce IFNβ production (47), chimeric mice were orogastrically inoculated with Stm to assess the roles of epithelial and hematopoietic compartments in resistance to infection (Fig. 7A). Strikingly, mice with KO epithelia and WT bone marrow were relatively resistant to oral Stm infection with reduced weight loss and distal organ bacterial loads compared to mice that had WT epithelia and bone marrow (Fig. 7B and SI Appendix, Fig. S6B), suggesting that IEC IFN-I signaling enhances Stm pathogenicity during infection. We also observed a similar phenotype in mice with WT epithelia and KO bone marrow (Fig. 7B), consistent with previous observations that immune cell IFN-I signaling also promotes Stm pathogenesis (23). Mice that had both KO epithelia and bone marrow were more protected than either chimera (Fig. 7B), further supporting the idea that Stm takes advantage of IFN-I signaling in both the gut epithelium as well as in bone marrow-derived cells.
Fig. 7.
IFN-I signaling in intestinal epithelial cells promotes Stm pathogenesis. (A) Experimental timeline for generation of Ifnar1 chimeras (Top) and oral Stm infection protocols for poly(I:C)-primed chimeras or primed/unprimed Villin-Cre transgenic mice (Bottom). (B) Liver and spleen Stm CFU burdens from chimeric mice 5-d post-Stm infection. n = 12 mice per group. (C) Representative hematoxylin/eosin-stained ileal sections from chimeric mice 5-d post-Stm infection. (Scale bars, 100 μm.) (D) Average histological scores of chimeric mice 5-d post-Stm infection from eight fields per group (n = 2 to 3 mice per group). (E) Representative images of ileal sections from chimeric mice 5-d post-Stm infection. IECs were identified with E-cadherin (red), dying cells with TUNEL (green), and nuclei with DAPI (blue). The white dashed line marks the epithelial surface. (Scale bar, 100 μm.) (F) Quantification of TUNEL+/E-cadherin–positive (red) or TUNEL+/E-cadherin–negative (blue) cells per field from eight fields per group (n = 2 to 3 mice per group). (G) Liver and spleen Stm CFU burdens from Villin-Cre/Ifnar1fl/fl mice 5-d post-Stm infection. n = 9 mice in each PBS treatment group and n = 8 in each poly(I:C) treatment group. Statistical analysis was performed by two-tailed Student’s t test in D. Statistical analysis was performed by two-tailed Mann–Whitney U test in B and G. Data shown are means ± SD from the indicated sample sizes. (*P < 0.05, **P < 0.01, and ***P < 0.001). See also SI Appendix, Fig. S6.
Although histological analyses revealed similar levels of tissue damage in both chimeras (Fig. 7 C and D), finer-scale immunofluorescence studies with terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining to quantify cell death showed that TUNEL+ (dying) cells tracked with the WT compartment. In chimeric mice with WT epithelia, a significantly greater proportion of TUNEL+ cells (76.3 ± 9.6%) also stained positive for the IEC marker E-cadherin than in chimeric mice with WT bone marrow where TUNEL staining was primarily represented in the E-cadherin–negative cellular compartment in the lamina propria (18.5 ± 6.1%) (P = 0.0002) (Fig. 7 E and F).
To bypass the complexities of the chimeric mouse system, and to more precisely decouple the contributions of the hematopoietic and intestinal epithelial cell compartments to host defense against Stm, we also generated transgenic Villin-Cre/Ifnar1fl/fl mice bearing an IEC-specific deletion of Ifnar1 to probe the role of IEC IFN-I signaling in Stm pathogenesis. The burden of Stm in distal organs was significantly lower in orally infected poly(I:C)-treated Villin-Cre/Ifnar1fl/fl mice than in Ifnar1fl/fl mice, similar to observations in the WT bone marrow/KO epithelial chimeric mice (Fig. 7G). Importantly, identical trends were observed in IFN-I–dependent pathogen dissemination to the liver and spleen in non-poly(I:C)–treated mice (Fig. 7G), ruling out potential confounding effects of systemic IFN-I induction by poly(I:C) administration. Together, the chimeric and transgenic in vivo studies demonstrate that IFN-I exacerbates Stm dissemination and suggest that intestinal epithelial IFN-I signaling and downstream IEC death is a key determinant of efficient Stm spread to distal organs.
Discussion
Our findings underscore the utility of model intracellular pathogens, such as Stm as probes for the investigation of fundamental cell processes. The top hits in the genome-scale CRISPR/Cas9 screen that initiated this study were remarkably coherent and revealed that IFN-I signaling sensitizes epithelial cells to Stm cytotoxicity. IFN-I–dependent lysosome acidification in IECs was associated with elevated Stm virulence gene expression, heightened SCV damage, and exacerbated cell death, offering a plausible molecular pathway that explains the results of the screen. Importantly, our paper uncovered a fundamental role for IFN-I signaling. We discovered that this canonical antiviral signaling pathway, which has been studied for more than five decades (48), controls the subcellular localization, protein content, pH, and protease activity of lysosomes. Several ISGs, including IFITM3, SLC15A3, and CNP, that were found to localize to lysosomes, were shown to contribute to lysosomal acidification. Thus, IFN-I signaling controls the function of an organelle—the lysosome—in addition to directly or indirectly modulating the expression and activities of hundreds of ISGs and their interaction partners.
The disparate functions of the three ISGs we identified as participants in IFN-I–mediated lysosomal acidification suggest that more than one mechanism accounts for this phenotype. It seems likely that putative v-ATPase–associated proteins, such as IFITM3 contribute to acidification processes by directly modulating the lysosomal proton concentration gradient. However, ISGs with known non-ATPase–related functions, such as SLC15A3, a proton-coupled histidine and a di-tripeptide transporter, may not play similar roles. While we cannot exclude v-ATPase–mediated mechanisms for such proteins, we speculate that SLC15A3 may show transport preferences for nonneutral dipeptides that could influence lysosome luminal pH. Previous studies have linked CNP with not only lysosomal, but also mitochondrial compartments (49), raising the possibility that ISG function in additional cell compartments could also indirectly contribute to lysosomal acidification. Interestingly, CNP, a membrane-bound protein, has additionally been linked to microtubule function, suggesting that it may also play a role in IFN-I–mediated lysosomal repositioning (50). Understanding the molecular bases of and potential interactions between IFN-I–dependent lysosomal acidification and relocalization will be an interesting future topic of study.
Although a virtually universal antiviral immune signal, the consequences of IFN-I signaling during bacterial infection is less clear and is in many cases detrimental to the host (19). Several intracellular bacterial pathogens, including Listeria monocytogenes, Mycobacterium tuberculosis, and Stm, are less virulent in Ifnar1-deficient mice. Studies of the bases for these phenotypes have primarily focused on immune-mediated explanations (51–53). For Stm, we propose that IFN-I signaling contributes to negative infection outcomes, at least, in part, by inducing epithelial cell lysosome remodeling, stimulating the Stm virulence program, and SCV breakage, leading to cytosolic escape and host cell death. Our findings show that IFN-I signaling can modify innate defense in the epithelial as well as the immune compartments and are complementary to and compatible with previously proposed mechanisms for IFN-I–enhanced Stm infection in bone marrow-derived immune cells. Such mechanisms include elevated macrophage necroptosis (23, 54) and transcriptional reprogramming (55), altered dendritic cell homeostasis (56), and increased neutrophil-mediated inflammation (57). The role of IFN-I modulation of lysosome function in Stm infection in nonepithelial cells, such as macrophages, requires further study.
The results from our screen suggest that besides IFN-I signaling, diverse cellular pathways ranging from cytoskeletal maintenance to control of posttranslational modifications may influence Stm-induced cytotoxicity in IECs. These pathways may directly or indirectly impact this phenotype; for example, host genes required for Stm invasion reduce host cell death due to lack of infection. This global approach complements existing literature that indicates Stm can induce canonical and noncanonical cell death pathways in IECs, including apoptosis, necroptosis, and pyroptosis (11–15). The role that cell death at the intestinal epithelial surface plays is complex and likely to be context dependent. Previous reports have proposed that inflammasome activation and associated IEC cell death/expulsion are a generally protective mechanism during infection (58–60). However, it is also known that cytosolic Stm in epithelial cells can be hyper-replicative and primed for infection, readily reinvading neighboring cells upon primary cell lysis (14, 61). By selectively elevating the frequency of Stm entry into the cytosol, IFN-I signaling could promote pathogenesis even if cell death is, in part, a protective mechanism due to the increase in hyperinfectious bacteria that are released upon cell lysis. IFN-I’s role in promoting Stm-infected IEC death may also negatively affect the integrity of the intestinal epithelial barrier, aiding pathogen dissemination (62). Our paper does not conclusively define the mechanism(s) of death in IFN-I-primed Stm-infected IECs and suggests that multiple pathways, including apoptosis and pyroptosis, may be active at the same time. Additional work is needed to untangle the relative contributions, kinetics, and potentially hierarchical nature of infection-induced cytotoxicity pathways. Furthermore, it remains an open question whether Stm purposely stimulates IFN-I signaling as part of its pathogenic strategy.
Although IFN-I–induced lysosomal acidification sensitizes cells to an intracellular bacterial pathogen, our finding that several known ISGs with antiviral properties, such as IFITM3, SLC15A3, and CNP, participate in this process leads us to speculate that this mechanism may be protective against viral threats. IFN-I–mediated lysosomal remodeling may also play a role in noninfectious pathologies, such as lysosomal cholesterol accumulation (63) and other lysosome-related disorders. It remains unclear whether these effects might be driven by the tonic IFN-I signaling that occurs in many tissues (35) or instead require pathogenic elevations of IFN-I.
Our finding that IFN-I signaling governs the composition and function of the lysosome provides a cell biologic perspective for understanding IFN-I function. It will be of interest to determine whether other immune signals (i.e., other cytokines) can also direct remodeling of lysosomes and other organelles under homeostasis and in diverse pathogenic contexts. Such activities may constitute a broadly applicable lens through which to view and enhance our understanding of the cell biology of innate defense.
Materials and Methods
Core methods for this study are described below. Detailed methods for the remaining assays are specified in SI Appendix, Supplementary Methods. Bacterial strains (Dataset S3), plasmids (Dataset S4), oligos (Dataset S5), antibodies (Dataset S6), and cell lines; pharmacologic inhibitors and IFNβ priming; genomic DNA preparation, sequencing, and analyses of screen results; lentivirus preparation and transductions; construction of cell lines with targeted gene disruptions; cell survival assays; infection of primary human small intestinal organoids; Stm invasion assays; immunofluorescence microscopy of tissue cultured cells; gentamicin sensitivity assay; cathepsin D activity assay; endocytosis and lysosome function assays; immunoblot analyses; qRT-PCR quantification of Stm virulence gene expression; histology and tissue immunofluorescence.
Stm Infections.
All tissue culture infections were performed according to the following procedure unless otherwise indicated. WT and mutant Stm were grown for ∼16 h at 37 °C with shaking and then subcultured (1:33) in lysogeny broth (LB) without antibiotics for 3 h until the cultures reached an optical density at 600 nm of 0.8. To prepare the inoculum, cultures were first pelleted at 5,000 × g for 5 min. The pellets were resuspended in Dulbecco’s modified Eagle’s medium without fetal bovine serum (FBS), and an appropriate volume of bacterial solution was added to cells to reach a multiplicity of infection (MOI) of 100 bacteria per eukaryotic cell. The cells were then incubated with bacteria for 30 min at 37 °C with 5% CO2. Extracellular bacteria were removed by extensive washing with phosphate-buffered saline (PBS) (Gibco, Catalog No. 14190250) and addition of 50 μg/mL gentamicin to the medium. At 2 hpi, the gentamicin concentration was decreased to 5 µg/mL.
CRISPR/Cas9 Stm Infection Screen.
HT29-Cas9 CRISPR libraries were constructed as described previously (20) using the Avana sgRNA library, which contains four different sgRNAs targeting each human protein-coding gene (64). For each library, two sets of four T225 flasks (Corning, Catalog No. 14–826-80) were seeded with 15 × 106 cells per flask and then incubated for 48 h. At the time of the screen, there were ∼150 × 106 cells per experimental condition, corresponding to ∼2,000 × coverage per sgRNA. Cells were at ∼70% confluence at the time of infection. The infection was performed as described above with minor modifications. Briefly, HT29 libraries were infected with WT Stm at a MOI of 300 for 30 min. After infection, the libraries were expanded in McCoy's 5A + FBS containing 5 µg/mL gentamicin to both permit intracellular bacterial cytotoxicity and minimize the intracellular gentamicin concentration to allow Stm invasion during the next round of infection. Flasks were checked daily to monitor recovery of survivor cells; when 70% confluency was achieved, cells were trypsinized, pooled, and reseeded for the next round of infection. In total, four rounds of infection were conducted. Surviving cells from the last round of infection were used for preparation of genomic DNA.
Annexin V Staining and Fluorescence-Activated Cell Sorter Analysis.
Cell death was detected with the FITC Annexin V Apoptosis Detection kit (BioLegend, Catalog No. 640922). Infections were performed as described above with mCherry-Stm at a MOI of 100. At 20 hpi, both suspended and attached cells were collected, resuspended in 100 μL of Annexin V binding buffer at 1 × 107 cells/mL, and mixed with 5 μL of FITC-conjugated Annexin V. After incubation at room temperature for 15 min in the dark, 400 μL of Annexin V binding buffer was added, and stained cells were immediately analyzed by flow cytometry as described above.
Quantification of Lysosome Distribution.
Lysosome distribution was analyzed as previously described (65); the area occupied by nuclei was excluded from analyses. Average LAMP1 intensities were measured for the area within 5 μm of the nucleus (Iperinuclear) and the area >10 μm from the nucleus (Iperipheral). The average intensities were calculated and normalized to cell areas. The perinuclear index was defined as Iperinuclear/Iperipheral. Quantifications were carried out on 10 cells per group with ImageJ.
Measurement of Lysosomal Acidity.
Cells with no treatment or with either 10 ng/mL IFNβ or 5 nM BfA1 (Santa Cruz, Catalog No. sc-201550) treatment for 16 h were stained with 75 nM Lysotracker or Lysosensor Green DND-189 (Thermofisher, Catalog No. L7535) for 15 min and washed with PBS. The fluorescence intensity of the stained cells was determined by flow cytometry. Ratiometric measurement of lysosomal pH was performed with LysoSensor Yellow/Blue DND-160 (Invitrogen, Catalog No. L7545). Cells were labeled with 2 µM LysoSensor Yellow/Blue DND-160 for 30 min at 37 °C in regular medium after which ratiometric fluorescence imaging was performed by confocal microscopy. Images were analyzed in ImageJ where background was subtracted, regions of interest identified by finding circular spots, fluorescence intensity measured in the 540- and 440-nm channels, and the intensity ratio of corresponding regions of interests calculated. Absolute quantification of lysosomal pH was performed using a previously described fluorescence-activated cell sorter-based FITC-dextran (Sigma-Aldrich, Catalog No. FD40S) labeling assay where cells are labeled for 72 h with 1 µM FITC-dextran and then subjected to quantitative flow cytometry against a standard pH curve of FITC-dextran fluorescence at specific pHs (27).
LysolP.
LysoIP was performed largely as described (36). Briefly, pLJC5-3×hemagglutinin (HA)-TMEM192 was used to introduce a lysosomal tag protein in WT and IFNAR2 KO HeLa cells. Cells (15 million per replicate) were rinsed twice with prechilled PBS and then scraped in 1 mL of PBS-containing protease and phosphatase inhibitors and pelleted at 100 × g for 2 min at 4 °C. Cells were resuspended in 950 µL of the same buffer, and 25 µL (equivalent to 2.5% of the total number cells) was reserved for further processing to generate the whole-cell sample. The remaining cells were gently homogenized with 25 strokes of a 2 mL Dounce-type homogenizer. The homogenate was then centrifuged at 100 × g for 2 min at 4 °C to pellet the cell debris and intact cells, while cellular organelles including lysosomes remained in the supernatant. The supernatant was incubated with 150 µL of anti-HA magnetic beads preequilibrated with PBS on a rotator shaker for 3 min. Immunoprecipitates were then gently washed three times with PBS on a DynaMag spin magnet. Beads with bound lysosomes were resuspended in 100 µL prechilled 1% Triton-X lysis buffer to extract proteins. After 10-min incubation on ice, the beads were removed with the magnet. Samples (5 μL each) were subjected to 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunodetected using antibodies listed in Dataset S6, while the remainder was submitted to the Thermo Fisher Center for Multiplexed Proteomics of Harvard Medical School, Boston, MA for isobaric tandem mass tag-based quantitative proteomics.
Flow Cytometric Analysis of Stm Virulence Gene Expression.
HeLa cells were infected with mCherry- and sifB-GFP–expressing Stm as described above. Cell lysis was performed 4 hpi by washing three times with PBS and subsequent incubation for 10 min with PBS containing 0.1% Triton X-100. Cell lysates were then analyzed by flow cytometry. Stm were first identified by gating on the mCherry signal, and sifB expression was quantified by gating on the mCherry+/GFP+ population. LB-cultured Stm served as a negative control.
CHQ Protection Assay.
To quantify the proportion of cytosolic bacteria in the total population, we used the previously described CHQ protection assay (66). HeLa cells were infected in 24-well plates as described above. Two wells were incubated in the presence of 400 µM CHQ and 10 µg/mL gentamicin for 1 h (CHQ-resistant, cytosolic bacteria) and another two wells were incubated with 10 µg/mL gentamicin only (total bacteria). Infected cells were solubilized in lysis buffer, and the numbers of viable bacteria were determined by plating serial dilutions on LB agar. The proportion of cytosolic Stm was determined by dividing the colony-forming units (cfu) from wells treated with CHQ and gentamicin by the cfu from wells treated with gentamicin alone.
Mice.
C57BL/6, Ifnar1−/−, and Ifnar1flox/flox mice were purchased from The Jackson Laboratory, Bar Harbor, ME, Villin-Cre transgenic mice were a generous gift from Richard Blumberg, Brigham and Women’s Hospital, Boston, MA. These mice were maintained on a 12-h light–dark cycle and a standard chow diet at the Harvard Institute of Medicine specific pathogen-free animal facility. Animal experiments were performed according to guidelines from the Center for Animal Resources and Comparative Medicine at Harvard Medical School. All protocols and experimental plans were approved by the Brigham and Women’s Hospital Institutional Animal Care and Use Committee (Protocol No. 2016N000416). Littermate control mice were randomly assigned to each group, and experiments were performed blinded with respect to treatment.
Bone Marrow Chimeras.
Recipient mice were irradiated two times with 600 rad 1 d before injection of bone marrow from WT or Ifnar1−/− mice. Bone marrow was extracted from femurs of donor mice by flushing with PBS and then washed once in PBS; 1 × 106 cells were injected into the tail vein of recipient mice. Mice were monitored for 4 wk at which point engraftment was evaluated by flow cytometry.
In Vivo Stm Infection.
Poly(I:C) (Sigma-Aldrich, Catalog No. P1530) (20 μg per mouse) was given i.p. to chimeric and Villin-Cre Ifnar1fl/fl mice 1 d before Stm infection and every other day for a total of three doses to stimulate IFN production. Mice that did not receive poly(I:C) received injections of PBS. Food was withdrawn for 4 h before infection. Stm inocula were prepared as described above. Mice without or with poly(I:C) treatment were infected orogastrically with 5 × 108 Stm in 100 μL PBS. Food was returned to the cages 2 hpi. Infected mice were killed 5 d after infection. Tissue samples of the small intestine, spleen, and liver were collected for histological analysis and enumeration of cfu. cfu were quantified by serial-dilution plating of homogenized tissue samples on LB plates containing 100 μg/mL streptomycin.
Statistical Methods.
Statistical analyses were carried out using the two-tailed Student’s t test or one-way analysis of variance with Dunnett’s postcorrection on GraphPad Prism 5.
Supplementary Material
Acknowledgments
We thank members of the Waldor Laboratory for helpful discussions on all aspects of this project, Dr. David Breault at the Harvard Digestive Diseases Center (HDDC) Organoid Core for the primary human small intestine organoids, Dr. Jonathan Kagan for insightful suggestions, and Michal Pyzik from Dr. Richard Blumberg’s Laboratory for assistance in creation of bone marrow chimeric mice. Research in the M.K.W. Laboratory was supported by HHMI and NIH Grant R01 AI-042347. A.Z. was supported by an European Molecular Biology Organization long-term fellowship (Award ALTF 1514-2016) and by a HHMI Fellowship of the Life Sciences Research Foundation.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010723117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Geoghegan J. L., Holmes E. C., The phylogenomics of evolving virus virulence. Nat. Rev. Genet. 19, 756–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribet D., Cossart P., How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 17, 173–183 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Hybiske K., Stephens R. S., Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 6, 99–110 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lee H. C., Chathuranga K., Lee J. S., Intracellular sensing of viral genomes and viral evasion. Exp. Mol. Med. 51, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omotade T. O., Roy C. R., Manipulation of host cell organelles by intracellular pathogens. Microbiol. Spectr. 7, 199–214 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Welch M. D., Why should cell biologists study microbial pathogens? Mol. Biol. Cell 26, 4295–4301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo E. K., Interplay between host and pathogen: Immune defense and beyond. Exp. Mol. Med. 51, 1–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurley D., McCusker M. P., Fanning S., Martins M., Salmonella-host interactions–Modulation of the host innate immune system. Front. Immunol. 5, 481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuli A., Sharma M., How to do business with lysosomes: Salmonella leads the way. Curr. Opin. Microbiol. 47, 1–7 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Hernandez L. D., Hueffer K., Wenk M. R., Galán J. E., Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304, 1805–1807 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Hefele M., et al. , Intestinal epithelial Caspase-8 signaling is essential to prevent necroptosis during Salmonella Typhimurium induced enteritis. Mucosal Immunol. 11, 1191–1202 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Ingram J. P., et al. , A nonpyroptotic IFN-γ-triggered cell death mechanism in nonphagocytic cells promotes Salmonella clearance in vivo. J. Immunol. 200, 3626–3634 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J. M., et al. , Apoptosis of human intestinal epithelial cells after bacterial invasion. J. Clin. Invest. 102, 1815–1823 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knodler L. A., et al. , Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U.S.A. 107, 17733–17738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broz P., et al. , Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207, 1745–1755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess C. B., Niesel D. W., Klimpel G. R., The induction of interferon production in fibroblasts by invasive bacteria: A comparison of Salmonella and Shigella species. Microb. Pathog. 7, 111–120 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Negishi H., Taniguchi T., Yanai H., The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb. Perspect. Biol. 10, a028423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubel P., et al. , A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 20, 493–502 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Kovarik P., Castiglia V., Ivin M., Ebner F., Type I., Type I interferons in bacterial infections: A balancing act. Front. Immunol. 7, 652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blondel C. J., et al. , CRISPR/Cas9 screens reveal requirements for host cell sulfation and fucosylation in bacterial type III secretion system-mediated cytotoxicity. Cell Host Microbe 20, 226–237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unsworth K. E., Way M., McNiven M., Machesky L., Holden D. W., Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cell. Microbiol. 6, 1041–1055 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Yeung A. T. Y., et al. , A genome-wide knockout screen in human macrophages identified host factors modulating Salmonella infection. MBio 10, e02169-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson N., et al. , Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 13, 954–962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Velden A. W., Lindgren S. W., Worley M. J., Heffron F., Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype typhimurium. Infect. Immun. 68, 5702–5709 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galán J. E., Lara-Tejero M., Marlovits T. C., Wagner S., Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjardins M., et al. , Molecular characterization of phagosomes. J. Biol. Chem. 269, 32194–32200 (1994). [PubMed] [Google Scholar]
- 27.Eriksson I., Öllinger K., Appelqvist H., Analysis of lysosomal pH by flow cytometry using FITC-dextran loaded cells. Methods Mol. Biol. 1594, 179–189 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Reis R. C., Sorgine M. H., Coelho-Sampaio T., A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur. J. Cell Biol. 75, 192–197 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Butor C., Griffiths G., Aronson N. N. Jr, Varki A., Co-localization of hydrolytic enzymes with widely disparate pH optima: Implications for the regulation of lysosomal pH. J. Cell Sci. 108, 2213–2219 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y., Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 (1991). [PubMed] [Google Scholar]
- 31.Bao S., Beagley K. W., France M. P., Shen J., Husband A. J., Interferon-gamma plays a critical role in intestinal immunity against Salmonella typhimurium infection. Immunology 99, 464–472 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalkhali-Ellis Z., et al. , IFN-gamma regulation of vacuolar pH, cathepsin D processing and autophagy in mammary epithelial cells. J. Cell. Biochem. 105, 208–218 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spence J. S., et al. , IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 15, 259–268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wee Y. S., Roundy K. M., Weis J. J., Weis J. H., Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 18, 834–845 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Schoggins J. W., et al. , Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abu-Remaileh M., et al. , Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song F., et al. , Regulation and biological role of the peptide/histidine transporter SLC15A3 in Toll-like receptor-mediated inflammatory responses in macrophage. Cell Death Dis. 9, 770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty S., Mizusaki H., Kenney L. J., A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol. 13, e1002116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prost L. R., et al. , Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26, 165–174 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Garmendia J., Beuzón C. R., Ruiz-Albert J., Holden D. W., The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology (Reading) 149, 2385–2396 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Gunn J. S., Alpuche-Aranda C. M., Loomis W. P., Belden W. J., Miller S. I., Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177, 5040–5047 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy D., et al. , A process for controlling intracellular bacterial infections induced by membrane injury. Science 304, 1515–1518 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Xu Y., et al. , A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell 178, 552–566(2019). [DOI] [PubMed] [Google Scholar]
- 44.Myrdal S. E., Johnson K. C., Steyger P. S., Cytoplasmic and intra-nuclear binding of gentamicin does not require endocytosis. Hear. Res. 204, 156–169 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thurston T. L., Wandel M. P., von Muhlinen N., Foeglein A., Randow F., Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Zhao Y., Shi J., Shao F., Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 110, 14408–14413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauterbach H., et al. , Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J. Exp. Med. 207, 2703–2717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.González-Navajas J. M., Lee J., David M., Raz E., Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12, 125–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McFerran B., Burgoyne R., 2′,3′-Cyclic nucleotide 3′-phosphodiesterase is associated with mitochondria in diverse adrenal cell types. J. Cell Sci. 110, 2979–2985 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Bifulco M., Laezza C., Stingo S., Wolff J., 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: A membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc. Natl. Acad. Sci. U.S.A. 99, 1807–1812 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Connell R. M., et al. , Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 200, 437–445 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boxx G. M., Cheng G., The roles of type I interferon in bacterial infection. Cell Host Microbe 19, 760–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kernbauer E., Maier V., Rauch I., Müller M., Decker T., Route of infection determines the impact of type I interferons on innate immunity to Listeria monocytogenes. PLoS One 8, e65007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hos N. J., et al. , Type I interferon enhances necroptosis of Salmonella Typhimurium-infected macrophages by impairing antioxidative stress responses. J. Cell Biol. 216, 4107–4121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins D. J., et al. , Salmonella typhimurium co-opts the host type I IFN system to restrict macrophage innate immune transcriptional responses selectively. J. Immunol. 195, 2461–2471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stefan K. L., Fink A., Surana N. K., Kasper D. L., Dasgupta S., Type I interferon signaling restrains IL-10R+ colonic macrophages and dendritic cells and leads to more severe Salmonella colitis. PLoS One 12, e0188600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson R. P., et al. , STAT2 dependent type I Interferon response promotes dysbiosis and luminal expansion of the enteric pathogen Salmonella Typhimurium. PLoS Pathog. 15, e1007745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rauch I., et al. , NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46, 649–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crowley S. M., et al. , Intestinal restriction of Salmonella Typhimurium requires caspase-1 and caspase-11 epithelial intrinsic inflammasomes. PLoS Pathog. 16, e1008498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knodler L. A., et al. , Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe 16, 249–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malik-Kale P., Winfree S., Steele-Mortimer O., The bimodal lifestyle of intracellular Salmonella in epithelial cells: Replication in the cytosol obscures defects in vacuolar replication. PLoS One 7, e38732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labarta-Bajo L., et al. , Type I IFNs and CD8 T cells increase intestinal barrier permeability after chronic viral infection. J. Exp. Med. 217, e20192276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kühnl A., et al. , Late endosomal/lysosomal cholesterol accumulation is a host cell-protective mechanism inhibiting endosomal escape of influenza A virus. MBio 9, e01345-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X., et al. , A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein J. A., Powers T. R., Knodler L. A., Measurement of Salmonella enterica internalization and vacuole lysis in epithelial cells. Methods Mol. Biol. 1519, 285–296 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.